Abstract
C-type lectin domain family 5, member A (CLEC5A), also known as myeloid DNAX activation protein 12 (DAP12)-associating lectin-1 (MDL-1), is a cell surface receptor strongly associated with activation and differentiation of myeloid cells. CLEC5A associates with its adaptor protein DAP12 to activate a signaling cascade resulting in activation of downstream kinases in inflammatory responses. Currently, little is known about the transcriptional regulation of CLEC5A. We identified CLEC5A as one of the most highly induced genes in a microarray gene profiling experiment of PU.1 restored myeloid PU.1-null cells. We further report that CLEC5A expression is significantly reduced in several myeloid differentiation models upon PU.1 inhibition during monocyte/macrophage or granulocyte differentiation. In addition, CLEC5A mRNA expression was significantly lower in primary acute myeloid leukemia (AML) patient samples than in macrophages and granulocytes from healthy donors. Moreover, we found activation of a CLEC5A promoter reporter by PU.1 as well as in vivo binding of PU.1 to the CLEC5A promoter. Our findings indicate that CLEC5A expression in monocyte/macrophage and granulocytes is regulated by PU.1.
Keywords: CLEC5A, PU.1, myeloid differentiation, innate immunity
1. Introduction
CLEC5A is a cell surface receptor involved in the activation of myeloid cells. It associates non-covalently with the adaptor and signaling molecule DAP12 (also called KARAP and TYROBP) and was the first DAP12-associating receptor described in myeloid cells (Bakker et al., 1999). Signaling via this complex constitutes a significant activation pathway in myeloid cells and has an important role in immune defense. DAP12 is expressed on several immune cells, contains an immunotyrosine-based activation motif (ITAM) and associates with more than 20 different cell surface activating receptors to regulate immune responses. Upon ligand binding by a DAP12-associated receptor, the DAP12 cytoplasmic ITAM is phosphorylated by Src kinases and interacts with the Syk cytoplasmic tyrosine kinase, initiating a cascade of events that lead to macrophage activation (Campbell and Colonna, 1999; Lanier and Bakker, 2000; Lanier et al., 1998). Furthermore, recent studies have demonstrated that CLEC5A is a receptor for Dengue virus on the surface of macrophages. CLEC5A interacts directly with the Dengue virion, thereby causing phosphorylation of DAP12 and triggering a signaling cascade, which results in the release of proinflammatory cytokines (Chen et al., 2008).
In myeloid development CLEC5A expression is associated with mature stages of myeloid differentiation (Gingras et al., 2002). Furthermore, CLEC5A is expressed constitutively at very low levels but is highly induced in activated macrophages during infections (Aoki et al., 2009; Aoki et al., 2004; Bakker et al., 1999). In addition, CLEC5A is also implicated in osteoclastogenesis and associates with DAP10 in osteoclasts and bone marrow-derived macrophages. This association seems to be dependent on the presence of DAP12, and signaling through this trimolecular complex stimulates osteoclastogenesis and bone remodeling (Inui et al., 2009). CLEC5A expression is also induced upon neutrophil differentiation and activation of mouse 32Dcl3 myeloid cells (Aoki et al., 2009). In general, CLEC5A has an important function in innate immunity due to its role in macrophage and neutrophil differentiation as well as activation.
The CLEC5A gene is located on human chromosome 7q33 and murine chromosome 6B2, two loci that have not been associated with any disease relation to date. The molecular mechanisms that are responsible for the transcriptional regulation of the CLEC5A gene remain mostly unidentified. We discovered strong induction of CLEC5A in myeloid PU.1 knockout cells where PU.1 had been restored and provide evidence that CLEC5A is a direct PU.1 target gene in AML cells. Our results suggest that PU.1 is a major regulator of CLEC5A expression during myeloid differentiation.
2. Material and methods
2.1 Cell lines, primary patient samples and culture conditions
The human acute myeloid leukemia (AML) cell lines HL60, HT93, U937 and THP1 were maintained in RPMI-1640 or Dulbecco’s modified Eagle’s medium (DMEM) (Sigma-Aldrich, Buchs, Switzerland) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin. Cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C. For differentiation experiments, cells were treated with 1µM all-trans retinoic acid (ATRA; Sigma-Aldrich, Buchs), 10−7 M vitamin D3 (1,25-(OH)2D3) or 10 ng/ml phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) for days indicated.
Successful granulocyte or monocyte differentiation was evidenced by FACS analysis of CD11b and CD14 (BD Pharmingen), respectively. Successful macrophage differentiation was assessed morphologically. Isolation and differentiation of primary myeloid cells was done as described (Tschan et al., 2003; Tschan et al., 2001). Protocols and the use of all human samples were approved by the Cantonal Ethical Committee at the Inselspital.
2.2 Microarray analysis
Chip assays and analysis of the myeloid 503 PU.1 null cell line and PU.1 restored cells were utilized as described (Jenal et al., 2010).
2.3 Human CLEC5A promoter reporter assay
The CLEC5A promoter region was PCR amplified from genomic DNA of HL60 AML cells using the GC-RICH PCR system (Roche Diagnostics, Rotkreuz, Switzerland) and cloned into pCR-XL vector using the TOPO XL cloning kit (Invitrogen). The CLEC5A KpnI/HindIII promoter fragment was further subcloned into the pGL4-basic luciferase vector (Promega, Madison, WI, USA) using standard cloning techniques. PU.1 binding site mutations were introduced using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA).
For reporter assays, 293T cells were transfected in triplicate with 100 ng reporter, 300 ng effectors and 10 ng of pRL-TK plasmid per 24-wll using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cells were lysed 24 hours after transfection and Luciferase activity was measured using the Dual-Luciferase Reporter Plasmid System (Promega, Madison, WI). Results, expressed relative to a value of 1.0 for cells transfected with empty vector, are the means of two replications, and error bars represent standard deviations.
2.4 Chromatin immunoprecipitation assay (ChIP)
U937 cells were collected and ChIP assay performed as described (Weinberg et al., 2005). Antibodies used were using anti-PU.1, anti-C/EBPA (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-RNA pol II (Active Motif, Carlsbad, CA) antibodies. The following primers were used to amplify the CLEC5A genomic region containing the proximal PU.1 binding site by SYBR® Green based Quantitative PCR: Fw 5’-GGAAGTCTGCTCTTGCCACCACTag-‘3 and Rev 5’-CTGCCTTGGTAGCATCCCCAAG-‘3. Results were normalized to an IgG control and are given as % input chromatin.
2.5 TaqMan Low Density Arrays (LDA) and quantitative real-time RT-PCR (RQ-PCR)
RQ-PCR for LDAs and the 96-well format were performed using the ABI 7900HT Fast Real-Time PCR System or the ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Rotkreuz, Switzerland), respectively. Taqman® Gene Expression Assays for HMBS, ABL1, PU.1 and CLEC5A preloaded on LDAs were Hs00609297_m1, Hs00245445_m1, Hs00231368_m1, and Hs00183780_m1 (Applied Biosystems), respectively. For the 96-well format, we used the CLEC5A Taqman® Gene Expression Assay Hs00183780_m1. Primers and probe for PU.1, ABL1 and HMBS as well as data analysis have been described (Tschan et al., 2008).
2.6 Lentiviral knockdown constructs
pLKO.1 lentiviral vectors expressing short hairpin RNAs targeting PU.1 (shPU.1_256: NM_003120.1-256s1c1/TRCN0000020536 and_ shPU.1_928: NM_003120.1-928s1c1/ TRCN0000020538) and the non-targeting control shRNA vector (SHC002) were purchased from Sigma-Aldrich. Lentivirus production and transduction were done as described (Tschan et al., 2003).
2.7 Western Blot Analysis
Immunoblotting and protein extraction have been described previously (Andrews and Faller, 1991; Radziwill et al., 2003). Primary antibodies used were anti-PU.1 (Cell Signaling) and anti-GAPDH (MAB374; Millipore). Secondary antibodies used were donkey anti-rabbit and sheep anti-mouse horseradish peroxidase-conjugated IgG (Amersham, Zurich, Switzerland).
2.8 Statistical analysis
Nonparametric Mann-Whitney-U tests were applied to compare the difference between two groups using the program GraphPad Prism 4. P-values <0.05 were considered to be statistically significant.
3. Results and discussion
3.1 Silencing PU.1 significantly impairs CLEC5A induction during monocyte/macrophage differentiation
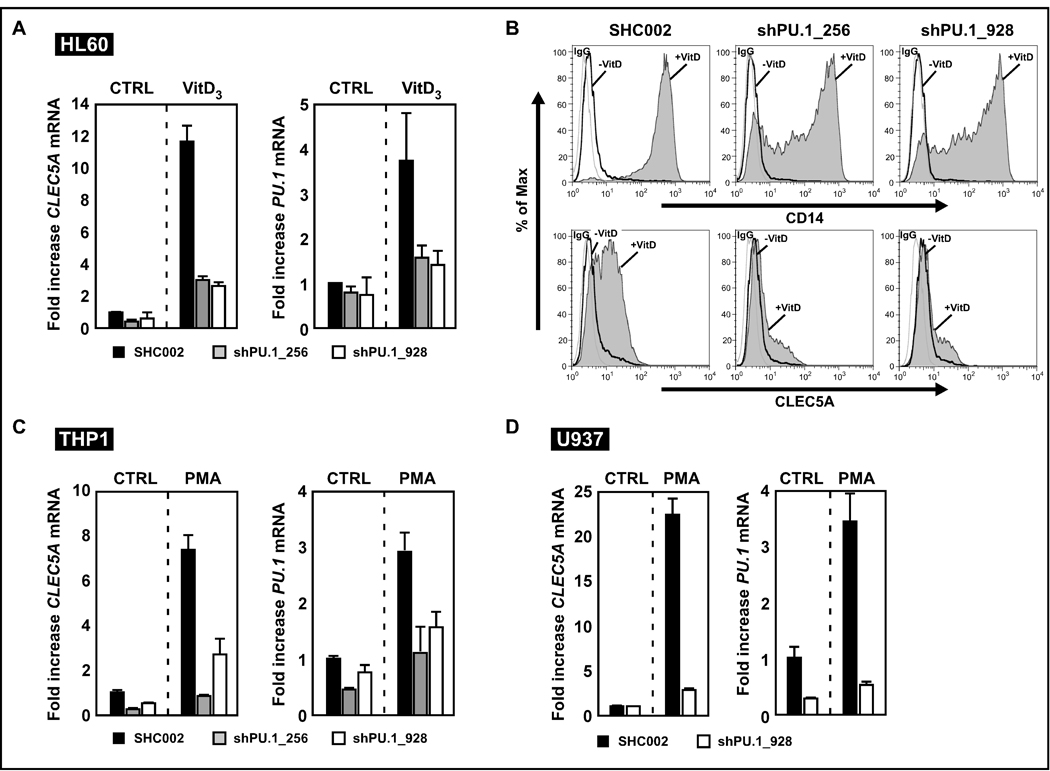
To identify novel PU.1-regulated genes, we introduced PU.1 into murine 503 PU.1-null myeloid cells using lentiviral gene transfer. We subsequently assessed gene expression profiles of PU.1 restored versus 503 PU.1-null myeloid cells. Several known PU.1 target genes were upregulated in the PU.1 rescued 503 cell line thereby confirming successful restoration of PU.1 (Jenal et al., 2010). CLEC5A was among the most highly induced genes in PU.1-restored 503 cells. To validate this finding we decreased PU.1 expression in HL60 AML cells, which can be differentiated into monocytes by vitamin D3 (VitD3) treatment. HL60 cells were stably transduced with non-targeting shRNA (SHC002) or with two different short hairpin RNAs targeting PU.1. Both shRNA constructs significantly reduced PU.1 expression in the two knockdown cell lines (Fig. 1A, right panel and Supplementary Fig. 1). We observed significantly reduced CLEC5A mRNA expression levels in VitD3-differentiated HL60 cells from 11.5-fold in parental cells to 2.3- and 2-fold in the two PU.1 knockdown cell lines, respectively (Fig. 1A). We further assessed CLEC5A protein surface expression by flow cytometry. During monocytic differentiation of HL60 cells induction of CLEC5A in the two knockdown cell lines was significantly impaired compared to control cells (CD11b MFI in control cells 422, compared to 110 and 140 in the two PU.1 knockdown lines, respectively). In parallel, CD14 expression, a marker for monocytic differentiation, was reduced in the two PU.1 knockdown cell lines compared to the control cells upon VitD3 treatment (Fig. 1B). Next, we included two macrophage differentiation models to assess CLEC5A regulation by PU.1. THP1 or U937 AML cells were differentiated into macrophages using phorbol 12-myristate 13-acetate (PMA). Both cell lines displayed a markedly attenuated increase in CLEC5A mRNA expression in PU.1 knockdown cells (Figs. 1C and D).
Figure 1. PU.1 knockdown attenuates CLEC5A induction during monocyte/macrophage differentiation.
(A) CLEC5A and PU.1 real-time quantitative polymerase chain reaction (RQ-PCR) analysis of untreated or monocyte (VitD3) differentiated HL60 cells expressing non- (SHC002) or PU.1-targeting (shPU.1_256 and shPU.1_928) shRNA. Results are given as n-fold induction compared to untreated cells and normalized to the housekeeping gene HMBS. (B) CD14 and CLEC5A flow cytometry analysis of HL60 control (SHC002) and PU.1 knockdown (shPU.1_256 and shPU.1_928) cells. Cells were treated as in A. (C) THP1 control (SHC002) and PU.1 knockdown (shPU.1_256 and shPU.1_928) AML cells were differentiated with PMA for two days to adherent macrophages. CLEC5A as well as PU.1 mRNA was determined by RQ-PCR. (D) U937 control (SHC002) and PU.1 knockdown (shPU.1_928) AML cells. Cells were treated as in C.
Overall, our results indicate that PU.1 is a crucial positive regulator of CLEC5A during monocyte/macrophage differentiation of AML cells.
3.2 PU.1 regulates CLEC5A expression during neutrophil differentiation of APL cells
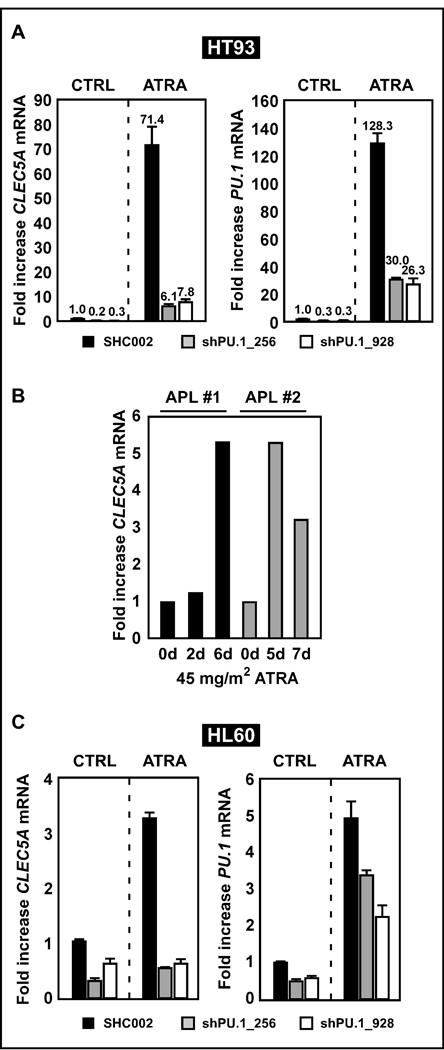
Recent findings by Aoki et al. (Aoki et al., 2009) showing CLEC5A induction during neutrophil differentiation of mouse myeloid cells, prompted us to analyze CLEC5A expression during all-trans retinoic acid (ATRA)-induced neutrophil differentiation of acute promyelocytic leukemia (APL) cell lines and patient samples. APL is characterized by the t(15;17) translocation resulting in the PML-RARA fusion protein. The block in differentiation caused by PML-RARA can be reversed by treating the patients with pharmacological doses of ATRA. PU.1 is a PML-RARA repressed gene and degradation of the PML-RARA fusion allows restoration of PU.1 expression (Mueller et al., 2006). Here, we investigated whether ATRA treatment induces CLEC5A expression via PU.1 expression. We utilized ATRA-differentiated parental as well as PU.1 knockdown HT93 APL cells and then monitored CLEC5A and PU.1 expression. CLEC5A expression was decreased in both PU.1 knockdown cell lines (Fig. 2A). To further provide insights into CLEC5A regulation two diagnosed APL patients undergoing ATRA-therapy were evaluated for CLEC5A mRNA expression at indicated days after treatment. Both APL patients displayed a 3- to 5-fold induction of CLEC5A at days 5–6 after treatment (Fig. 2B). Lastly, we investigated PU.1-dependent CLEC5A regulation in HL60, a second neutrophil differentiation model without the t(15;17) translocation. We confirmed impaired CLEC5A induction upon ATRA-induced granulocytic differentiation in HL60 cells and disruption of CLEC5A expression in PU.1 knockdown HL60 cells (Fig. 2C and Supplemental Fig. 1).
Figure 2. Silencing PU.1 inhibits CLEC5A mRNA induction upon all-trans retinoic acid (ATRA)-induced neutrophil differentiation of AML cells.
(A) HT93 acute promyelocytic leukemia cells stably expressing non- (SHC002) or PU.1-targeting (shPU.1_256, shPU.1_928) shRNAs were differentiated towards neutrophils with 1 µM ATRA for 4 days. Total RNA was extracted, and CLEC5A and PU.1 mRNA were measured by RQ-PCR. Values are given as n-fold induction compared to untreated cells and normalized to the housekeeping gene HMBS. (B) CLEC5A mRNA levels during a short-term follow up of two newly diagnosed APL-patients treated with orally administered ATRA (45mg/m2) were determined by RQ-PCR. CLEC5A mRNA levels are given as n-fold induction compared to day 0 and to ABL1 mRNA as control gene. (C) CLEC5A RQ-PCR analysis of control (SHC002) and PU.1 knockdown (shPU.1_256 and shPU.1_928) HL60 AML cells upon ATRA-induced neutrophil differentiation. PU.1 knockdown efficiency is shown by RQ-PCR.
Taken together, we clearly show that CLEC5A expression is restored in APL cells upon ATRA-induced neutrophil differentiation in a PU.1-dependent manner.
3.3 CLEC5A is repressed in AML patient samples
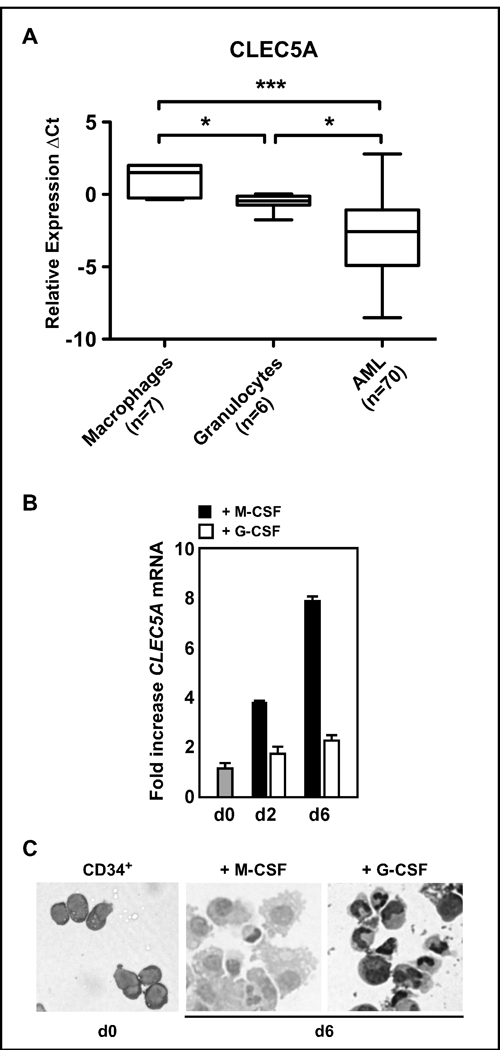
Earlier reports showed a positive correlation of PU.1 expression with the mature stages of normal myeloid cells, and that the gene is repressed in AML. We asked whether CLEC5A expression patterns would be altered in leukemic myeloid cells as compared to normal myeloid cells. CLEC5A mRNA expression levels were determined by quantitative RT-PCR in 70 AML blasts from patients and in 7 macrophages and in 6 granulocytes samples from healthy donors. CLEC5A mRNA expression was significantly lower in primary AML patient samples than in macrophages (p<0.001) and granulocytes (p<0.05) from healthy donors, suggesting that PU.1 and CLEC5A may be co-regulated during differentiation (Fig. 3A). In concordance with our cell line findings, primary macrophages expressed significantly more CLEC5A than granulocytes (p<0.05).
Figure 3. Repression of CLEC5A in primary AML patient samples.
(A) CLEC5A RQ-PCR analysis in macrophages and granulocytes from healthy donors as well as in undifferentiated AML patient cells. Values are the differences in Ct-values between CLEC5A mRNA and the levels of the two housekeeping genes HMBS and ABL1. Mann-Whitney U-test: *p<0.05; ***p<0.001 (B) Cord blood-derived CD34+ cells from healthy donors were expanded in vitro and differentiated into macrophages and neutrophils using M-CSF and G-CSF, respectively. RQ-PCR for CLEC5A mRNA was done as in A. Data are expressed as n-fold induction compared to untreated, control cells. (C) Morphological assessment of macrophage and granulocytic differentiation of CD34+ cells.
To further establish a role for CLEC5A in macrophage and granulocyte development, we isolated CD34+ progenitor cells from cord blood samples obtained from two healthy donors. CD34+ cells were differentiated in vitro to macrophages and neutrophils using M-CSF and G-CSF, respectively. CD34+ cells differentiated to monocytes/macrophages showed an 8-fold induction of CLEC5A mRNA expression at day 6. During neutrophil differentiation, CLEC5A mRNA levels increased up to 2-fold at day 6 (Fig. 3B). Successful macrophage and neutrophil differentiation of CD34+ was shown by morphology (Fig. 3C).
Our data demonstrate that CLEC5A expression is high in mature myeloid cells and that its expression is downregulated in AML.
3.4 Human CLEC5A is a direct transcriptional target of PU.1
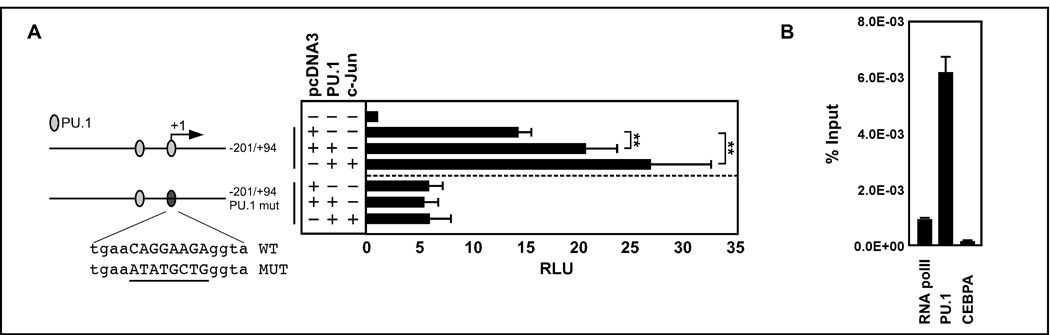
In a global ChIP-chip screen to identify PU.1 transcriptional targets in mouse macrophages, Weigelt et al. (Weigelt et al., 2008) identified two putative PU.1 binding sites in the proximal CLEC5A promoter region that are conserved in humans. We thus cloned a 295 bp fragment of the proximal promoter encompassing the two potential PU.1 sites into a luciferase reporter plasmid to analyze activation of the CLEC5A promoter by PU.1. Although the CLEC5A promoter reporter showed strong basal level activity, co-transfection with a PU.1 expression vector alone or in combination with the PU.1 co-factor c-Jun resulted in a further, significant activation. Mutating the proximal potential PU.1 binding site abolished PU.1-mediated activation of the CLEC5A promoter (Fig. 4A). This indicates that only the proximal PU.1 binding site is needed for CLEC5A induction.
Figure 4. PU.1 activates and directly binds to the human CLEC5A promoter.
(A) Left panel: Schematic representation of the 295 bp proximal human CLEC5A promoter. Two potential PU.1 binding sites (open circles) as determined by MatInspector analysis are indicated. Numbering indicates the relative position to exon 1. Right panel: Transactivation assays of 293T cells transiently transfected with empty or PU.1 and c-Jun expression vectors (300 ng) together with wild-type or PU.1 mutated CLEC5A promoter reporter constructs (100 ng). The promoter activity is shown as relative light units (RLU) normalized to the co-transfected Renilla Luciferase expression vector (phRL-TK, 10 ng). Results are the mean ± s.d. of five transfections. Mann-Whitney U-test: **p<0.01 (B) Chromatin immunoprecipitation (ChIP) assays using untreated U937 cells and antisera against PU.1 and. Anti-RNA pol II and anti-CEBPA served as positive and negative ChIP controls, respectively. Primers were used to amplify a CLEC5A promoter fragment encompassing the proximal PU.1 binding site. Results from two pull down experiments were normalized to an IgG control and are given as % chromatin input.
Next, we performed PU.1 chromatin immunoprecipitation assays (ChIP) in U937 cells to test if PU.1 binds to the human CLEC5A promoter in vivo. Precipitated chromatin was used to amplify the active PU.1 binding site in the proximal CLEC5A promoter. As a positive ChIP control we used an RNA pol II pull-down. As a negative control we performed a CCAAT/enhancer binding protein alpha (CEBPA) pulldown, because the CLEC5A promoter does not have any CEBPA binding sites. We found marked enrichment of PU.1 on the CLEC5A promoter in human U937 cells confirming data from the global ChIP-chip screen in mouse macrophages (Fig. 4B). Together, the reporter and ChIP assays strongly indicate that CLEC5A is a novel, direct transcriptional target of PU.1.
In summary, we identified CLEC5A as novel PU.1 target gene by gene expression profiling of PU.1-restored 503 PU.1-null myeloid cells. By decreasing PU.1 levels in several monocyte/macrophage and neutrophil differentiation models, we show that CLEC5A induction depends primarily on PU.1 during myeloid differentiation. Moreover, we found that CLEC5A is a direct transcriptional target of PU.1. Therefore, PU.1 contributes to the myeloid expression of CLEC5A. Further, we provide evidence that CLEC5A expression is regulated by PU.1 during neutrophil differentiation of APL cell lines and primary patient samples as well as in G-CSF differentiated CD34+ cells. These findings indicate that CLEC5A receptors are both part of the monocytic maturation process but also present in neutrophil differentiation consistent with an earlier report showing CLEC5A expression in mature neutrophils and macrophages.
Our study provides insight into the PU.1 regulatory network in inflammatory responses and points to a central role of PU.1 in this process. Moreover, CLEC5A is one of the many PU.1 regulated genes involved in myeloid activation and inflammatory responses including the triggering receptor expressed on myeloid cells (TREM-1) and DAP12 (Henkel et al., 2002; Weigelt et al., 2007). TREM-1 is a receptor that associates with the DAP12 adaptor molecule, similar to CLEC5A, to initiate a signaling cascade and thereby is involved in activation of immune cells (Tessarz and Cerwenka, 2008).
Our results support the importance of PU.1 as a mediator of immune responses by directly activating C-type lectin and triggering receptors in myeloid cells.
Supplementary Material
Western blot analysis of non-, ATRA- (neutrophil differentiation) or VitD3-treated (monocyte differentiation) HL60 cells expressing non- (SHC002) or PU.1-targeting (shPU.1_256, shPU.1_928) shRNA showing efficient PU.1 knockdown.
Acknowledgements
Deborah Shan and Gustav Arvidsson are gratefully acknowledged for excellent technical support. This work was supported by grants from the Swiss National Science Foundation 3100A0-118276 (to MPT), the Bern University Research Foundation (to MPT), the Werner and Hedy Berger-Janser Foundation of Cancer Research (to MFF and MPT), the NIH, HL091219 and DK54938, and the Joyce Klein Stock Gift (to BET), the Marlies-Schwegler Foundation, the Ursula-Hecht-Foundation for Leukemia Research, and the Bernese Foundation of Cancer Research (to MFF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship
JB: project design, performed experiments, collected and analyzed all data, drafted the manuscript; MMM: performed expression analysis in CD34 cells; MJ: PU.1 ChIP assays; VAR: performed CHIP arrays; MFF and BET: project planning and design, reviewed manuscript; MPT: project design, data analysis and manuscript writing.
References
- Andrews NC, Faller DV. A Rapid Micropreparation Technique for Extraction of DNA-Binding Proteins from Limiting Numbers of Mammalian-Cells. Nucleic Acids Research. 1991;19:2499–2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki N, Kimura Y, Kimura S, Nagato T, Azumi M, Kobayashi H, Sato K, Tateno M. Expression and functional role of MDL-1 (CLEC5A) in mouse myeloid lineage cells. J Leukoc Biol. 2009;85:508–517. doi: 10.1189/jlb.0508329. [DOI] [PubMed] [Google Scholar]
- Aoki N, Zganiacz A, Margetts P, Xing Z. Differential regulation of DAP12 and molecules associated with DAP12 during host responses to mycobacterial infection. Infect Immun. 2004;72:2477–2483. doi: 10.1128/IAI.72.5.2477-2483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker AB, Baker E, Sutherland GR, Phillips JH, Lanier LL. Myeloid DAP12-associating lectin (MDL)-1 is a cell surface receptor involved in the activation of myeloid cells. Proc Natl Acad Sci U S A. 1999;96:9792–9796. doi: 10.1073/pnas.96.17.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KS, Colonna M. DAP12: a key accessory protein for relaying signals by natural killer cell receptors. Int J Biochem Cell Biol. 1999;31:631–636. doi: 10.1016/s1357-2725(99)00022-9. [DOI] [PubMed] [Google Scholar]
- Chen ST, Lin YL, Huang MT, Wu MF, Cheng SC, Lei HY, Lee CK, Chiou TW, Wong CH, Hsieh SL. CLEC5A is critical for dengue-virus-induced lethal disease. Nature. 2008;453:672–676. doi: 10.1038/nature07013. [DOI] [PubMed] [Google Scholar]
- Gingras MC, Lapillonne H, Margolin JF. TREM-1, MDL-1, and DAP12 expression is associated with a mature stage of myeloid development. Mol Immunol. 2002;38:817–824. doi: 10.1016/s0161-5890(02)00004-4. [DOI] [PubMed] [Google Scholar]
- Henkel GW, McKercher SR, Maki RA. Identification of three genes up-regulated in PU.1 rescued monocytic precursor cells. Int Immunol. 2002;14:723–732. doi: 10.1093/intimm/dxf040. [DOI] [PubMed] [Google Scholar]
- Inui M, Kikuchi Y, Aoki N, Endo S, Maeda T, Sugahara-Tobinai A, Fujimura S, Nakamura A, Kumanogoh A, Colonna M, Takai T. Signal adaptor DAP10 associates with MDL-1 and triggers osteoclastogenesis in cooperation with DAP12. Proc Natl Acad Sci U S A. 2009;106:4816–4821. doi: 10.1073/pnas.0900463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal M, Batliner J, Reddy VA, Haferlach T, Tobler A, Fey MF, Torbett BE, Tschan MP. The anti-apoptotic gene BCL2A1 is a novel transcriptional target of PU.1. Leukemia. 2001;24:1073–1076. doi: 10.1038/leu.2010.26. [DOI] [PubMed] [Google Scholar]
- Lanier LL, Bakker AB. The ITAM-bearing transmembrane adaptor DAP12 in lymphoid and myeloid cell function. Immunol Today. 2000;21:611–614. doi: 10.1016/s0167-5699(00)01745-x. [DOI] [PubMed] [Google Scholar]
- Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- Mueller BU, Pabst T, Fos J, Petkovic V, Fey MF, Asou N, Buergi U, Tenen DG. ATRA resolves the differentiation block in t(15;17) acute myeloid leukemia by restoring PU.1 expression. Blood. 2006;107:3330–3338. doi: 10.1182/blood-2005-07-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radziwill G, Erdmann RA, Margelisch U, Moelling K. The Bcr kinase downregulates ras signaling by phosphorylating AF-6 and binding to its PDZ domain. Molecular and Cellular Biology. 2003;23:4663–4672. doi: 10.1128/MCB.23.13.4663-4672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz AS, Cerwenka A. The TREM-1/DAP12 pathway. Immunol Lett. 2008;116:111–116. doi: 10.1016/j.imlet.2007.11.021. [DOI] [PubMed] [Google Scholar]
- Tschan MP, Fischer KM, Fung VS, Pirnia F, Borner MM, Fey MF, Tobler A, Torbett BE. Alternative splicing of the human cyclin D-binding Myb-like protein (hDMP1) yields a truncated protein isoform that alters macrophage differentiation patterns. J Biol Chem. 2003;278:42750–42760. doi: 10.1074/jbc.M307067200. [DOI] [PubMed] [Google Scholar]
- Tschan MP, Reddy VA, Ress A, Arvidsson G, Fey MF, Torbett BE. PU.1 binding to the p53 family of tumor suppressors impairs their transcriptional activity. Oncogene. 2008;27:3489–3493. doi: 10.1038/sj.onc.1211004. [DOI] [PubMed] [Google Scholar]
- Tschan MP, Vonlanthen S, Cajot JF, Peters UR, Oppliger E, Betticher DC, Yarbrough WG, Fey MF, Tobler A. Different p16INK4a and p14ARF expression patterns in acute myeloid leukaemia and normal blood leukocytes. Leuk Lymphoma. 2001;42:1077–1087. doi: 10.3109/10428190109097728. [DOI] [PubMed] [Google Scholar]
- Weigelt K, Ernst W, Walczak Y, Ebert S, Loenhardt T, Klug M, Rehli M, Weber BH, Langmann T. Dap12 expression in activated microglia from retinoschisin-deficient retina and its PU.1-dependent promoter regulation. J Leukoc Biol. 2007;82:1564–1574. doi: 10.1189/jlb.0707447. [DOI] [PubMed] [Google Scholar]
- Weinbert MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen Z-X, Riggs AD, Rossi JJ, Morris KV. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt K, Moehle C, Stempfl T, Weber B, Langmann T. An integrated workflow for analysis of ChIP-chip data. Biotechniques. 2008;45:131–132. doi: 10.2144/000112819. 134, 136 passim. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western blot analysis of non-, ATRA- (neutrophil differentiation) or VitD3-treated (monocyte differentiation) HL60 cells expressing non- (SHC002) or PU.1-targeting (shPU.1_256, shPU.1_928) shRNA showing efficient PU.1 knockdown.