Abstract
Halophilic enzymes function optimally at high salt concentrations and are active at low water availability. Such conditions are encountered at elevated concentrations of solutes such as salts and sugars, and at high concentrations of organic solvents. However, expression in heterologous hosts such as Escherichia coli can cause problems, since halophilic proteins typically misfold and aggregate in conditions of low ionic strength. We have harnessed the sophisticated genetic tools available for the haloarchaeon Haloferax volcanii, to develop a system for the overexpression and purification of halophilic proteins under native conditions.
Key words: protein overexpression, His-tag, archaea, Haloferax volcanii, halophile
Halophilic archaea offer an unparalleled resource of enzymes that are tolerant of high concentrations of salt and organic solvents.1–3 Halophilic microorganisms found in hypersaline lakes such as the Dead Sea are able to grow in the presence of molar concentrations of salt.4 There are two biological strategies to cope with a high salt environment: halophilic bacteria maintain an osmotic balance of their cytoplasm with the medium by accumulating organic solutes, whereas halophilic archaea (haloarchaea) accumulate high cytoplasmic concentrations of salt (typically potassium). The latter strategy mandates that intracellular enzymes are able to function in high salt concentrations; this is achieved by a reduction in overall hydrophobicity and an increase in acidic residues on the protein surface.5–7 The high number of negative charges coordinates a network of hydrated cations, thereby maintaining the protein in solution via a salt-enriched solvation shell.
Since halophilic enzymes are adapted to function at high salt concentrations, they are active at low water availability. Such conditions are encountered at elevated concentrations of solutes such as salts and sugars, and at high concentrations of organic solvents. It is well known that halophilic enzymes have significantly greater activity and stability in organic solvents than equivalent enzymes from non-halophilic organisms.1 For example, catalase from Halobacterium cutirubrum exhibits up to threefold stimulation of activity by 0.5–1.5 M monovalent salts and by 2–4 M organic solvents, such as ethylene glycol, glycerol and dimethylsulfoxide.8
Halophilic proteins usually feature an excess of acidic amino acids, the negative surface charge is critical to solubility in high salt.6,7 This can pose problems for expression in heterologous hosts such as Escherichia coli, since halophilic proteins typically misfold and aggregate in conditions of low ionic strength. Purification of halophilic enzymes from E. coli has often relied on the recovery of insoluble protein from inclusion bodies, followed by denaturation and refolding in hypersaline solutions.9 To circumvent this problem, we have developed a system for the expression of halophilic proteins in the haloarchaeon Haloferax volcanii.10
Hfx. volcanii is the organism of choice for haloarchaeal genetics.11,12 It has simple growth requirements (aerobic conditions, optimum temperature 45°C) and is easy to culture in the laboratory.12 As a halophile, it is extremely resilient to contamination—few organisms can survive in 2.5 M NaCl. This is a significant advantage for industry, since it circumvents the need for sterile growth conditions. Another feature of haloarchaea that make them attractive to industry is the ease of lysis upon addition of water, reducing the cost of protein purification.12 Amongst halophiles, Hfx. volcanii can grow across a wide range of salinities, from 12–24% salt (w/v), approximately 1.8–3.5 M NaCl. It is therefore ideally placed for investigating halophilic proteins from other organisms, which might grow at different salt concentrations.
Hfx. volcanii can grow in defined media, which has enabled the development of a wide range of selectable markers.11 The genetic toolbox for Hfx. volcanii also includes gene knockout methods, plasmids with high and low copy-number replication origins, and the tryptophan-inducible tnaA promoter.13–16 We have made significant improvements to transformation protocols, by generating strains deficient in the Mrr restriction endonuclease.10 Mrr cleaves foreign DNA that is methylated at 5′-GATC and reduces the efficiency of transformation by 50-fold. The Δmrr strains allow direct and efficient transformation of Hfx. volcanii (>107 transformants/µg DNA) without the need to passage DNA through an E. coli dam mutant, they are ideal for generating expression libraries.
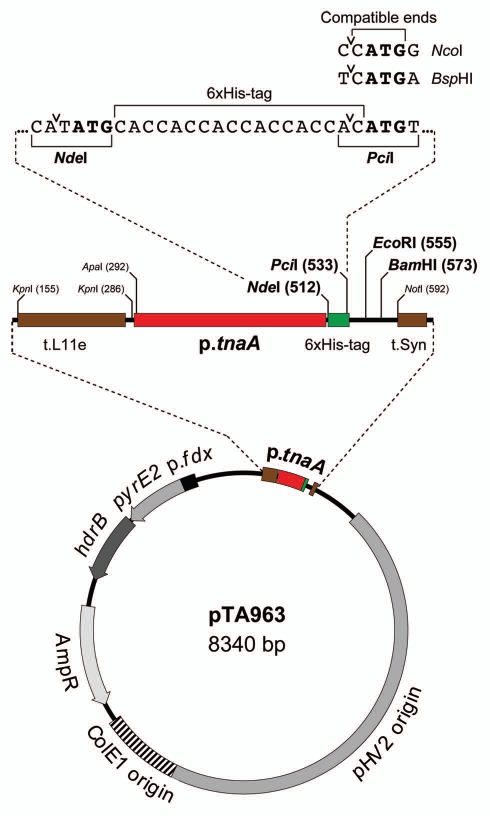
The overexpression system we have developed consists of a highly-engineered host strain H1209 (ΔpyrE2 ΔhdrB Δmrr Nph-pitA), and a corresponding plasmid pTA963 for inducible overexpression of His-tagged proteins.10 pTA963 utilizes the p.tnaA tryptophanase promoter of Hfx. volcanii, genes under control of the p.tnaA promoter show rapid and strong induction of expression upon addition of ≥1 mM tryptophan.16 For expression of proteins with an N-terminal hexahistidine (6xHis) tag, a (CAC)6 tract was incorporated in pTA963 (Fig. 1), and a polylinker ensures that cloning is facile. To insulate the gene from read-through transcription, the expression cassette was flanked by two transcriptional terminators (t.L11e and t.Syn). The vectors are based on pTA230 and use the pHV2 replication origin that maintains the plasmid at a copy number of ∼6 per genome equivalent.13 Selection is based on the pyrE2 and hdrB markers, which allow growth of the host strain H1209 on media lacking uracil and thymidine, respectively; the latter marker allows the plasmid to be maintained in rich media (Hv-YPC), thus maximizing protein overexpression.13,14
Figure 1.
Conditional overexpression vector pTA963. pTA963 utilizes the tryptophan-inducible promoter p.tnaA. For N-terminally 6xHis tagged proteins, the 5′ end of the gene is ligated with the PciI site located downstream of a (CAC)6 tract. PciI-compatible ends are generated by NcoI and BspHI, and are used where the second codon starts with G and A, respectively. For expression of native proteins, the 5′ end of the gene is ligated with the NdeI site downstream of the promoter.10
In Hfx. volcanii H1209 the pitA gene is replaced with its orthologue from Natronomonas pharaonis.10 PitA is a fusion of chlorite dismutase-like and monooxygenase-like domains, it is unique to haloarchaea and in almost all species features a histidine-rich linker.17 Owing to this histidine-rich region, PitA is the major contaminant of His-tagged recombinant proteins purified from H. volcanii by immobilized metal affinity chromatography. We were unable to delete pitA gene by standard techniques, indicating that it is essential.13,14 Instead, we replaced pitA with its orthologue from the haloalkaliphile N. pharaonis, which does not feature a high number of histidines in the central linker region. The H. volcanii strain H1209 with Nph-pitA gene replacement shows no contamination by PitA of His-tagged recombinant proteins, thus improving protein yield and purity.
We have used this overexpression system to purify several halophilic proteins from Hfx. volcanii, including the DNA repair proteins RadA and RadB.10 The protocol is simple and robust, and compatible with the molar concentrations of salt required for the stability of halophilic proteins.
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/11794
References
- 1.Margesin R, Schinner F. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles. 2001;5:73–83. doi: 10.1007/s007920100184. [DOI] [PubMed] [Google Scholar]
- 2.Sellek GA, Chaudhuri JB. Biocatalysis in organic media using enzymes from extremophiles. Enzyme and Microbial Technology. 1999;25:471–482. [Google Scholar]
- 3.Danson MJ, Hough DW. Structure, function and stability of enzymes from the Archaea. Trends Microbiol. 1998;6:307–314. doi: 10.1016/s0966-842x(98)01316-x. [DOI] [PubMed] [Google Scholar]
- 4.Oren A. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Systems. 2008;4:2. doi: 10.1186/1746-1448-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanyi JK. Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol Rev. 1974;38:272–290. doi: 10.1128/br.38.3.272-290.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danson MJ, Hough DW. The structural basis of protein halophilicity. Comp Biochem Physiol A Physiol. 1997;117:307–312. [Google Scholar]
- 7.Mevarech M, Frolow F, Gloss LM. Halophilic enzymes: proteins with a grain of salt. Biophys Chem. 2000;86:155–164. doi: 10.1016/s0301-4622(00)00126-5. [DOI] [PubMed] [Google Scholar]
- 8.Lanyi JK, Stevenson J. Effect of salts and organic solvents on the activity of Halobacterium cutirubrum catalase. J Bacteriol. 1969;98:611–616. doi: 10.1128/jb.98.2.611-616.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connaris H, Chaudhuri JB, Danson MJ, Hough DW. Expression, reactivation and purification of enzymes from Haloferax volcanii in Escherichia coli. Biotechnol Bioeng. 1999;64:38–45. [PubMed] [Google Scholar]
- 10.Allers T, Barak S, Liddell S, Wardell K, Mevarech M. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl Environ Microbiol. 2010;76:1759–1769. doi: 10.1128/AEM.02670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allers T, Mevarech M. Archaeal genetics—the third way. Nat Rev Genet. 2005;6:58–73. doi: 10.1038/nrg1504. [DOI] [PubMed] [Google Scholar]
- 12.Soppa J. From genomes to function: haloarchaea as model organisms. Microbiology. 2006;152:585–590. doi: 10.1099/mic.0.28504-0. [DOI] [PubMed] [Google Scholar]
- 13.Allers T, Ngo HP, Mevarech M, Lloyd RG. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol. 2004;70:943–953. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bitan-Banin G, Ortenberg R, Mevarech M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J Bacteriol. 2003;185:772–778. doi: 10.1128/JB.185.3.772-778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norais C, Hawkins M, Hartman AL, Eisen JA, Myllykallio H, Allers T. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 2007;3:77. doi: 10.1371/journal.pgen.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Large A, Stamme C, Lange C, Duan Z, Allers T, Soppa J, et al. Characterization of a tightly controlled promoter of the halophilic archaeon Haloferax volcanii and its use in the analysis of the essential cct1 gene. Mol Microbiol. 2007;66:1092–1106. doi: 10.1111/j.1365-2958.2007.05980.x. [DOI] [PubMed] [Google Scholar]
- 17.Bab-Dinitz E, Shmuely H, Maupin-Furlow J, Eichler J, Shaanan B. Haloferax volcanii PitA: an example of functional interaction between the Pfam chlorite dismutase and antibiotic biosynthesis monooxygenase families? Bioinformatics. 2006;22:671–675. doi: 10.1093/bioinformatics/btk043. [DOI] [PubMed] [Google Scholar]