Summary
Autophagy influences numerous cellular processes, including innate and adaptive immunity against intracellular pathogens. However, some viruses, including dengue virus (DENV), usurp autophagy to enhance their replication. The mechanism for a positive role of autophagy in DENV infection is unclear. We present data that DENV induction of autophagy regulates cellular lipid metabolism. DENV infection leads to an autophagy-dependent processing of lipid droplets and triglycerides to release free fatty acids. This results in an increase in cellular β-oxidation, which generates ATP. These processes are required for efficient DENV replication. Importantly, exogenous fatty acids can supplant the requirement of autophagy in DENV replication. These results define a role for autophagy in DENV infection and provide a mechanism by which viruses can alter cellular lipid metabolism to promote their replication.
Introduction
Autophagy is a cellular process by which cytoplasmic components are sequestered in double membrane vesicles (DMVs) and degraded to maintain cellular homeostasis (Mizushima, 2007). It has also been implicated as an important component of the innate and adaptive immune response against a variety of viral and bacterial pathogens, reviewed in (Deretic, 2005; Deretic and Levine,2009;Kirkegaard et al.,2004;Schmid et al.,2006). Intact autophagocytic machinery has been reported to contribute to defense against infection by herpes simplex virus-1 (HSV-1) (Orvedahl et al., 2007), vesicular stomatitis virus (VSV) (Shelly et al., 2009), and Sindbis virus (Liang et al., 1998).
Some viruses, however, require components of the autophagocytic machinery for robust replication, reviewed in (Deretic and Levine, 2009). In particular, many positive-stranded RNA viruses require autophagy for efficient replication. Members of the family Picornaviridae, such as poliovirus (Jackson et al., 2005; Taylor and Kirkegaard, 2007) and coxsackieviruses B3 (Wong et al., 2008) and B4 (Yoon et al., 2008), have reduced replication in the presence of autophagy inhibitors. Further, markers of poliovirus replication and autophagosomes co-localize in infected cells, which has led to the hypothesis that polio virus replication structures may be at least in part derived from autophagosomes (Jackson et al., 2005). In addition to the Picornaviridae, members of the Flaviviridae, hepatitis C virus (HCV) (Dreux et al., 2009; Sir et al., 2008) and dengue virus (DENV), have been shown to require autophagy (or a component of autophagic machinery)for efficient replication. Several reports have shown that DENV infection induces autophagy and that the inhibition of autophagy leads to a significant reduction in DENV replication and the release of viral particles (Heaton et al., 2010; Khakpoor et al., 2009; Lee et al., 2008; Panyasrivanit et al., 2009).
Following virion endocytosis and uncoating, DENV proteins are translated and establish membranous replication complexes in the cytosol that facilitate efficient viral RNA replication (Miller and Krijnse-Locker, 2008). A proteolytically processed form of DENV NS4A protein, termed NS4A-2K, appears to be sufficient for DENV membrane remodeling, perhaps performing a structural role by inducing membrane curvature (Miller et al., 2007). It was previously postulated that DENV-2 replicates on amphisomes, which are fused endosomes and autophagosomes (Khakpoor et al., 2009; Panyasrivanit et al., 2009). However, this hypothesis is inconsistent with the cryo-EM tomography data, which shows viral RNA replication complexes are contiguous with the ER and thus are not distinct structures (Welsch et al., 2009). Therefore, although several groups have characterized autophagy as an important determinant for DENV replication, the role of autophagy in DENV infection is unclear.
Recently, autophagy has been show to regulate cellular lipid metabolism by modifying lipid droplets (Singh et al., 2009), which are cellular stores of triglycerides and cholesterol esters (Martin and Parton, 2006). The triglycerides are processed by lipases which results in the release of fatty acids (Haemmerle et al., 2003), which are then imported into mitochondria via the carnitine carrier system (McGarry and Brown, 1997) where they undergo β-oxidation (Ren and Schulz, 2003) that eventually leads to ATP production (Jormakka et al., 2003). The role of autophagy in maintaining normal cellular physiology is well established; yeast deficient in autophagy rapidly die under low nutrient conditions (Tsukada and Ohsumi, 1993) and mice with impaired autophagy have abnormal cellular membrane inclusions and protein aggregates (Komatsu et al., 2005). In addition, many human diseases are associated with deregulated autophagosome formation including neurodegenerative diseases, myopathies, and liver injury (Mizushima et al., 2002; Perlmutter, 2002).
In this study, we investigated whether DENV-induced autophagy regulates lipid metabolism. We found that the induction of autophagy in DENV-infected cells correlates with decreases in lipid droplet area and triglyceride levels and increases in β-oxidation. These alterations in lipid metabolism are autophagy-dependent and are required for efficient DENV replication. Complementation studies demonstrated that although autophagy regulates many cellular processes, its modulation of lipid metabolism is the requirement for efficient DENV replication.
Results
Induction and localization of autophagosomes in DENV-infected cells
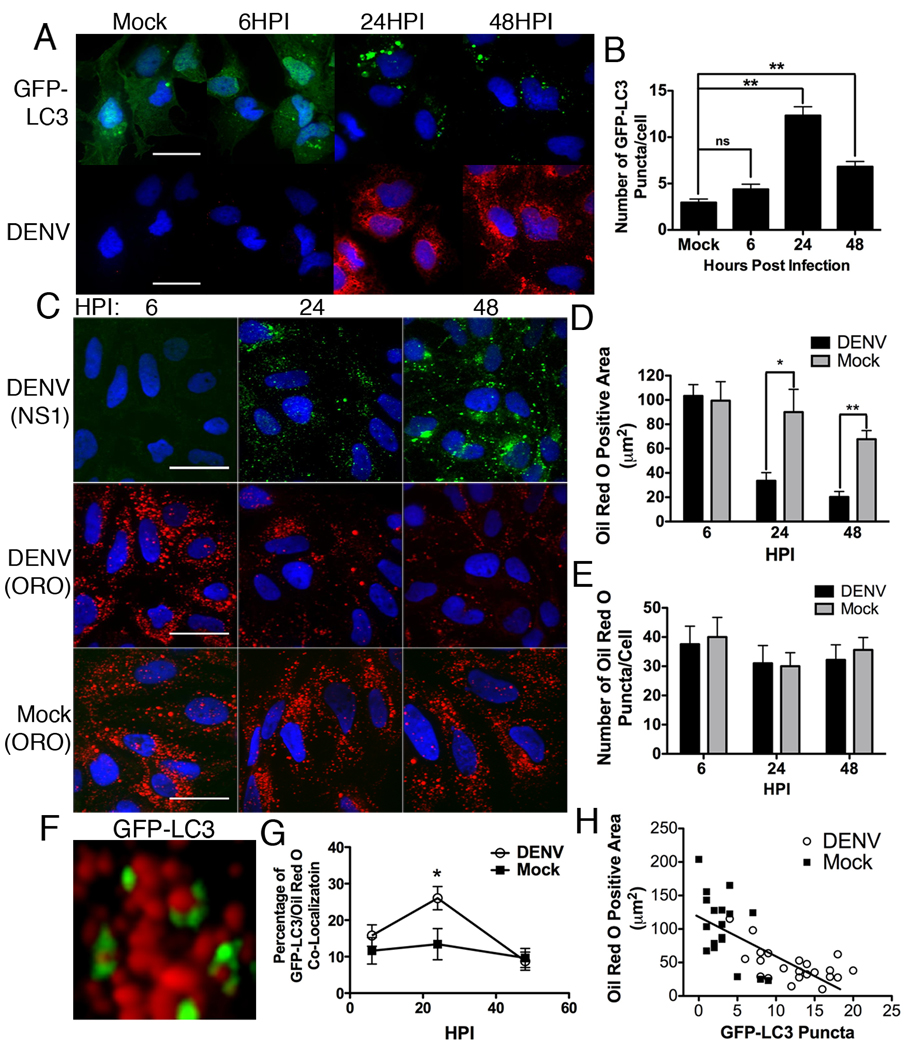
We first examined the sub-cellular localization of autophagosomes in DENV-infected cells. In order to visualize autophagosomes, we fused GFP to LC3, which specifically associates with autophagosomal membranes. Huh-7.5 cells were transfected with GFP-LC3 for 24 hours, then infected with DENV for 6, 24, or 48 hours, fixed, probed with DENV2-specific monoclonal antibody D1-4G2-4-15 and visualized using confocal microscopy. Consistent with previous reports, the number of GFP-LC3 puncta per DENV-infected cell significantly increased at 24 and 48 HPI, as compared with mock-infected cells (p<0.001, Fig. 1A,B).
Figure 1. Lipid droplet area decreases as the number of GFP-LC3 puncta increase in DENV-infected cells.
(A) Huh-7.5 cells were transfected with a GFP-LC3 construct, then DENV- or mock-infected at an MOI of 2 for the indicated times, fixed, and probed with specific monoclonal antibody D1-4G2-4-15. Nuclei were visualized by DAPI staining. Scale bar = 30µm. (B) ImageJ quantification of the number of GFP-LC3 puncta per cell in (A) **p<0.001. (C) Huh-7.5 cells were DENV- or mock-infected at an MOI of 2 for the indicated times and then stained with oil red O to visualize lipid droplets and antibodies to NS1 were used to verify cells were infected. Scale bar = 30µm. (D) Quantification of the total oil red O positive area per cell representative of three independent experiments. *p<0.05, **p<0.001. (E) Quantification of the number of oil red O puncta per cell. (F) Huh-7.5 cells were transfected with a GFP-LC3 construct and then DENV infected for, a high magnification image of GFP-LC3 association with lipid droplets is shown. (G) Huh7.5 cells were DENV- or mock-infected for the indicated times. The association of GFP-LC3 positive structures with oil red O are quantified, *p<0.05. (H) Scatter plot of the number of lipid droplets and GFP-LC3 puncta in individual DENV- or mock-infected cells from random fields of view, 36HPI. The trend-line indicates a statistically significant (p<0.05) linear regression showing a negative correlation between the two variables.
We then interrogated whether GFP-LC3 co-localizes with markers of DENV replication complexes using antibodies that recognize NS1, NS3 or dsRNA, the positive strand RNA virus replication intermediate. We were unable to identify conditions in which LC3-GFP positive structures co-localized with markers of dengue virus replication (Fig. S1). In some cases, the GFP-LC3 localization was reminiscent of lipid droplet distributions in Huh-7.5 cells. Therefore, we examined co-localization of GFP-LC3 with lipid droplets in DENV-infected cells. Huh-7.5 cells were transfected with GFP-LC3, infected with DENV for 24 hours, fixed, and stained with oil red O. We observed a subset of GFP-LC3 positive autophagosomes associating with lipid droplets in DENV-infected cells (Figs. 1F and 5D). A kinetic analysis of autophagosome association with lipid droplets showed maximal association 24 hours after DENV infection (Fig. 1G).
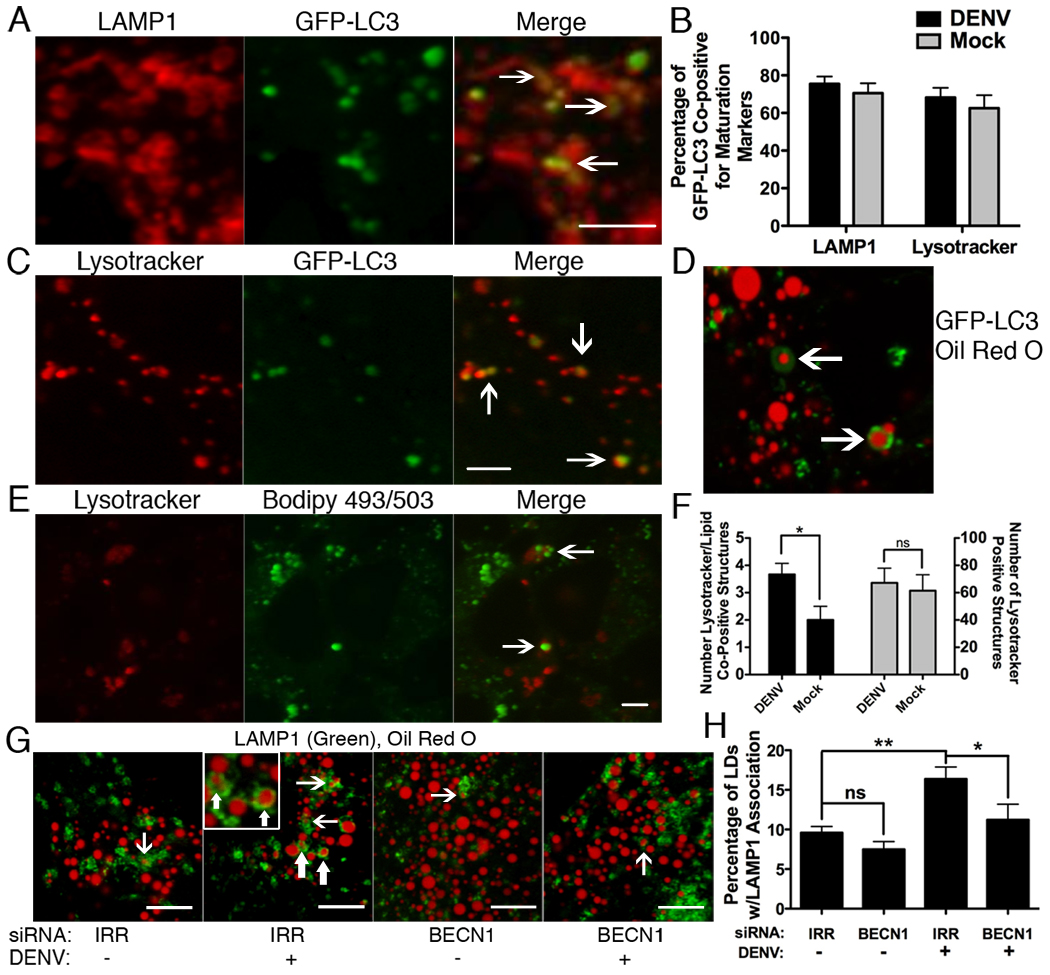
Figure 5. Increased delivery of lipids to acidified lysosomes in DENV-infected cells.
Huh-7.5 cells were transfected with GFP-LC3, then DENV-infected at an MOI of 3–5 for 24 hours, then (A) fixed and probed with antibodies against LAMP1 or (C) stained with Lysotracker. (B) ImageJ quantitation of co-localization of GFP-LC3 with LAMP1 or Lysotracker. (D) GFP-LC3 transfected cells were DENV infected for 24 hours and the association of Oil Red O and GFP-LC3 was visualized. (E) Huh-7.5 cells were DENV infected (MOI=2) for 24 hours before treatment with Lysotracker and Bodipy 493/503 to stain lipid droplets. Arrows indicate examples of Lysotracker positive structures containing lipids and are quantified in (F). The numbers of in (F) are per field of view. (G) Huh-7.5 cells were treated with indicated siRNAs and maintained for 48 hours, then mock- or DENV-infected at an MOI=2 for 24 hours, then fixed and stained with oil red O and antibodies against LAMP1. Arrows depict examples of co-positive structures. (H) Image J quantitation of 10 randomly selected fields of view from part (E), *p<0.05, **p<0.001. Scale bars = 5µm.
DENV-induced autophagy reduces cellular lipid droplet area
We next examined whether lipid droplets were modified in DENV-infected cells. Huh-7.5 cells were infected as before, fixed, probed with antibody to NS1, and stained with oil red O. Lipid droplet area was reduced by ~70% in DENV-infected cells at 48 hours post infection, as compared with mock-infected cells (p<0.001, Fig. 1C,D). Despite this decrease in total oil red O staining area per cell, the total number of oil red O foci per cell remained relatively unchanged (Fig. 1E). This indicated that lipid droplets were not eliminated in DENV infection, but that their size, and possibly content, was reduced. The results were reproduced in BHK, Huh7 and HepG2 cells to confirm that effects were not cell type specific (Fig. S2).
Lipid droplets were generally depleted in cells with high numbers of autophagosomes. To test for a direct correlation, the number of autophagosomes and lipid droplet area per mock- and DENV-infected cells were quantified and linear regression analysis was performed. There was a statistically significant (p<0.05) negative correlation between the number of autophagosomes and the area of lipid droplets per cell (Fig. 1H).
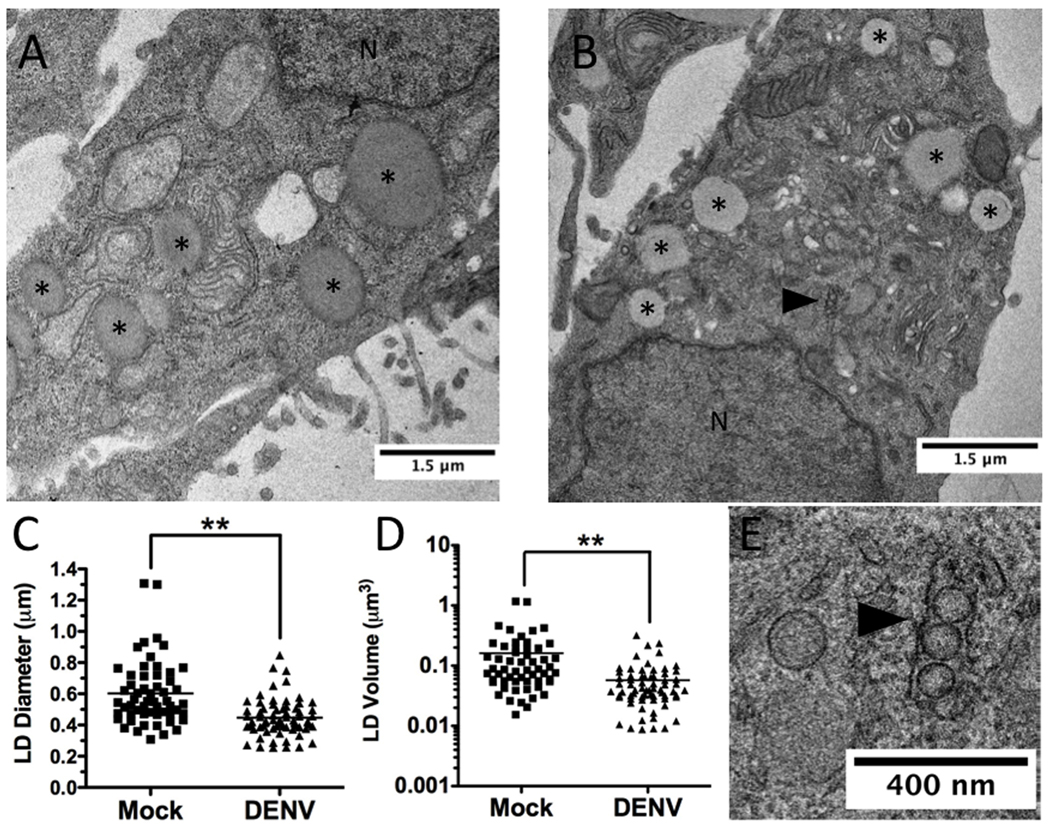
Since oil red O staining is an indirect measurement of lipid droplet area, we next quantified the size of lipid droplets in DENV and mock-infected cells following visualization by EM (Fig. 2A and B). Huh-7.5 cells were mock- or DENV-infected for 48 hours, then fixed and processed for EM. The lipid droplet diameters in these cells were then measured. DENV infection was verified by the presence of virally induced membrane structures (Fig. 2B,E). Lipid droplet diameter in DENV-infected cells was decreased ~35% as compared to mock-infected cells, which translated to an ~70% reduction in lipid droplet volume (p<0.001, Fig. 2C,D).
Figure 2. EM analysis of lipid droplet size during DENV infection.
Huh-7.5 cells were DENV- or mock-infected (MOI=2) for 48 hours, fixed, and pelleted for EM staining and sectioning. Representative images of (A) mock- and (B) DENV-infected cells. *denotes lipid droplets, N denotes the nucleus, and the black arrowhead denotes DENV induced membrane structures. (B). Plot of the (C) average diameter and (D) calculated volume of lipid droplets (n>50) in EM images. **p<0.001. (E) High magnification image of B showing DENV induced membrane re-organization.
Autophagy is required for lipid droplet depletion during DENV infection
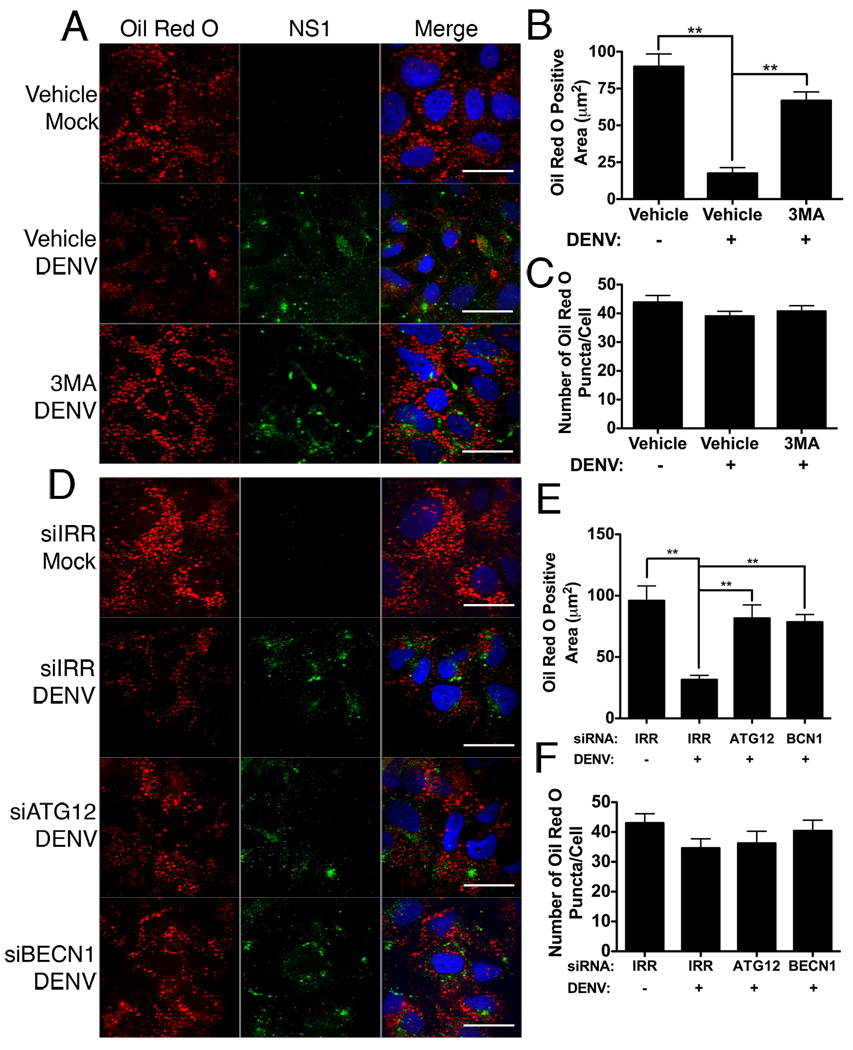
We next tested whether autophagy was required for the lipid droplets alterations in DENV-infected cells. Huh-7.5 cells were infected as before and treated with 3-methyl adenine (3MA), a well-characterized inhibitor of autophagy. In addition, we inhibited autophagy by silencing key components of the autophagy pathway (ATG12 and BECN1) as compared to an irrelevant siRNA (IRR) that targets HCV. In IRR siRNA or vehicle treated cells, lipid droplet area was again depleted in DENV-infected cells as compared to mock-infected cells (p<0.001, Fig. 3A,B) with only minor changes to lipid droplet number (Fig. 3C). Inhibition of autophagy significantly increased the area of lipid droplets in DENV-infected cells (Fig. 3D–F). In parallel, we confirmed that the siRNAs specifically reduced accumulation of the target gene mRNAs (Table S1) and protein (Fig. S3A), and that treatment with 3MA or siRNAs targeting autophagy genes precluded the induction of GFP-LC3 puncta during DENV infection (Fig. S3B).
Figure 3. Inhibition of autophagy prevents depletion of lipid droplets in DENV-infected cells.
(A) Huh-7.5 cells were mock- or DENV-infected at an MOI of 2 for two hours, then treated with 2.5mM 3MA or vehicle control. Cells were fixed 48HPI and stained with Oil Red O to visualize lipid droplets and an antibody against DENV NS1 (green). (C) Alternatively, Huh-7.5 cells were treated with siRNAs targeting an irrelevant HCV sequence (IRR), ATG12, or beclin1 (BECN1) for 24 hours, and then infected with DENV for 48 hours at an MOI of 2. Respective Image J quantification of total area staining positive for oil red O (B,E) and the total number of oil red O puncta per cell (C,F). Values represent the average of at least eight cells per treatment. **p≤0.001. Scale bar = 30µm.
Inhibition of autophagy leads to a defect in viral RNA replication
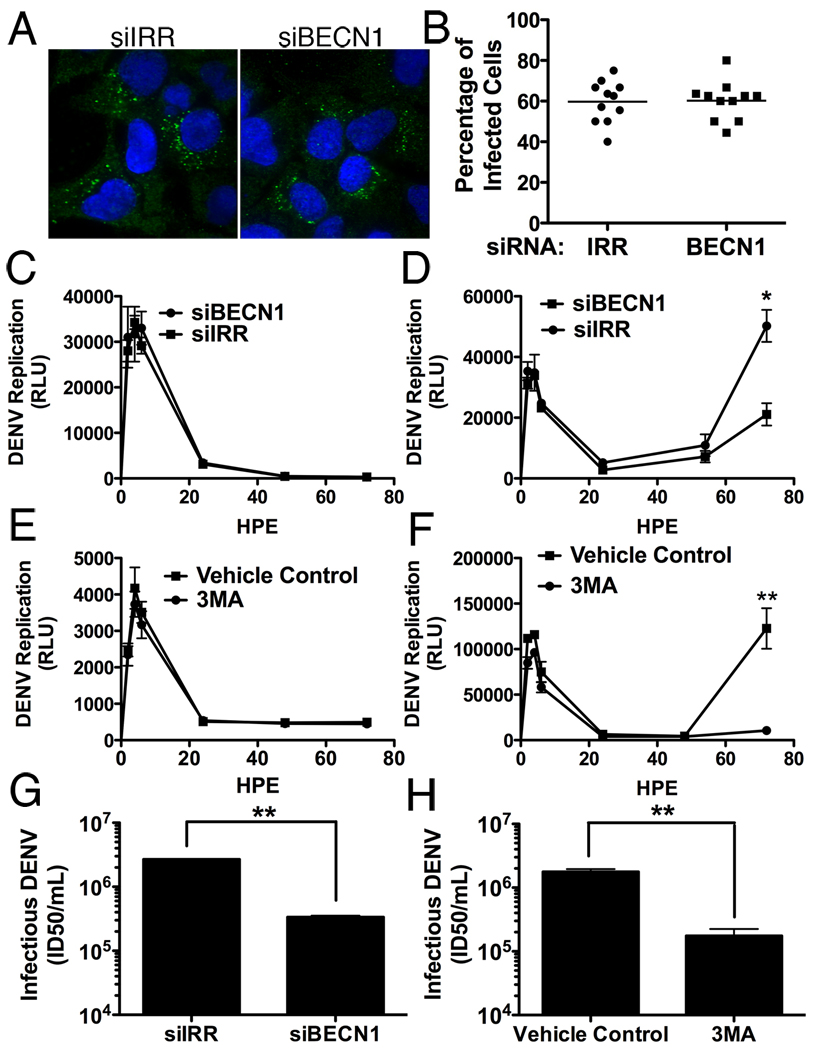
We next investigated the stage of the viral life cycle that requires autophagy. To determine if autophagy plays a role in viral entry, we transfected IRR or BECN1 siRNAs into Huh-7.5 cells, then infected the cells (MOI=0.5) for 15 hours, fixed and stained cells for dsRNA as a marker of viral infection (Fig 4. A,B). Approximately 60% of cells (11 random fields of view, n>100 cells) in both treatments stained positive for dsRNA, indicating that under the conditions tested, autophagy has no apparent role in viral entry.
Figure 4. DENV replication, but not entry or translation, is dependent upon autophagy.
(A) Huh-7.5 cells silenced for BECN1 or an irrelevant control were infected (MOI=0.5) for 15hrs before fixation and staining for dsRNA and DAPI. Random fields of view were imaged (A) and quantified for the percentage of cells containing DENV-specific dsRNA. (B). A polymerase defective DENV-luciferase replicon was electropotated into Huh-7.5 cells silenced for BECN1 or treated with 3MA to measure input RNA translation (C and E respectively). In parallel, the wild-type replicon was introduced under the same conditions to determine the effect on viral RNA replication corresponding to time points after 24hrs (D and F). HPE is hours post electroporation. Autophagy was inhibited in Huh7.5 cells via siRNAs targeting BECN1 (G) or treatment with 3MA (H) and the release of infectious DENV was quantified. *p≤0.05, **p<0.001.
It has been reported that autophagy plays a role in translation of HCV RNA (Dreux et al., 2009). To determine if this was the case during DENV infection, we introduced a DENV luciferase reporter replicon construct with a polymerase mutation (GDD→GΔΔ) and measured luciferase over time. In cells either silenced for BECN1 or treated with 3MA, there was no significant reduction in RNA translation compared to their respective controls (Fig. 4C,E). In parallel, we introduced the wild-type replicon and again observed no defect in the early translation peak but significant inhibition of replication at later time points (p<.05, Fig. 4D,F). The decreases in DENV replication under conditions of inhibited autophagy are also reflected in infectious DENV production. Treatment of Huh-7.5 cells with 3MA or siRNAs targeting BECN1 inhibit infectious DENV production, as has been reported previously (Fig. 4G,H) (Khakpoor et al., 2009; Lee et al., 2008; Panyasrivanit et al., 2009). 3MA inhibition of DENV replication and infectious virus production is consistent in all cell lines tested, including Huh-7.5, Huh7, HepG2, and BHK cells (Fig. S4). We therefore conclude that autophagy is primarily required for DENV replication, although a secondary role in assembly or egress cannot be excluded.
Lipid delivery to the lysosomal compartment is increased during DENV infection
We next characterized autophagosomal maturation during DENV infection. Huh-7.5 cells were DENV- or mock-infected for 24 hours, fixed and analyzed for GFP-LC3 association with the late endosomal and lysosomal marker LAMP1. The majority of GFP-LC3 positive structures are co-positive for LAMP1. (Fig. 5A,B). Autophagosomal acidification was investigated under the same conditions by examining GFP-LC3 co-localization with Lysotracker Red, a fluorescent probe that selectively accumulates in low pH compartments. In mock-and DENV-infected cells, we observed that the majority of GFP-LC3 puncta also stained positive for Lysotracker Red, indicative of autophagolysosomal acidification (Fig. 5B,C). Thus, DENV does not appear to perturb autophagosomal maturation.
We next interrogated whether lipids were delivered to the lysosomal compartment in an autophagy-dependent manner. DENV infected cells expressing GFP-LC3 were analyzed for co-localization of Oil Red O and LC3 (Fig. 4D). As previously observed, there is an association of lipid droplet markers with autophagosome markers during infection. To test whether lipids were being delivered to acidified, degradative compartments, Huh-7.5 cells were mock- or DENV-infected for 24 hours and stained with Lysotracker and Bodipy 493/503 to label acidic compartments and lipid droplets, respectively. DENV infection increased the number of structures were co-positive for Lysotracker and the Bodipy lipid droplet marker, without altering the total number of Lysotracker positive structures (Fig. 5E,F). Thus, DENV infection increased the delivery of lipids to acidified lysosomes. To determine the requirement of autophagy for delivery of lipids to LAMP1 compartments, Huh-7.5 cells were treated with IRR or BECN1 siRNAs, then mock- or DENV-infected for 24 hours. Cells were then fixed and probed for the association of LAMP1 with oil red O stained lipids. LAMP1 structures surrounding oil red O lipids were detectable in all samples (Fig. 5G). DENV-infection increased the accumulation of these LAMP1/oil red O co-positive structures and this effect was significantly precluded by treatment with Beclin1 siRNAs (p<0.05, Fig. 5G,H). Thus, DENV infection enhances the delivery of lipids to the lysosome in an autophagy-dependent manner.
Triglycerides are specifically depleted in DENV-infected cells
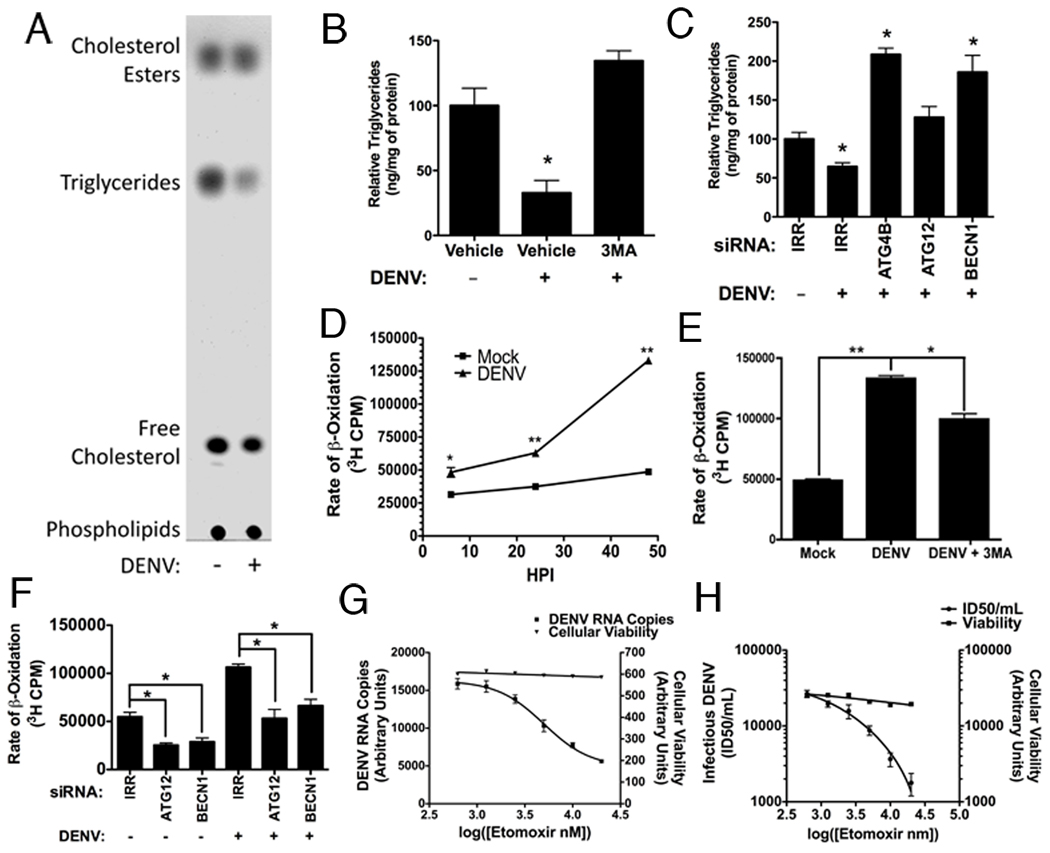
Triglycerides (TGs)are a major component of lipid droplets. Since lipid droplet area decreases in DENV-infected cells, we investigated whether TG levels were also reduced. Huh-7.5 cells were DENV- or mock-infected for 48 hours and total cellular lipids were extracted and separated via thin layer chromatography (TLC). DENV infection caused a specific depletion of TGs with minimal effect on other major lipid classes. Cholesterol and cholesterol esters, which are also components of lipid droplets, are relatively unchanged by DENV infection at the whole cell level (Fig. 6A). This may reflect a selective processing of triglycerides; or alternatively, it is possible that decreases in lipid droplet cholesterols are offset by enhanced cholesterol synthesis at sites of dengue virus replication, as has been proposed for West Nile virus (Mackenzie et al., 2007). We then quantified TGs in DENV-infected cells using a colorimetric assay. DENV-infected cells had ~65% less TGs than mock-infected cells (Fig. 6B). We also tested the requirement of autophagy for triglyceride reduction using 3MA and siRNAs targeting critical autophagy genes (Fig. 6B,C). As was the case with lipid droplet degradation, inhibiting autophagy in DENV-infected cells prevented TG depletion.
Figure 6. DENV infection depletes triglycerides and increases β-oxidation in an autophagy-dependent manner.
(A–C) DENV infection depletes triglyceride in an autophagy dependent manner. (A) DENV- or mock-infected cells (MOI=2) were harvested 48 HPI and total cellular lipids were extracted. Several major lipid classes were then resolved by TLC in a non-polar mobile phase. A representative TLC image (n=6) is shown. (B) Huh-7.5 cells were either mock-, DENV, or DENV+3MA-infected (MOI=2) for 48 hours. Cellular lipids were then extracted and the amount of triglycerides was quantified (n=3) relative to a protein loading control by colorimetric assay. *p≤0.05. (C) Huh-7.5 cells were maintained for 24 hours after introduction of irrelevant (IRR), ATG4B, ATG12 or BECN1 siRNAs, then mock- or DENV-infected (MOI=2) for 48 hours. Triglycerides were quantified as in (B). (D–G) DENV infection increases and requires β-oxidation in an autophagy-dependent manner. (D) Huh-7.5 cells were DENV- or mock-infected (MOI=2) for the indicated times, then pulsed for two hours with 3H palmitic acid to determine the rate of β-oxidation by measuring 3H2O (a byproduct of β-oxidation) via scintillation counting. (E) Huh-7.5 cells were either mock-, DENV, or DENV+3MA-infected (MOI=2) for 48 hours. β-oxidation was measured as in (D). (F) Huh-7.5 cells were transfected with the indicated siRNA (or irrelevant control) for 48 hours, then DENV infected for 32 hours. β-oxidation was measured as in (D). Huh-7.5 cells were DENV-infected for four hours, and then treated with the indicated concentrations of Etomoxir to inhibit β-oxidation. 24 hours post infection, (G) DENV RNA or (H) infectious DENV production was quantified. *p≤0.05, **p≤0.001
β-oxidation is increased by and required for DENV replication
During normal cellular metabolism, lipid droplet TGs are processed by lipases and released as free fatty acids (FFAs), which are transported into the mitochondria to undergo β-oxidation for ATP production. We tested whether the decrease in TGs in DENV-infected cells was associated with increased cellular β-oxidation. Huh-7.5 cells were DENV- or mock-infected for 6, 24, or 48 hours, then 3H-labeled palmitic acid was added to the cells and 3H2O levels (an end product of β-oxidation) were quantified. Over time, the rate of β-oxidation was significantly increased in DENV-infected cells (Fig. 6D) as compared to mock-infected cells (p<0.001), with kinetics similar to the lipid droplet depletion. To determine if autophagy was contributing to the increase in cellular β-oxidation, we assayed β-oxidation in the presence of 3MA or siRNAs targeting ATG12 and BECN1 (Fig. 6E,F). Inhibition of autophagy by 3MA significantly (p<0.05) reduced the levels of β-oxidation in DENV-infected cells, although not to the level of mock-infected cells. Silencing components of the autophagy machinery inhibited the rate of β-oxidation in both mock- and DENV- infected cells, suggesting that autophagy is required for basal and virally enhanced levels of cellular β-oxidation.
We tested the dependence of DENV replication on β-oxidation using Etomoxir, which inhibits β-oxidation by preventing transport of FFAs into the mitochondria. We first verified that Etomoxir treatment significantly inhibited β-oxidation in our assay (Fig S5A). Huh-7.5 cells were then infected with DENV for four hours and then treated with the indicated concentrations of Etomoxir to inhibit β-oxidation. 24 hours later, viral titer was determined using limiting dilution titer and RNA was harvested to measure viral RNA by quantitative RT-PCR. DENV RNA replication and the release of extracellular virus were inhibited in a dose-dependent manner, while no significant changes in cellular viability occurred (Fig. 6G,H). Thus, optimal DENV replication requires robust β-oxidation. It was recently predicted that cellular enzymes involved in β-oxidation may be regulated in HCV infection, based on cellular proteomic, lipidomic and microarray data (Blackham et al., 2010; Diamond et al., 2010). Therefore, we also examined the effects of Etomoxir on HCV replication (Fig. S5B). Although higher concentrations of Etomoxir led to an inhibition of HCV replication, HCV does not appear to be as sensitive to the inhibition of β-oxidation as is DENV. Thus, the replication of multiple viruses may be influenced by rates of cellular β-oxidation; however, their sensitivities to this process appear to differ.
Regulation of lipid metabolism is the critical function of autophagy for DENV replication
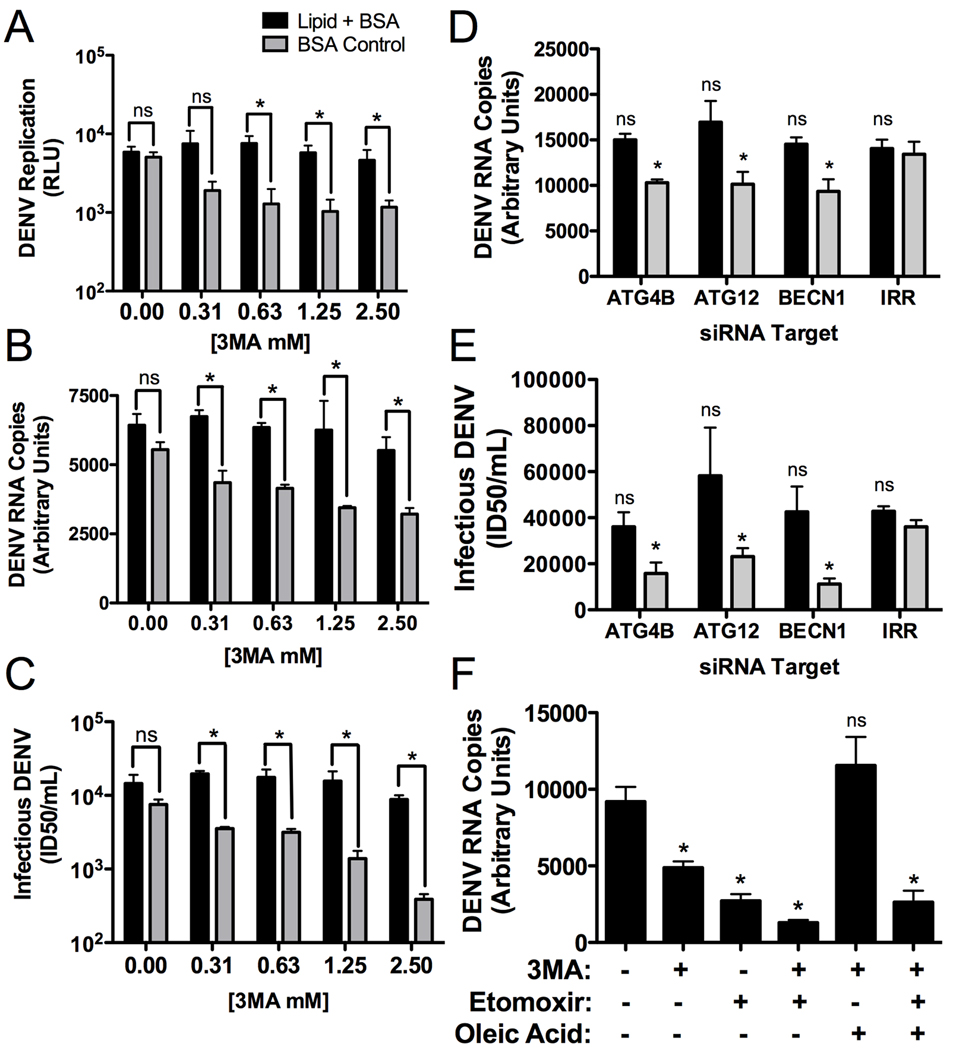
Autophagy regulates multiple cellular processes. In order to test whether its regulation of lipid metabolism was the critical function for DENV replication, we developed a lipid complementation assay. If DENV replication requires autophagosomal processing of triglycerides to release FFAs, then we should be able to complement defects in autophagy by adding exogenous FFAs to DENV-infected cells. DENV replication was quantified in the presence of 3MA with an exogenously added fatty acid (oleic acid) conjugated to BSA, as compared to BSA-only treated control cells, using the DENV luciferase reporter replicon. We first titrated the levels of BSA-oleic acid to define concentrations that did not alter DENV replication and then repeated the 3MA inhibition assay in the presence of BSA or BSA-oleic acid. In the control BSA-treated cells, 3MA inhibited DENV replication in a dose-dependent manner. In contrast, BSA-oleic acid treated cells maintained wild type levels of replication in the presence of increasing doses of 3MA (Fig. 7A). The complementation assay was repeated with infectious DENV and exogenous lipids could complement autophagy inhibition for both DENV replication and infectious virus production (Fig. 7B,C) Similarly, inhibiting autophagy with siRNAs leads to a decrease in viral RNA and titer, which can be complemented with the addition of exogenous fatty acid (Fig. 7D,E). Cellular viability was unaltered in these assays (Fig. S6). Thus, the requirement of autophagy for DENV replication can be supplanted by the addition of fatty acids.
Figure 7. Defects in DENV replication caused by autophagy inhibition can be complimented by exogenous free fatty acids.
(A) Huh-7.5 cells were electroporated with DENV luciferase reporter replicon RNAs. 24 hours post-electroporation, media was replaced with 3MA supplemented with oleic acid conjugated to BSA or BSA alone. 24 hours after addition of the drug, cells were lysed and the amount of DENV replication was assayed via luciferase assay. (B & C) Huh-7.5 cells were infected for four hours (MOI=0.5), then virus was removed and 3MA was applied at the indicated concentrations. The media was supplemented with an oleic acid-BSA conjugate or BSA alone. 48 hours post infection, (B) total RNA or (C) infectious DENV production was quantified. (D & E) Huh-7.5 cells were treated with the indicated siRNAs, maintained for 24 hours, then mock- or DENV-infected (MOI=2) for 24 hours, then (D) DENV RNA or (E) infectious DENV was quantified. (F). Huh-7.5 cells were infected for four hours (MOI=0.5), then virus was removed and the indicated inhibitors were applied. 24 hours after addition of the drug, DENV RNA was quantified. *p≤0.05.
Treatment with 3MA and Etomoxir led to an additive inhibition of DENV replication, as is expected of drugs that inhibit different steps of the same pathway (the modulation of lipid metabolism for energy production) (Fig. 7F). We next tested whether effective complementation of autophagy inhibitors with exogenous fatty acids required β-oxidation. The complementation assay was repeated in the absence or presence of Etomoxir. We observe, as before, that we can complement the 3MA DENV replication defect by adding fatty acids. However, with the addition of Etomoxir prevents complementation of the viral RNA replication defect (Fig. 7F). We conclude that the complementation of autophagy by exogenous lipids requires functional β-oxidation. Taken together, the data indicate that the primary function of autophagy in DENV replication is the regulation of lipid metabolism.
Discussion
Autophagy is critical for the replication of numerous viruses. In many cases, autophagosomes have been proposed to be sites of viral RNA replication, reviewed by (Wileman, 2006) and (Deretic and Levine, 2009). In this study, we define a novel function for autophagy in viral RNA replication: the regulation of lipid metabolism. We show that DENV infection led to the processing of cellular lipid droplets and triglycerides. The depletion of lipid droplet area correlates with an increase in autophagosomes. DENV infection stimulates β-oxidation, which is required for DENV replication. All of these DENV-induced cellular alterations require autophagy. To confirm that fatty acid release from lipid droplets is the major role of autophagy in DENV infection, we demonstrated that the requirement of autophagy for DENV replication could be supplanted by adding exogenous fatty acids to infected cells. These data suggest a model wherein DENV infection induces autophagy, which leads to the depletion of lipid droplet triglyceride stores. This results in the release of free fatty acids, which are imported into the mitochondria where they undergo β-oxidation, which generates ATP, and facilitates robust viral RNA replication. Since ATP is the critical molecule for cellular energy and an important enzymatic cofactor, increased ATP levels would enhance numerous functions in DENV replication. All mammalian cell types examined thus far have the ability to generate lipid droplets, reviewed by (Martin and Parton, 2006). Monocytes, which are thought to be a primary target of DENV, may have additional roles for autophagy including antigen presentation and innate immunity.
DENV has been proposed to assemble capsids at the ER in membranes directly adjacent to replication complexes, based on cryo-EM tomography studies (Welsch et al., 2009). Alternatively, DENV capsid co-localizes with lipid droplets under certain fixation conditions, leading to speculation that lipid droplets may be sites of virion assembly (Samsa et al., 2009). Despite the possibility that DENV may assemble at lipid droplets, it is clear that the autophagy-dependent lipid droplet modifications do not impede infectious virus assembly and release. There is a positive correlation between autophagy induction, lipid droplet depletion, robust DENV replication, and infectious virus release.
It is well established that a variety of intracellular pathogens modulate the metabolic state of the host cell. There are clear examples of both eukaryotic and bacterial pathogens that manipulate host lipid metabolism, as reviewed in (van der Meer-Janssen et al.). Trypanosoma cruzi replication vacuoles are found in close proximity to, attached to, or even containing lipid droplets (Melo et al., 2006). Similarly, Chlamydia trachomatis recruits and internalizes lipid droplets into their resident cellular vacuoles for nutrient acquisition (Kumar et al., 2006).
Other viral infections also manipulate cellular lipid metabolism. Early work on poliovirus and Semliki Forest virus showed a requirement for phospholipid biosynthesis during viral infection (Guinea and Carrasco, 1990, 1991; Perez et al., 1991). Brome mosaic virus replication is dependent on specific localized lipid compositions (Lee and Ahlquist, 2003; Lee et al., 2001). Cytomegalovirus infection profoundly alters cellular metabolism, including increases in fatty acid synthesis, glycolysis, the citric acid cycle, and nucleotide biosynthesis (Munger et al., 2006; Munger et al., 2008). West Nile virus induces cholesterol biosynthesis and redistributes cholesterol to viral RNA replication membranes (Mackenzie et al., 2007). Dengue virus RNA replication also requires cholesterol synthesis (Rothwell et al., 2009) and the DENV non-structural protein 3 recruits and stimulates fatty acid synthase activity at sites of viral replication (Heaton et al., 2010). Hepatitis B virus induces accumulation of a specific cholesterol precursor (Rodgers et al., 2009).
HCV globally alters cellular cholesterol synthesis and lipid metabolism, as shown in a recent proteomic and lipidomic study (Diamond et al., 2010). Following HCV infection, there is an early increase in host catabolic and biosynthetic activities, later followed by compensatory metabolic changes. HCV lterations in lipid metabolism appear to be mediated in part by an inhibition of the AMP-activated protein kinase (Mankouri et al., 2010). In addition to alterations in lipid metabolism, HCV and some enteroviruses also depend on phospholipid signaling for replication (Berger et al., 2009; Berger and Randall, 2009; Borawski et al., 2009; Hsu et al., 2010; Li et al., 2009; Tai et al., 2009; Trotard et al., 2009; Vaillancourt et al., 2009). HIV-1 infection stimulates similar changes, with alterations in glycolysis, the tricarboxylic acid (TCA) cycle, and cholesterol synthesis (Chan et al., 2007; Chan et al., 2009; Ringrose et al., 2008).
Lipids also play an important role in virion secretion and infectivity. HCV assembles virions at lipid droplets and secretion of infectious virus is dependent upon the very low-density lipid secretion pathway (Chang et al., 2007; Huang et al., 2007). A number of viruses manipulate the lipid content of viral envelopes. HCV virions associate with lipids and apolipoproteins and this is thought to alter the physical properties of virions, in addition to greatly enhancing infectivity (Andre et al., 2002; Andre et al., 2005). Finally, HIV-1 Nef alters cellular lipid microdomains and the lipid composition of virions (Rodgers et al., 2009).
It is clear that many intracellular pathogens have a dependence on cellular lipids. Viruses appear to have this requirement at multiple stages of the viral life cycle and frequently manipulate lipid metabolism to enhance viral production. The induction of autophagy is one mechanism by which viruses can alter cellular lipid metabolism.
Experimental Procedures
Cells, Virus, Plasmids
Huh-7.5 cells, a subline derived from the hepatocyte Huh7 cell line (Blight et al., 2002) were primarily used in this study. In addition, the human hepatocyte cell lines Huh-7, HepG2 and the baby hampster kidney cell line BHK were also used. Cells were maintained in DMEM-high glucose supplemented with 0.1mM non-essential amino acids, 5% FBS, and penicillin-streptomycin (Invitrogen). Infectious DENV-2 16681 and a DENV luciferase reporter replicon containing only the non-structural proteins (Heaton et al., 2010) were used. GFP-LC3 was constructed by PCR amplification of LC3 from a cDNA clone (Open Biosystems), which was cloned into a c-terminal monomeric GFP fusion vector, generously provided by Benjamin Glick (The University of Chicago).
Pharmacological Inhibitor Studies
3-Methyladenine (MP Biomedicals), Etomoxir (US Biological) and BSA-Oleic Acid (Sigma) were used. Huh-7.5 cells were infected at an MOI of 0.5 for 4 hours, the inoculum was removed and fresh media containing the inhibitor or vehicle alone were added to the cells and maintained for indicated times. Virus in cellular supernatants was quantified by titration, while intra-cellular RNA was isolated at 24 or 48 HPI using an RNeasy kit (Qiagen) and quantified by real time RT-PCR, as previously described (Heaton et al., 2010), see supplemental experimental procedures. For lipid complementation assays, 3-methyladenine or siRNAs were applied as described above with the addition of a 1:100 dilution of BSA-Oleic Acid (Sigma) or the equivalent concentration of BSA alone. For DENV-replicon assays, cells were treated with 3MA ± lipid or BSA control 24 hours after electroporation. 24 hours after drug addition, DENV replication was determined via use of the Renilla Luciferase Assay System per the manufacturer’s instruction (Promega). For the combination drug studies, cells were infected at an MOI=0.5 for 4 hours, then the inoculum was replaced with 2.5mM 3MA, 20µM Etomoxir, or a combination of the two. BSA-lipid was used at a 1:100 dilution as before. RNA was quantified ~24HPI. Cell viability was assayed using a MTT or CellTiter-Glo assay kit (Promega).
Immunofluorescence
Cells were infected at an MOI of 1–5 for 24–48 hours in DMEM supplemented with either 1% or 5% FBS before methanol or paraformaldehyde plus saponin fixation and antibody staining. Antibodies used included dsRNA (English and Scientific Consulting Bt.), NS1 (Abcam), LAMP1 (Abcam), NS3 (gift of Richard Kuhn, Purdue University), the DENV2-specific monoclonal antibody D1-4G2-4-15 (ATCC) and Alexa-fluor 488 or 594 secondary antibodies (Invitrogen). Stained cover slips were mounted with Prolong Gold with DAPI (Invitrogen). Oil red O (MP Biomedicals) was used per the manufacturer’s instructions. 50nM Lysotracker and 20µg/mL Bodipy 20 493/503 (Invitrogen) were visualized in live cells. Images were acquired using an Olympus DSU confocal microscope with a 100X oil objective or a Leica SP5 tandem scanner two-photon spectral confocal system using a 100X oil objective with digital zoom. Digital images were taken using Slidebook 4.1 software and processed using ImageJ (National Institutes of Health) and Adobe Photoshop. Quantification of images was performed with ImageJ using a set of defined intensity thresholds that were applied to all images.
Electron Microscopy
Huh-7.5 cells were maintained in 1% DMEM and DENV infected (MOI=5) for 48HPI and processed for EM using standard procedures (see supplemental experimental procedures). Images were taken at 300 kV with an FEI Tecnai F30 electron microscope equipped with a high-performance Gatan CCD camera. The average diameters of the observed lipid droplets were calculated by averaging multiple diameter measurements using NIH Image J software. The volume of the lipid droplets were calculated by using the average diameter in the equation v=4/3Πr3.
siRNA Inhibition
siRNAs were introduced into cells using RNAiMax (Invitrogen) according to the manufacturers instructions for optimal periods, depending on gene. The siRNA sequences and the Taqman assays used to verify knockdown relative to 18S control are described in Table S1.
Cellular lipid analysis
Huh-7.5 cells in 2% DMEM were DENV- or mock-infected at an MOI of 2 for 48 hours, washed in PBS, then scraped, pelleted, and resuspended in a glass tube in 400µL of methanol:chloroform (1:2 v/v). Lipids were extracted via the Folch procedure (Folch et al., 1957) and processed by standard thin layer chromatography, as described in supplemental experimental procedures.
β-oxidation assay
Huh-7.5 cells were infected at an MOI of 3 in 2% FBS DMEM. 10mM unlabeled palmitic acid and 10µCi/mL of 3H-palmitic acid were conjugated to 7.5µM free fatty acid free BSA (1:450 dilution). At the indicated time point, the DMEM was replaced with 500µL of the palmitic acid-BSA mixture in Kreb’s buffer (5µCi/6 well), incubated at 37°C for two hours, the Kreb’s buffer was removed from the cells and added to equal volume 10% trichloroacetic acid (TCA). After incubation at RT for 5 min, solution was spun at 13,000×g for 5 minutes, 500µL of the supernatant was removed and added to 100µL 6N NaOH, mixed, then applied to 90µm pore columns (Spectrum Labs) that were packed with a Dowex 1×8 100–200 mesh ion exchange resin (Acros Organics). The 3H2O was eluted with 1mL ddH20 and assayed for 3H using a scintillation counter.
Statistical Analysis
All statistical significance was determined using either a paired or an un-paired student’s T-test depending on the experimental design. Statistical analysis of microscopy images was based on ImageJ quantification of randomly selected fields of view or cells (n>5) for each treatment or time point. Each study shown is representative of at least two independent experiments. The level of significance is denoted in each figure, *p≤0.05, **p≤0.001.
Highlights.
DENV-induced autophagosomes partially co-localize with lipid droplets.
DENV-induced autophagy depletes lipid droplets and triglycerides.
Autophagy increases cellular β-oxidation, which is required for DENV replication.
Exogenous fatty acids supplant the requirement of autophagy for DENV replication.
Supplementary Material
Acknowledgements
We thank Benjamin Glick (University of Chicago) for providing pmGFP-C1, Charles Rice (Rockefeller University) for providing Huh-7.5 cells, Rushika Perrera and Richard Kuhn (Purdue University) for providing the DENV replicon and NS3 antibody, and Claire Huang (CDC, Fort Collins CO) for providing the DENV-2 16681 infectious clone. We thank Ernst Lengyel, Songul Dogan, and Kristin Nieman for technical expertise with the β-oxidation studies. We thank Kristi Berger, Rushika Perrera, and Richard Kuhn for critical reading of the manuscript. The authors wish to acknowledge membership within and support from the Region V 'Great Lakes' RCE (NIH award 1-U54-AI-057153). N.H. is funded by NIH training grant T32 AI065382-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andre P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Brechot C, Paranhos-Baccala G, Lotteau V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76:6919–6928. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre P, Perlemuter G, Budkowska A, Brechot C, Lotteau V. Hepatitis C virus particles and lipoprotein metabolism. Semin Liver Dis. 2005;25:93–104. doi: 10.1055/s-2005-864785. [DOI] [PubMed] [Google Scholar]
- Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KL, Randall G. Potential roles for cellular cofactors in hepatitis C virus replication complex formation. Commun Integr Biol. 2009;2:471–473. doi: 10.4161/cib.2.6.9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackham S, Baillie A, Al-Hababi F, Remlinger K, You S, Hamatake R, McGarvey MJ. Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus. J Virol. 2010;84:5404–5414. doi: 10.1128/JVI.02529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borawski J, Troke P, Puyang X, Gibaja V, Zhao S, Mickanin C, Leighton-Davies J, Wilson CJ, Myer V, Cornellataracido I, et al. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J Virol. 2009;83:10058–10074. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Qian WJ, Diamond DL, Liu T, Gritsenko MA, Monroe ME, Camp DG, 2nd, Smith RD, Katze MG. Quantitative analysis of human immunodeficiency virus type 1-infected CD4+ cell proteome: dysregulated cell cycle progression and nuclear transport coincide with robust virus production. J Virol. 2007;81:7571–7583. doi: 10.1128/JVI.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Sutton JN, Jacobs JM, Bondarenko A, Smith RD, Katze MG. Dynamic host energetics and cytoskeletal proteomes in human immunodeficiency virus type 1-infected human primary CD4 cells: analysis by multiplexed label-free mass spectrometry. J Virol. 2009;83:9283–9295. doi: 10.1128/JVI.00814-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol. 2005;26:523–528. doi: 10.1016/j.it.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R, et al. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Guinea R, Carrasco L. Phospholipid biosynthesis and poliovirus genome replication, two coupled phenomena. EMBO J. 1990;9:2011–2016. doi: 10.1002/j.1460-2075.1990.tb08329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinea R, Carrasco L. Effects of fatty acids on lipid synthesis and viral RNA replication in poliovirus-infected cells. Virology. 1991;185:473–476. doi: 10.1016/0042-6822(91)90802-i. [DOI] [PubMed] [Google Scholar]
- Haemmerle G, Zimmermann R, Zechner R. Letting lipids go: hormone-sensitive lipase. Curr Opin Lipidol. 2003;14:289–297. doi: 10.1097/00041433-200306000-00009. [DOI] [PubMed] [Google Scholar]
- Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Jr, Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci U S A. 2007;104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jormakka M, Byrne B, Iwata S. Protonmotive force generation by a redox loop mechanism. FEBS Lett. 2003;545:25–30. doi: 10.1016/s0014-5793(03)00389-2. [DOI] [PubMed] [Google Scholar]
- Khakpoor A, Panyasrivanit M, Wikan N, Smith DR. A role for autophagolysosomes in dengue virus 3 production in HepG2 cells. J Gen Virol. 2009;90:1093–1103. doi: 10.1099/vir.0.007914-0. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Y, Cocchiaro J, Valdivia RH. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr Biol. 2006;16:1646–1651. doi: 10.1016/j.cub.2006.06.060. [DOI] [PubMed] [Google Scholar]
- Lee WM, Ahlquist P. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J Virol. 2003;77:12819–12828. doi: 10.1128/JVI.77.23.12819-12828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WM, Ishikawa M, Ahlquist P. Mutation of host delta9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J Virol. 2001;75:2097–2106. doi: 10.1128/JVI.75.5.2097-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Lei HY, Liu MT, Wang JR, Chen SH, Jiang-Shieh YF, Lin YS, Yeh TM, Liu CC, Liu HS. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374:240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JM, Khromykh AA, Parton RG. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe. 2007;2:229–239. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Mankouri J, Tedbury PR, Gretton S, Hughes ME, Griffin SD, Dallas ML, Green KA, Hardie DG, Peers C, Harris M. Enhanced hepatitis C virus genome replication and lipid accumulation mediated by inhibition of AMP-activated protein kinase. Proc Natl Acad Sci U S A. 2010;107:11549–11554. doi: 10.1073/pnas.0912426107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- McGarry JD, Brown NF. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur J Biochem. 1997;244:1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- Melo RC, Fabrino DL, Dias FF, Parreira GG. Lipid bodies: Structural markers of inflammatory macrophages in innate immunity. Inflamm Res. 2006;55:342–348. doi: 10.1007/s00011-006-5205-0. [DOI] [PubMed] [Google Scholar]
- Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K–regulated manner. J Biol Chem. 2007;282:8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Panyasrivanit M, Khakpoor A, Wikan N, Smith DR. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. J Gen Virol. 2009;90:448–456. doi: 10.1099/vir.0.005355-0. [DOI] [PubMed] [Google Scholar]
- Perez L, Guinea R, Carrasco L. Synthesis of Semliki Forest virus RNA requires continuous lipid synthesis. Virology. 1991;183:74–82. doi: 10.1016/0042-6822(91)90119-v. [DOI] [PubMed] [Google Scholar]
- Perlmutter DH. Liver injury in alpha1-antitrypsin deficiency: an aggregated protein induces mitochondrial injury. J Clin Invest. 2002;110:1579–1583. doi: 10.1172/JCI16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Schulz H. Metabolic functions of the two pathways of oleate beta-oxidation double bond metabolism during the beta-oxidation of oleic acid in rat heart mitochondria. J Biol Chem. 2003;278:111–116. doi: 10.1074/jbc.M209261200. [DOI] [PubMed] [Google Scholar]
- Ringrose JH, Jeeninga RE, Berkhout B, Speijer D. Proteomic studies reveal coordinated changes in T-cell expression patterns upon infection with human immunodeficiency virus type 1. J Virol. 2008;82:4320–4330. doi: 10.1128/JVI.01819-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers MA, Saghatelian A, Yang PL. Identification of an overabundant cholesterol precursor in hepatitis B virus replicating cells by untargeted lipid metabolite profiling. J Am Chem Soc. 2009;131:5030–5031. doi: 10.1021/ja809949r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell C, Lebreton A, Young Ng C, Lim JY, Liu W, Vasudevan S, Labow M, Gu F, Gaither LA. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology. 2009;389:8–19. doi: 10.1016/j.virol.2009.03.025. [DOI] [PubMed] [Google Scholar]
- Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Dengjel J, Schoor O, Stevanovic S, Munz C. Autophagy in innate and adaptive immunity against intracellular pathogens. J Mol Med. 2006;84:194–202. doi: 10.1007/s00109-005-0014-4. [DOI] [PubMed] [Google Scholar]
- Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J Virol. 2007;81:12543–12553. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotard M, Lepere-Douard C, Regeard M, Piquet-Pellorce C, Lavillette D, Cosset FL, Gripon P, Le Seyec J. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J. 2009;23:3780–3789. doi: 10.1096/fj.09-131920. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Vaillancourt FH, Pilote L, Cartier M, Lippens J, Liuzzi M, Bethell RC, Cordingley MG, Kukolj G. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology. 2009;387:5–10. doi: 10.1016/j.virol.2009.02.039. [DOI] [PubMed] [Google Scholar]
- van der Meer-Janssen YP, van Galen J, Batenburg JJ, Helms JB. Lipids in host-pathogen interactions: pathogens exploit the complexity of the host cell lipidome. Prog Lipid Res. 49:1–26. doi: 10.1016/j.plipres.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wileman T. Aggresomes and autophagy generate sites for virus replication. Science. 2006;312:875–878. doi: 10.1126/science.1126766. [DOI] [PubMed] [Google Scholar]
- Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol. 2008;82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SY, Ha YE, Choi JE, Ahn J, Lee H, Kweon HS, Lee JY, Kim DH. Coxsackievirus B4 uses autophagy for replication after calpain activation in rat primary neurons. J Virol. 2008;82:11976–11978. doi: 10.1128/JVI.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.