Abstract
Misfolded, luminal endoplasmic reticulum (ER) proteins are retro-translocated into the cytosol and degraded by the ubiquitin/proteasome system. This ERAD-L pathway requires a protein complex consisting of the ubiquitin ligase Hrd1p, which spans the ER membrane multiple times, and the membrane proteins Hrd3p, Usa1p, and Der1p. Here, we show that Hrd1p is the central membrane component in ERAD-L; its overexpression bypasses the need for the other components of the Hrd1p-complex. Hrd1p function requires its oligomerization, which in wild type cells is facilitated by Usa1p. Site-specific photocrosslinking indicates that, at early stages of retro-translocation, Hrd1p interacts with a substrate segment close to the degradation signal. This interaction follows the delivery of substrate through other ERAD components, requires the presence of transmembrane segments of Hrd1p, and depends on both the ubiquitin ligase activity of Hrd1p and the function of the Cdc48p ATPase complex. Our results suggest a model for how Hrd1p promotes polypeptide movement through the ER membrane.
Introduction
Misfolded proteins in the lumen or membrane of the endoplasmic reticulum (ER) are ultimately retro-translocated into the cytosol, poly-ubiquitinated, and degraded by the proteasome (for review, see Hirsch et al., 2009; Xie and Ng, 2010). The process is called ERAD (for ER-asociated protein degradation) and is conserved in all eukaryotes. In S. cerevisiae, substrates use three ERAD pathways (ERAD-L, -M, and -C), depending on whether their misfolded domain is located in the ER lumen, ER membrane, or on the cytoplasmic side of the ER membrane (Carvalho et al., 2006; Taxis et al., 2003; Vashist and Ng, 2004). ERAD-L requires a hetero-tetrameric membrane protein complex, the Hrd1p-complex, comprised of the ubiquitin ligase Hrd1p, as well as Hrd3p, Usa1p, and Der1p (Bays et al., 2001a; Bordallo et al., 1998; Carvalho et al., 2006). ERAD-M requires only a subset of these components. Finally, ERAD-C uses a different ubiquitin ligase (Doa10p) (Swanson et al., 2001). Following poly-ubiquitination, these pathways converge at an ATPase complex, consisting of the ATPase Cdc48p and two cofactors (Ufd1p and Npl4p) (Bays et al., 2001b; Braun et al., 2002; Carvalho et al., 2006; Jarosch et al., 2002; Rabinovich et al., 2002; Ye et al., 2001).
Although most, if not all, components have been identified, the molecular mechanisms of the ERAD pathways remain unclear. Some insight exists into the events that occur during ERAD-L on the luminal and cytoplasmic sides of the ER membrane. Misfolded, glycosylated ERAD-L substrates are initially recognized in the ER lumen. Their prolonged residence time in the ER results in the processing of their carbohydrate moiety to generate a terminal α1–6 mannose residue (Bhamidipati et al., 2005; Clerc et al., 2009; Kim et al., 2005; Quan et al., 2008; Szathmary et al., 2005). This sugar, together with the unfolded polypeptide segment surrounding the carbohydrate attachment site, constitutes the degradation signal (Xie et al., 2009). The signal is then recognized through a luminal domain of Hrd3p as well as the lectin Yos9p (Denic et al., 2006; Gauss et al., 2006a). Once the substrate appears on the cytoplasmic side of the ER membrane, the RING finger domain of Hrd1p, together with the ubiquitin conjugating enzymes Ubc7p or Ubc1p, catalyze poly-ubiquitination (Bays et al., 2001a; Bordallo et al., 1998). The modified substrate is then recognized by the Cdc48p/Ufd1p/Npl4p ATPase complex and moved into the cytosol (Bays et al., 2001b; Braun et al., 2002; Jarosch et al., 2002; Rabinovich et al., 2002; Ye et al., 2001). Finally, the substrate is delivered to the proteasome and degraded.
The events of ERAD-L occurring inside the ER membrane are unknown, particularly the mechanism by which substrates are moved through the ER membrane. It has been proposed that a retro-translocation channel is required, and several channel candidates have been considered (for review, see Hirsch et al., 2009). Among the components of the Hrd1p-complex, the most attractive candidates are Der1p and its mammalian homologs, the Derlins (Knop et al., 1996; Lilley and Ploegh, 2004; Ye et al., 2004), as well as Hrd1p, simply because they possess the largest number of transmembrane segments. In addition, some studies suggest a role for the Sec61 channel, which is normally involved in the translocation of proteins from the cytosol into the ER (Pilon et al., 1997; Schafer and Wolf, 2009; Wiertz et al., 1996; Willer et al., 2008; Zhou and Schekman, 1999). However, there is no direct evidence in support of any of these candidates and it is not even clear whether a channel exists. In fact, it has been suggested that translocation is instead linked to the formation of lipid droplets (Ploegh, 2007). How the driving force for retro-translocation is provided is also unclear. While it is conceivable that the Cdc48p ATPase pulls on a poly-ubiquitinated substrate once it appears on the cytosolic side of the membrane, it is mysterious how energy would be provided for moving the polypeptide through the membrane to make it accessible to the ubiquitination machinery. Finally, and more generally, the specific functions of the individual components of the Hrd1p-complex are unknown and it is unclear whether they act in a temporal order during retro-translocation of ERAD-L substrates.
Here, we show that Hrd1p is the central membrane component in the ERAD-L process and propose a model for polypeptide movement through the ER membrane.
Results
Bypassing ERAD components by Hrd1p overexpression
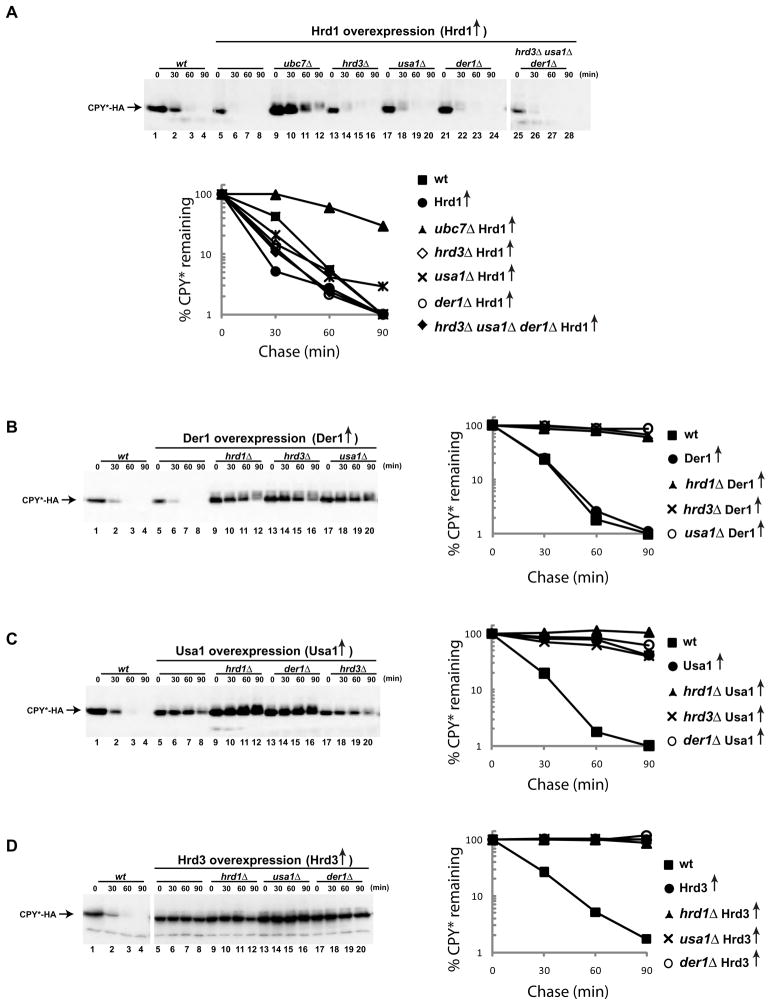
In wild type S. cerevisiae cells, all four components of the Hrd1p complex (Hrd1p, Hrd3p, Usa1p, and Der1p) are essential for the degradation of ERAD-L substrates (Knop et al., 1996; Taxis et al., 2003; Gauss et al., 2006a; Carvalho et al., 2006). However, we reasoned that the overexpression of one component may make other components dispensable, a result that would indicate a functional hierarchy among these factors. These experiments were also motivated by the previous observation that overexpression of Hrd1p compensates for the absence of Hrd3p (Gardner et al., 2000; Plemper et al., 1999), although this result was interpreted as simply restoring the levels of Hrd1p, which becomes unstable in a HRD3 deletion mutant.
We tested the effect of overexpression of Hrd1p on the degradation of a well-characterized ERAD-L substrate, a misfolded version of carboxypeptidase Y (CPY*) that is tagged with a C-terminal hemagglutinin (HA) epitope (Finger et al., 1993; Ng et al., 2000). In wild type cells CPY*-HA is degraded with a half-life of ~30 min (Figure 1A, lanes 1–4 and graph). When the endogenous promoter for Hrd1p was replaced by the strong, galactose-inducible GAL1 promoter and the cells were grown in the presence of galactose, CPY*-HA degradation was accelerated (lanes 5–8). In the presence of glucose, when the Gal promoter is repressed, CPY*-HA was stable, as expected from the depletion of Hrd1p (Figure S1A). Although overexpressed Hrd1p is unstable, the steady state levels are increased by a factor of ~10 (Figure S1B). CPY*-HA degradation by overexpressed Hrd1p was much slower when the ubiquitin-conjugating enzyme Ubc7p was absent (Figure 1A, lanes 9–12) and was completely abrogated when an essential cysteine in the RING finger domain of Hrd1p was mutated (Figure S1C). In addition, degradation was attenuated in a cdc48 mutant (Figure S1C). Thus, the requirements for ubiquitin-ligase activity by Hrd1p and for the function of the Cdc48 ATPase are maintained when Hrd1p is overexpressed.
Figure 1. Bypassing ERAD components by Hrd1p overexpression.
(A) The degradation of the misfolded luminal ER protein CPY*-HA was followed after inhibition of protein synthesis by cycloheximide in wild type (wt) cells or in cells overexpressing Hrd1p under the GAL promoter in the presence of galactose. Where indicated, genes for ERAD components were deleted. The graph shows quantification of the data.
(B) As in (A), but with overexpression of Der1p. The overexpression was confirmed by immunoblotting with specific antibodies (not shown).
(C) As in (B), but with overexpression of Usa1p.
(D) As in (B), but with overexpression of Hrd3p.
Next, we tested CPY*-HA degradation in cells that overexpressed Hrd1p, but lacked other components of the Hrd1p-complex. As reported (Plemper et al., 1999), degradation was not affected when Hrd3p was absent (Figure 1A, lanes 13–16). Interestingly, the process was also not affected by the absence of Usa1p or Der1p (lanes 17–20 and 21–24). Moreover, even in cells that simultaneously lacked Hrd3p, Usa1p, and Der1p, the degradation of CPY*-HA was unimpeded (lanes 25–28). Degradation in the triple-deletion mutant required Hrd1p expression (Figure S1A) and ubiquitin ligase activity (Figure S1D). These experiments indicate that Hrd1p overexpression bypasses the need for the other components of the Hrd1p-complex. Similar results were obtained with KHN-HA (a soluble protein) and KWW-HA (a membrane-bound protein), both containing a misfolded luminal domain (Figure S2) (Vashist and Ng, 2004).
The bypass effect observed with Hrd1p was not seen when the other components of the Hrd1p complex were overexpressed. Der1p under the Gal promoter did not accelerate CPY*-HA degradation and did not alleviate the requirement for the other Hrd1p complex components (Figure 1B). Usa1p or Hrd3p overexpression blocked degradation of CPY*-HA (Figures 1C and 1D), indicating that excess of these components interferes with the normal function of the Hrd1p complex. Taken together, our data suggest that Hrd1p is the key component of the Hrd1p-complex, while the other subunits may have ancillary roles.
ERAD-L substrate degradation requires Hrd1p oligomers
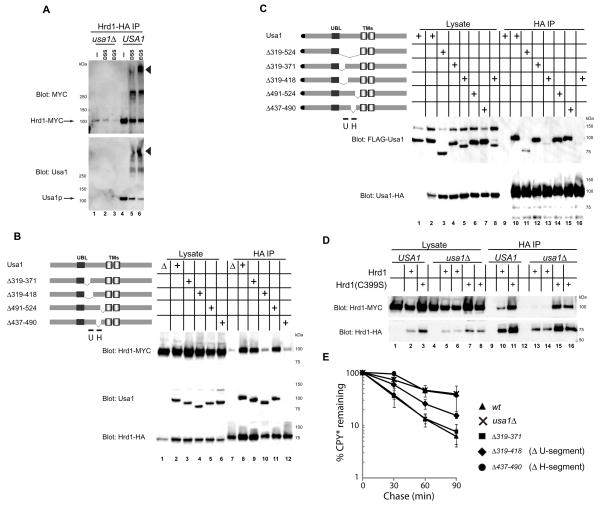
We hypothesized that Hrd1p overexpression bypasses the need of the other Hrd1p complex components because Hrd1p must oligomerize to be active in ERAD-L; in wild type cells, the oligomerization would be regulated by other components of the Hrd1p complex, whereas Hrd1p overexpression would force its spontaneous oligomerization. Both our own previous experiments and more recent results indicate that endogenous Hrd1p forms high-molecular weight complexes whose size depends on the presence of Usa1p, but not Hrd3p or Der1p (Carvalho et al., 2006; Horn et al., 2009). To investigate directly the oligomerization of Hrd1p, we expressed Myc- and HA-tagged versions of Hrd1p in the same cell, both under the endogenous promoter. Detergent-solubilized membrane extracts were then treated with bifunctional crosslinking reagents and subjected to immunoprecipitation with HA-antibodies. With extracts from wild type cells that were not treated with crosslinker, HA-antibodies precipitated Hrd1p-Myc (Figure 2A, upper panel, lane 4), as previously described (Horn et al., 2009). Upon treatment with crosslinkers, several high-molecular weight bands were observed (lanes 5 and 6). The highest molecular weight band appeared with multiple crosslinkers and contains not only Hrd1p, but also Usa1p (Figure 2A, lower panel, lanes 5 and 6). With extracts lacking Usa1p, HA-antibodies precipitated only negligible amounts of Hrd1p-Myc or the crosslinked bands (lanes 1–3). Thus, Usa1p facilitates Hrd1p oligomerization.
Figure 2. Usa1p-mediated oligomerization of Hrd1p is required for ERAD-L.
(A) Hrd1p-HA and Hrd1p-Myc were co-expressed under the endogenous promoter in either wild type (wt) or usaΔ cells. Detergent-solubilized membranes were treated with the bifunctional crosslinkers DSS or EGS, as indicated. Following quenching of the crosslinking reaction, immunoprecipitation with HA-antibodies was performed. Bound proteins were analyzed by SDS-PAGE followed by immunoblotting with Myc or Usa1p antibodies. The arrowhead indicates the position of a crosslinked species containing both Hrd1p and Usa1p.
(B) Hrd1p-HA and Hrd1p-Myc were co-expressed in cells containing either wild type Usa1p or Usa1p mutants with the indicated deletions. Detergent-solubilized membranes were subjected to immunoprecipitation (IP) with HA-antibodies and bound proteins were analyzed by SDS-PAGE and immunoblotting with HA-, Myc-, or Usa1p- antibodies. H and U indicate the segments in Usa1p that are responsible for interaction with Hrd1p and Usa1p, respectively.
(C) Usa1p-HA was co-expressed with FLAG-Usa1p or with the indicated FLAG-tagged deletion mutants of Usa1p. Detergent-solubilized membranes were subjected to immunoprecipitation (IP) with HA-antibodies and bound proteins were analyzed by SDS-PAGE and immunoblotting with HA- or FLAG- antibodies. Lanes 5 and 8 and the corresponding lanes 13 and 16 show the results with two independent clones of the same construct.
(D) Hrd1p-HA and Hrd1p-Myc or Hrd1(C399S)-HA and Hrd1(C399S)-Myc were co-expressed in cells containing or lacking Usa1p. Detergent-solubilized membranes were subjected to immunoprecipitation (IP) with HA-antibodies and bound proteins were analyzed by SDS-PAGE and immunoblotting with HA- or Myc- antibodies. For cells lacking Usa1p, two independent clones co-expressing Hrd1p-HA and Hrd1p-Myc (Lanes 5 and 6 and the corresponding lanes 13 and 14) or Hrd1(C399S)-HA and Hrd1(C399S)-Myc (Lanes 7 and 8 and the corresponding lanes 15 and 16) are shown.
(E) Kinetics of CPY*-HA degradation in cells expressing either wild type Usa1p or the indicated deletion mutants. The levels of CPY*-HA were determined by immunoblotting at different time points after cycloheximide addition (Figure S3). Shown are the means and standard deviations of three independent experiments.
See also Figure S3.
Next we identified the regions in Usa1p that are responsible for Hrd1p interaction and oligomerization. Usa1p contains two transmembrane segments flanked by long cytoplasmic domains (Figure 2B). Preliminary experiments showed that the cytoplasmic region preceding the first transmembrane segment was required for both Hrd1p oligomerization and Usa1p interaction (not shown). For a more precise analysis, we introduced deletions into Usa1p (Figure 2B). We found a segment (437–490), called segment H, whose deletion abolished both the binding of Usa1p to Hrd1p and the oligomerization of Hrd1p (lane 12). Segment H likely interacts with the C-terminal 34 residues of Hrd1p (Horn et al., 2009). Another deletion (residues 319–418) left the interaction between Usa1p and Hrd1p intact, but significantly reduced Hrd1p oligomerization (lane 10). The region from 371–418, called segment U, appears to be important for Hrd1p oligomerization, because a deletion mutant lacking residues 319–371 behaves like wild type Usa1p (lane 9). A recent report suggested that the segment from 259–312 encompassing the Ubl domain is responsible for inducing oligomers of Hrd1p (Horn et al., 2009). However, we did not observe any defects of Hrd1p oligomerization upon complete deletion of the Ubl domain (259–318) (Figure S3A).
A simple model for Usa1p-dependent Hrd1p oligomerization is that Usa1p itself forms oligomers through segment U and binds Hrd1p through segment H (see scheme in Figure 7A). To test this idea, we expressed HA-tagged full-length Usa1p together with FLAG-tagged versions of Usa1p mutants, all under the endogenous promoter. HA-antibodies precipitated all Usa1p-FLAG constructs, with the exception of the one lacking segment U (Figure 2C). Similar results were obtained with hrd1Δ cells (data not shown). These results support a model in which Usa1p oligomers facilitate Hrd1p oligomerization. Based on our overexpression experiments it appears that Hrd1p has an intrinsic propensity to form oligomers, which are stabilized by Usa1p. Indeed, crosslinking experiments show that overexpressed Hrd1p can form high molecular weight species even in the absence of Usa1p (Figure S3B). This is further supported by experiments in which ERAD was inactivated by a mutation of an essential cysteine in the RING finger of Hrd1p (C399S). As expected (Horn et al., 2009), in the presence of Usa1p, this mutant protein formed oligomers (Figure 2D, lane 11). Interestingly, however, Hrd1p(C399S) oligomerized even in the absence of Usa1p (Figure 2D, lanes 15 and 16 versus lanes 13 and 14). These data support the idea that Hrd1p has an intrinsic propensity to oligomerize and also suggest that substrate flux through the Hrd1p complex counteracts Usa1p-dependent oligomerization.
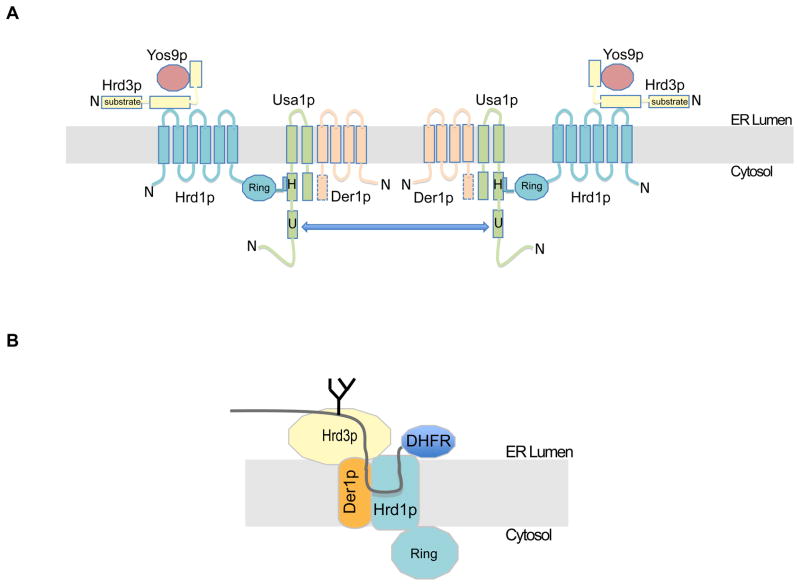
Figure 7. Organization of the Hrd1 complex and its interaction with substrate.
(A) The scheme summarizes interactions between Hrd1p complex components, identified in the present study and elsewhere (for references, see text). For each protein, the N-terminus is indicated. Although not demonstrated, the C-terminal domain of Der1p (dotted) likely interacts with the C-terminal segment of Usa1p. The arrow indicates interaction between two Usa1p molecules, which facilitates oligomerization of the complex.
(B) The scheme shows the ERAD-L substrate at an early stage of retro-translocation, as analyzed by photo-crosslinking.
Finally, we tested whether Hrd1 oligomerization is required for ERAD-L. The Usa1p mutant lacking segment H was completely inactive in the degradation of CPY* (Figure 2E; Figure S3C). The deletion of segment U also significantly impaired ERAD, but did not completely block it, consistent with the residual Hrd1p oligomerization activity (see Figure 2B). Thus, in contrast to previous suggestions (Horn et al., 2009), our results indicate that Hrd1p oligomerization is required for ERAD-L.
Crosslinking of an ERAD-L substrate to the Hrd1p complex
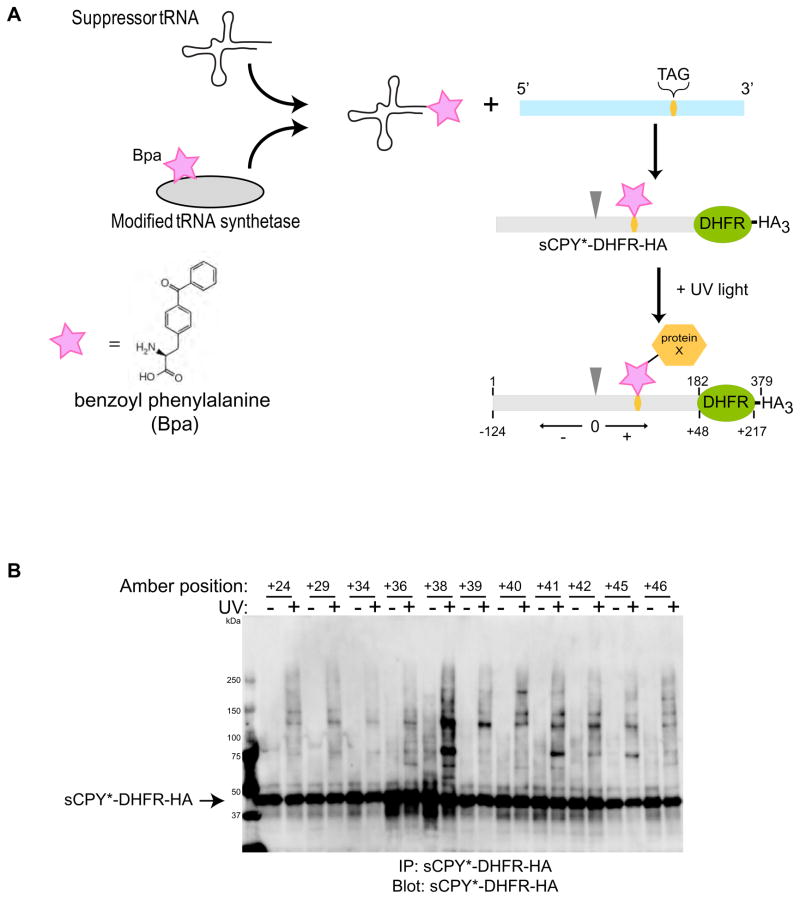
Next we tested whether an ERAD-L substrate undergoing retro-translocation would interact with components of the Hrd1p complex, particularly with the crucial membrane component Hrd1p. To test substrate interactions, we employed a site-specific in vivo photo-crosslinking method (Figure 3A) (Chen et al., 2007; Chin et al., 2003). An ERAD-L substrate with an amber stop codon at a selected site was expressed together with a suppressor tRNA and a modified tRNA-synthetase. The synthetase charges the tRNA with a phenylalanine derivative that carries a photoreactive benzophenone (Bpa), and the photo-reactive amino acid analog is then incorporated at the position specified by the stop-codon. Irradiation of the cells leads to crosslinks between the ERAD-L substrate and any protein that is in close proximity to the photoreactive probe.
Figure 3. Site-specific in vivo crosslinking of an ERAD-L substrate.
(A) A shortened version of CPY* containing the last 180 amino acids, including the glycosylation site, was fused to dihydrofolate reductase (DHFR) and three HA tags (sCPY*-DHFR-HA). Single amber stop codons (TAG) were introduced at different sites of the coding sequence. The stop codon was suppressed in vivo by expression of a suppressor tRNA that is charged with the photoreactive amino acid analog benzoyl phenylalanine (BPA) by a modified amino acyl tRNA synthetase. UV irradiation leads to crosslinks with proteins in close proximity of the photoreactive probe. The position of the probe is defined relative to the glycosylation site (position 0; arrow head; corresponds to position 124 in sCPY*-DHFR-HA), with amino acid residues upstream and downstream given negative and positive numbers, respectively.
(B) Photoreactive probes were placed at the indicated positions and the cells were irradiated with UV light, as indicated. Detergent-solubilized membranes were subjected to immunoprecipitation with HA antibodies, and bound proteins were analyzed by SDS-PAGE and immunoblotting with HA antibodies.
See also Figure S4.
To reduce the number of crosslinking positions to be tested, we used a shortened version of CPY* that contains the last 180 amino acids. This segment includes the glycosylation site that is part of the degradation signal. In addition, we fused dihydrofolate reductase (DHFR) to the C-terminus whose folding in the ER lumen slows the degradation of the substrate (Bhamidipati et al., 2005). Finally, three HA tags were added at the C-terminus. Degradation of this substrate (sCPY*-DHFR-HA) requires the normal ERAD-L components, i.e. Yos9p, Hrd1p, Hrd3p, Usa1p, and Der1p (Figure S4A). Next, we introduced single amber stop codons at various positions of sCPY*-DHFR-HA (Figure 3A; positions downstream and upstream of the glycosylation site are given positive and negative numbers, respectively). Yeast cells harboring a plasmid coding for tRNA and tRNA synthetase, as well as a plasmid coding for one of the sCPY*-DHFR-HA amber mutants, were grown in the presence of Bpa and then irradiated with UV light. Cell extracts were subjected to immunoprecipitation with HA antibodies, followed by SDS-PAGE and immunoblotting with HA-antibodies (Figure 3B). In the absence of the plasmid coding for tRNA and tRNA synthetase, no HA-reactive protein was detected (data not shown), demonstrating that suppression of the amber codon is required to generate sCPY*-DHFR-HA. Upon UV irradiation, crosslinked products were observed for most sCPY*-DHFR-HA mutants (Figure 3B); no crosslinks were seen in the absence of an amber codon (Figure S4B). The crosslinking pattern varied with the position of the photoreactive probe (Figure 3B). These results show that our approach can detect site-specific interactions of sCPY*-DHFR-HA. However, the most prominent crosslinked products could be extracted with alkali (data not shown), indicating that they do not contain any of the integral membrane proteins of the Hrd1p-complex. In fact, one may expect that, at any given time, only a small percentage of the luminal substrate is moving through the membrane (Gauss et al., 2006b).
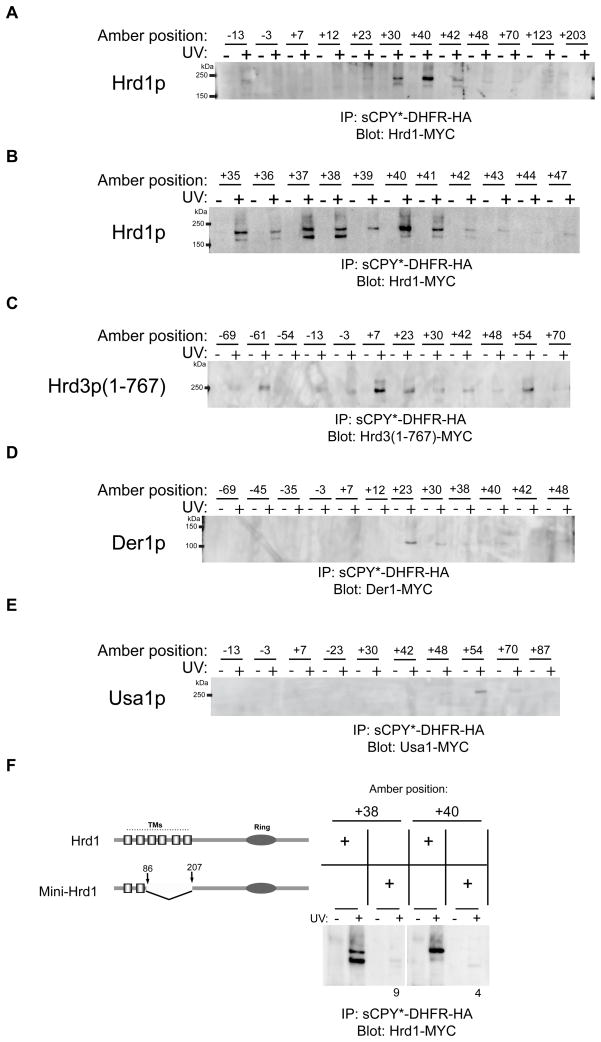
To test whether Hrd1p is among the less prominent crosslinking partners, the experiments were repeated in a strain that expresses under the endogenous promoter Hrd1p with 13 Myc tags at its C-terminus (Hrd1-Myc), a modification that maintains functionality of Hrd1p (Figure S4C). After irradiation, the samples were subjected to immunoprecipitation with HA-antibodies, followed by immunoblotting with Myc-antibodies (Figure 4A). Crosslinks to Hrd1p were observed with positions +30, +40 and +42, but not other positions. A more refined screen indicated that all positions between +30 and +42 crosslinked to Hrd1p, with the strongest interaction seen at positions +35 to +41 (Figure 4B). Although the crosslinking efficiency was low (Supplemental Table S1), these data attest to the specificity of substrate interaction with Hrd1p. Most positions gave two crosslinked bands in SDS gels, which might represent crosslinks to different sites in Hrd1p, as differences in mobility have been observed in other crosslinking experiments (Plath et al., 1998). Similar results were obtained when Hrd1p-Myc was first immunoprecipitated with Myc antibodies and the crosslinked products were analyzed by immunoblotting for sCPY*-DHFR-HA with HA-antibodies (data not shown). Taken together, these data show that Hrd1p interacts directly with a specific region of the substrate that starts ~30 amino acids downstream of the glycosylation site in the degradation signal and extends for ~12 amino acids almost to the DHFR domain (see scheme in Figure 7B).
Figure 4. Site-specific in vivo crosslinking of an ERAD-L substrate to Hrd1p complex components.
(A) Photoreactive probes were placed at the indicated positions of the substrate sCPY*-DHFR-HA. The constructs were expressed in cells that harbor under the endogenous promoter Hrd1p fused to 13 Myc tags (Hrd1-Myc). After immunoprecipitation with HA antibodies, the bound material was analyzed by SDS-PAGE and immunoblotting with Myc antibodies.
(B) As in (A), but with a narrower range of positions of the photoreactive probes.
(C) As in (A), but in cells that express under the endogenous promoter the luminal domain of Hrd3p (amino acids 1–767) fused to 13 Myc tags (Hrd3(1–767)-Myc).
(D) As in (A), but in cells that express under the endogenous promoter Der1p fused to 13 Myc tags (Der1-Myc).
(E) As in (A), but in cells that express under the endogenous promoter Usa1p fused to 13 Myc tags (Usa1-Myc).
(F) As in (A), but in cells expressing Myc-tagged Hrd1 or mini-Hrd1p. As indicated in the diagram, mini-Hrd1p was generated by deletion of the last four transmembrane segments (TMs) of Hrd1p. Ring refers to the Ring finger domain essential for ubiquitin ligase activity. The intensity of the crosslinks to mini-Hrd1p was normalized to that of wild type Hrd1p. The numbers are the average of three experiments.
Next we tested substrate interaction with the other components of the Hrd1p complex in similar photo-crosslinking experiments. We first analyzed the crosslinking of sCPY*-DHFR-HA with a functional version of the luminal domain of Hrd3p (amino acids 1–767) containing 13 Myc tags at the C-terminus (Hrd3(1–767)-Myc) (Figure S4C). The most prominent crosslinks were observed with probes at positions close to the degradation signal (position +7), although other positions also showed interactions (Figure 4C). These results are consistent with the proposal that the luminal domain of Hrd3p interacts with misfolded segments of ERAD-L substrates (Denic et al., 2006; Gauss et al., 2006a). Similar experiments were performed with Der1p containing 13 Myc tags at its C-terminus (Der1-Myc), which partially supports degradation of CPY* (Figure S4D). Substrate interaction was seen mostly for position +23 (Figure 4D). Finally, a functional fusion of Usa1p to 13 Myc tags (Usa1-Myc) did not show any significant crosslinks, except for a weak interaction with position +54 in the DHFR domain (Figure 4E; Figure S4C). Together, these data suggest that the crosslinks correspond to an early intermediate during retro-translocation, where the substrate interacts with the Hrd1p complex on the luminal side of the ER membrane. The degradation signal of the substrate is in contact with Hrd3p, a short segment downstream contacts Der1p, and ~12 residues immediately following interact with Hrd1p (Figure 7B). The C-terminal DHFR moiety is also in the ER lumen (Bhamidipati et al., 2005). The slow degradation of the substrate that is caused by the DHFR domain appears to allow the accumulation of this early translocation intermediate. Indeed, when the DHFR domain was deleted, the resulting substrate (sCPY*-HA) was degraded much faster than sCPY*-DHFR-HA, and photoreactive probes incorporated at equivalent positions did not give crosslinks to Hrd1p (data not shown). Taken together, these results suggest that the substrate inserts as a loop into the ER membrane from the luminal side, contacting primarily Hrd1p. The length of the interacting substrate segment indicates that no part of the polypeptide has reached the cytosolic side of the ER membrane (Figure 7B).
To provide further evidence that the substrate interacts with Hrd1p inside the membrane, we generated a mutant that lacks four of the six transmembrane segments. The resulting construct (mini-Hrd1p) contains the first two transmembrane segments directly fused to the C-terminal cytoplasmic domain (Figure 4F). Mini-Hrd1p is expressed at only slightly lower levels than wild type Hrd1p (Figure S5A), but it is totally inactive in degrading CPY* (Figure S5B) and two ERAD-M substrates (Sec61-2p and Hmg2p-Myc; data not shown). However, it interacts with the same ERAD components as wild type Hrd1p (Figure S5C) and forms oligomers, although these appear to be less dependent on the presence of Usa1p (Figure S3A). In addition, it is unstable in the absence of Hrd3p (Figure S5A), indicating that Hrd3p stabilizes Hrd1p by interacting with the luminal domain between transmembrane segments 1 and 2. Mini-Hrd1p no longer gave photo-crosslinks to sCPY*-DHFR-HA (Figure 4F). Because transmembrane segments 3–6 are not required for binding to ERAD components, they appear to be involved in substrate interaction. Thus, in the early translocation intermediate analyzed, the substrate appears to form a loop that contacts Hrd1p in the ER membrane, although it remains unclear how deeply the loop inserts.
Because of the proposed role of the Sec61 channel in ERAD, we also tested whether sCPY*-DHFR-HA would crosslink to a functional Sec61p fusion containing 13 Myc tags at its C-terminus (Sec61p-Myc). Very weak crosslinks were seen at positions +30, +37, +38, +40, and +54 (Figure S6A). The crosslinking yields were at least two orders of magnitude lower than seen with components of the Hrd1p complex (Table S1). In addition, they were not dependent on the presence of other ERAD components (Figure S6B), in contrast to those observed with Hrd1p (see below). These results argue against a role of Sec61p in the retro-translocation of the ERAD-L substrate.
Components required for substrate interaction with Hrd1p
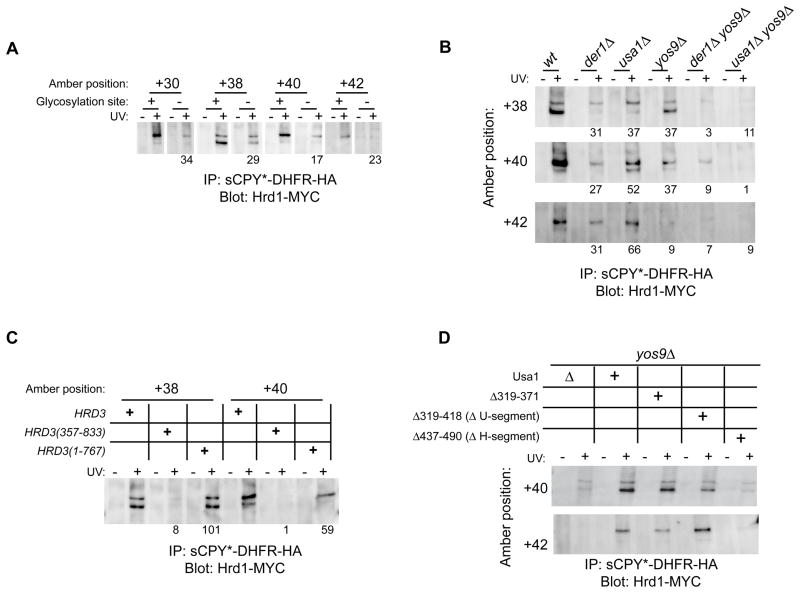
Because our data suggest that Hrd1p is the key membrane component in the ERAD-L pathway, we analyzed the interactions between substrate and Hrd1p in more detail. When N-glycosylation at the critical site was prevented by a mutation in sCPY*-DHFR-HA, crosslinking to Hrd1p was reduced (shown for positions +30, +38, +40, and +42 in Figure 5A). Deletion of YOS9, had a similar effect (Figure 5B), indicating that carbohydrate recognition by Yos9p affects substrate transfer to Hrd1p. The Hrd1p crosslinks completely disappeared in a HRD3 deletion mutant (not shown). In principle, this might be explained by a reduction of the steady state levels of Hrd1p in this strain (Gardner et al., 2000; Plemper et al., 1999). However, the Hrd1 crosslinks were also absent in cells expressing a fragment (residues 357–833) of the luminal domain of Hrd3p (Figure 5C), even though this fragment partially restored the levels of Hrd1p (data not shown and Gardner et al., 2000). Thus, substrate interaction with Hrd1p appears to critically depend on Hrd3p function. The recognition of the carbohydrate moiety by Yos9p, although essential for the overall ERAD process, is less important.
Figure 5. Effect of luminal ERAD events on substrate-Hrd1p crosslinking.
(A) sCPY*-DHFR-HA or a mutant lacking the glycosylation site with photoreactive probes at the indicated positions were expressed in cells together with Hrd1-Myc. Following UV irradiation, detergent-solubilized membranes were subjected to immunoprecipitation with HA antibodies and bound proteins were analyzed by SDS-PAGE and immunoblotting with Myc antibodies. The intensity of the crosslinked band for the glycosylation mutant is given as a percentage of the intensity obtained with sCPY*-DHFR-HA. The numbers are the average of two to four experiments.
(B) As in (A), but with sCPY*-DHFR-HA expressed in wild type (wt) cells or cells lacking the indicated ERAD components. The numbers give percentage of crosslinking intensity relative to wild type and are the average of two to four experiments.
(C) As in (A), but with sCPY*-DHFR-HA expressed in cells harboring either wild type Hrd3p or the indicated Hrd3p deletion mutants. The numbers are the average of two experiments.
(D) As in (A), but with sCPY*-DHFR-HA expressed in yos9Δusa1Δ cells harboring the indicated Usa1p deletion mutants.
See also Figure S7.
Substrate crosslinking to Hrd1p was only moderately reduced in the absence of Usa1p (Figure 5B). This could be due to the role of Usa1p in recruiting Der1p (Carvalho et al., 2006). Consistent with this assumption, deletion of DER1 had reproducibly a stronger effect on substrate-Hrd1p crosslinking than deletion of USA1 (Figure 5B). When cells lacked Yos9p and Usa1p, or Yos9p and Der1p, the crosslinks to Hrd1p completely disappeared (Figure 5B). These data suggest that there are two parallel pathways for substrate delivery to Hrd1p, one involving Yos9p and the other Der1p and Usa1p (Gauss et al., 2006b). Consistent with this model, when cells lacked Usa1p and Der1p, the crosslinks were reduced to approximately the same level as seen in the absence of Usa1p alone (Figure S7A). Both substrate delivery pathways to Hrd1p appear to require Hrd3p. Our data also confirm that the Hrd1p-substrate crosslinks represent an early stage of translocation, because in usa1Δder1Δ cells substrate still interacts with the Hrd1p complex, but is not translocated across the membrane. To determine the domain of Usa1p that is required for substrate-Hrd1p interaction, we expressed Usa1p mutants lacking either the H- or U-segment in usa1Δyos9Δ cells. Hrd1p crosslinks were restored as long as Usa1p contained the H-segment (Figure 5D). These data are consistent with the idea that Usa1p-Hrd1p interaction is required to allow substrate delivery from Der1p to Hrd1p.
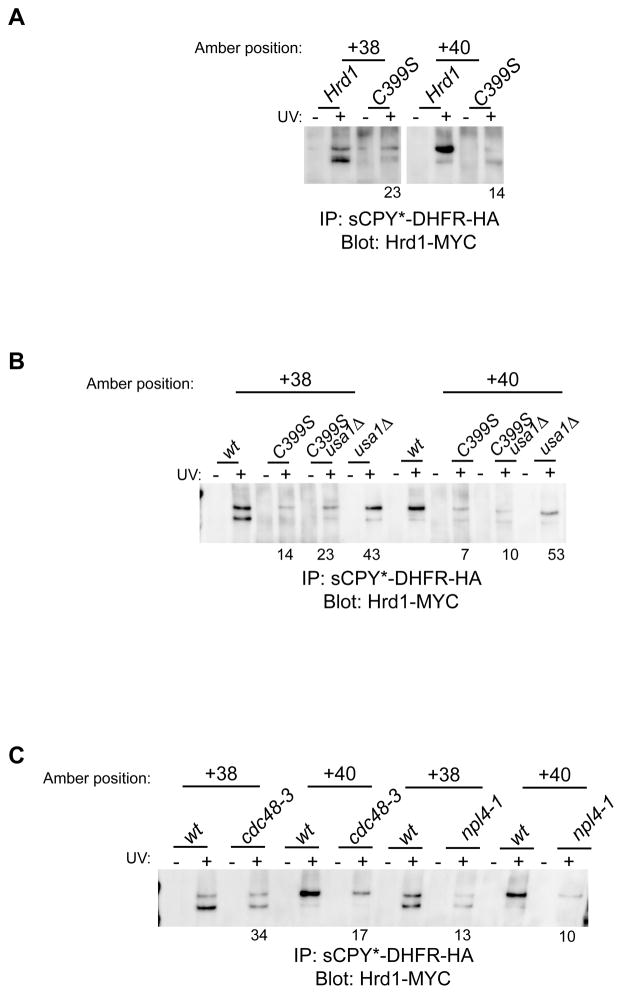
Although the Hrd1-substrate crosslinks correspond to an early retro-translocation intermediate, they were dependent on the ubiquitin ligase activity of Hrd1p; mutation of a critical Cys residue in the RING domain of Hrd1p led to a substantial reduction of the Hrd1p crosslinks (Figure 6A). The same results were obtained when the Cys mutation in Hrd1p was combined with a deletion of USA1 (Figure 6B). The absence of Ubc7p also reduced the Hrd1p crosslinking yields (Figure S7B); residual crosslinking may be explained by the fact that Ubc1p can also serve as an ubiquitin conjugating enzyme in ERAD-L (Bays et al., 2001a). Finally, Hrd1p crosslinks were also reduced in cells expressing a cdc48 or a npl4 temperature-sensitive mutant (cdc48-3 or npl4-1; Figure 6C). Taken together, these data indicate that the ubiquitination activity of Hrd1p and the function of the Cdc48 ATPase, i.e. events that occur on the cytosolic side of the ER membrane, are required for Hrd1p-substrate interaction on the luminal side of the membrane.
Figure 6. Mutations of ERAD components on the cytosolic side affect substrate-Hrd1p crosslinking.
(A) sCPY*-DHFR-HA with photoreactive probes at the indicated positions was expressed in cells together with Myc-tagged Hrd1 or a mutant defective in its ubiquitin ligase activity (C399S). Following UV irradiation, detergent-solubilized membranes were subjected to immunoprecipitation with HA antibodies, and bound proteins were analyzed by SDS-PAGE and immunoblotting with Myc antibodies. The numbers give percentage of crosslinking intensity relative to wild type and are the average of four experiments.
(B) As in (A), with either wild type (wt) or usa1Δ cells. The numbers are the average of two experiments.
(C) As in (A), with either wild type (wt) cells or cells bearing the indicated temperature-sensitive alleles of components of the Cdc48 ATPase complex analyzed after 1hr incubation at the restrictive temperature of 37°C. The numbers are the average of two experiments.
See also Figure S7.
Discussion
Our results provide important insight into the mechanism of ERAD-L. We show that the ubiquitin ligase Hrd1p is the key membrane component for moving a misfolded protein across the ER membrane. This conclusion is based on the observation that the overexpression of Hrd1p bypasses the need for its interaction partners Hrd3p, Usa1p, and Der1p, whereas all downstream cytosolic components are still required. To function in ERAD-L, Hrd1p needs to form homo-oligomers, a process that is normally dependent on Usa1p, but can be induced in the absence of Usa1p by the overexpression of Hrd1p. Using a site-specific photocrosslinking approach, we demonstrate that endogenous Hrd1p interacts directly with a substrate undergoing ERAD. This interaction requires the presence of transmembrane segments of Hrd1p and is dependent on the delivery of substrate through other ERAD components. Unexpectedly, substrate interaction with Hrd1 on the luminal side of the ER membrane is also dependent on the ubiquitination activity of Hrd1p and on the cytosolic Cdc48p ATPase complex. As discussed below, these results suggest a model for the mechanism by which a misfolded luminal protein is moved through the membrane.
Our results indicate that Hrd3p, Usa1p, and Der1p are all regulators of Hrd1p function. Hrd3p and Der1p are involved in substrate delivery to Hrd1p, whereas Usa1p serves both to recruit Der1p to Hrd1p and to induce Hrd1p oligomerization (Figure 7A). Usa1p facilitates Hrd1p oligomerization by interacting with Hrd1p through one domain (segment H) and interacting with another Usa1p molecule through another domain (segment U). Our data show that Usa1p does not significantly interact with substrate, consistent with it being a scaffolding protein (Horn et al., 2009). Taken together with results in the literature (Carvalho et al., 2006; Denic et al., 2006; Gardner et al., 2000; Gauss et al., 2006a; Gauss et al., 2006b; Horn et al., 2009), we have now a fairly comprehensive picture of the domain structure of the Hrd1p complex components and their interactions (Figure 7A), although it is possible that some of these associations are not permanent, but rather induced by substrate or regulated in other ways. Upon overexpression of Hrd1p, none of the regulatory components is required, indicating that Hrd1p can spontaneously oligomerize and bind substrates on its own. Under these conditions, substrate selection is less specific (Denic et al., 2006).
In wild type cells, Hrd3p is a crucial component for substrate delivery to Hrd1p. Our photo-crosslinking experiments indicate that the luminal domain of Hrd3p interacts with substrate and that in the absence of Hrd3p there is no transfer of substrate to Hrd1p. Hrd3p appears to collaborate with two alternative components to recruit substrate, either with Yos9p or Der1p, because only the deletion of both components abolishes all Hrd1p-substrate crosslinking. Dual delivery of substrate to Hrd1p was suggested before on the basis of co-immunoprecipitation experiments (Gauss et al., 2006b). Yos9p recognizes a terminal α1,6-mannose residue on a carbohydrate chain attached to the substrate (Clerc et al., 2009; Quan et al., 2008). We found that Der1p directly interacts with substrate, but its precise role remains unclear. Because both Yos9p and Der1p are essential in ERAD-L, they must have non-redundant functions in addition to providing parallel pathways of substrate recruitment.
The photo-crosslinking experiments give us a snapshot of an early translocation intermediate that follows substrate recognition (Figure 7B). The data show that Hrd1 interacts with a ~12 amino acid region of the sCPY*-DHFR-HA substrate. The interacting segment starts about 30 amino acid residues downstream of the glycosylation site in the degradation signal and immediately precedes the DHFR domain (Figure 7B). The polypeptide likely interacts with Hrd1p close to the luminal side of the membrane, because the degradation signal is in contact with the luminal domain of Hrd3p, and the DHFR moiety is also in the ER lumen (Bhamidipati et al., 2005). Thus, following recognition of the degradation signal and while still bound to Hrd3p, the adjacent C-terminal segment of the polypeptide chain interacts with Hrd1p. Given that the last four transmembrane segments of Hrd1p are required for substrate interaction, it appears that the polypeptide chain is inserted into the membrane-embedded parts of Hrd1p, likely as a loop. However, considering the length of the crosslinking region in the substrate, the polypeptide cannot be inserted deeply into Hrd1p, certainly not completely across the membrane. The N-terminal part of the substrate loop contacts Der1p (Figure 7B), suggesting that Der1p may play a role in inserting the polypeptide into Hrd1p.
Our data would be consistent with the assumption that the substrate interacts with a Hrd1p monomer, because the crosslinking yields were only moderately reduced in the absence of Usa1p, the component required for efficient Hrd1p oligomerization. Since the crosslinking efficiency is rather low, it is also possible that the substrate crosslinks to a small population of spontaneously generated Hrd1p oligomers. However, dissociation of the Hrd1p oligomer upon substrate binding would be consistent with the observation that blocking substrate flux through Hrd1p, either by mutation of its critical cysteine or by deletion of four of its transmembrane segments, makes Hrd1p oligomerization less dependent on the presence of Usa1p. We therefore propose that substrate and Usa1p have opposing effects on the oligomerization of Hrd1p.
Surprisingly, we found that the ubiquitin ligase activity of Hrd1p is required for an early interaction of substrate with Hrd1p. Because no part of the substrate has yet emerged on the cytoplasmic side of the membrane to become accessible to the ubiquitination machinery, these results suggest that Hrd1p modifies a target that is different from the substrate. The simplest possibility is that Hrd1p ubiquitinates another Hrd1p molecule, although this modification would not be expected to result in poly-ubiquitination since Hrd1p is stable in wild type cells. We do not have direct evidence for self-ubiquitination of endogenous Hrd1p, but it is well established that Hrd1p can modify itself upon overexpression or in the absence of Hrd3p (Bays et al., 2001a; Carroll and Hampton, 2009). Furthermore, the retro-translocation of two substrates that are not ubiquitinated themselves still requires the ubiquitination activity of the ligase (Bernardi et al., 2010; Hassink et al., 2006).
Our results also show that the activity of the Cdc48p ATPase complex is involved at an early stage of retro-translocation, likely the same that is dependent on Hrd1p ligase activity, given that the Cdc48p complex is generally recruited to ubiquitinated targets (Ye, 2006). An attractive possibility is that the Cdc48p ATPase remodels the Hrd1p complex following self-ubiquitination, either changing its conformation or its oligomeric state, a model that would be consistent with known activities of Cdc48p in other processes (Ramadan et al., 2007; Rape et al., 2001; Ye, 2006).
Based on our results, we propose a simple model for how a polypeptide is moved through the membrane. Because Hrd1p is the crucial component of the Hrd1p complex and needs to form oligomers, it may surround a polypeptide chain during its movement through the membrane. The polypeptide loop that is inserted into Hrd1p at the beginning of retro-translocation might simply be extended, with the transmembrane segments of the Hrd1p oligomer offering transient binding sites for the substrate inside the membrane. Because substrate and Usa1p have opposing effects on the oligomerization of Hrd1p, Hrd1p oligomers appear to be destabilized by substrate binding. The self-ubiquitination of Hrd1p and subsequent Cdc48p ATPase activity may be required for conformational changes of the Hrd1p-substrate complex. One possibility is that Hrd1p undergoes repeated cycles of Cdc48p- and ATP-dependent dissociation and Usa1p-dependent association, which may be coupled to cycles of substrate binding and release. Once the substrate loop has emerged on the cytosolic side of the membrane, it can be poly-ubiquitinated by Hrd1p and pulled out of the membrane by the Cdc48p ATPase complex. According to this model, Hrd1p alone would provide the conduit for a polypeptide through the ER membrane. Our photo-crosslinking data argue against the proposed role for the Sec61 channel in retro-translocation, but the participation of this or other components can only be formally excluded upon reconstitution of the process with purified proteins.
The proposed model could also apply to the degradation of proteins that have their misfolded domains inside the ER membrane (ERAD-M). Because Usa1p is not required for all ERAD-M substrates (Carroll and Hampton; Carvalho et al., 2006; Horn et al., 2009), one might assume that the binding of Hrd1p to different regions of the membrane-embedded substrate would promote Hrd1p oligomerization. Although many of the details of ERAD-L and -M remain to be elucidated, it is now clear that future work has to concentrate on Hrd1p and its regulation by the Cdc48p ATPase complex.
Experimental Procedures
Yeast strains and Plasmids
Tagging of proteins and individual gene deletions were performed by standard PCR-based homologous recombination (Longtine et al., 1998). Strains with multiple gene deletions and/or genomically encoded fusion proteins were made by PCR-based homologous recombination (Longtine et al., 1998) or by crossing haploid cells of opposite mating types, followed by sporulation and tetrad dissection using standard protocols (Guthrie and Fink, 1991). The strains used are isogenic either to BY4741 (Mata ura3Δ0 his3Δ1 leu2Δ0 met15Δ0) or to FY251 (Mata ura3-52 his3Δ200 leu2Δ1 trp1Δ63) and are listed in the Supplemental Table S2. Plasmids used in this study are listed in Supplemental Table S3 and described in Supplemental Experimental Procedures.
ERAD-substrate degradation experiments
Cycloheximide shut-off experiments were performed in exponentially growing cells, as described (Gardner et al., 2000). For experiments in which one of the ERAD components was expressed from the GAL1 promoter, cells were grown for 3 to 5 doubling times in medium containing 3% raffinose for de-repression of the promoter. ERAD components were then overexpressed in medium containing 3% galactose for 5 to 16 hours before performing cycloheximide shut-off experiments.
Chemical crosslinking and immunoprecipitations
A membrane fraction derived from 40–80 OD600 units of cells was isolated as previously described (Carvalho et al., 2006) and solubilized in 50 mM Hepes/KOH pH 7.9, 200 mM NaCl, 2 mM MgCl2, containing 1% digitonin or 1% Nonidet P-40. For chemical crosslinking experiments, the extracts were incubated with 0.2 mM disuccinimidyl suberate (DSS) or 0.2 mM ethylene glycol bis[succinimidylsuccinate] (EGS) for 30 min at room temperature. The reaction was quenched with 50 mM Tris/HCl pH 7.4 for 10 min. Immunoprecipitation, SDS-PAGE with 4–20% gradient gels, and immunoblotting were performed as described previously (Carvalho et al., 2006). In all immunoprecipitation experiments, 5% of the lysate was used directly for SDS-PAGE and immunoblotting.
Site-specific in vivo photocrosslinking
Cells were co-transformed with two plasmids, one coding for a tRNA that suppresses the amber stop codon and a modified tRNA-synthetase that charges the tRNA with the photoreactive amino acid analog benzoyl phenylalanine (Bpa), and another one coding for sCPY*-DHFR-HA with an amber stop codon at a selected position (Chen et al., 2007; Chin et al., 2003). The cells were grown overnight at 30°C in 150 ml of minimal medium. At OD600 of 0.3–0.5, 0.2mM of Bpa was added (from a stock solution of 0.2M Bpa in 1M NaOH) for 3 to 5 hrs at 25°C. The cells were harvested, washed with cold water, and resupended in 1ml of water. One half of the cells was transferred to a 12-well plate and exposed to long-range UV irradiation for ~45min using a B-100AP lamp (UVP, CA); the other half of the cells was kept on ice and used as control. Cells were lysed in LB buffer (50mM Tris/HCl pH 7.4, 200mM NaCl, 1mM EDTA, 2 mM phenylmethylsulfonyl fluoride (PMSF), and a protease inhibitor cocktail (Roche)) using glass beads in a bead beater (Biospec). The lysates were cleared by a 10 min centrifugation at ~600 x g, and a crude membrane fraction was obtained from the supernatant by a 20 min centrifugation at 100,000 x g. The membranes were solubilized at 65°C in urea buffer (50mM Tris/HCl pH7.4, 1mM EDTA, 1%SDS, 2M urea). Following dilution with LB buffer containing 1% Nonidet P-40, the extracts were incubated with HA antibodies (1:2,000; rat monoclonal clone 3F10, Roche) and protein G Sepharose (GE Healthcare). Bound proteins were eluted with SDS buffer and analyzed by SDS-PAGE and immunoblotting.
Supplementary Material
Acknowledgments
We thank R. Hampton, D. Ng, D. Pellman, P. Schultz, J. Weissman, and Y. Ye for reagents. We thank D. Pellman, Y. Ye, A. Stein, and D. Finley for critical reading of the manuscript. P.C. was supported by the Jane Coffin Child Memorial fund and is currently a special fellow of the Leukemia and Lymphoma Society, and A. M.S. is supported by a Ruth L. Kirschstein National Research Service Award postdoctoral fellowship. T.A.R. is supported by NIH grant GM052586 and is a Howard Hughes Medical Institute Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001a;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. HRD4/NPL4 Is Required for the Proteasomal Processing of Ubiquitinated ER Proteins. Mol Biol Cell. 2001b;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi KM, Williams JM, Kikkert M, van Voorden S, Wiertz EJ, Ye Y, Tsai B. The E3 ubiquitin ligases Hrd1 and gp78 bind to and promote cholera toxin retro-translocation. Mol Biol Cell. 2010;21:140–151. doi: 10.1091/mbc.E09-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. Embo J. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SM, Hampton RY. Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase. J Biol Chem. 2009;285:5146–5156. doi: 10.1074/jbc.M109.067876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Chen S, Schultz PG, Brock A. An improved system for the generation and analysis of mutant proteins containing unnatural amino acids in Saccharomyces cerevisiae. J Mol Biol. 2007;371:112–122. doi: 10.1016/j.jmb.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Finger A, Knop M, Wolf DH. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006a;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- Gauss R, Sommer T, Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. Embo J. 2006b;25:1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink G. Guide to yeast genetics and molecular biology. Vol. 194. Academic Press; 1991. [Google Scholar]
- Hassink GC, Barel MT, Van Voorden SB, Kikkert M, Wiertz EJ. Ubiquitination of MHC class I heavy chains is essential for dislocation by human cytomegalovirus-encoded US2 but not US11. J Biol Chem. 2006;281:30063–30071. doi: 10.1074/jbc.M602248200. [DOI] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- Horn SC, Hanna J, Hirsch C, Volkwein C, Schutz A, Heinemann U, Sommer T, Jarosch E. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell. 2009;36:782–793. doi: 10.1016/j.molcel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- Kim W, Spear ED, Ng DT. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell. 2005;19:753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth K, Wolf DH. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. Embo J. 1996;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Ng DT, Spear ED, Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Schekman R, Romisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Bordallo J, Deak PM, Taxis C, Hitt R, Wolf DH. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J Cell Sci. 1999;112:4123–4134. doi: 10.1242/jcs.112.22.4123. [DOI] [PubMed] [Google Scholar]
- Ploegh HL. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 2007;448:435–438. doi: 10.1038/nature06004. [DOI] [PubMed] [Google Scholar]
- Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, Weissman JS. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a Cytosolic Chaperone Required for Endoplasmic Reticulum-Associated Protein Degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan K, Bruderer R, Spiga FM, Popp O, Baur T, Gotta M, Meyer HH. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Mobilization of Processed, Membrane-Tethered SPT23 Transcription Factor by CDC48(UFD1/NPL4), a Ubiquitin-Selective Chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- Schafer A, Wolf DH. Sec61p is part of the endoplasmic reticulum-associated degradation machinery. Embo J. 2009;28:2874–2884. doi: 10.1038/emboj.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Taxis C, Hitt R, Park SH, Deak PM, Kostova Z, Wolf DH. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J Biol Chem. 2003;278:35903–35913. doi: 10.1074/jbc.M301080200. [DOI] [PubMed] [Google Scholar]
- Vashist S, Ng DTW. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz EJHJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- Willer M, Forte GM, Stirling CJ. Sec61p is required for ERAD-L: genetic dissection of the translocation and ERAD-L functions of Sec61P using novel derivatives of CPY. J Biol Chem. 2008;283:33883–33888. doi: 10.1074/jbc.M803054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Kanehara K, Sayeed A, Ng DT. Intrinsic conformational determinants signal protein misfolding to the Hrd1/Htm1 endoplasmic reticulum-associated degradation system. Mol Biol Cell. 2009;20:3317–3329. doi: 10.1091/mbc.E09-03-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Ng DT. ERAD substrate recognition in budding yeast. Semin Cell Dev Biol. 2010 doi: 10.1016/j.semcdb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Ye Y. Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J Struct Biol. 2006;156:29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Zhou M, Schekman R. The engagement of Sec61p in the ER dislocation process. Mol Cell. 1999;4:925–934. doi: 10.1016/s1097-2765(00)80222-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.