Abstract
Epidermal growth factor receptor-2 (EGFR-2) has been implicated in the pathogenesis of breast and other carcinomas. In this report, we tested the ability of a bacteriophage selected peptide KCCYSL, radiolabeled with 64Cu, to image EGFR-2 expressing breast tumors in vivo by positron emission tomography (PET). We evaluated and compared the in vivo tissue distribution and imaging properties of 64Cu-X-(Gly-Ser-Gly)-KCCYSL peptide (X = 1,4,7,10, tetraazacyclododecane-N,N’,N’’,N’’’-tetracetic acid, [DOTA] 1,4,8,11-tetraazabicyclo[6.6.2]hexadecane-4,11-diacetic acid [CB-TE2A], and 1,4,7-triazacyclononane-1,4,7-triacetic acid [NOTA] chelators) in a human breast cancer xenograft mouse model using dual modality PET and computed tomography (CT). The synthesized peptides DO3A-GSG-KCCYSL, CB-TE2A-GSG-KCCYSL, and NO2A-GSG-KCCYSL were purified, radiolabeled with 64Cu, and evaluated for human breast cancer cell (MDA-MB-435) binding, 50% inhibitory concentration, and serum stability. In vivo pharmacokinetic and small animal PET/CT imaging studies were performed using SCID mice bearing MDA-MB-435 xenografts. The radiolabeled peptides bound with an 50% inhibitory concentration of 42.0 ± 10.2 nM (DO3A), 31 ± 7.9 nM (CB-TE2A), and 44.2 ± 6.6 nM (NO2A) to cultured MDA-MB-435 cells. All of the radiolabeled peptides were stable in vitro. The tumor uptake of DO3A, CB-TE2A, and NO2A peptides were 0.73 ± 0.15 percent injected dose per gram (%ID/g), 0.64 ± 0.08%ID/g, and 0.52 ± 0.04%ID/g, respectively at 1 hour. Liver uptake for the 64Cu-DO3A-peptide (1.68 ± 0.42%ID/g) was more than that of the 64Cu-CB-TE2A-peptide (0.52 ± 0.02% ID/g) and 64Cu-NO2A-peptide (0.48 ± 0.05%ID/g) at 2 hours. PET/CT studies revealed successful tumor uptake of 64Cu-peptides at 2 hours postinjection. In vivo kidney retention was observed with all of the radiolabeled peptides. The optimization of bifunctional chelators improves the pharmacokinetic properties of the 64Cu-labeled GSG-KCCYSL peptide, which enables the selection of a suitable peptide homolog as a PET imaging agent for EGFR-2 expressing breast carcinomas.
Key words: breast cancer, 64Cu-chelators, EGFR-2, peptide, PET-imaging
Introduction
Epidermal growth factor receptor-2 (EGFR-2, erythroblastic leukemia viral oncogene homolog 2 [ErbB-2]) is a member of the EGFR family of transmembrane receptor tyrosine kinases.1,2 EGFR-2 expression has been reported to be minimal in normal tissues,3 however, amplification of this receptor occurs in approximately 30% of invasive breast cancers and is associated with poor patient outcome.4–6 Hence, EGFR-2 has been identified as a target for cancer imaging and therapy. In the past few years, several groups have developed antibodies, scFV fragments, and peptides7–10 that bind to EGFR-2.
Bacteriophage (phage) display libraries have been extensively used to identify peptides that bind to a variety of targets including EGFR-2.11–13 Such phage-derived peptides, once radiolabeled, hold promise as effective imaging agents for identifying specific human tumors and evaluating response to therapy. Although many imaging agents have been developed using antibodies and proteins, they often demonstrate low specificity, poor accumulation in target tissue, sensitivity to reaction conditions, and slow clearance properties in vivo.14,15 Accordingly, the number of radiopharmaceutials utilizing bifunctional chelator-conjugated peptides for use in imaging has grown considerably over the past few decades.16,17 Previously, we reported an 111In-labeled phage display selected peptide, KCCYSL, for single photon emission computed tomography (SPECT) imaging of EGFR-2 expressing human breast tumor xenografts in mice.18 However, due to the inherent limited sensitivity and resolution of SPECT imaging, we were interested in evaluating KCCYSL peptide for the positron emission tomography (PET) targeted imaging of EGFR-2 expressing tumor(s).
PET imaging depends on the delivery of a targeting ligand containing a positron emitting radionuclide to a tissue or organ of interest. Copper-64 (t1/2= 12.7 hours) is an attractive radionuclide for PET imaging that can be used for both diagnostic imaging and radionuclide therapy due to its dual decay characteristics.19 The positron emissions (β+:17.8%, E β+max= 653 keV; β−: 38.4%, E β-max= 578 keV) would allow clinicians to image a patient using PET and provide therapeutic treatment using the same 64Cu-targeted pharmaceutical. Unfortunately, the application of 64Cu-based radiopharmaceuticals has been hindered by the lack of stable copper radiotracers in vivo. The high uptake and retention of copper-containing compounds in the blood and liver are well known.20,21 Therefore, more stable chelator-compounds are necessary for in vivo studies using 64Cu.
Macrocyclic polyamine complexes of 1,4,8,11-tetraazacyclotetradecane-N,N’,N’’,N’’’-tetracetic acid (TETA) and 1,4,7,10, tetraazacyclododecane-N,N’,N’’,N’’’-tetracetic acid (DOTA) with copper have been shown to exhibit greater kinetic inertness than acyclic analogs, but dissociation of copper from both the chelators occurs in vivo.22–24 Weisman, Wong, and Anderson designed and characterized the novel cross-bridged tetraaza-macrocyclic chelator 1,4,8,11-tetraazabicyclo[6.6.2]hexadecane-4,11-diacetic acid (CB-TE2A) and demonstrated enhanced in vivo stability with significantly less transchelation in the liver compared with 64Cu-TETA. Thus, the rigidity of the cross-bridge system was credited for the greater kinetic stability of the copper (II)-CB-TE2A complex.25,26
Previous studies have also demonstrated the use of 1,4,7-triazacyclononane-1,4,7-triacetic acid (NOTA), a 9-member cyclic chelating agent for divalent copper.27–29 NOTA was shown to have the capacity to form stable metal complex with Cu2+ and other divalent and trivalent metals,29 which overcome demetallation and tracer uptake in tissues such as liver. Functionalized NOTA derivatives suitable for conjugation with proteins or peptides have been reported by different groups.29,30 In this study, we evaluated and compared the in vitro and in vivo properties of DO3A-GSG-KCCYSL, CB-TE2A-GSG-KCCYSL, and NO2A-GSG-KCCYSL radiolabeled with 64Cu. The radiolabeled peptides were tested for cell binding, stability, pharmacokinetics, and tumor imaging in MDA-MB-435 human breast carcinoma xenografted mice using small-animal PET/CT.
Materials and Methods
Reagents, cell lines, and animals
Reverse phase-high performance liquid chromatography (RP-HPLC)–grade acetonitrile, and trifluoroacetic acid were purchased from Fischer Scientific for HPLC analysis. Copper-64 was purchased from Nuclear Reactor Laboratory, University of Wisconsin. RPMI (Roswell Park Memorial Institute) 1640 and human mammary epithelial cell media were purchased from Invitrogen/Gibco. Bovine serum albumin (BSA) and trypsin were obtained from Sigma Chemical Company. DOTA was purchased from Macrocyclics, and CB-TE2A was synthesized as previously described.31 NOTA was procured from Chematech.
Normal mammary 184A1 cell line was obtained from American Type Tissue Culture. The MDA-MB-435 cells were maintained as monolayer cultures in RPMI-1640 medium supplemented with 10% fetal bovine serum, sodium pyruvate, nonessential amino acids, and l-glutamine. 184A1 cells were grown in (human mammary epithelial cell) media supplemented with bovine pituitary extract. Cell cultures were maintained at 37°C in a 5% CO2 humidified incubator. Sub-culturing was performed using standard trypsinization procedures.
Four- to 5-week-old ICR-SCID (Institute of Cancer Research severely compromised immunodeficient) female mice were obtained from Taconic Farms. The mice were supplied with sterilized water and irradiated rodent chow ad libitum. Under gas anesthesia, each SCID mouse was subcutaneously inoculated in the right shoulder with 1 × 107 MDA-MB-435 cells. The tumors were allowed to grow for 3–4 weeks before studies were performed. Animal studies were conducted in accordance with the protocols approved by the institutional animal care and use committee in accordance with U.S. Public Health Service guidelines.
Solid-phase peptide synthesis
Peptides used in this study were synthesized using an Advanced Chem Tech 396 multiple peptide synthesizer (AAPPTEC) using standard Fmoc chemistry. Briefly, the resin (50 μmol) was deprotected using piperidine, resulting in the formation of a primary amine from which the C-terminus of the growing peptide was anchored. The Fmoc protected amino acids (225 μmol) with appropriate orthogonal protection were activated using O-benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluoro-phosphate, N-hydroxy-benzotriazole, and N,N–diisopropylethylamine and sequentially added to the resin. The metal chelators DOTA or NOTA were coupled to the amino terminus of the linear GSG-KCCYSL. In the case of CB-TE2A, the chelator was coupled to the peptide on the resin through an intermediate preparation of the activated mixed anhydride by use of diisopropylcarbodiimide/N,N-diisopropylethylamine/dimethylformamide as described earlier.31 The resulting peptide was deprotected and cleaved from the resin using a cocktail consisting of thioanisol, water, 1,2-ethanedithiol, phenol, triisopropylsilane (2.5 ratio each), and trifluoroacetic acid, respectively. The cleaved peptide was then precipitated and washed using ether. The crude conjugate was dried, weighed, and analyzed by Electrospray Ionization mass-spectrometry (Thermo Finnigan). A NovaPak (HR-C18, 7.8 × 300 mm, 6 μM, 60 Å) (Waters) column was used for the purification of bulk amounts of peptides. The peptides were peak purified to ≥98% purity before in vitro cell binding assays.
Radiolabeling of peptides with 64Cu
Radiolabeling of DO3A-, CB-TE2A-, or NO2A-(GSG)-KCCYSL with 64Cu was performed as follows: For DO3A-(GSG)-KCCYSL, the peptide (50 μg) was radiolabeled with 29.6 MBq 64Cu in 80 μL ammonium acetate (0.1 M) pH 5.5, at 85°C for 90 minutes. Similarly, CB-TE2A-(GSG)-KCCYSL (50 μg) was radiolabeled with 29.6 MBq 64Cu in ammonium acetate (0.4 M) pH 7.0, at 85°C for 90 minutes, and NO2A-(GSG)-KCCYSL (50 μg) was radiolabeled with 29.6 MBq 64Cu in ammonium acetate (0.4 M) pH 6.8, at 75°C for 60 minutes. The reaction buffer was purged with nitrogen before radiolabeling to inhibit or minimize peptide oxidation. Also, as a precautionary measure, the reaction buffers contained 0.05 mM Tris- (2-carboxyethyl) phosphine hydrochloride (TCEP.HCl) to maintain cysteine reduction in the wake of radiolytic oxidation during the peptide metal chelation process. The resulting radiolabeled peptides were peak purified using a Phenomenex Jupiter 5u C18 300 Å 250 × 4.6 mm column and RP-HPLC (10%–95% acetonitrile/0.1% TFA) for 30 minutes to separate them from their nonradiolabeled counterparts. The peak-purified peptides were concentrated using Empore high efficiency (C18) extraction disk cartridges and eluted with 400 μL of an 8:2 ethanol/sterile saline solution. The ethanol was evaporated under a stream of nitrogen and was diluted to the appropriate volume with sterile saline. Radiochemical yields for 64Cu-DO3A-, 64Cu-CB-TE2A-, and 64Cu-NO2A-GSG-KCCYSL averaged 55%, 45%, and 57%, respectively. Radiochemical purity of all 64Cu-labeled peptides was found to be ≥98% pure.
Preparation of native Cu-peptides (natCu)
For nonradioactive Cu-labeling, 0.5 mg samples of DO3A-(GSG)-KCCYSL, CB-TE2A-(GSG)-KCCYSL, and 64Cu-NO2A-GSG-KCCYSL peptides were dissolved separately in 500 μL of ammonium acetate buffer (purged with nitrogen and contained TCEP) with appropriate pH (as mentioned above) containing 0.8 mM copper(II) sulfate-pentahydrate solution. The solutions were heated at identical conditions used for radiolabeling procedure and allowed to cool to room temperature. natCu-peptides were then peak purified by RP-HPLC. The native (natCu)-peptides were determined to be ≥98% pure by RP-HPLC analyses. Electrospray Ionization mass-spectrometry was performed to check the integrity of the peptides.
In vitro cell binding of radiolabeled KCCYSL peptides
Experiments were performed to test the ability of the radiolabeled peptides to bind cell lines that express low to moderate levels of EGFR-232 (MDA-MB-435) or normal mammary epithelial cells (184A1) with negligible EGFR-2 expression. Cells grown in culture flasks were trypsinized, released, and washed once in cell binding media (RPMI 1640 with 25 mM HEPES,8 pH 7.4, 0.2% BSA, 3 mM 1, 10-phenanthroline). Cells (1 × 105 cells/tube) were transferred to microcentrifuge tubes containing 0.3 mL cell binding medium and were incubated at 37°C for different times (15 minutes, and 0.5, 0.75, 1, 1.5 and 2 hours) with 1 × 105 counts per minute (cpm) of radiolabeled peptides. After incubation, the media were removed, and the cells were rinsed with ice-cold 0.01 M PBS, pH 7.4, 0.2% BSA, and centrifuged. This process was repeated twice. Radioactivity bound to the cells was quantified in a Wallac γ counter (PerkinElmer Life and Analytical Sciences Inc.). Cell binding ability was reported as total radioactivity in cpm that was bound to the cells.
Fifty-percent inhibitory concentration (IC50) binding studies for radiopeptides were performed using cultured MDA-MB-435 breast carcinoma cells. For competitive binding assays, MDA-MB-435 cells (2.5 × 106 cells/tube) were incubated with 2.5 × 104 cpm 64Cu-DO3A or 64Cu-CB-TE2A or 64Cu-NO2A peptide and increasing concentrations of their respective nonradioactive natCu-peptide counterparts (10−12–10−5 M) (n = 3 each). The cell-associated radioactivity was measured in a Wallac γ counter, and the binding affinity was determined by the Grafit software program (Erithacus Software Limited).
In vitro serum stability of the radiolabeled peptides was tested by incubating 29.6 MBq of the respective peptide in 0.3 mL of mouse serum at 37°C for 0.5, 1, 2, 4, and 24 hours, respectively. At various time points, 40 μL aliquots were removed, and the proteins were precipitated with 40 μL acetonitrile. The samples were centrifuged at 12,500 rpm for 5 minutes, and the cleared lysate (∼40 μL aliquots) was analyzed by RP-HPLC with a 0%–95% gradient acetonitrile in 30 minutes to assess the integrity of the radiolabeled peptides.
Pharmacokinetic studies of 64Cu-labeled peptides in MDA-MB-435 xenograft SCID mice
SCID mice bearing MDA-MB-435 tumors (3 per time point) were injected with approximately 0.185 MBq of different radiolabeled peptides and were sacrificed by cervical dislocation at 30 minutes and 1, 2, 4, and 24 hours postinjection (p.i.), after which tissues and organs of interest were collected. Gastrointestinal tract contents were not removed. The animal, tissue, and organ samples were weighed, and counts were determined with an automated γ counter. Uptake of radioactivity in the tumor and normal tissues and organs was expressed as a percentage of the injected radioactive dose per gram (%ID/g). Whole blood %ID or %ID/g was determined assuming the blood accounted for 6.5% of the body weight of the mouse. For blocking experiments, the MDA-MB-435 breast tumor-bearing mice (n = 3) were preinjected with 100 μg of natCu-DO3A or natCu-CB-TE2A or natCu-NO2A peptides. After 10 minutes p.i., of the nonradiolabeled peptide, 0.185 MBq of radiolabeled counterpart was injected and the blocking efficiency was evaluated after 2 hours.
For reducing the kidney retention of the radiolabeled peptides in mice, inhibition experiments with albumin fragments were performed.33 Briefly, 335 mg of trypsin was added to 3.0 g of BSA in 50 mM ammonium carbonate (15 mL). The mixture was incubated at 37°C for 24 hours. The trypsin-digested sample was filtered using 50 kDa Centripep (YM-50) centrifugal filters (Millipore). The filtrate containing albumin fragments <50 kDa was further fractionated using a YM-3 filter (3 kDa cutoff), which yielded albumin fragments <3 kDa (filtrate) and fragments with molecular weight range of 3–50 kDa (residue). The effect of inhibition on renal uptake of the radiolabeled peptides was tested in the presence of the 3–50 kDa albumin fraction.
In separate experiments, the 3–50 kDa albumin fragment (200 μg) sample in normal saline (100 μL) was preinjected into the MDA-MB-435 tumor-bearing mice (n = 3) via the tail vein. After 5 minutes, 0.185 MBq of 64Cu-DO3A-(GSG)-KCCYSL or 64Cu-CB-TE2A-(GSG)-KCCYSL or 64Cu-NO2A-(GSG)-KCCYSL was administered into the mice. The mice were sacrificed after 2 hours p.i., and the kidney uptake of the radiolabeled peptides was measured in a γ-counter. Control experiments (n = 3) were run in parallel with preinjection of normal saline instead of albumin fragments.
MicroPET/CT imaging studies
In vivo microPET/CT of MDA-MB-435 tumor-bearing mice with the radiolabeled peptides were performed as follows. MDA-MB-435 tumor-bearing mice were injected in the tail vein with 12.0 MBq of 64Cu-DO3A- or 64Cu-CB-TE2A- or 64Cu-NO2A- (GSG)-KCCYSL and imaged at 2 hours p.i., in a small animal PET scanner (MOSAIC small animal PET unit [Philips]). This PET scanner has a transverse field of view of 12.8 cm and a gantry diameter of 21 cm. The scanner operates in a three-dimensional (3D) mode. The mouse to be imaged was laser aligned at the center of the scanner field of view for imaging. The microPET image reconstruction was performed with a 3D row-action maximum-likelihood algorithm. Small animal CT was performed for the purpose of anatomic/molecular data fusion, and concurrent image reconstruction was performed with a Fanbeam (Feldkamp) filtered-backprojection algorithm. Reconstructed Digital Imaging and Communication in Medicine PET images were imported into AMIRA 3.1 software (Visage Imaging, Inc.) for subsequent image fusion with microCT and 3D visualization.
Statistical analysis
The data are presented as mean ± standard deviation. For statistical analysis, a Student's t-test was performed using Prism software (Graphpad Software). A p-value ≤0.05 was considered significant.
Results
Preparation and characterization of different chelator-KCCYSL peptide
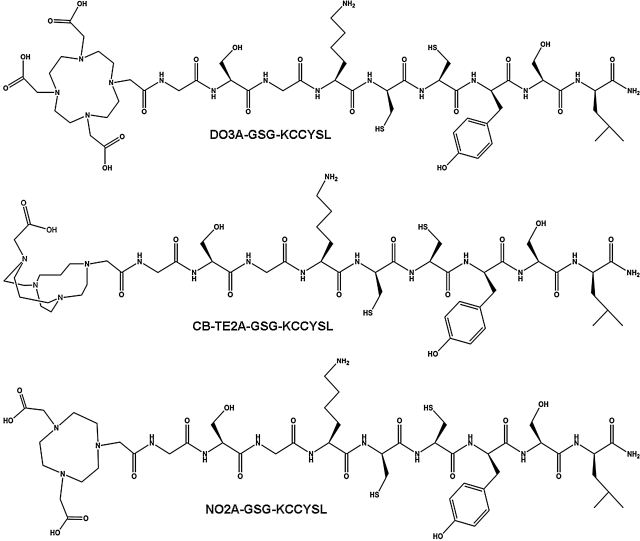
The peptides DO3A-(GSG)-KCCYSL, CB-TE2A-(GSG)-KCCYSL, and NO2A-(GSG)-KCCYSL were synthesized by solid-phase synthesis with GSG spacers between the chelators and the amino termini of the peptides to avoid possible steric hindrances. The chemical structures of different chelator-peptides are shown in Figure 1. Nonradioactive Cu-chelator-peptides were prepared by heating aqueous solutions of the peptides with CuSO4.5H2O at temperatures similar to those described for radioactive peptides. Nonradiolabeled copper-labeled peptides were purified and analyzed by mass spectrometry before cell binding assays. The calculated and observed molecular weight of natCu-peptide was 1365.0, 1364.7.0 (DO3A-peptide), 1302.0, 1301.8 (CB-TE2A–peptide), and 1326.0, 1325.8 (NO2A-peptide), respectively.
FIG. 1.
Chemical structures of DO3A-(GSG)-KCCYSL, CB-TE2A-(GSG)-KCCYSL, and NO2A-(GSG)-KCCYSL.
For radiolabeling, DO3A-(GSG)-KCCYSL was dissolved in 0.1 M ammonium acetate buffer (pH 5.5), CB-TE2A-(GSG)-KCCYSL in 0.4 ammonium acetate (pH 7.0) buffer, and NO2A-(GSG)-KCCYSL in ammonium acetate (0.4 M) pH 6.8. 64CuCl2 was added to the solutions and heated at different temperatures (85°C, 90 minutes) for DO3A and CB-TE2A peptides and (75°C for 60 minutes) for NO2A-peptide, respectively. The radiolabeled peptides were purified by RP-HPLC in order to separate them from their respective nonradiolabeled counterparts. The retention times of 64Cu-DO3A-, 64Cu-CB-TE2A-, and 64Cu-NO2A-GSG-KCCYSL were 12.8, 13.3, and 13.0 minutes, respectively, under identical RP-HPLC conditions. All the peptides were obtained at high radiochemical purity with a reasonable radiochemical yield (Table 1).
Table 1.
Characterization of 64Cu Labeled Peptides
| Peptide | RP-HPLC (tr-min) | Labeling yield (%) | Radiochemical purity | Receptor affinity (IC50, nM) |
|---|---|---|---|---|
| 64Cu-DO3A-GSG-KCCYSL | 12.8 | 55 | ≥98 | 42 ± 10.2 |
| 64Cu-CB-TE2A-GSG-KCCYSL | 13.3 | 45 | ≥98 | 31 ± 7.9 |
| 64Cu-NO2A-GSG-KCCYSL | 13.0 | 57 | ≥98 | 44 ± 6.6 |
The IC50 was determined using corresponding nonradiolabeled counterparts. Details are given in Materials and Methods section.
IC50, 50% inhibitory concentration; RP-HPLC, Reverse phase-high performance liquid chromatography.
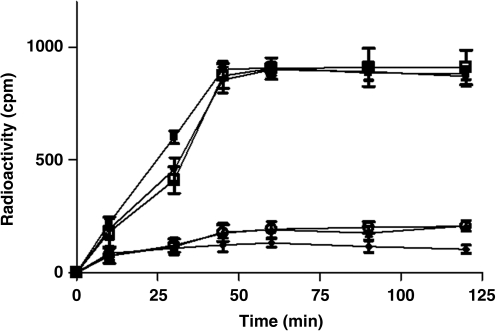
Studies to test the ability of the 64Cu-radiolabeled peptides to bind MDA-MB-435 breast carcinoma cells known to express EGFR-232 or normal mammary epithelial cells (184A1) with negligible expression of EGFR-2 were performed. The 64Cu-DO3A, 64Cu-CB-TE2A, and 64Cu-NO2A peptides were able to bind EGFR-2 expressing MDA-MB-435 cells in a time-dependent manner. The peptides exhibited a similar cell binding profile up to 45 minutes, beyond which binding of the peptides to MDA-MB-435 cells appeared to be saturated. Further increase in cell binding was not observed up to 2 hours (Fig. 2). The peptides demonstrated less or no binding to 184A1 cells, indicating the specificity of the radiolabeled peptides for EGFR-2 expressing cells.
FIG. 2.
Epidermal growth factor receptor-2 receptor binding properties of the radiolabeled 64Cu-DO3A-(GSG)-KCCYSL, 64Cu-CB-TE2A-(GSG)-KCCYSL, and 4Cu-NO2A-(GSG)-KCCYSL peptides. Approximately, 1.0 × 105 MDA-MB-435 breast carcinoma or normal mammary epithelial cells/tube were incubated at 37°C for different time intervals with 100,000 counts per minute radioligand. The radiolabled peptides bound to human MDA-MB-435 breast carcinoma cells. 64Cu-DO3A- peptide (•); 64Cu-CB-TE2A-peptide (▪) and 64Cu-NO2A- peptide (□).Very minimal or no binding was observed with normal mammary epithelial (184A1) cells using 64Cu-DO3A-peptide (▴) or 64Cu-CB-TE2A-peptide (♦) or 64Cu-NO2A- peptide (○), respectively.
Receptor binding affinity studies with radiolabeled peptides in the presence of their respective nonradioactive counterparts were carried out using cultured human MDA-MB-435 breast carcinoma cells. The radiolabeled peptides demonstrated nanomolar affinity with IC50 values of 42 ± 10.2 nM (64Cu-DO3A-peptide), 31 ± 7.9 nM (64Cu-CB-TE2A-peptide), and 44 ± 6.6 nM (64Cu-NO2A-peptide) (mean ± standard deviation, n = 3, Table 1), respectively. The IC50 values suggest that the radiolabeled peptides with different chelators possessed comparable EGFR-2 binding affinities.
To determine their metabolic stability, the radiolabeled peptides were incubated in mouse serum for various time points in vitro, at 37°C. As determined by RP-HPLC, the radiolabeled peptides were stable in serum for periods up to 2 hours. However, an additional peak was observed with all three radiolabeled peptides, which eluted very close to the original radiolabeled peak, indicating degradation of peptide with passage of time at 4 and 24 hours (data not shown).
Biodistribution and imaging studies
Detailed in vivo pharmacokinetic studies with all the radiolabeled peptides in SCID mice bearing MDA-MB-435 xenografts were performed and are summarized in Tables 2–4. At 2 hours p.i., 89.4 ± 3.6%ID had been excreted from the mice using 64Cu-DO3A-peptide conjugate in comparison with 95.2 ± 0.48%ID of 64Cu-CB-TE2A-peptide and 96.05 ± 0.15%ID of 64Cu-NO2A-peptide conjugate, respectively. Overall clearance of all the 64Cu-chelated peptides proceeded primarily through the renal system, with ∼2.4 ± 0.36%ID (64Cu-DO3A-peptide), 0.54 ± 0.05%ID (64Cu-CB-TE2A-peptide), and 0.48 ± 0.07%ID (64Cu-NO2A-peptide) being excreted through the hepatobiliary system.
Table 2.
Pharmacokinetic Studies of 64Cu-DO3A-GSG-KCCYSL Using MDA-MB-435 Tumor-Bearing SCID Mice
| Tissues | 0.5 hour | 1 hour | 2 hours | 2 hours block | 4 hours | 24 hours |
|---|---|---|---|---|---|---|
| Percent injected dose/g (%ID/g)a | ||||||
| Tumor | 0.91 ± 0.12 | 0.73 ± 0.15 | 0.54 ± 0.06 | 0.29 ± 0.09b | 0.45 ± 0.20 | 0.27 ± 0.02 |
| Blood | 1.08 ± 0.22 | 0.35 ± 0.14 | 0.20 ± 0.08 | 0.18 ± 0.04 | 0.16 ± 0.12 | 0.15 ± 0.02 |
| Brain | 0.05 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.03 ± 0.01 |
| Heart | 0.57 ± 0.03 | 0.34 ± 0.17 | 0.35 ± 0.09 | 0.24 ± 0.02 | 0.36 ± 0.21 | 0.30 ± 0.05 |
| Lung | 1.40 ± 0.07 | 0.96 ± 0.15 | 0.79 ± 0.07 | 0.70 ± 0.09 | 0.64 ± 0.20 | 0.56 ± 0.12 |
| Liver | 2.30 ± 0.21 | 1.72 ± 0.94 | 1.68 ± 0.42 | 1.47 ± 0.53 | 1.65 ± 1.01 | 1.07 ± 0.11 |
| Spleen | 0.51 ± 0.07 | 0.40 ± 0.14 | 0.57 ± 0.31 | 0.52 ± 0.40 | 0.68 ± 0.44 | 0.54 ± 0.03 |
| Stomach | 0.54 ± 0.20 | 0.56 ± 0.26 | 0.36 ± 0.11 | 0.29 ± 0.10 | 1.14 ± 1.00 | 0.14 ± 0.08 |
| Kidneys | 6.37 ± 1.44 | 4.32 ± 0.57 | 4.27 ± 0.84 | 4.12 ± 1.23 | 2.96 ± 0.31 | 1.49 ± 0.24 |
| Muscle | 0.23 ± 0.02 | 0.08 ± 0.03 | 0.06 ± 0.01 | 0.05 ± 0.01 | 0.07 ± 0.04 | 0.05 ± 0.01 |
| Pancreas | 0.31 ± 0.10 | 0.14 ± 0.09 | 0.23 ± 0.05 | 0.18 ± 0.07 | 0.18 ± 0.09 | 0.17 ± 0.08 |
| Bone | 0.26 ± 0.09 | 0.12 ± 0.05 | 0.15 ± 0.08 | 0.12 ± 0.05 | 0.12 ± 0.06 | 0.03 ± 0.01 |
| Percent injected dose (%ID) | ||||||
| Intestines | 2.05 ± 0.17 | 2.34 ± 1.3 | 3.1 ± 1.50 | 3.4 ± 0.40 | 3.3 ± 1.94 | 1.11 ± 0.14 |
| Urine | 81.11 ± 1.20 | 88.4 ± 4.5 | 89.4 ± 3.60 | 87.8 ± 0.57 | 90.0 ± 5.40 | 94.9 ± 0.50 |
| Uptake ratio of tumor/normal tissue | ||||||
| Tumor/blood | 0.84 | 2.08 | 2.70 | — | 2.40 | 1.80 |
| Tumor/muscle | 3.95 | 9.12 | 9.00 | 6.40 | 5.40 | |
| Tumor/liver | 0.39 | 0.42 | 0.32 | 0.27 | 0.20 | |
Data are presented as %ID/g ± SD except for intestines and urine, values for which are expressed as%ID ± standard deviation (n = 3).
p = 0.03, significance comparison between the tumor uptake of radiolabeled peptide in the absence and presence of its nonradiolabeled counterpart at 2 hours p.i.
p.i., postinjection; SD, standard deviation.
Table 4.
Pharmacokinetic Studies of 64Cu-NO2A-GSG-KCCYSL Using MDA-MB-435 Tumor-Bearing SCID Mice
| Tissues | 0.5 hour | 1 hour | 2 hours | 2 hours block | 4 hours | 24 hours |
|---|---|---|---|---|---|---|
| Percent injected dose/g (%ID/g)a | ||||||
| Tumor | 0.78 ± 0.03 | 0.64 ± 0.04 | 0.61 ± 0.06 | 0.32 ± 0.09b | 0.43 ± 0.02 | 0.1 ± 0.01 |
| Blood | 1.29 ± 0.09 | 0.33 ± 0.11 | 0.08 ± 0.01 | 0.10 ± 0.04 | 0.05 ± 0.02 | 0.01 ± 0.01 |
| Brain | 0.05 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.001 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Heart | 0.40 ± 0.05 | 0.16 ± 0.06 | 0.04 ± 0.01 | 0.08 ± 0.02 | 0.04 ± 0.02 | 0.07 ± 0.02 |
| Lung | 1.21 ± 0.08 | 0.49 ± 0.07 | 0.22 ± 0.08 | 0.34 ± 0.07 | 0.15 ± 0.06 | 0.10 ± 0.03 |
| Liver | 0.90 ± 0.09 | 0.55 ± 0.07 | 0.48 ± 0.05 | 0.57 ± 0.20 | 0.34 ± 0.10 | 0.27 ± 0.05 |
| Spleen | 0.41 ± 0.07 | 0.19 ± 0.11 | 0.11 ± 0.05 | 0.11 ± 0.05 | 0.08 ± 0.05 | 0.07 ± 0.02 |
| Stomach | 0.37 ± 0.04 | 0.18 ± 0.03 | 0.14 ± 0.04 | 0.20 ± 0.10 | 0.13 ± 0.03 | 0.10 ± 0.05 |
| Kidneys | 7.87 ± 0.96 | 6.51 ± 0.63 | 4.23 ± 1.00 | 6.70 ± 0.89 | 1.37 ± 0.28 | 0.68 ± 0.07 |
| Muscle | 0.20 ± 0.05 | 0.06 ± 0.02 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.00 |
| Pancreas | 0.34 ± 0.09 | 0.10 ± 0.08 | 0.09 ± 0.03 | 0.05 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 |
| Bone | 0.08 ± 0.05 | 0.04 ± 0.05 | 0.04 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.01 |
| Percent injected dose (%ID) | ||||||
| Intestines | 1.2 ± 0.13 | 1.1 ± 0.27 | 1.06 ± 0.14 | 1.30 ± 0.60 | 1.33 ± 0.35 | 0.5 ± 0.10 |
| Urine | 84.4 ± 1.93 | 90.1 ± 4.04 | 96.0 ± 0.15 | 93.9 ± 1.06 | 96.7 ± 0.13 | 98.1 ± 0.14 |
| Uptake ratio of tumor/normal tissue | ||||||
| Tumor/blood | 0.60 | 1.94 | 7.62 | — | 8.60 | 3.00 |
| Tumor/muscle | 4.00 | 10.70 | 10.71 | 17.00 | 14.20 | |
| Tumor/liver | 0.86 | 1.16 | 1.27 | 1.26 | 0.11 | |
Data are presented as %ID/g ± standard deviation except for intestines and urine, values for which are expressed as %ID ± standard deviation (n = 3).
p = 0.027, significance comparison between the tumor uptake of radiolabeled peptide in the absence and presence of its nonradiolabeled counterpart at 2 hours p.i.
The disappearance of different radiolabeled peptides from the blood was comparable at initial time points (0.5, 1, and 2 hours) tested. For instance, at the end of 2 hours p.i., peptide retention in the blood was 0.20 ± 0.08%ID/g, 0.05 ± 0.01%ID/g, and 0.08 ± 0.01%ID/g for 64Cu-DO3A-, 64Cu-CB-TE2A-, and 64Cu-NO2A-peptides, respectively. Despite lower blood values observed for CB-TE2A and NO2A peptides compared with DO3A-peptide at or beyond 2 hours time point, the values were not statistically different (p > 0.05).
Unstable copper compounds are known to exhibit higher retention in the liver in vivo. In this study, the retention of 64Cu in the liver with 64Cu-DO3A-peptide was observed to be higher than 64Cu-CB-TE2A- or 64Cu-NO2A-peptide at all time points, suggesting the relative stable chelation of 64Cu with CB-TE2A and NO2A. The difference in radiolabled uptake in the liver between 64Cu-DO3A- and 64Cu-CB-TE2A-peptide (n = 3, p = 0.001), 64Cu-DO3A-, and 64Cu-NO2A-peptide (n = 3, p = 0.03) was statistically different, whereas the uptake between 64Cu-NO2A- and 64Cu-CB-TE2A-peptide was comparable and not statistically significant (n = 3, p > 0.05). The tumor uptake of the 64Cu-labeled peptides was comparable at 0.5 hours p.i. (0.91 ± 0.12%ID/g, 64Cu-DO3A-peptide), (0.98 ± 0.31%ID/g, 64Cu-CB-TE2A-peptide), and (0.78 ± 0.03%ID/g, 64Cu-NO2A-peptide). Also, in the later time points, there were no significant differences in the tumor uptake between the three radiolabeled peptides. For instance, at the 2 hours time point, the tumor uptake of 64Cu-DO3A-peptide was 0.54 ± 0.06%ID/g compared with that of 0.32 ± 0.08%ID/g for 64Cu-CB-TE2A- and 0.61 ± 0.06%ID/g for 64Cu-NO2A-peptide, respectively.
The tumor-to-blood ratio was found to be significantly different between 64Cu-DO3A/64Cu-CB-TE2A peptides (n = 3, p = 0.04) and 64Cu-DO3A/64Cu-NO2A peptides (n = 3, p = 0.045), whereas there was no significant difference between 64Cu-CB-TE2A/64Cu-NOTA peptides for all time points (p > 0.05). Similarly, tumor-to-liver ratios were also significantly different between 64Cu-DO3A/64Cu-CB-TE2A (n = 3, p = 0.027) and 64Cu-DO3A/64Cu-NO2A (n = 3, p = 0.01) peptides, respectively. These results suggest that 64Cu-DO3A peptide uptake in the liver was high compared with 64Cu-CB-TE2A and 64Cu-NO2A peptides. However, no major difference was observed between 64Cu-CB-TE2A and 64Cu-NO2A peptides, suggesting their uptake was comparable in liver tissue at all time points. No variation in tumor-to-muscle ratio was observed between the three radiolabeled peptides (Tables 2–4), and the differences in values were not statistically significant (n = 3, p > 0.05). Increased tumor-to-liver tissue ratio of both 64Cu-CB-TE2A and 64Cu-NO2A peptides compared with 64Cu-DO3A peptide suggests less transchelation of 64Cu from the former two chelator-peptides.
The retention of radiolabeled peptides was observed to be high in kidneys. Radioactivity in the kidneys decreased from 6.37 ± 1.44%ID/g at 0.5 hours to 4.27 ± 0.84%ID/g at 2 hours and 1.49 ± 0.24%ID/g at 24 hours, for 64Cu-DO3A-peptide. Similarly, the radioactivity declined from 7.11 ± 0.75%ID/g after 0.5 hours p.i., to 6.43 ± 1.56%ID/g at 2 hours, and 3.2 ± 0.23%ID/g at 24 hours for 64Cu-CB-TE2A-peptide. 64Cu-NO2A-peptide uptake in kidneys was 7.87 ± 0.96 at 0.5 hours, which declined to 4.23 ± 1.00 at 2 hours, and 0.68 ± 0.07 at 24 hours. Further statistical comparison of kidney uptake values for earlier time points (0.5, 1, and 2 hours) was comparable and not statistically significant between 64Cu-CB-TE2A-/64Cu-DO3A-peptide, 64Cu-DO3A-/64Cu-NO2A-peptide, or 64Cu-NO2A-/64Cu-CB-TE2A-peptide (n = 3, p > 0.05). With regard to the radioactivity retention in lungs, 64Cu-DO3A-peptide was relatively higher at all time points compared with 64Cu-CB-TE2A and 64Cu-NO2A peptides, respectively (Tables 2–4). Significant difference was noted in the values between 64Cu-DO3A-/64Cu-CB-TE2A-peptide (n = 3, p = 0.001) and 64Cu-DO3A-/64Cu-NO2A-peptide (n = 3, p = 0.02), suggesting more lung retention of 64Cu-DO3A-peptide at all time points. The lung uptake values between 64Cu-NO2A- and 64Cu-CB-TE2A-peptides were comparable and did not significantly differ (p > 0.05).
Blocking studies were performed with 100 μg of respective natCu-peptides 15 minutes before the injection of the radiolabeled peptides in tumor mice (n = 3). As a positive control, another set of mice (n = 3) was injected with respective radiolabeled peptides only. The results demonstrated that tumor uptake of radiolabeled peptide was blocked by 46% (DO3A-peptide, p = 0.03), 50% (CB-TE2A-peptide, p = 0.024), and 48% (NO2A-peptide, p = 0.027), respectively (Tables 2–4).
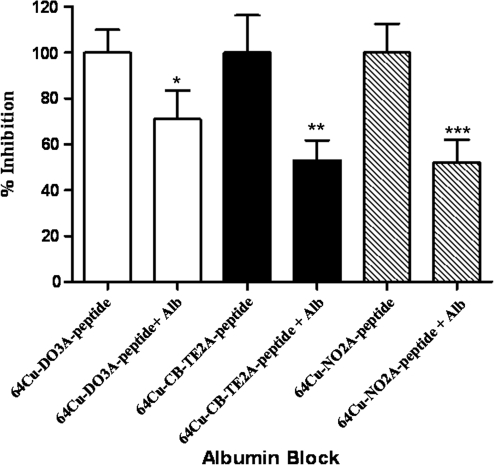
It is well known that high and persistent renal retention of radiolabeled peptides is a concern when designing receptor-targeted radiopharmaceuticals. In an attempt to study an agent that could interfere with kidney retention of the radiolabeled peptides, albumin fragments33 were used as inhibiting agents. The renal uptake of the radiolabeled peptides with and without albumin fragments (3–50 kDa) is shown in (Fig. 3). The kidney retention of the radiolabeled peptides with 64Cu-DO3A-, 64Cu-CB-TE2A-, and 64Cu-NO2A-(GSG)-KCCYSL was blocked in the presence of albumin fragments by 29%, 47%, and 47.5% (n = 3 each, p ≤ 0.05), respectively.
FIG. 3.
Renal blocking studies with bovine serum albumin fragments. Albumin fragments (200 μg) were preinjected in the mice 5 minutes before the injection of radiolabeled peptides (0.185 MBq). The kidney uptake of the radioligands in mice with and without albumin fragments was measured in a γ-counter. Inhibition in the uptake of 64Cu-DO3A-(GSG)-KCCYSL (29%, *p = 0.037), 64Cu-CB-TE2A-(GSG)-KCCYSL (47%, **p = 0.02), and 64Cu-NO2A-(GSG)-KCCYSL (48%, ***p = 0.032) in the presence of albumin fragments are shown.
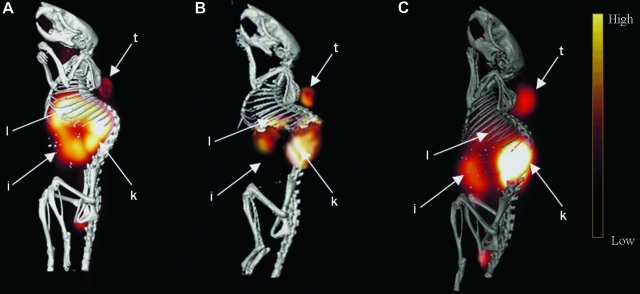
Observations from small animal microPET experiments performed in SCID mice bearing MDA-MB-435 tumors at different times after intravenous injection of 12.0 MBq of each 64Cu-DO3A-, 64Cu-CB-TE2A, and 64Cu-NO2A-(GSG)-KCCYSL are depicted in Figure 4. All the images were acquired at 2 hours p.i., of the radiolabeled peptides. Tumors could be easily visualized with radiolabeled peptides. For 64Cu-DO3A-peptide, the abdominal uptake and retention appeared high compared with 64Cu-CB-TE2A- and 64Cu-NO2A-peptides, which showed relatively less retention in the gastrointestinal tract and liver. A more notable difference in retention was observed in liver tissue for the 64Cu-DO3A-peptide,which was higher compared with 64Cu-CB-TE2A- or 64Cu-NO2A-KCCYSL, which agrees with the biodistribution data. Substantial uptake of all radiolabeled peptides in the kidneys was evident, indicating that theses tracers are mainly excreted through the renal route.
FIG. 4.
MicroPET/CT of MDA-MB-435 tumor-bearing mice. 64Cu-DO3A-(GSG)-KCCYSL, 64Cu-CB-TE2A-(GSG)-KCCYSL, or 64Cu-NO2A-(GSG)-KCCYSL 12.0 (MBq) each, was injected into the tail vein of SCID mice bearing MDA-MB-435 tumor xenograft. Imaging was acquired 2 hours postinjection, of the radiolabeled peptides in a PET scanner. The PET images were fused with conventional microCT images to validate regions of increased radiolabeled peptide uptake. The figures depict PET/CT images with (A) 64Cu-DO3A-(GSG)-KCCYSL; (B) 64Cu-CB-TE2A-(GSG)-KCCYSL; and (C) 64Cu-NO2A-(GSG)-KCCYSL (t, tumor; l, liver; k, kidney; i, intestine). CT, computed tomography; PET, positron emission tomography.
Discussion
Our study demonstrates that a phage derived peptide, KCCYSL, that binds the extracellular domain of EGFR-2,11,18 once synthesized with a bi-functional chelator and radiolabeled with 64Cu, can be developed into a PET imaging agent for detection of breast tumors in vivo. Though other groups have reported peptides derived from phage display that bind EGFR-2,13,34,35 our studies are the first to demonstrate in vivo in a animal model that an EGFR-2 targeting peptide selected by phage display can be exploited as an SPECT or PET radioimaging agent.18
DOT3A-(GSG)-KCCYSL, CB-TE2A-(GSG)-KCCYSL, and NO2A-(GSG)-KCCYSL were synthesized and efficiently radiolabeled with 64Cu. One potential problem with cysteine-containing peptide(s) in aqueous buffers especially at neutral or alkaline pH is the oxidation of its thiol group to form disulfide bonds during radiolabeling reactions. Though the oxidation may be minimal in acidic pH (as in DO3A labeling), the chances are relatively high in near neutral pH such as the reaction conditions used in CB-TE2A and NO2A. To overcome this situation, the reaction buffers were extensively purged with nitrogen to keep oxygen levels minimal and also contained reducing agent TCEP to prevent the formation of disulfide bonds.
All the radiolabeled peptides bound to EGFR-2 expressing MDA-MB-435 breast carcinoma cells, whereas minimal binding was observed with normal mammary cells (184A1). MDA-MB-435 human breast carcinoma cells were originally isolated from a pleural effusion of a woman with metastatic breast adenocarcinoma.36 Rae at al. speculated in 2004 that the original MDA-MB-435 cancer cells were lost early after their establishment and found to be identical to the M-14 melanoma cell line.37 However, a recent report based on the different origin of these cell lines indicated that both MDA-MB-435 and M-14 cell lines are consistent with MDA-MB-435 breast cancer origin38 and that some poorly differentiated breast tumors can exhibit lineage infidelity and express markers associated with both epithelial and melanomas. Given this, the MDA-MB-435 breast carcinoma cell line has been widely used as a breast carcinoma model and remains one of the most reliable in vivo models of human breast cancer.
In vitro competition studies with unlabeled peptides demonstrated specificity of the radiolabeled peptide for breast carcinoma cells. Comparable binding affinities were noted with the three radiolabeled peptides. One would anticipate that binding affinity differences between the peptides may arise due to the use of different chelators. Previous studies with somatostatin receptor targeting DOTA-[Tyr3]-octreotide peptide analogs suggested that relatively small changes in a radiolabeled peptide, such as altering the chelator, can markedly change the receptor binding profile.39 On the contrary, we did not observe any marked changes in the binding affinity profile of all three radiolabeled peptides tested to bind the breast carcinoma cells.
Comparative in vivo studies of 64Cu-DO3A-, 64Cu-CB-TE2A-, and 64Cu-NO2A- (GSG)-KCCYSL indicated reasonable tumor uptake of these peptides. 64Cu-CB-TE2A-(GSG)-KCCYSL conjugate demonstrated significant disappearance from the tumor, at 24 hours p.i., compared with 64Cu-DO3A-(GSG)-KCCYSL, which had slower disappearance for the same time period (Tables 2 and 3). The delayed tumor tissue disappearance pattern of the 64Cu-DO3A-radioconjugate could be due to a trapping mechanism, such as transchelation of the radiometal from the chelator to intracellular protein(s).22,26,40 Moreover, significant transchelation of the 64Cu-DO3A-peptide to serum proteins occurs in blood,23,41 and the protein-bound 64Cu form may be transported back into the blood, which over time decreases target tissue:blood ratios.
Table 3.
Pharmacokinetic Studies of 64Cu-CB-TE2A-GSG-KCCYSL Using MDA-MB-435 Tumor-Bearing SCID Mice
| Tissues | 0.5 hour | 1 hour | 2 hours | 2 hours block | 4 hours | 24 hours |
|---|---|---|---|---|---|---|
| Percent injected dose/g (%ID/g)a | ||||||
| Tumor | 0.98 ± 0.31 | 0.64 ± 0.08 | 0.32 ± 0.17 | 0.16 ± 0.07b | 0.17 ± 0.05 | 0.10 ± 0.03 |
| Blood | 1.18 ± 0.20 | 0.28 ± 0.12 | 0.05 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.011 ± 0.01 |
| Brain | 0.04 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.01 | 0.007 ± 0.001 |
| Heart | 0.42 ± 0.31 | 0.15 ± 0.03 | 0.06 ± 0.02 | 0.05 ± 0.03 | 0.07 ± 0.01 | 0.012 ± 0.001 |
| Lung | 1.13 ± 0.16 | 0.46 ± 0.09 | 0.24 ± 0.02 | 0.20 ± 0.05 | 0.18 ± 0.02 | 0.05 ± 0.002 |
| Liver | 0.68 ± 0.11 | 0.52 ± 0.02 | 0.45 ± 0.05 | 0.37 ± 0.10 | 0.40 ± 0.03 | 0.29 ± 0.05 |
| Spleen | 0.36 ± 0.11 | 0.20 ± 0.08 | 0.12 ± 0.06 | 0.10 ± 0.05 | 0.10 ± 0.01 | 0.01 ± 0.003 |
| Stomach | 0.25 ± 0.08 | 0.18 ± 0.10 | 0.13 ± 0.06 | 0.15 ± 0.02 | 0.21 ± 0.12 | 0.03 ± 0.001 |
| Kidneys | 7.11 ± 0.75 | 6.72 ± 0.91 | 6.43 ± 1.56 | 6.94 ± 1.47 | 5.96 ± 0.89 | 3.18 ± 0.23 |
| Muscle | 0.21 ± 0.07 | 0.06 ± 0.01 | 0.03 ± 0.01 | 0.05 ± 0.01 | 0.02 ± 0.01 | 0.007 ± 0.001 |
| Pancreas | 0.32 ± 0.11 | 0.14 ± 0.04 | 0.05 ± 0.01 | 0.02 ± 0.00 | 0.05 ± 0.01 | 0.04 ± 0.001 |
| Bone | 0.20 ± 0.05 | 0.07 ± 0.02 | 0.04 ± 0.01 | 0.03 ± 0.01 | 0.04 ± 0.01 | 0.03 ± 0.001 |
| Percent injected dose (%ID) | ||||||
| Intestines | 1.02 ± 0.19 | 2.0 ± 2.10 | 0.75 ± 0.07 | 0.73 ± 1.27 | 1.3 ± 1.90 | 1.0 ± 0.14 |
| Urine | 78.2 ± 9.20 | 91.2 ± 3.74 | 95.2 ± 0.48 | 93.7 ± 4.30 | 95.3 ± 1.40 | 98.0 ± 2.40 |
| Uptake ratio of tumor/normal tissue | ||||||
| Tumor/blood | 0.83 | 2.20 | 6.40 | — | 8.50 | 10.00 |
| Tumor/muscle | 4.00 | 10.70 | 10.71 | 17.00 | 14.20 | |
| Tumor/liver | 1.40 | 1.20 | 0.70 | 0.42 | 0.34 | |
Data are presented as %ID/g ± standard deviation except for intestines and urine, values for which are expressed as%ID ± SD (n = 3).
p = 0.024, significance comparison between the tumor uptake of radiolabeled peptide in the absence and presence of its nonradiolabeled counterpart at 2 hours p.i.
All the radiolabeled peptides demonstrated significant blood disappearance rates, whereas the liver clearance was modest with relatively more retention of 64Cu-DO3A- compared with 64Cu-CB-TE2A and 64Cu-NO2A peptides. Unstable copper compounds in vivo (e.g., DO3A-peptides) are known to exhibit high retention in the liver that could be likely due to transchelation.42 The use of the 64Cu-DO3A chelation system can result in release of uncoordinated 64Cu by decomposition in the blood or transchelation in the liver, resulting in elevated uptake in liver tissue.23,41 On the other hand, CB-TE2A and NO2A chelator-peptides demonstrated less tranchelation of the bound metal. Boswell et al. observed that the 64Cu-labeled chelator CB-TE2A during biodistribution studies in normal rats underwent significantly less transchelation in liver.25 In this study, it was shown that transchelation of 64Cu from 64Cu-DOTA was 39% ± 1% at 1 hour, whereas transchelation of 64Cu from 64Cu-CB-TE2A was 12% ± 7% for the same time period.25 Also, previous studies indicated that the injection of [64Cu]Cu2+ alone in mice resulted in half the injected dose (∼54%) concentrated in the liver after 2 hours.43 Also, oral administration of copper acetate in tumor-bearing mice revealed the accumulation of copper in liver (∼62.1 μg/g tissue) and kidneys (∼4.8 μg/g tissue).43 Studies with 64Cu-NO2A-RGD-bombesin peptide uptake resulted in less uptake in the liver, compared to 64Cu tracers42 suggesting the high kinetic stability of the radiotracer. Another study with 64Cu-NO2A-bombesin analogs also reported the stability of NO2A peptides in vivo.28
Nonspecific accumulation of radioactivity in the kidneys is often associated with the use of radiolabeled peptides in vivo.44 The kidney retention of copper KCCYSL peptides was found to be substantially high. Similar kidney retention of peptides with 64Cu-labeled azamacrocycles, including that of 64Cu-CB-TE2A-Y3-TATE and 64Cu-CB-TE2A-somatostatin receptor subtype 2 selective antagonist (sst2-ANT), was previously reported.25,45 Recent studies indicated that administering albumin fragments could reduce the renal uptake of radiolabeled peptides.33 Since only a small fraction of total albumin passes through the glomerular membrane, it was hypothesized that the use of albumin fragments could competitively inhibit the uptake of radiolabeled peptides in the kidneys.33 In the present study, biodistribution studies with an attempt to block the in vivo kidney uptake of 64Cu-DO3A-, 64Cu-CB-TE2A-, and 64Cu-NO2A-(GSG)-KCCYSL in the presence of albumin fragments resulted in a partial inhibition in the uptake of radiotracers by the kidneys.
Similar to radiolabeled GSG-KCCYSL peptides, corresponding nonradiolabeled peptides presumably undergo faster blood clearance, and demetallation from these peptides may also occur in the liver. This presumably could have been attributed to partial blocking of tumoral uptake of the radiolabeled peptides in the presence of their nonradiolabeled counterparts during biodistribution studies. Thus, lack of longer residence time of the competing molecule and its uptake in nontarget tissues including kidney could result in less efficient blocking of the radioligand uptake in tumor unlike large molecules such as antibodies, which have much longer circulation times in vivo.46
Small-animal PET/CT studies with all three radiolabeled peptides demonstrated reasonable tumor uptake and retention. In spite of similar tumor uptake by these peptides, differences in uptake and retention of these compounds in nontarget tissues were found. For the 64Cu-DO3A-peptide, the image revealed significant activity in the liver, gastrointestinal tract, and the kidneys. The image obtained with 64Cu-CB-TE2A and 64Cu-NO2A-peptides, however, demonstrated relatively reduced activity in the liver and the gastrointestinal tract with comparable retention in the kidneys. Likewise, biodistribution and imaging studies with 64Cu-CB-TE2A-Y3-TATE, a somatostatin receptor targeting peptide, showed low uptake in liver and blood and more in the kidneys.31 Recent studies with 64Cu-CB-TE2A-ReCCMSH(Arg11), a melanocortin receptor targeting peptide, demonstrated reduced uptake in liver and blood and a modest uptake in the kidneys compared with 64Cu-DOTA-ReCCMSH(Arg11).47 Taken together, 64Cu-CB-TE2A- and 64Cu-NO2A-peptides demonstrated better tumor-to-background ratios and nontarget tissue clearance compared with the DO3A-peptide. Overall, the cross-bridged cyclam chelator CB-TE2A and NOTA exhibit comparable stability, thus enabling efficient 64Cu-radiopharmaceutical applications with biological targeting molecules such as the KCCYSL peptide.
Conclusions
The peptides 64Cu-DO3A-(GSG)-KCCYSL, 64Cu-CB-TE2A-(GSG)-KCCYSL, and 64Cu-NO2A-(GSG)-KCCYSL were evaluated as agents for PET imaging of breast carcinoma. The 64Cu-stabilization properties of CB-TE2A and NOTA compared with those of DOTA resulted in improved blood disappearance and lower retention in liver and the gastrointestinal tract, with improved tumor-to-nontarget tissue ratios for 64Cu-CB-TE2A- and 64Cu-NO2A-GSG-KCCYSL. Thus, CB-TE2A and NO2A-peptides appear to be better candidates for application as diagnostic PET agents for EGFR-2 expressing breast cancers.
Acknowledgments
This work was supported by awards from the National Institutes of Health, (P50CA103130-01, 1R-21CA137239-01A1, and 2 R01 CA093375) and in part by a Merit Review Award from the Veterans Administration (SLD). We thank Dr. Said Figueroa for his help with imaging studies. We also thank the VA Biomolecular Imaging Center (BIC) at the Harry S. Truman Memorial Veteran's Hospital. We acknowledge Lisa Watkinson and Terry Carmack for performing animal experiments and Marie Dickerson for technical help.
Disclosure Statement
No competing financial interests exist.
References
- 1.Ullrich A. Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 2.Hung MC. Lau YK. Basic science of HER2/neu: A review. Semin Oncol. 1999;26(Suppl):51. [PubMed] [Google Scholar]
- 3.Press MF. Cordon-Cardo C. Slamon DJ. Expression of the Her2/neuproto-oncogene in normal human and adult and fetal tissues. Oncogene. 1990;5:53. [PubMed] [Google Scholar]
- 4.Ross J. Fletcher JA. The Her2/neu oncogene in breast cancer: Prognostic factor, predictive factor, and target for therapy. Stem Cells. 1998;16:413. doi: 10.1002/stem.160413. [DOI] [PubMed] [Google Scholar]
- 5.Hynes NE. Stern DF. The biology of ErbB-2/neu/Her-2 and its role in cancer. Biochimica et Biophysica Acta. 1994;1198:165. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 6.Holbro T. Hynes NE. ErbB receptors: Directing key signaling networks throughout life. Ann Rev of Pharmacol Toxicol. 2004;44:195. doi: 10.1146/annurev.pharmtox.44.101802.121440. [DOI] [PubMed] [Google Scholar]
- 7.Hudziak RM. Lewis GD. Winget M, et al. p185HER2 monoclonal antibody has antiproliferative effects in vitro and sensitizes human breast tumor cells to tumor necrosis factor. Mol Cell Biol. 1989;9:1165. doi: 10.1128/mcb.9.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harwerth IM. Wels W. Schlegel J, et al. Monoclonal antibodies directed to the ErbB-2 receptor inhibit in vivo tumour cell growth. Br J Cancer. 1993;68:1140. doi: 10.1038/bjc.1993.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houimel M. Schneider P. Terskikh A, et al. Selection of peptides and synthesis of pentameric peptabody molecules reacting specifically with ErbB-2 receptor. Int J Cancer. 2001;92:748. doi: 10.1002/1097-0215(20010601)92:5<748::aid-ijc1258>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 10.Li X. Stuckert P. Bosch I, et al. Single-chain antibody-mediated gene delivery into ErbB2-positive human breast cancer cells. Cancer Gene Ther. 2001;8:555. doi: 10.1038/sj.cgt.7700337. [DOI] [PubMed] [Google Scholar]
- 11.Karasseva NG. Glinsky VV. Chen NX, et al. Identification and characterization of peptides that bind human ErbB-2 selected from a bacteriophage display library. J Protein Chem. 2002;21:287. doi: 10.1023/a:1019749504418. [DOI] [PubMed] [Google Scholar]
- 12.Nilsson F. Tarli L. Viti F, et al. The use of phage display for the development of tumour targeting agents. Adv Drug Deliv Rev. 2000;43:165. doi: 10.1016/s0169-409x(00)00068-5. [DOI] [PubMed] [Google Scholar]
- 13.Pero SC. Shukla GS. Armstrong AL, et al. Identification of a small peptide that inhibits the phosphorylation of ErbB2 and proliferation of ErbB2 overexpressing breast cancer cells. Int J Cancer. 2004;111:951. doi: 10.1002/ijc.20306. [DOI] [PubMed] [Google Scholar]
- 14.Blok D. Feitsma RI. Vermeij P, et al. Peptide radiopharmaceuticals in nuclear medicine. Eur J Nucl Med. 1999;26:1511. doi: 10.1007/s002590050488. [DOI] [PubMed] [Google Scholar]
- 15.Signore A. Annovazzi A. Chianelli M, et al. Peptide radiopharmaceuticals for diagnosis and therapy. Eur J Nucl Med. 2001;28:1555. doi: 10.1007/s002590100583. [DOI] [PubMed] [Google Scholar]
- 16.Reubi JC. Maecke HR. Peptide-based probes for cancer imaging. J Nucl Med. 2008;49:1735. doi: 10.2967/jnumed.108.053041. [DOI] [PubMed] [Google Scholar]
- 17.Wadas TJ. Anderson CJ. Radiolabeling of TETA- and CB-TE2A-conjugated peptides with copper-64. Nat Protoc. 2006;1:3062. doi: 10.1038/nprot.2006.431. [DOI] [PubMed] [Google Scholar]
- 18.Kumar SR. Quinn TP. Deutscher SL. Evaluation of an 111In- radiolabeled peptide as a targeting and imaging agent for ErbB-2 receptor expressing breast carcinomas. Clin Cancer Res. 2007;13:6070. doi: 10.1158/1078-0432.CCR-07-0160. [DOI] [PubMed] [Google Scholar]
- 19.Wadas TJ. Wong EH. Weisman GR, et al. Copper chelation chemistry and its role in copper radiopharmaceuticals. Curr Pharm Des. 2007;13:3. doi: 10.2174/138161207779313768. [DOI] [PubMed] [Google Scholar]
- 20.Bearn AG. Kunkel HG. Localization of Cu64 in serum fractions following oral administration: An alteration in Wilson's disease. Proc Soc Exp Biol Med. 1954;85:44. doi: 10.3181/00379727-85-20780. [DOI] [PubMed] [Google Scholar]
- 21.Owen CA., Jr Hazelrig JB. Metabolism of cu-64-labeled copper by the isolated rat liver. Am J Physiol. 1966;210:1059. doi: 10.1152/ajplegacy.1966.210.5.1059. [DOI] [PubMed] [Google Scholar]
- 22.Jones-Wilson TM. Deal KA. Anderson CJ, et al. The in vivo behavior of copper-64- labeled azamacrocyclic complexes. Nucl Med Biol. 1998;25:523. doi: 10.1016/s0969-8051(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 23.Cole WC. DeNardo SJ. Meares CF, et al. Serum stability of 67Cu chelates: Comparison with 111In and 57Co. Int J Rad Appl Instrum B. 1986;13:363. doi: 10.1016/0883-2897(86)90011-5. [DOI] [PubMed] [Google Scholar]
- 24.Smith SV. Molecular imaging with copper-64. J Inorg Biochem. 2004;98:1874. doi: 10.1016/j.jinorgbio.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Boswell CA. Sun X. Niu W, et al. Comparative in vivo stability of copper-64-labeled cross-bridged and conventional tetraazamacrocyclic complexes. J Med Chem. 2004;47:1465. doi: 10.1021/jm030383m. [DOI] [PubMed] [Google Scholar]
- 26.Sun X. Wuest M. Weisman GR, et al. Radiolabeling and in vivo behavior of copper-64-labeled cross-bridged cyclam ligands. J Med Chem. 2002;45:469. doi: 10.1021/jm0103817. [DOI] [PubMed] [Google Scholar]
- 27.Prasanphanich AF. Nanda PK. Rold TL, et al. [64Cu-NOTA-8-Aoc-BBN(7-14)NH2] targeting vector for positron-emission tomography imaging of gastrin-releasing peptide receptor-expressing tissues. Proc Natl Acad Sci USA. 2007;104:12462. doi: 10.1073/pnas.0705347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y. Zhang X. Xiong Z, et al. MicroPET imaging of glioma integrin {alpha}v{beta}3 expression using (64)Cu-labeled tetrameric RGD peptide. J Nucl Med. 2005;46:1707. [PubMed] [Google Scholar]
- 29.Studer M. Meares CF. Synthesis of novel 1,4,7-triazacyclononane-N,N',N”-triacetic acid derivatives suitable for protein labeling. Bioconjug Chem. 1992;3:337. doi: 10.1021/bc00016a013. [DOI] [PubMed] [Google Scholar]
- 30.Brechbiel MW. McMurry TJ. Gansow OA. A direct synthesis of bifunctional chelating agent for radiolabeling protein. Tetrahedron Lett. 1993;34:3691. [Google Scholar]
- 31.Sprague JE. Peng Y. Sun X, et al. Preparation and biological evaluation of copper-64- labeled tyr3-octreotate using a cross-bridged macrocyclic chelator. Clin Cancer Res. 2004;10:8674. doi: 10.1158/1078-0432.CCR-04-1084. [DOI] [PubMed] [Google Scholar]
- 32.Tommasi S. Fedele V. Lacalamita R, et al. Molecular and functional characteristics of erbB2 in normal and cancer breast cells. Cancer Lett. 2004;209:215. doi: 10.1016/j.canlet.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Vegt E. van Eerd JE. Eek A, et al. Reducing renal uptake of radiolabeled peptides using albumin fragments. J Nucl Med. 2008;49:1506. doi: 10.2967/jnumed.108.053249. [DOI] [PubMed] [Google Scholar]
- 34.Houimel M. Schneider P. Terskikh A, et al. Selection of peptides and synthesis of pentameric peptabody molecules reacting specifically with ErbB-2 receptor. Int J Cancer. 2001;92:748. doi: 10.1002/1097-0215(20010601)92:5<748::aid-ijc1258>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 35.Urbanelli L. Ronchini C. Fontana L, et al. Targeted gene transduction of mammalian cells expressing the HER2/neu receptor by filamentous phage. J Mol Biol. 2001;313:965. doi: 10.1006/jmbi.2001.5111. [DOI] [PubMed] [Google Scholar]
- 36.Price JE. Zhang FA. Radinsky RD, et al. Characterization of variants of a human breast cancer cell line isolated from metastases in different organs of nude mice. Int J Oncol. 1994;5:459. doi: 10.3892/ijo.5.3.459. [DOI] [PubMed] [Google Scholar]
- 37.Rae JM. Ramus SJ. Waltham M, et al. Common origins of MDA-MB-435 cells from various sources with those shown to have melanoma properties. Clin Exp Metastasis. 2004;21:543. doi: 10.1007/s10585-004-3759-1. [DOI] [PubMed] [Google Scholar]
- 38.Chambers AF. MDA-MB-435 and M14 cell lines: Identical but not M14 melanoma? Cancer Res. 2009;69:5292. doi: 10.1158/0008-5472.CAN-09-1528. [DOI] [PubMed] [Google Scholar]
- 39.Rolleman EJ. Valkema R. de Jong M, et al. Safe and effective inhibition of renal uptake of radiolabelled octreotide by a combination of lysine and arginine. Eur J Nucl Med Mol Imaging. 2003;30:9. doi: 10.1007/s00259-002-0982-3. [DOI] [PubMed] [Google Scholar]
- 40.Bass LA. Wang M. Welch MJ, et al. In vivo transchelation of copper-64 from TETA-octreotide to superoxide dismutase in rat liver. Bioconjug Chem. 2000;11:527. doi: 10.1021/bc990167l. [DOI] [PubMed] [Google Scholar]
- 41.Moi MK. Meares CF. McCall MJ, et al. Copper chelates as probes of biological systems: Stable copper complexes with a macrocyclic bifunctional chelating agent. Anal Biochem. 1985;148:249. doi: 10.1016/0003-2697(85)90653-0. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z. Yan Y. Liu S, et al. 18F, 64Cu, and 68Ga lableled RGD-bombesin heterodimeric peptides for PET imaging of breast cancer. Bioconjug Chem. 2009;20:1016. doi: 10.1021/bc9000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu M. Qing H. Guojian H, et al. Biodistribution of [64Cu]Cu2+ and variance of metallothionein during tumor treatment by copper. Nucl Med Biol. 1998;25:111. doi: 10.1016/s0969-8051(97)00169-8. [DOI] [PubMed] [Google Scholar]
- 44.Reubi JC. Schär JC. Waser B, et al. Affinity profiles for human somatostatin receptor subtypes SST1-SST5 of somatostatin radiotracers selected for scintigraphic and radiotherapeutic use. Eur J Nucl Med. 2002;27:273. doi: 10.1007/s002590050034. [DOI] [PubMed] [Google Scholar]
- 45.Wadas TJ. Eiblmaier M. Zheleznyak A, et al. Preparation and biological evaluation of 64Cu-CB-TE2A-sst2-ANT, a somatostatin antagonist for PET imaging of somatostatin receptor-positive tumors. J Nucl Med. 2008;49:1819. doi: 10.2967/jnumed.108.054502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perik PJ. Lub-De Hooge MN. Gietema JA, et al. Indium-111-labeled trastuzumab scintigraphy in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer. J Clin Oncol. 2006;24:2276. doi: 10.1200/JCO.2005.03.8448. [DOI] [PubMed] [Google Scholar]
- 47.Wei L. Butcher C. Miao Y, et al. Synthesis and biologic evaluation of 64Cu-labeled rhenium-cyclized alpha-MSH peptide analog using a cross-bridged cyclam chelator. J Nucl Med. 2007;48:64. [PubMed] [Google Scholar]