Abstract
The infrequently feeding Burmese python (Python molurus) experiences significant and rapid postprandial cardiac hypertrophy followed by regression as digestion is completed. To begin to explore the molecular mechanisms of this response, we have sequenced and assembled the fasted and postfed Burmese python heart transcriptomes with Illumina technology using the chicken (Gallus gallus) genome as a reference. In addition, we have used RNA-seq analysis to identify differences in the expression of biological processes and signaling pathways between fasted, 1 day postfed (DPF), and 3 DPF hearts. Out of a combined transcriptome of ∼2,800 mRNAs, 464 genes were differentially expressed. Genes showing differential expression at 1 DPF compared with fasted were enriched for biological processes involved in metabolism and energetics, while genes showing differential expression at 3 DPF compared with fasted were enriched for processes involved in biogenesis, structural remodeling, and organization. Moreover, we present evidence for the activation of physiological and not pathological signaling pathways in this rapid, novel model of cardiac growth in pythons. Together, our data provide the first comprehensive gene expression profile for a reptile heart.
Keywords: Illumina, RNA-Seq
an understanding of the molecular mechanisms that regulate cardiac hypertrophy is of clinical relevance, as cardiac hypertrophy is one of the leading predictors of mortality (27). In mammals, cardiac hypertrophy occurs via an increase in cardiomyocyte size rather than by hyperplasia and can arise from either pathological or physiological stimuli. In the former case, hypertrophy can be caused by hypertension, maladaptive responses to diseases, or mutations within the sarcomere, ultimately leading to heart failure. On the other hand, physiological hypertrophy is beneficial, completely reversible, and caused by normal stimuli such as postnatal growth, exercise, and pregnancy (10). Research leading to a more thorough understanding of the molecular mechanisms that underlie physiological cardiac growth in animal models could lead to therapies aimed at counteracting pathological hypertrophy.
Gene expression studies in mammalian models of cardiac hypertrophy have contributed critical information to our mechanistic understanding of these processes. For example, experiments characterizing the expression patterns of the two cardiac isoforms of myosin heavy chain (MyHC) in both physiological and pathological models of hypertrophy have provided a means to separate these conditions by their gene expression profiles (29). Additionally, the induction of a fetal program of gene expression has been widely used to distinguish pathological hypertrophy from physiological hypertrophy, where no such markers are induced. Elucidating the molecular pathways that control enlargement of the heart by different stimuli remains a focus of cardiovascular research, and the identification of key physiological vs. pathological effectors may provide novel therapeutic targets to prevent, reverse, or modify the pathological hypertrophic phenotype.
Expression array analysis has been the method of choice for quantitative analysis of cardiac gene expression in varying states, but more recently whole transcriptome shotgun sequencing (RNA-Seq) has been used in a number of experimental systems (4). In fact, recent comparisons of these two techniques have determined that expression data generated through RNA-Seq analysis is at least as reproducible as data from expression array analysis (18). Rapidly emerging next generation genomics tools including low-cost, high-throughput sequencing of genomes, exomes, and transcriptomes are changing the face of inquiry into adaptations found in novel systems where the genome sequence resources required to perform expression arrays are unavailable (13, 28).
One such model, the infrequently feeding Burmese python, undergoes numerous extreme physiological responses to cope with the stress of digesting meals of at least 25% its body mass, including a 40-fold increase in standard metabolic rate and significant systemic organ hypertrophy (25). Specifically, organs involved in digestion, substrate handing, and energy utilization such as the intestine, liver, kidneys, and heart experience a substantial postprandial increase in dry tissue mass (24). Blood flow must increase to these organs to satisfy the energy needs of their increased metabolic demand, a requirement fulfilled by the hypertrophic heart. After digestion is complete, the heart and other organs experience a complete reversal of hypertrophy and a complete regression of all other systemic postprandial changes to their respective fasting levels (25). Accordingly, we hypothesize that the postprandial heart growth exhibited by the Burmese python is a potentially novel model for physiological cardiac hypertrophy.
A current critical obstacle to studying the molecular mechanisms of the unique form of heart growth exhibited by Burmese pythons is the inability to perform whole transcriptome analysis and gene expression experiments due to the lack of available gene-specific nucleotide sequences for pythons. To address this limitation, we have used Illumina sequencing to generate a python heart transcriptome for three states during digestion: fasted, 1 day postfed (DPF), and 3 DPF, the two postprandial states being when systemic metabolic rate is highest (25) and heart size is the largest compared with fasted (3), respectively. In addition, we have used RNA-Seq in conjunction with gene ontological analyses to create a comparative gene expression profile for several biological processes that underlie cardiac hypertrophy in each of these three states. In this article, we will present 1) data from sequencing the python heart transcriptome, 2) the assembly and annotation of the transcriptome using the phylogenetically related Gallus sequence assemblies, and 3) RNA-Seq, gene ontological, and Ingenuity pathway analyses showcasing the postprandial increase in transcripts of genes that regulate biological processes in pythons that are characteristic of physiological cardiac hypertrophy in mammals.
METHODS
Experimental animals.
Juvenile Burmese pythons were obtained from Strictly Reptiles and housed at the University of Colorado for 3 mo prior to experimentation. These animals were kept at 27 ± 3°C in individual containers with free access to water and under a 12 h light/dark photoperiod. All animals were fed rats equivalent to 25% of their body mass every other week until experimentation. At the time of tissue collection, the snakes weighed between 300 and 350 g. Prior to tissue collection, animals were fasted for 30 days and then fed as previously described. Heart ventricles were obtained from euthanized snakes at three experimental time points: fasted, 1 DPF, and 3 DPF. For sequencing, two snakes were killed per time point, while four snakes were killed per time point for the quantitative real-time PCR (qPCR) validation. Ventricles from each of these animals were quickly excised, sectioned into ∼100 mg pieces with a sterile razor blade, snap-frozen in liquid nitrogen, and stored at −80°C until RNA isolation.
RNA isolation and reverse transcription.
Total RNA was isolated from ∼50 mg of snap-frozen python ventricles using TRI Reagent (MRC). Isolated RNA was quantified using spectroscopy and tested for integrity by denaturing electrophoresis (data not shown). Total RNA samples for Illumina sequencing were poly(A+) purified using MPG Streptavidin Complex kit (Pure Biotech LLC). Subsequent mRNA quality and yields were assessed using the Agilent Bioanalyzer at the University of Colorado Health Sciences Center Microarray Facility. Purified mRNA samples were then chemically fragmented and reverse transcribed into double-stranded cDNA libraries using a modified version of the Paired-End Sample Preparation Kit (Illumina) optimized for sequencing. Total RNA samples for qPCR were reverse transcribed into cDNA using SuperScript III (Invitrogen). Specifically, 2 μg total RNA from each sample was primed with 2.5 μM random hexamers and 500 μM dNTPs.
Illumina sequencing and assembly.
cDNA samples were sequenced on an Illumina Genome Analyzer platform using the version 1.3 pipeline. Reads for each time point (fasted, 1 DPF, and 3 DPF) were assembled using ABySS version 1.0.15, which implements a short-read de novo sequence assembly method using de Bruijn digraphs (26). Reads are then decomposed into subreads of length k (k-mers) that are then processed for read errors and used to construct contigs in stages. Multiple assembly trials were performed to determine the parameters for this analysis: for example, a k-mer length of 21 was determined to produce the best N50 length contigs. Because the library was selected at 175−250 bp, an inner mate distance of 178 was used. A final assembly was then produced, combining the assemblies for each experimental animal from each time point after feeding. The ABySS assembler source code and binaries are available for download at http://www.bcgsc.ca/platform/bioinfo/software/abyss.
RNA-Seq analysis.
Assembled python contigs were compared with the available reference genomes for genus Anolis, Gallus, and Homo. Basic Local Alignment Search Tool (BLAST) hits with >80% identity (near the noise boundary between distantly related taxa) were selected for analysis. TopHat version 1.0.10 and high-throughput short-read aligner Bowtie were used to align and produce a gene expression profile for reads from each time point (16). An indexed reference genome is required for TopHat to perform its analysis, and since there is no established reference genome for the Burmese python phylogenetically similar Gallus assemblies [mRNA, protein, and expressed sequence tags (ESTs)] were selected for this study. The 2.1 version (May 2006) of the Gallus genome is ∼1.05 gb in size and has a Q20 base redundancy of 7.1× with 600,000 available ESTs (6). The assembled reads for each time point were independently aligned to the Gallus reference genome using TopHat, producing a list of genes with expression levels measured by reads per kilobase of exon model per million-mapped reads (RPKM). In order for a gene to be classified as differentially expressed between states, the log2 of its fold change needed to be >2. The two-sample homoscedastic, independent t-test was used for comparing the pair-wise combinations of the states described herein. The significance threshold was set at P ≤ 0.05 to test the hypothesis that mean 1 = mean 2. These tests were done on log2 transformed data, a common variance reduction method.
Primer design and qPCR.
Gene-specific primers were selected for qPCR analysis based on sequences from the Illumina assembly obtained using homologous Gallus genes as a reference. Primers were designed for 50–200 bp amplicons and optimized for an annealing temperature of 60°C with the assistance of Primer3 (23) (Supplemental Table S1).1 Primer specificity was determined using conventional PCR and gel electrophoresis with ethidium bromide staining. These PCR products were cloned using the TOPO cloning kit (Invitrogen), and purified DNA was subsequently sent to the DNA sequencing facility at the University of Colorado for sequencing. qPCR was performed for each python gene using the absolute quantification with a standard curve on the Applied Biosystems 7500 Fast system as follows: 10 μl reactions in triplicate containing 5 μl 2× SYBR Green Master Mix (ABI), 0.2 μl forward primer at 12.5 μM, 0.2 μl reverse primer at 12.5 μM, 0.6 μl nuclease-free water, and 4 μl template cDNA at 1.25 ng/μl. Template cDNA from fasted, 1 DPF, and 3 DPF python ventricles with four biological replicates for each time point were used for this study. The cycling conditions for the qPCR reaction were as follows: 95°C for 10 min (polymerase activation), 40 cycles of 95°C for 15 s and 60°C for 1 min (amplification and quantification), with a melting curve from 65 to 95°C to confirm primer specificity. Absolute quantities of transcript were determined based on the cycle threshold of each reaction in reference to the standard curve. Expression data was normalized using the Pfaffl method (21) with the reference gene hypoxanthine phosphoribosyl transferase (HPRT1), which was selected based on its utility in qPCR experiments conducted in other highly metabolic systems like the postprandial python heart, where the expression of most conventional reference genes would change (8). Statistical analyses were conducted using the student's t-test for each postprandial state in reference to fasting levels, with P ≤ 0.05 considered to be significant.
Gene Ontology analysis of RNA-Seq expression data.
Gene Ontology (GO) analysis was conducted by importing the list of Gallus corresponding to python contigs in the assembly to the DAVID functional GO annotation program (9, 15). GO terms corresponding to biological processes (BP-FAT) were exclusively selected for this study. To sort the extensive lists of biological processes obtained using GO analysis into descriptive general categories, GO terms were grouped into the following broad processes using these criteria (detailed analysis available in Supplemental S2, available at http://mcdb.colorado.edu/files/S2_Python-Transcriptome-Gene-Ontology.xls/view):
Metabolism and Energy - Processes involving the metabolism of nucleic acids, proteins, lipids, and glucose or mitochondrial processes resulting in the production of ATP.
Gene Expression - Any process that is involved in or regulates transcription, translation, or posttranslational modification of genes and gene products.
Signaling and Response to Stimuli - Processes regulating how cells respond to extracellular stimuli and relay these responses using intracellular signaling mechanisms.
Differentiation, Cycle, and Death - Processes that significantly impact the cellular life cycle: differentiation, proliferation, DNA replication, mitosis, and death.
Biogenesis, Morphogenesis, and Organization - Processes involved in the generation, organization, and localization of complex, functional cellular components including ribosomes and organelles.
Structure and Movement - Any process that involves the assembly and/or organization of structural proteins, the cytoskeleton, or movement. This group includes processes responsible for cardiac function, including contraction and myofibril assembly.
Other Processes - Any process not covered by the previous categories.
For GO analysis of the whole transcriptome, only biological processes with 50 or more genes were selected to be grouped into the aforementioned categories. For GO analysis comparing states, genes that were considered to be differentially expressed between time points (log2 fold-change of ≥ 2 and P ≤ 0.05 by t-test) were selected, and all biological processes considered. All biological processes identified by DAVID were statistically significant (P ≤ 0.05).
Ingenuity Pathway Analysis.
Python genes determined to be differentially expressed between states by TopHat were uploaded into the Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, http://www.ingenuity.com). IPA uses a human-curated knowledge base containing genes, proteins, and RNAs to construct association graphs for mammalian canonical signaling pathways, and additionally attempts to build new associations using gene expression data. Associations are computed using the right-tailed Fisher exact test to test the significance within the context of new data. All signaling pathways identified by IPA with a P value ≤0.05 have a statistically significant, nonrandom association.
RESULTS
Illumina sequencing of the postprandial python heart transcriptome.
To obtain the cardiac transcriptome of the python heart, samples were selected for sequencing from the following states: pythons that had been fasted for 1 mo (fasted), pythons that had been fed 24 h previously (1 DPF), and pythons that had been fed 72 h previously (3 DPF). In preparation for sequencing, cDNAs from the three states were sequenced using the Illumina platform. Approximately 15 million to 30 million paired-end reads of 36 bp in length were obtained for each sample as summarized in Table 1. We subsequently compiled each of the separate sequencing reactions according to state and used the ABySS assembler to generate a series of contiguous nucleotide sequences (contigs) of no less than 100 bp in length using homologous 36 bp reads. A summary of all the contigs from the assemblies pertaining to each state as well as from the comprehensive, combined assembly is shown in Table 2.
Table 1.
Number of 36 bp sequencing reads for each time point by Illumina
| Fasted | 1 DPF | 3 DPF | |
|---|---|---|---|
| Sequencing run 1 | 20,419,196 | 21,269,929 | 20,424,488 |
| Sequencing run 2 | 14,751,134 | 31,643,800 | 15,547,198 |
DPF, days postfed.
Table 2.
Summary of ABySS contigs assembled from each time point postfeeding
| Fasted | 1 DPF | 3 DPF | Combined | |
|---|---|---|---|---|
| Contigs, n | 32,719 | 39,441 | 36,861 | 76,371 |
| Cutoff contig length | 100 | 100 | 100 | 100 |
| Max. contig length | 8,603 | 7,796 | 8,405 | 12,967 |
| Avg. contig length | 280 | 324 | 280 | 325 |
| N50 | 355 | 459 | 352 | 461 |
| Total bases | 9,189,005 | 12,806,707 | 10,327,026 | 24,843,421 |
| Min. GC composition, % | 25.00 | 23.00 | 24.00 | 22.00 |
| Max. GC composition, % | 85.00 | 91.00 | 89.00 | 92.00 |
| Avg. GC composition, % | 48.00 | 47.00 | 49.00 | 47.00 |
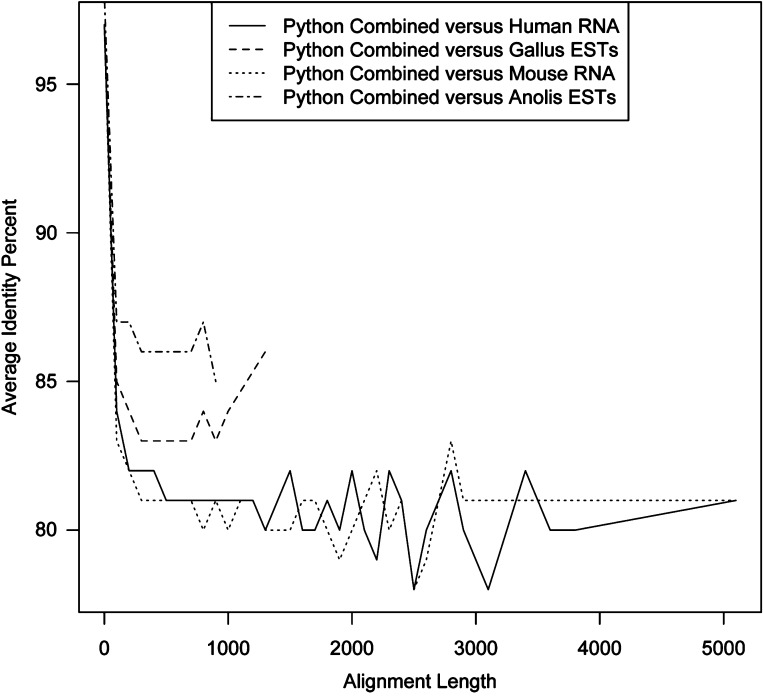
Since no reference genome exists for the Burmese python, we used BLAST to reference the contigs from each assembly against annotated assemblies for the genera Homo, Mus, Gallus, and Anolis from the UCSC Genome Bioinformatics website (5–7). Figure 1 shows a graphical comparison of the python transcriptome to the best candidate assemblies to use as references. The Anolis and Gallus assemblies had the highest average identity percent per total alignment length. Although the assemblies for Mus and Homo are much more complete and have a much larger total alignment length, the average identity percent of the python contigs to these assemblies was below the acceptable background level for this analysis. Despite the fact that the Anolis EST assembly produced the highest percent similarity compared with the python assembly, we selected the collective Gallus EST, RNA, and protein assemblies to use as a reference with which to match each python contig to its corresponding gene. Because the Anolis assembly is not sufficiently annotated to function as a reference database, the annotated Gallus assemblies provide more thorough coverage for the homologous python contigs. Approximately 4,300 Gallus genes with genomic positions were matched to python contigs from our assemblies.
Fig. 1.
Sequence similarity between Python, Gallus, Anolis, Mus, and Homo. The average identity percent per alignment length of the python assembly compared with assemblies from other potential reference assemblies from organisms. An average identity percent <80% was considered to be below the acceptable background noise level for these alignments. EST, expressed sequence tag.
RNA-Seq analysis comparing transcript levels between postprandial states.
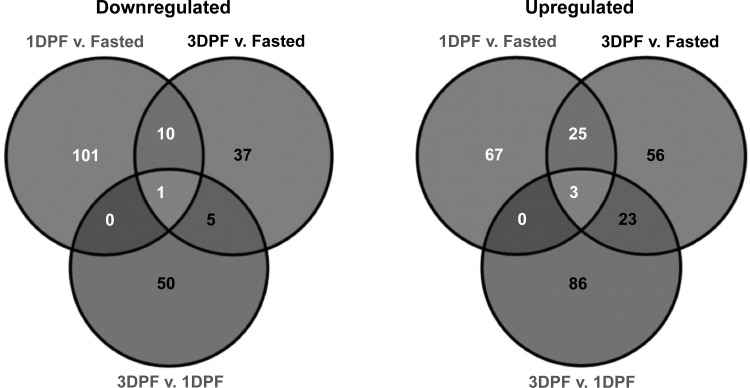
After annotating the python transcriptome using the WUGSC2.1/galGal3 version of the Gallus genome, we used the fast-splice junction mapper TopHat to perform RNA-Seq analysis comparing expression patterns of the annotated contigs between each of the three aforementioned states. This analysis comprised 4,407 gene annotations in a single GTF file, which was then converted into GFF3 format for input into TopHat. Of those 4,407 genes, 88 were rejected by TopHat due to errors such as incorrect transcript annotations. Approximately 2,774 of the remaining genes were measured by TopHat in at least one state in the python heart using the number of RPKMs. The range of RPKM values varied on a log2 scale from a minimum of 0 to a maximum of 18.3, the average being 3.53 and the median 3.19. Through this analysis, we observed that 464 genes exhibited significant differential expression between the fasted and postprandial states with 260 genes upregulated, 204 genes downregulated, and 45 genes overlapping. Paired-state comparisons were made between up- and downregulated genes between 1 and 3 DPF (Fig. 2).
Fig. 2.
The number of genes significantly downregulated and upregulated between time points as determined by TopHat. Comparing 1 day postfed (DPF) vs. fasted, 112 genes were down; 53 down, comparing 3 DPF vs. fasted, and 56 down comparing 3 DPF vs. 1 DPF. Comparing 1 DPF vs. fasted, 95 genes were up; 107 up comparing 3 DPF vs. fasted, and 112 up, comparing 3 DPF vs. 1 DPF. The expression of each gene was changed by at least 2-fold with a P value ≤ 0.05 by t-test.
In addition, we used qPCR to validate the expression patterns of 10 genes demonstrated by TopHat to be upregulated, downregulated, or unchanged at each postprandial state versus fasted (Table 3). We observed that the expression patterns were directionally similar for 7 out of 10 genes at 1 DPF vs. fasted, and for 9 out of 10 genes at 3 DPF vs. fasted, for a total similarity of 80% in expression data using fractional fold changes between states from TopHat and qPCR (Table 3). However, differences in expression as determined by qPCR for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ribosomal protein, large, P1 (RPLP1), heat shock 90 kDa protein 1, beta (HSPCB), and Jumonji, AT-rich interactive domain 2 (JARID2) at 1 DPF and for GAPDH, RPLP1, and JARID2 at 3 DPF were not statistically significant (identified in the table by an asterisk), meaning that the expression of these genes should be considered unchanged between fasting and postfed. To further validate the accuracy of the Illumina sequencing and contig assembly, each of these 10 cDNA amplicons was cloned and sequenced. Subsequently, we determined that these cloned sequences had in total a 95% identity to their respective contigs.
Table 3.
Comparison of fold changes in gene expression between TopHat and qPCR data
| 1 DPF vs. Fasted |
3 DPF vs. Fasted |
|||
|---|---|---|---|---|
| Gene Name | TopHat | qPCR | TopHat | qPCR |
| ACTN1 | −0.95 | 0.57 | 0.40 | 1.21 |
| COL1A | 0.92 | 2.55 | 3.79 | 4.14 |
| PCNA | 0.77 | 1.99 | 1.64 | 2.09 |
| VDAC2 | 0.30 | 1.46 | 0.74 | 1.48 |
| CTNNB1 | 0.29 | 1.68 | 0.81 | 1.82 |
| TPM1 | 0.02 | 3.76 | 0.63 | 1.94 |
| RPLP1 | −1.12 | 0.01* | −0.41 | −0.32* |
| GAPDH | −1.36 | 0.30* | −1.11 | −0.07* |
| HSPCB | −2.52 | −0.64* | −2.50 | −1.63 |
| JARID2 | −4.30 | −0.15* | −2.64 | 0.58* |
Unchanged between fasting and postfed states.
Gene ontological pathways for physiological hypertrophy are postprandially represented in the transcriptome.
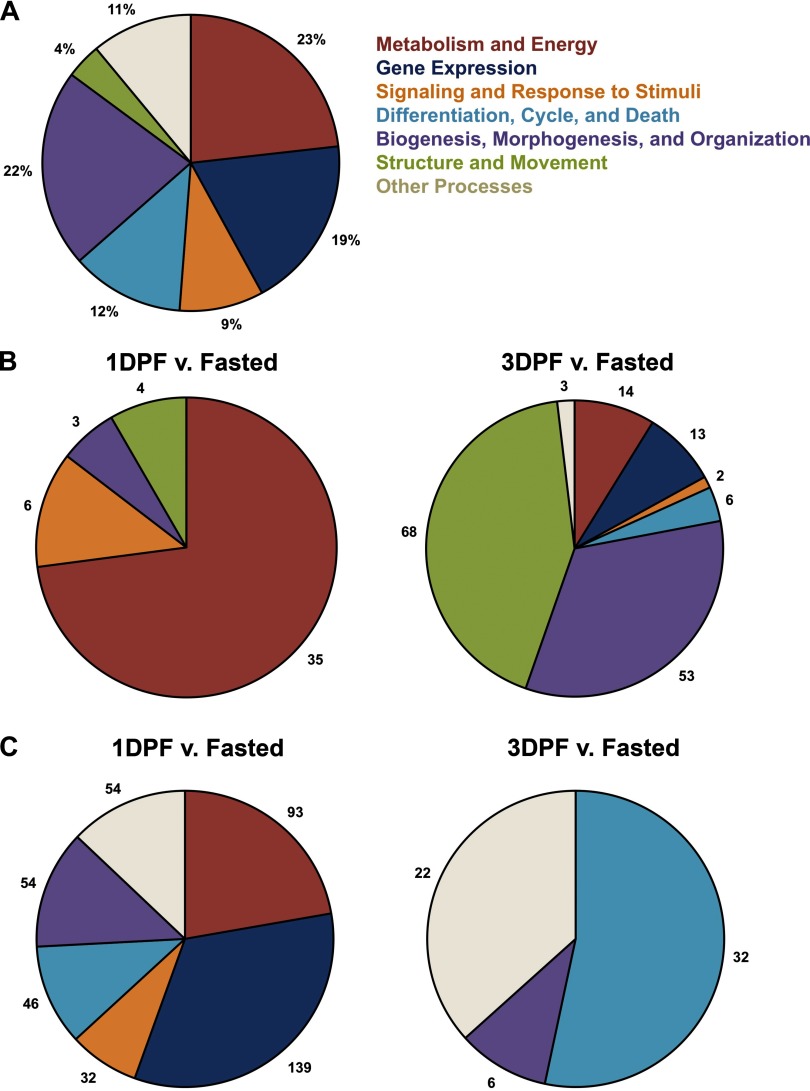
In an effort to understand the biological processes that are represented in the python heart and to define which processes were differentially expressed between the fasted and fed states, we used the GO annotation program DAVID to group all the genes expressed in the python transcriptome into specific biological processes. After grouping the top biological processes for the entire transcriptome (any gene with an RPKM value from at least one state) as determined by DAVID (with 50 or more genes per specific process) into the broad, descriptive ontological groups as described in methods, we found that the top biological processes in the python heart are metabolism and energy and gene expression, covering nearly half of the top biological processes (Fig. 3A) (Supplemental S1 and S2, available at http://mcdb.colorado.edu/files/S1_Python-Transcriptome-Gene-Lists.xls/view and http://mcdb.colorado.edu/files/S2_Python-Transcriptome-Gene-Ontology.xls/view, respectively). Signaling and response to stimuli, other processes, differentiation, cycle, and death, and biogenesis, morphogenesis, and organization were each less represented than the previous two, and structure and movement was least represented.
Fig. 3.
Top-represented Gene Ontology (GO) biological processes in the whole transcriptome and the regulation of GO biological processes at the postprandial states compared with fasted. A: this chart shows the percent of genes (out of the total number of genes grouped into biological processes with 50 genes or more by DAVID) sorted into each of the descriptive broad categories for the comprehensive transcriptome containing combined data from fasted, 1 DPF, and 3 DPF. B: each graph depicts the number of upregulated genes for each state compared with fasted as determined by TopHat that have been annotated into specific biological processes by DAVID and then grouped into the described broad categories. C: each graph depicts the number of downregulated genes for each state compared with fasted as determined by TopHat that have been annotated into specific biological processes by DAVID and then grouped into the described broad categories.
In addition to identifying the predominant biological processes in the python heart, we were also interested in how these processes might be regulated at the two postprandial states compared with fasted. To answer this question, we also used DAVID to annotate the lists of genes that were found to be significantly differentially expressed at these time points by TopHat. While the data are shown as total genes annotated into each broad category, GO biological processes often display considerable overlap with individual genes, meaning that the total number of genes graphed is in fact the total number of genes assigned to a biological process by DAVID. Processes not depicted for each chart were not represented in the GO analysis at that state. Figure 3B shows that out of a total 46 upregulated genes at 1 DPF compared with fasted, the majority of genes grouped into metabolism and energy with a few grouping into other processes, structure and movement, and signaling and response to stimuli. Moreover, out of a total 158 upregulated genes at 3 DPF compared with fasted, nearly half of these genes grouped into structure and movement, with a notable amount of genes also grouping into biogenesis, morphogenesis, and organization and gene expression, with the other processes being much less represented.
We were able to use this same strategy to identify downregulated processes at these time points compared with fasted. Out of a total of 418 genes downregulated at 1 DPF compared with fasted, over half of these genes grouped into gene expression and metabolism and energy with a considerable amount of genes also grouping into other processes, biogenesis, morphogenesis, and organization, and differentiation, cycle, and death. Signaling and response to stimuli was the least downregulated descriptive group at this state. Out of a total 60 genes downregulated at 3 DPF compared with fasted, over half of these genes grouped into differentiation, cycle, and death, with a considerable amount also grouping into other processes. Biogenesis, morphogenesis, and organization was only slightly represented compared with these other two processes (Fig. 3C).
IPA reveals a preferential activation of physiological signaling pathways over pathological pathways.
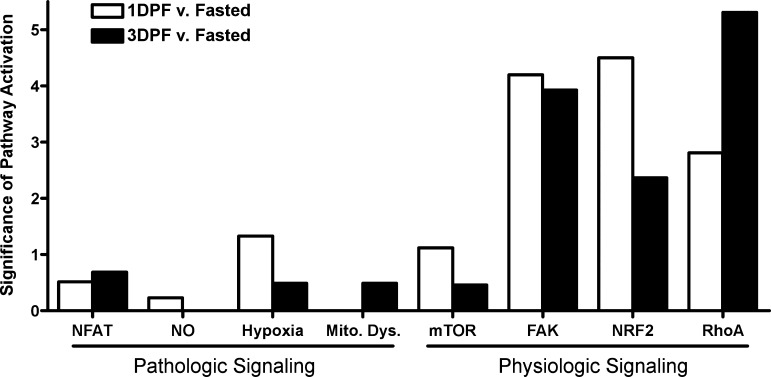
While GO analysis provides a general overview of the biological processes that are heavily regulated and highly represented at each state, we were further interested in the signaling pathways that might be activated in the postprandial python heart and whether those pathways were more similar to physiological or pathological models in mammals. To accomplish this, we used IPA to group the lists of genes that were significantly differentially expressed at each state as determined by TopHat into hypertrophic signaling pathways. Out of eight canonical hypertrophic signaling pathways selected by IPA grouping of genes that change between states in the python, three pathways involved in physiological hypertrophy in mammals: FAK (22), NRF2 (17), and RhoA (20) were shown to be significantly activated compared with four pathways that have been implicated in pathological hypertrophy in mammals: NFAT (14), nitric oxide (30), hypoxia (11), and mitochondrial dysfunction (1) (Fig. 4). The downstream target genes of FAK, NRF2, and RhoA that were significantly upregulated are listed in Supplemental Table S2.
Fig. 4.
Canonical signaling pathways shown to be activated through Ingenuity Pathways Analysis (IPA). IPA highlighted 8 signaling pathways involved in 2 types of hypertrophic responses. The data are plotted as the −log2 of the P value given during the analysis, meaning larger values correspond to more significant pathway activation. The pathological pathways are: nuclear factor of activated T-cell signaling (NFAT), nitric oxide signaling (NO), cardiac hypoxia signaling (Hypoxia), and mitochondrial dysfunction signaling (Mito. Dys.). The physiological pathways are: mammalian target of rapamycin signaling (mTOR), focal adhesion kinase-1 signaling (FAK), NF-E2 related factor 2 signaling (NFR2), and ras homolog member A signaling (RhoA).
DISCUSSION
In this report, we describe the sequencing and assembly of the fasted and postprandial Burmese python heart transcriptome using the Illumina platform. We have used RNA-Seq to generate and qPCR to validate expression data from this assembly and have annotated these data through GO and IPA to show the highly represented biological processes and signaling pathways that are likely activated in the postprandial python heart. We have shown that biological processes common to mammalian models of physiological cardiac hypertrophy are activated in the postprandial heart using GO analysis and that physiological signaling pathways are preferentially expressed over pathological pathways in the python heart using IPA. The sequencing data described in this study will provide a valuable resource to researchers interested in conducting detailed investigation into the molecular mechanisms of postprandial cardiac hypertrophy in pythons and provides a preliminary representation of the general processes and pathways that facilitate the rapid morphological changes exhibited by the hypertrophic python heart.
The python transcriptome was sequenced using the Illumina platform with ∼15 million reads generated per state with high (≥350) N50 values and almost 25 million nucleotides of unique python sequence. These sequences also validated with a 95% similarity compared with actual python sequences cloned from cDNA amplicons, which further establishes the robustness of the Illumina platform. The python heart transcriptome consists of ∼2,800 unique genes with expression levels at least one state homologous to Gallus genes, many of which have functional roles in physiological heart processes. Although ∼4,400 python genes were originally annotated to the Gallus genome, only 60% of these genes were detected by TopHat in at least one state in the python heart. Reads that fail to align can do so for a number of reasons including the differences between the Gallus reference genome and the python, low coverage of transcripts, artifacts such as contamination due to the total RNA protocol being used, stringency of the allowed mismatches, and others. In addition to validating the expression data generated by TopHat through qPCR, we also considered a previous study by Andersen et al. (3) that examined the expression of two transcripts (atrial and ventricular MyHC) in the postprandial python heart. When comparing our transcriptome to their study, we found that the expression patterns for individual myosin isoforms did not match. We believe this inconsistency to be largely attributable to the inability of short-read sequencing to effectively distinguish between highly similar isoforms (in mammals, the two cardiac isoforms are 93% identical), as assembling 36 bp reads into separate isoforms in this instance would likely yield chimeric contiguous sequences.
When all three states are considered, there is a clear representation of genes that are involved in metabolism and energy, gene expression, and signaling and response to stimuli. Interestingly, a similar study utilizing Illumina technology also found that GO processes involved in metabolism and energy and signaling and response to stimuli also accounted for the majority of the mouse heart transcriptome (19). While GO analysis cannot effectively provide a detailed representation of how specific biological processes are represented in the transcriptome given that many genes have overlapping functions, and processes concerning specific mechanisms of regulation are often too complicated to characterize based solely on transcript levels, the GO analysis of the whole transcriptome shows that genes controlling these common cardiac biological processes are overrepresented compared with other genes.
In terms of changes in representations of genes and biological processes between the postprandial states, each time point seems to have its own unique expression profile. TopHat was able to identify at least 50 genes that are both up- and downregulated comparing fasted, 1 DPF, and 3 DPF. The charts showing genes that change in expression between the states differ from each other and also from the profile of the whole transcriptome, suggesting that the molecular changes that underlie hypertrophy in pythons are not as simple as “hypertrophic” and “nonhypertrophic.” Specifically, processes clustering to metabolism and energy were persistently upregulated at 1 and 3 DPF compared with fasted, while processes clustering to differentiation, cycle, and death were persistently downregulated at these states. Yet the changes in gene expression at each state still mirror known mechanisms of hypertrophy. For example, the highly represented upregulation of metabolic processes at 1 DPF is in support of work showing a 40-fold increase in metabolic rate at the same state in pythons (25). This observation is also consistent with the fact that the heart itself is a highly metabolic organ and is likely working much harder to circulate the necessary nutrients to satisfy this systemic demand. The highly represented increase in the expression of processes that regulate structure and movement as well as biogenesis, morphogenesis, and organization at 3 DPF also supports this idea, as this state is the apex of growth for the python heart where the organ likely undergoes its highest level of structural remodeling (3, 25), yet we still see that processes such as gene expression are also highly represented, suggesting that the genes upstream of these structural changes are highly regulated. These data suggest that the hypertrophic python heart is similar to mammalian models of physiological cardiac hypertrophy where hypertrophic hearts are more highly metabolic to support their energy requirements, are regulating several mechanisms that control structure and movement to facilitate growth and cardiac function, and are activating several signaling pathways that are upstream of both of these processes (2, 12).
Interestingly, more genes involved in metabolic processes and gene expression are also downregulated at 1 DPF compared with fasted than are upregulated at the same state. While at first glance contradictory, this finding can be rationalized by the general manner through which genes and biological processes are categorized in GO studies. As previously mentioned, GO analysis does not necessarily provide specific information about how one particular process is changing, rather, it suggests that genes either directly involved in or that regulate a particular process, either positively or negatively, are experiencing considerable changes in expression. As such, this research aims primarily to narrow the focus of molecular research in the python to the descriptive categories used for GO analysis in this study at each state, as these are the processes that are experiencing the most regulation in terms of transcript levels. With this idea in mind, at 3 DPF processes that control differentiation, cycle, and death are strikingly downregulated compared with fasted, suggesting that these developmental processes are under stringent control as the heart undergoes its morphological changes.
Given the similarities in biological processes expressed by the hypertrophic python heart and in hypertrophic mammalian hearts, there could be important clinical implications of applying this rapid, reversible, and novel model of cardiac growth to pathological hypertrophy in mammals Using IPA, we found evidence that three pathways characteristic of physiological hypertrophy (FAK, NRF2, and RhoA) are activated in the postprandial python heart, while four pathways characteristic of pathological hypertrophy (NFAT, NO, hypoxia, and mitochondrial dysfunction) are activated to a much lesser extent. Collectively, this analysis suggests a direct link between cardiac hypertrophy in pythons and mammals in terms of common signaling mechanisms that regulate these processes. Further research into the nature of these signaling pathways in pythons could lead to a clinically applicable method to rapidly onset or regress hypertrophy in mammals. While cardiac hypertrophy in pythons likely harbors many similar traits to these models, many of the precise mechanisms that are responsible for initiating and regressing the changes in heart morphology are likely unique to this novel animal model.
GRANTS
We acknowledge Hiberna, Inc. and National Institutes of Health Grants 1S10RR-023702-01 to W. R. McCombie and HL-50560 to L. A. Leinwand for funding this project.
DISCLOSURES
Steven Cozza, Tom Marr, and Leslie Leinwand have a total financial interest worth more than US $10,000 in Hiberna, Inc., which also provided funding for this study.
Supplementary Material
ACKNOWLEDGMENTS
The short-read transcriptome sequences for each state are available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive under the accession SRA020826.7 and the study under the accession SRP002796.1. Validated sequences for qPCR contigs are available at NCBI GenBank under the accession numbers HM641801-HM641811.
The authors thank Jason Magida, Brooke Harrison, and Kristen Bjorkman for providing critical advice and support throughout this study.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Ago T, Kuroda J, Pain J, Fu C, Li H, Sadoshima J. Upregulation of Nox4 by hypertrophic stimuli promotes apoptosis and mitochondrial dysfunction in cardiac myocytes. Circ Res 106: 1253–1264, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allen DL, Harrison BC, Leinwand LA. Molecular and genetic approaches to studying exercise performance and adaptation. Exerc Sport Sci Rev 30: 99–105, 2002. [DOI] [PubMed] [Google Scholar]
- 3. Andersen JB, Rourke BC, Caiozzo VJ, Bennett AF, Hicks JW. Physiology: postprandial cardiac hypertrophy in pythons. Nature 434: 37–38, 2005. [DOI] [PubMed] [Google Scholar]
- 4. Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25: 169–193, 2000. [DOI] [PubMed] [Google Scholar]
- 5. Consortium HGS. Initial sequencing and analysis of the human genome. Nature 409: 860–921, 2001. [DOI] [PubMed] [Google Scholar]
- 6. Consortium ICGS. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716, 2004. [DOI] [PubMed] [Google Scholar]
- 7. Consortium MGS. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562, 2002. [DOI] [PubMed] [Google Scholar]
- 8. De Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, Swinkels DW, Span PN. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest 85: 154–159, 2005. [DOI] [PubMed] [Google Scholar]
- 9. Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: P3, 2003. [PubMed] [Google Scholar]
- 10. Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol 65: 45–79, 2003. [DOI] [PubMed] [Google Scholar]
- 11. Giordano FJ. Oxygen, oxidative stress, hypoxia, and heart failure. J Clin Invest 115: 500–508, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harrison BC, Leinwand LA. Fighting fat with muscle: bulking up to slim down. Cell Metab 7: 97–98, 2008. [DOI] [PubMed] [Google Scholar]
- 13. Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet 11: 476–486, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Houser SR, Molkentin JD. Does contractile Ca2+ control calcineurin-NFAT signaling and pathological hypertrophy in cardiac myocytes? Sci Signal 1: pe31, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009. [DOI] [PubMed] [Google Scholar]
- 16. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J, Ichikawa T, Villacorta L, Janicki JS, Brower GL, Yamamoto M, Cui T. Nrf2 protects against maladaptive cardiac responses to hemodynamic stress. Arterioscler Thromb Vasc Biol 29: 1843–1850, 2009. [DOI] [PubMed] [Google Scholar]
- 18. Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matkovich SJ, Zhang Y, Van Booven DJ, Dorn GW., 2nd Deep mRNA sequencing for in vivo functional analysis of cardiac transcriptional regulators: application to Galphaq. Circ Res 106: 1459–1467, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miyamoto S, Del Re DP, Xiang SY, Zhao X, Florholmen G, Brown JH. Revisited and revised: is RhoA always a villain in cardiac pathophysiology? J Cardiovasc Transl Res 3: 330–343, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pham CG, Harpf AE, Keller RS, Vu HT, Shai SY, Loftus JC, Ross RS. Striated muscle-specific beta(1D)-integrin and FAK are involved in cardiac myocyte hypertrophic response pathway. Am J Physiol Heart Circ Physiol 279: H2916–H2926, 2000. [DOI] [PubMed] [Google Scholar]
- 23. Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386, 2000. [DOI] [PubMed] [Google Scholar]
- 24. Secor SM. Gastric function and its contribution to the postprandial metabolic response of the Burmese python Python molurus. J Exp Biol 206: 1621–1630, 2003. [DOI] [PubMed] [Google Scholar]
- 25. Secor SM, Diamond J. A vertebrate model of extreme physiological regulation. Nature 395: 659–662, 1998. [DOI] [PubMed] [Google Scholar]
- 26. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res 19: 1117–1123, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vakili BA, Okin PM, Devereux RB. Prognostic implications of left ventricular hypertrophy. Am Heart J 141: 334–341, 2001. [DOI] [PubMed] [Google Scholar]
- 28. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weiss A, Leinwand LA. The mammalian myosin heavy chain gene family. Annu Rev Cell Dev Biol 12: 417–439, 1996. [DOI] [PubMed] [Google Scholar]
- 30. Wollert KC, Drexler H. Regulation of cardiac remodeling by nitric oxide: focus on cardiac myocyte hypertrophy and apoptosis. Heart Fail Rev 7: 317–325, 2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.