Abstract
Understanding the prevailing mutational mechanisms responsible for human genome structural variation requires uniformity in the discovery of allelic variants and precision in terms of breakpoint delineation. We develop a resource based on capillary end-sequencing of 13.8 million fosmid clones from 17 human genomes and characterize the complete sequence of 1,054 large structural variants corresponding to 589 deletions, 384 insertions, and 81 inversions. We analyze the 2,081 breakpoint junctions and infer potential mechanism of origin. Three mechanisms account for the bulk of germline structural variation: microhomology-mediated processes involving short (2–20 bp) stretches of sequence (28%), non-allelic homologous recombination (NAHR) (22%) and L1 retrotransposition (19%). The high quality and long-range continuity of the sequence reveals more complex mutational mechanisms including repeat-mediated inversions and gene conversion that are most often missed by other methods including comparative genomic hybridization, SNP microarrays and next-generation sequencing.
Introduction
Despite significant advances in the discovery and genotyping of human genome structural variation, only a small fraction of common structural variation has been resolved at the sequence level (Conrad et al., 2010b; Freeman et al., 2006; Itsara et al., 2009; Kidd et al., 2008; Lam et al., 2010; McCarroll et al., 2008b; Redon et al., 2006). The majority of human genome structural variation has been discovered using SNP microarrays and array comparative genomic hybridization (arrayCGH), approaches that provide limited information about the precise structure and location of identified variants. Due to their dependence on the reference genome, array-based approaches preferentially detect deletions over insertions and are unable to directly detect copy-number neutral events such as inversions. Higher-density array platforms give a better estimation of variant sizes but most breakpoints cannot be resolved at a scale finer than 50-bp regions (Conrad et al., 2010b), while targeted next-generation sequencing approaches have difficulty resolving breakpoints within homologous segments (Conrad et al., 2010a). These methodological biases threaten to skew our understanding of the underlying mechanisms responsible for the formation of structural variation and limit our ability to comprehensively discover and genotype this form of genetic variation.
We resolve the breakpoints of 1,054 structural variants based on capillary sequencing of clone inserts. The high-quality sequence of contiguous variant haplotypes allows alternative structures to be included in future human genome assemblies and provides the breakpoint resolution necessary to accurately genotype these variants in sequence data generated from next-generation sequencing platforms. The sequences and the associated clones also provide a resource for assessing future methods for structural variation discovery.
Results
The Human Genome Structural Variation Clone Resource
The high quality of the reference human genome is due, in large part, to the fact that it was assembled based on capillary sequencing of individual large-insert clones whose complete sequence was resolved prior to final genome assembly. This strategy allowed complex duplicated and repetitive regions to be incorporated that were missed by other approaches (Istrail et al., 2004; She et al., 2004). Since genome structural variation is similarly biased to these regions, we proposed that developing clone libraries for a modest number of additional genomes would serve as a valuable resource for characterizing complex and difficult-to-assay regions of genome structural variation (Eichler et al., 2007). The overall strategy involved the construction of individual genome libraries using a fosmid cloning vector (40-kbp inserts) and capillary sequencing of the ends of the inserts to generate a high-quality end-sequence pair. Discrepancies in the length and orientation of these mapped end-sequence pairs with respect to the reference genome serve as signatures of copy-number variation and inversion, respectively. Since the underlying clones can be retrieved, the complete sequence context of the discovered structural variant can also be obtained. Previously, we discovered and cloned 1,695 structural variants using fosmid libraries derived from nine individuals and presented sequence of 261 structural variants (Kidd et al., 2008; Tuzun et al., 2005). We expand this resource to include capillary end-sequencing of 4.1 million additional fosmid clones from eight additional human genomes (Supplementary Table 1). The combined set includes 13.8 million clones derived from the genomes of six Yoruba Nigerians, five CEPH European, three Japanese, two Han Chinese and one individual of unknown ancestry.
Structural Variant Alleles
Using this resource, we searched for clusters of clones that suggest a structural difference when compared to the reference. We discovered a total of 2,051 discordant regions (Supplementary Table 1) having support from multiple clones for a structure different from the reference genome. The size distribution of the fosmid clone inserts limited us to the detection of structural variants greater than 5 kbp in length. Inversions also tend to be biased to larger events due to the probability of capturing a breakpoint by a pair of end-sequences. While there is no upper bound in the detection of deletions and inversions the direct capturing of insertions larger than the insert size of the clone (40 kb) requires specialized approaches. For example, new tandem duplications may be identified using an everted clone mapping signature (Supplementary Figure 1) (Cooper et al., 2008) and insertions of novel human sequence may be identified by read-pairs for which only one end maps (Kidd et al., 2010).
We targeted 1,054 structural variants (Supplementary Table 1) from nine human genomes and completely sequenced the inserts of 1,167 fosmid clones (46.4 Mb of sequence). We identified 81 loci for which breakpoints could not be resolved due to difficulty in clone assembly and the limits of 40-kb fosmid inserts (see Supplementary Experimental Procedures). We defined breakpoints relative to the reference genome assembly following a two-stage procedure (Kidd et al., 2010) (Figure 1 and Supplementary Table 2). We initially distinguished copy-number changes (n = 973 insertion/deletions) from balanced genome structural variants (81 inversions) (Figure 2). The analyzed variants altered 95 gene structures. We estimate that 1.04% (11/1,054) of the sequenced alleles are already known risk factors for common and rare human diseases (Figure 3, Supplementary Table 3).
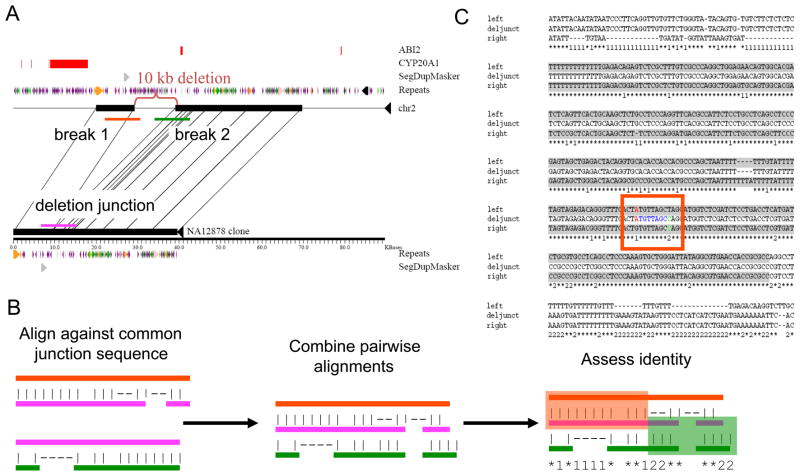
Figure 1. Sequence and breakpoint analyses.
Variant breakpoints were defined based on alignments of sequences from the sequenced insertion and deletion alleles. For example, (A) the sequence of fosmid clone AC207429 is compared with sequence from the corresponding region on chr2. A 10-kb deletion, relative to the reference sequence, is readily apparent (indicated by the red bracket). The position of segmental duplications, common repeats (LINEs are green, SINEs are purple, and LTR elements are orange), and RefSeq exons are shown. Sequence segments corresponding to three different breakpoint regions (red, green and purple bars) are extracted for further analysis. (B) The sequence across the variant junction is aligned against each of the other two sequences and the resulting pairwise alignments are merged. The pattern of sequence identity is assessed to identify the positions where the junction sequence switches from being a better match to the first breakpoint to being a better match to sequence from the second breakpoint. The breakpoint coordinates correspond to the innermost positions that can be confidently assigned to be before and after the variant boundary. (C) The result of aligning the three segments depicted in (A). Alignment columns where the junction sequence matches the sequence from the first (left-most) breakpoint are indicated by a ‘1’ while alignment columns where the junction sequence matches the second (right-most) breakpoint are indicated by a ‘2’. Positions where all three sequences are the same are indicated by an asterisk ‘*’. The red square highlights the position of the breakpoint coordinates (highlighted in red and green text). The two breakpoints are separated by seven nucleotides found at both breakpoints with perfect identity (blue text). Highlighted in gray is a 293-bp segment present at both breakpoints with a sequence identity of 91%. See also Tables S2, and S7.
Figure 2. Sequenced structural variant alleles.
(A) Size distribution for 1,054 sequenced structural variants. Insertions, deletions, and inversions relative to the genome reference assembly are depicted separately. Note that the bins are not of equal sizes. The mean size of the sequenced variants is 14.9 kb for deletions, 6.1 kb for insertions, and 196 kb for inversions. Our variant selection methodology can only identify deletions larger than ~5 kb and insertions from ~5 kb to ~40 kb in size and is biased against inversions smaller than ~40 kb. (B) The relationship between the donor site of transduced sequences and LINE insertion position are given for 30 events with a match to hg18 using BLAT. Relationships are shown for 20 LINE insertions in library source individuals relative to the reference (blue lines) and for 10 insertions in the genome reference (red lines). The blue circles represent three different loci associated with multiple distinct LINE insertions. See also Figure S1 and Table S1.
Figure 3. Examples of sequenced variants.
Examples of the complete sequence of structural variant alleles that have been associated with disease risk, including (A) a 45.5-kb deletion upstream of NEGR1, (B) a 72-kb deletion of RHD, (C) a 3.9-kb and a 20.1-kb deletion upstream of IRGM, and (D) a 32-kb deletion of LCE3C. See also Table S3.
Breakpoint Features
Using the 40 kbp of clone-based sequence, we examined the sequence features and inferred potential mechanism of origin for these variants (Table 1). We identified 30 variants associated with the expansion or contraction of a variable number of tandem repeats (VNTRs) (Buard et al., 2000; Jeffreys et al., 1994; Richard et al., 2008). VNTR repeat units ranged from 17 bp to 6.5 kbp with copy numbers ranging from 1 to 319 copies. We identified 198 events (20% of the total insertion/deletions) that we classified as being the result of L1 retrotransposition. Each of the 198 L1 elements associated with the retrotransposition events has a sequence identity of at least 97.5% when compared to the L1.3 reference sequence and 152 are at least 6 kb in size, consistent with full-length elements that may be capable of subsequent retrotransposition (Beck et al., 2010). We find evidence for transduction of flanking sequence for 20% (40/198) of the sites, with the transduced segment size ranging from 45 to 968 nucleotides (median of 81.5) (Goodier et al., 2000; Moran et al., 1999; Pickeral et al., 2000). Using the transduced sequence as a marker, we identified the potential donor location for 30 of these retrotranspositions (20 insertions in the fosmid source sample and 10 insertions in the reference genome). We identified three positions that have each given rise to multiple, LINE insertions (Figure 2B) suggesting the presence of L1 donor hotspots. We note that 11 of the 20 L1 insertions in the fosmid source (including the three recurrent L1 donors) correspond to elements that have been functionally determined to represent ‘hot’ L1 according to assays performed by Beck et al., 2010. We found two events consistent with the insertion of an intact HERV-K element: one insertion in the reference sequence (as indicated by clone AC209281) and an insertion contained in clone AC226770. Both events showed less than 1% divergence from the HERV-K sequence (Dewannieux et al., 2006) and were flanked by long terminal repeats (Tristem, 2000). Our discovery size thresholds (>5 kbp) preclude the identification of smaller retrotransposition events arising from SVA or Alu repeats that are common when smaller structural variants are considered (Bennett et al., 2008; Korbel et al., 2007; Lam et al., 2010; Mills et al., 2006).
Table 1. Summary of events and inferred mechanisms.
The number of events that fall into each breakpoint class is given. See also Table S6.
| Event Classification | Insertions and Deletions | Inversions | Potential Mechanisms |
|---|---|---|---|
| Retroelements | |||
| L1 | 198 (20.3%) | NA | Retrotransposition |
| HERVK | 2 (0.2%) | NA | Retrotransposition |
| VNTR | 30 (3.1%) | Minisatellite, NAHR | |
| CLASS I (no additional sequence at breakpoint) | 590 (60.6%) | 74 (91.3%) | |
| 0 or 1 matching nucleotides | 82 (8.4%) | 10 (12.3%) | NHEJ |
| 2–20 matching nucleotides | 289 (29.7%) | 8 (9.9%) | NHEJ, MMEJ |
| 21–100 matching nucleotides | 28 (2.9%) | 0 | NAHR, other |
| 101–199 matching nucleotides | 14 (1.4%) | 0 | NAHR, other |
| >=200 (NAHR) | 177 (18.2%) | 56 (69.1%) | NAHR |
| CLASS 2 (additional sequence at breakpoint) | 153 (15.7%) | 7 (8.6%) | |
| 1 to 10 additional nucleotides | 76 (7.8%) | 2 (2.5%) | NHEJ |
| >10 additional nucleotides | 77 (7.9%) | 5 (6.2%) | NHEJ, FoSTeS,template switching |
| TOTAL | 973 | 81 | |
We divided the remaining 824 structural variants into two broad categories. Class I consists of variants with no additional sequence at the breakpoint junction (Figure 4,A–D, Supplementary Figure 2). Class II variants contain additional sequence found across the variant junction that is not present at either of the other variant breakpoints (Figure 4E–G). We also assessed the presence of extended sequence homology and the extent of matching sequence at the breakpoints. We note that ‘microhomology’ is a qualitative term without clear delineation as 1 or 2 bp matches are expected to occur often by chance (Figure 4) and a range of homologous match lengths is observed (Conrad et al., 2010a; Lam et al., 2010). Similarly, there is ambiguity in assigning events to potential mechanisms based solely on the length of homologous segments. Consequently, we bin events based on observed ranges of homology and consider assignment to specific mechanisms as speculative.
Figure 4. Variant breakpoint analyses.
Class-I variants (A–D) are defined as those without additional nucleotides at the breakpoint. (A) A histogram of the extent of matching breakpoint sequence (black) and extended breakpoint homology (gray) is shown for 590 class-I insertion/deletion events. The red line corresponds to the expected distribution of breakpoint match lengths found from 100 random permutations. Note that bin sizes are not equal. The increase in extended homology segments 250–299 bp in length corresponds to variants having Alus at their breakpoints. (B) As in (A) zoomed in to show variants having 20 bp or less matching sequence. (C) Box plot of variant size partitioned by length of extended breakpoint homology for 590 class-I insertion-deletion variants (red line : median; blue box : interquartile range; whiskers : within 1.5X interquartile range). (D) Breakpoint density map within a consensus Alu repeat sequence based on 269 copy-number variant events (blue box : RNA pol III promoter, black boxes : AT-rich segment between the two monomers that make up the Alu element and the poly A tail, purple box : position of motif (CCNCCNTNNCCNC) found in some Alus and associated with recombination hotspots (Myers et al., 2008)). Class-II variants (E–G) contain additional sequence across the breakpoint junction. (A) A class-II variant containing a 55 nucleotide-long stretch of additional sequence (in blue) that is not found at either breakpoint. (B) Histogram of the length of additional sequence found at variant breakpoints (black) and the length of detected extended homology between breakpoint sequences (gray) for 153 class-II insertion/deletion variants. (C) Genomic location for class-II unmatched sequences (>20 bp) associated with deletions. The black lines connect the positions of a class-II deletion variant (relative to the genome assembly) and the corresponding location where the additional sequence across the variant breakpoint can be found. The relationship for 31 deletion variants is depicted. One event involves a match to unlocalized sequence on chromosome 1 (chr1_rand). See also Figure S2, Table S4, and Table S5.
Among the class I events, 49% (289/590) of insertion-deletion variants contain 2–20 bp of matching sequence, indicating that microhomology-mediated mechanisms, such as microhomology-mediated end joining (MMEJ) contribute to a substantial fraction (30%) of human structural variation (Table 1) (Hastings et al., 2009; McVey and Lee, 2008; Payen et al., 2008; Roth and Wilson, 1986). Although there is large overlap in the variant size when broken down by extent of homologous sequence (Figure 4C), we find that, as a class, the mean size of events associated with microhomology (2–20 bp of matching sequence, n = 289, mean size 9.7 kb) is significantly smaller (p = 0.02926, two sample t-test) than those showing a hallmark of NAHR (>=200 bp of matching sequence, n = 177, mean size is 21.0 kbp). The analyzed inversions are overwhelming driven by large homologous segments with 69% (56/81) of all analyzed inversions containing stretches of matching sequence at least 200 bp in length. In contrast, only 30% (177/590) of the class-I insertion-deletion variants contain matching breakpoint sequences of at least that length. It is important to note, however, that our clone end-sequence mapping strategy is biased toward the detection of larger inversions when compared to insertion/deletions. This is a direct consequence of the probability of capturing a breakpoint that diminishes when inversions become smaller than the clone insert size. Overall, we find that younger Alu events and segmental duplications contribute most significantly to the process of NAHR (Supplementary Table 4) as expected due to their higher levels of sequence identity. The strongest enrichment is found for paired Alu repeats at each breakpoint (5.2-fold enrichment). If each breakpoint is treated separately, rather than requiring that an element of the same subfamily be present at both breakpoints of a variant, then AluY also shows a substantial degree of enrichment (2.6-fold, Supplementary Table 4). Since AluY is the most recently active Alu family, dispersed AluY elements are expected to have a higher degree of sequence identity than other Alu families (Batzer and Deininger, 2002; Cordaux and Batzer, 2009). Closer examination of the distribution of breakpoints within individual Alus reveals a non-uniform pattern of breakpoint density (Figure 3D). The highest density of breakpoints occurs near the position of a sequence motif (CCNCCNTNNCCNC) that has been associated with meiotic recombination hotspots and is found in some Alu elements (Myers et al., 2008) and has also been observed for rearrangements between human and chimpanzee (Han et al., 2007; Sen et al., 2006).
We find that 16% (153/973) of the insertion-deletion variants and 9% (7/81) of the inversions contain additional sequence at the variant breakpoints (class II events; Figure 4). Many of the additional insertion sequences are relatively short in length, consistent with non-template directed repair associated with non-homologous end-joining (Figure 4B). For these shorter sequences, no inference could be made as to the source of the additions. However, 41% of all class-II variants (66/160) contain additional sequence at the junction at least 20 bp in length. Of these longer fragments, 88% (58/66) map to another location within the human genome. Since we are limited in this study to directly capturing the breakpoints of insertions smaller than 40 kb, we repeated this comparison using only deletions relative to the assembly where we expect to have less of a bias in terms of variant size. We find that the additional junction sequences for 30 of 39 class-II deletion events at least 20 bp long map elsewhere in the genome. 73% (22/30) are found on the same chromosome as the variant. In fact, eight of the insertions map less than 1 kb away from the variant breakpoint (Figure 4G, Supplementary Table 5) and all 22 are less than 250 kbp from the breakpoint. This pattern suggests the action of a replication-associated process that involves template switching or strand invasion (Hastings et al., 2009; Lee et al., 2007; Smith et al., 2007). In contrast to the class I events, only 2% of the class II events (3/160) contained stretches of homologous sequence flanking the breakpoint insertion confirming they arose by mechanisms other than NAHR. Interestingly, if we examine the sequence context of these regions, we find that 20% (30/153) of class II events map within 5 kbp of a segmental duplication. This represents a significant enrichment for proximity to duplicated sequence (p <0.002 based on comparisons with randomly sampled sequences) indicating that regions flanking segmental duplications may be generally more unstable and susceptible to multiple mutational processes such as template switching during replication (Itsara et al., 2009; Lee et al., 2007; Payen et al., 2008).
Gene Conversion and Structural Variation
During our analysis of putative NAHR events, we identified 10 structural variants having a complex pattern of exchange inconsistent with a simple model of unequal crossover. The breakpoint region contains an interleaved pattern of alternating patches of sequences from flanking homologous segments (Figure 5). These patterns are reminiscent of multiple rounds of gene conversion, although each of these events was also associated with a copy-number variant event. Using paralogous sequence variants that distinguish the 5’ and 3’ homologous segments, we investigated the overall extent of this non-allelic exchange (referred to as the conversion tract length) and the number of switches before unambiguous homology to the 5’ or 3’ end was re-established. We determined that most (6/10) of the conversion tracts were relatively short (200–600 bp in length) with a relatively consistent number (4–6) and length (30–40 bp) of switches before clear boundaries at the 5’ and 3’ could be re-established (Supplementary Figure 4). Seven of these events have breakpoints that map within segmental duplications and the remaining three have breakpoints that map within LINEs. Three of the variants contained at least 10 switches. One variant (AC212911) showed the largest associated conversion tract with a remarkable 182 switches extending over 7.9 kb (Figure 5D). We sequenced the deletion allele using fosmids derived from three different individuals for one event (AC226182). Each of the three deletion haplotypes contained identical patterns of interleaved sequence, a finding that is consistent with the creation of the pattern at the time of variant formation, or shortly thereafter, rather than as a result of a continual conversion process between deletion and insertion alleles leading to a diverse set of related molecules over time (Supplementary Figure 3). It is also possible that the conversion pattern arose before the formation of the structural variant and that the pattern we observe in sequenced variants is merely incidental or the result of a series of mismatch-repair processes prior to variant formation. Nevertheless, the observed switch pattern is reminiscent of patterns of toggling previously observed at some LINE insertions (Gilbert et al., 2005; Gilbert et al., 2002; Symer et al., 2002) and suggests a mechanism of serial strand invasion/repair during the rearrangement process.
Figure 5. Breakpoint assessment using paralogous sequence variants.
(A) Schematic comparison of the structures of the insertion and deletion haplotypes of a putative NAHR variant. The blue and red boxes represent homologous sequences present at the breakpoints, which mediate the rearrangement. The blue and red vertical lines identify paralogous sequence variants that distinguish the 5’ and 3’ copy of the matching sequence. Scanning along the deletion allele, which is missing the intervening sequence, one observes single nucleotides specific with the 5’ breakpoint, followed by a stretch of sequence that matches both, then sequences that match the 3’ breakpoint. (B) Representation for three variants showing a ‘classic’ NAHR pattern. Each line represents the deletion allele corresponding to the indicated variant. We note a single unexpected paralogous sequence variant mismatch located 145 bp past the 3’ breakpoint, which could correspond to a SNP, short gene conversion, or alignment artifact due to the placement of indels between 5’ and 3’ segments. (C) Representation of four variants having breakpoints that show a pattern of alternating sequences that match the 3’ then 5’ breakpoints. (D) An extreme pattern of alternating matches that contains 182 switches spanning over a 7.9-kb interval. (E) Rearrangements associated with gene conversion. See also Figure S3.
Comparison with Other Genome-wide Studies and Ascertainment Biases
In this study we focused on systematically characterizing large structural variants at the single basepair level. In order to identify events that may have been missed by the fosmid end-sequence pair approach, we compared our set of structural variants to other studies that have discovered and genotyped copy-number variants in the same DNA samples. We focused on five individuals analyzed by fosmid end-sequencing (Kidd et al., 2008), Affymetrix 6.0 microarray (McCarroll et al., 2008b), and high-density oligonucleotide arrayCGH (Conrad et al., 2010b). A comparison of the three studies shows that 11–65% of discovered variants are unique to a single study and corresponding experimental platform (Figure 6). The limited overlap should not be surprising since each approach preferentially identifies a subset of the total collection of genomic variation. For example, the fosmid end-sequence pair (ESP) mapping approach can detect insertions of sequence not represented in the genome assembly (Kidd et al., 2008; Kidd et al., 2010), as well as balanced events such as inversions (not depicted in Supplementary Figure 6), whereas array approaches can more readily detect copy-number variation caused by large duplications.
Figure 6. Comparison of events detected from three studies.
Only variants estimated to be >5 kb are included. The Kidd et al. set includes sites of insertion or deletion in one of the five samples relative to the genome assembly; the Conrad et al. set includes gains and losses in at least one of the five samples relative to a reference arrayCGH sample; and the McCarroll et al. set includes CNVs that were successfully genotyped on the Affymetrix 6.0 platform and are variable among the five included samples. Prior to comparison, the variant sets within each study were merged into a single, nonredundant interval set, and any overlap among regions between studies was sufficient regardless of which sample a variant was detected in.
Differences in ascertainment extend to the resolution of breakpoint sequences. The sequenced variants described in this manuscript include 237 of the regions targeted for array capture and 454-sequencing (Conrad et al., 2010a). 70 of these targeted events were successfully resolved by breakpoint array-capture experiments (Supplementary Table 6), with none of the events containing extended breakpoint homology successfully resolved by next-generation sequencing.
We also reassessed regions discovered by other studies that were missed by the fosmid ESP approach. Using the standard fosmid analysis criteria (two or more discordant clones with sufficient quality) (Tuzun et al., 2005), an overlapping deletion site is only identified for 53% (631/1,193) of the corresponding deletion genotypes reported by Conrad et al. (Conrad et al., 2010b). The intersection rate increases to 75% (900/1,193 sample level genotypes) if individual deletion clones are considered with reduced quality thresholds. This suggests that much of the variation missed by the fosmid ESP approach is a result of random fluctuations in the level of clone coverage and the quality of individual sequencing reads (Cooper et al., 2008).
Experimental approaches to discover structural variation can have reduced sensitivity in regions of segmental duplication due to difficulty in uniquely mapping reads or designing array probes (Cooper et al., 2008; Kidd et al., 2008; Tuzun et al., 2005). We compared the validated structural variants from Kidd et al. with those found by read-depth approaches (Alkan et al., 2009). Alkan et al. identified 113 genes that differ in copy number among three individuals. Only 38% of the genes greater than 5 kb (26/69) identified as copy-number variable by read-depth intersect with a structural variant reported in Kidd et al. This result indicates that even the fosmid ESP approach has underascertained copy-number variation associated with the most variable duplicated sequences.
We identified 81 loci during our sequence analysis with evidence for a non-reference structure for which we could not unambiguously define the variant breakpoint (see Supplemental Experimental Procedures). Of these 81 loci, 63 are associated with segmental duplications, including 10 examples of tandem duplications. We note that 23 of these duplication-containing loci map near gaps in the NCBI build36 genome assembly or to sequences that have been assigned to a chromosome but not fully integrated into the genome reference sequence. Duplication-mediated copy-number variation remains underascertained in terms of sequence-level resolution of variant haplotypes and mutational mechanism analysis. If we adjust for these biases, we estimate that the fosmid ESP approach has minimally missed at least 106 structural variants associated with segmental duplications.
Discussion
We describe a clone resource from 17 human DNA samples that provides 135-fold physical coverage of the human genome. The corresponding catalogue and clones can be used to further characterize almost any segment of human euchromatin. We used this resource to assess breakpoint characteristics of 1,054 events. The nature of our experimental design permitted us to discover more events mediated by larger segments of homology providing a more complete assessment of human genetic variation. Of particular interest are complex events whose sequence features have been difficult to previously assess at a genome-wide level. The high quality and length of the sequenced fosmids combined with defined paralogous sequence events allowed us to quantify alternating sequence matches suggestive of interlocus gene conversion (Bayes et al., 2003; Lagerstedt et al., 1997; Reiter et al., 1997; Visser et al., 2005).
Using this resource, we obtained the complete structure of several alleles that have been associated with disease, including a deletion variant upstream of the NEGR1 gene associated with increased body mass index (Willer et al., 2009) (clone AC210916), two deletion polymorphisms upstream of the IRGM gene associated with Crohn’s disease (Barrett et al., 2008; Bekpen et al., 2009; McCarroll et al., 2008a) (clone AC207974), and the deletion of the LCE3B and LCE3C genes. In total, we conservatively estimate that 1.04% (11/1,054) of the discovered variants are associated with disease. This yield of disease-causing alleles rivals that found by genome wide association studies using SNPs, which has identified 779 genome-wide associations based on genotyping of at least 100,000 SNPs (http://www.genome.gov/multimedia/illustrations/GWAS2010-3.pdf). Although the functional significance of many of the other structural variants remains to be determined, the clone resource and availability of the complete sequence of variant haplotypes will facilitate future disease association through the rapid design of assays to test for association with disease (Abe et al., 2009; An et al., 2009; Kidd et al., 2007) or direct comparison with short sequencing reads from next-generation sequence platforms (Kidd et al., 2010; Lam et al., 2010).
We investigated this approach for 1,024 non-VNTR sequenced structural variants (Supplementary Table 7) and found that 71% (726/1,024) of the variants are uniquely identifiable with a read-length of 36 bp and uniqueness threshold permitting up to one substitution. This includes 32 inversions—balanced events that are invisible to array-based genotyping approaches. As read lengths increase to 100 bp, we estimate that 88% (902/1,024) of these variants could be genotyped. The construction of complete alternative haplotypes then facilitates the use of read-pair information to distinguish among distinct structural configurations (Antonacci et al., 2010).
Although, short-read technologies may miss some of the breakpoint sequences, there are many advantages to the application of short-read technology to genome structural variation. This includes the detection of thousands more events per individual genome, especially variants below the detection threshold of the fosmid ESP approach. The dynamic range response and the sequence specificity of next-generation sequencing allow absolute copy number and the identity of duplicated genes to be accurately predicted. One of the strengths of this clone resource, however, is that it permits the iterative assessment of predicted variants. Clones corresponding to structural variants discovered by other methods applied to these 17 individuals, including newly developed approaches such as methods for identifying transposon insertions (Huang et al., 2010; Witherspoon et al., 2010), may be retrieved providing complete sequence information for additional events and thereby provide a resource set of sequenced variant haplotypes. The availability of the underlying clones and potential location of the variant within a specific DNA sample provides an approach for more fully exploring the genetic architecture and mutational properties of these regions. Thus, we predict that such a resource will be a valuable complement for understanding the true complexity of human genetic variation as human genomes become routinely sequenced using short-read sequencing technology.
Experimental Procedures
Identifying and Sequencing Variant Clones
Sites of structural variation, relative to the reference genome assembly, were identified through fosmid end-sequence pair mapping. Briefly, genomic DNA was obtained from transformed lymphoblastoid cell lines (available from the Coriell Cell Repository) and approximately 1 million 40-kb fragments from each individual were cloned into fosmid vectors. Paired end-sequences were obtained from both ends of each fragment using standard capillary sequencing. The resulting end-sequence pairs were mapped onto the reference assembly to identify clusters of multiple clones from a single individual showing the same type of discordancy (Tuzun et al., 2005). We previously identified 1,695 structural variants that have been experimentally validated (Kidd et al., 2008). In this manuscript, we focus on 1,054 events for which complete, finished clone sequence is available. High-quality finished sequence was obtained for all fosmid inserts using capillary-based shotgun sequencing and assembly using the procedures established for sequencing clones as part of the Human Genome Project. Some sequenced clones contain gaps in simple sequence repeats that are not related to the detected structural variants. For one individual, NA18956, additional clones were selected using a relaxed threshold of two standard deviations larger or smaller than the observed mean insert. In some cases, multiple clones were sequenced for a single event, whereas in other loci a single clone sequence appeared to contain multiple distinct variants relative to the genome reference.
Identifying Variant Breakpoints
Sequences of individual fosmid inserts were initially compared to the NCBI build36 (UCSC hg18) genome reference assembly using the program miropeats (Parsons, 1995) with a match threshold of -s 400. Images summarizing these comparisons that included annotations of the repeat content, predicted and observed segmental duplications (using DupMasker (Jiang et al., 2008)), and RefSeq exons were prepared and examined to identify clones harboring a structural difference relative to the build36. Clones that mapped to unassigned or ‘random’ parts of the reference genome or that do not contain an entire event (such as clones that contain one edge of a tandem duplication) were omitted from analysis. Approximate variant breakpoints were determined utilizing the context provided by long stretches of contiguous matching sequence. In many cases, the pattern of common repeats or segmental duplications was a useful aid in this assessment.
For each variant, three sequences were extracted and aligned. In the case of a deletion, two sequences at the variant boundaries are extracted from the genome assembly and one sequence (termed the deletion junction sequence) is extracted from the clone. For insertions, the junction sequence is extracted from the genome assembly and two sequences corresponding to the variant boundaries in the fosmid clone are extracted. For inversions, a single breakpoint is directly captured in the sequenced clone. However, the position of the other breakpoint can be inferred based on a comparison with the genome assembly. Thus, for inversions, two sequences are extracted from the assembly at the edges of the inferred inversion and the third sequence is extracted from the clone. For inversion analysis, one of the chromosome-derived segments is reverse-complimented prior to alignment.
An alignment is then constructed from the extracted breakpoint segments (Kidd et al., 2010). First, an optimal global alignment is computed between the ‘junction’ fragment and each of the other two fragments using the program needle with default parameters (Rice et al., 2000). These alignments are then merged to yield a single, three-sequence alignment. From this alignment, the innermost positions that can be confidently assigned to be before and after the structural variant are identified. The resulting positions are used to define membership as a class-I or class-II variant and correspond to the ‘breakpoint match length’ depicted in Figure 4. Extended breakpoint homology was determined using both cross_match (http://www.phrap.org/, -minmatch 4 -maxmatch 4 -minscore 20 -masklevel 100 -raw -word_raw) without complexity-adjusted scoring (Chiaromonte et al., 2002) and bl2seq (-W 7 -g F -F F -S 1 -e 20) to identify the longest extent and identity of additional matching sequence (termed ‘extended breakpoint homology’) that included the two breakpoints. For putative NAHR events, we additionally determined the longest stretch of 100% perfect identity, as well a parsimonious matching metric to account for mutations after the time of variant formation (Supplementary Figure 2).
VNTR and Retroelement Analysis
Events associated with tandem repeats were characterized using the output from miropeats (Parsons, 1995), tandem repeats finder (Benson, 1999), DupMasker (Jiang et al., 2008), and RepeatMasker(Smit et al., 1996–2004). Potential L1 insertions were characterized using both the TSDfinder program (Szak et al., 2002) and the results of the breakpoint identification and characterization process.
Genotyping Structural Variants with Diagnostic K-mers
Diagnostic k-mers were identified for each variant (Supplementary Table 7) by extracting overlapping k-mers of the indicated size across each sequenced breakpoint. K-mers were then searched against the build36 genome sequence and a set of sequenced fosmids using mrsFAST (http://mrfast.sourceforge.net/). To be considered diagnostic, a k-mer must be unique (within the given edit distance threshold) to the allele variant from which it was derived (Kidd et al., 2010).
All sequence data have been deposited in GenBank under project ID 29893.
Supplementary Material
Acknowledgments
We thank Douglas Smith and the staff at Agencourt Biosciences for library production, Ewen Kirkness and staff of the J. Craig Venter Institute for end-sequence data from the JVCI library and Lin Chen for computational assistance in the mapping of end-sequence data. We thank S. Girirajan, J. Moran, and C. Payen for thoughtful discussion, T. Brown for manuscript preparation assistance, and members of the University of Washington and Washington University Genome Centers for assistance with data generation. J.M.K. is supported by a US National Science Foundation Graduate Research Fellowship. This work was supported by the US National Institutes of Health grant HG004120 to E.E.E. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interests statement
E.E.E is on the scientific advisory board for Pacific Biosciences. T.L.N. is an employee and founder of iGenix Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe H, Ochi H, Maekawa T, Hatakeyama T, Tsuge M, Kitamura S, Kimura T, Miki D, Mitsui F, Hiraga N, et al. Effects of structural variations of APOBEC3A and APOBEC3B genes in chronic hepatitis B virus infection. Hepatol Res. 2009;39:1159–1168. doi: 10.1111/j.1872-034X.2009.00566.x. [DOI] [PubMed] [Google Scholar]
- Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An P, Johnson R, Phair J, Kirk GD, Yu XF, Donfield S, Buchbinder S, Goedert JJ, Winkler CA. APOBEC3B deletion and risk of HIV-1 acquisition. J Infect Dis. 2009;200:1054–1058. doi: 10.1086/605644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonacci F, Kidd JM, Marques-Bonet T, Teague B, Ventura M, Girirajan S, Alkan C, Campbell CD, Vives L, Malig M, et al. A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat Genet. 2010;42:745–750. doi: 10.1038/ng.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Bayes M, Magano LF, Rivera N, Flores R, Perez Jurado LA. Mutational mechanisms of Williams-Beuren syndrome deletions. Am J Hum Genet. 2003;73:131–151. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 Retrotransposition Activity in Human Genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekpen C, Marques-Bonet T, Alkan C, Antonacci F, Leogrande MB, Ventura M, Kidd JM, Siswara P, Howard JC, Eichler EE. Death and resurrection of the human IRGM gene. PLoS Genet. 2009;5:e1000403. doi: 10.1371/journal.pgen.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, Devine SE. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovee D, Zhou Y, Haugen E, Wu Z, Hayden HS, Gillett W, Tuzun E, Cooper GM, Sampas N, Phelps K, et al. Closing gaps in the human genome with fosmid resources generated from multiple individuals. Nat Genet. 2008;40:96–101. doi: 10.1038/ng.2007.34. [DOI] [PubMed] [Google Scholar]
- Buard J, Shone AC, Jeffreys AJ. Meiotic recombination and flanking marker exchange at the highly unstable human minisatellite CEB1 (D2S90) Am J Hum Genet. 2000;67:333–344. doi: 10.1086/303015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaromonte F, Yap VB, Miller W. Scoring pairwise genomic sequence alignments. Pacific Symposium on Biocomputing; 2002. pp. 115–126. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010a doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010b;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet. 2008;40:1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Nickerson DA, Altshuler D, Bowcock AM, Brooks LD, Carter NP, Church DM, Felsenfeld A, Guyer M, Lee C, et al. Completing the map of human genetic variation. Nature. 2007;447:161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME, et al. Copy number variation: new insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH., Jr Transduction of 3'-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- Han K, Lee J, Meyer TJ, Wang J, Sen SK, Srikanta D, Liang P, Batzer MA. Alu recombination-mediated structural deletions in the chimpanzee genome. PLoS Genet. 2007;3:1939–1949. doi: 10.1371/journal.pgen.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CR, Schneider AM, Lu Y, Niranjan T, Shen P, Robinson MA, Steranka JP, Valle D, Civin CI, Wang T, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istrail S, Sutton GG, Florea L, Halpern AL, Mobarry CM, Lippert R, Walenz B, Shatkay H, Dew I, Miller JR, et al. Whole-genome shotgun assembly and comparison of human genome assemblies. Proc Natl Acad Sci U S A. 2004;101:1916–1921. doi: 10.1073/pnas.0307971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36:151–156. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Tamaki K, MacLeod A, Monckton DG, Neil DL, Armour JAL. Complex gene conversion events in germline mutation at human minisatellites. Nature Genet. 1994;6:136–145. doi: 10.1038/ng0294-136. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Hubley R, Smit A, Eichler EE. DupMasker: A tool for annotating primate segmental duplications. Genome Res. 2008;18:1362–1368. doi: 10.1101/gr.078477.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Sampas N, Antonacci F, Graves T, Fulton R, Hayden HS, Alkan C, Malig M, Ventura M, Giannuzzi G, et al. Characterization of missing human genome sequences and copy-number polymorphic insertions. Nat Methods. 2010;7:365–371. doi: 10.1038/nmeth.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerstedt K, Karsten SL, Carlberg BM, Kleijer WJ, Tonnesen T, Pettersson U, Bondeson ML. Double-strand breaks may initiate the inversion mutation causing the Hunter syndrome. Hum Mol Genet. 1997;6:627–633. doi: 10.1093/hmg/6.4.627. [DOI] [PubMed] [Google Scholar]
- Lam HY, Mu XJ, Stutz AM, Tanzer A, Cayting PD, Snyder M, Kim PM, Korbel JO, Gerstein MB. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat Biotechnol. 2010;28:47–55. doi: 10.1038/nbt.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008a doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008b;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, Devine SE. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008 doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- Parsons J. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickeral OK, Makalowski W, Boguski MS, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Gibbs RA, Lupski JR. The human COX10 gene is disrupted during homologous recombination between the 24 kb proximal and distal CMT1A-REPs. Hum Mol Genet. 1997;6:1595–1603. doi: 10.1093/hmg/6.9.1595. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen SK, Han K, Wang J, Lee J, Wang H, Callinan PA, Dyer M, Cordaux R, Liang P, Batzer MA. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Jiang Z, Clark RA, Liu G, Cheng Z, Tuzun E, Church DM, Sutton G, Halpern AL, Eichler EE. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2004. [Google Scholar]
- Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3:research0052. doi: 10.1186/gb-2002-3-10-research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristem M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol. 2000;74:3715–3730. doi: 10.1128/jvi.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- Visser R, Shimokawa O, Harada N, Kinoshita A, Ohta T, Niikawa N, Matsumoto N. Identification of a 3.0-kb major recombination hotspot in patients with Sotos syndrome who carry a common 1.9-Mb microdeletion. Am J Hum Genet. 2005;76:52–67. doi: 10.1086/426950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherspoon DJ, Xing J, Zhang Y, Watkins WS, Batzer MA, Jorde LB. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genomics. 2010;11:410. doi: 10.1186/1471-2164-11-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.