Abstract
The genetic basis of the regeneration process in cultured immature embryos of rye (Secale cereale L.) was analyzed. The experiments were designed to reveal differences between the in vitro culture responses of two inbred lines: L318 (a high regeneration ability) and L9 (a low potential for regeneration). The rye ortologues of plant genes previously recognized as crucial for somatic embryogenesis and morphogenesis in vitro were identified. Using oligonucleotide primers designed to conserved regions of the genes Somatic Embryogenesis Receptor-like Kinase (SERK), Leafy Cotyledon 1 (LEC1), Viviparous 1 (VP1) and NiR (encoding ferredoxin-nitrite reductase), it was possible to amplify specific homologous sequences from rye RNA by RT-PCR. The transcript levels of these genes were then measured during the in vitro culture of zygotic embryos, and the sites of expression localized. The expression profiles of these genes indicate that their function is likely to be correlated with the in vitro response of rye. In line L9, increased expression of the rye SERK ortologue was observed at most stages during the culture of immature embryos. The suppression of ScSERK expression appears to start after the induction of somatic embryogenesis and lasts up to plant regeneration. The rye ortologues of the LEC1 and VP1 genes may function in a complimentary manner and have a negative effect on the production of the embryogenic callus. The expression of the rye NiR ortologue during in vitro culture reveals its importance in the process of plant regeneration.
Keywords: Expression profile, Immature embryos, In situ RT-PCR, Real Time RT-PCR, Regeneration ability
Introduction
Efficient regeneration from cultured cells is a prerequisite for most aspects of plant biotechnology, especially for transformation experiments. Rye is known to be one of the most recalcitrant cereals with regard to its regeneration ability (Ma et al. 2003), which is highly dependent on genotype (Linacero and Vasquez 1990; Rakoczy-Trojanowska and Malepszy 1993, 1995). Immature embryos are the most widely used explants and have been used successfully to regenerate rye (Ward and Jordan 2001).
In vitro plant regeneration depends on numerous factors including the physiological status of the donor plant and the organ used as the explant (Rakoczy-Trojanowska and Malepszy 1990). However, the genotype is the factor that has the most influence on this process. Rakoczy-Trojanowska and Malepszy (1993, 1995) found significant differences between the tissue response (TCR) of several inbred rye lines, both in the case of immature inflorescences and immature embryos cultured. Genetic analysis at the Mendelian level showed that the in vitro response of immature inflorescences is controlled by a polygenic system with different gene interactions, and that the ability to regenerate plants is a recessive trait (Rakoczy-Trojanowska and Malepszy 1993). Results obtained in a study of immature embryos demonstrated that the production of embryogenic callus and the plant and root regeneration are determined by recessive genes, whereas the reduced ability to produce non-embryogenic callus most probably depends on dominant genes. The lack of response was found to be controlled by at least two interacting genes (Rakoczy-Trojanowska and Malepszy 1995).
Recently, we have identified nine putative QTLs for rye TCR: two loci (eci-1, eci-2) for percentage of immature embryos producing callus, four loci for percentage of immature embryos producing embryogenic callus (ece-1, ese-2, ese-3, ese-4), two loci for percentage of immature inflorescences producing callus (ici-1, ici2) and one locus for percentage of immature inflorescences producing embryogenic callus (ise-1) (Bolibok et al. 2007). These loci were mapped to chromosomes 1R, 4R, 5R, 6R and 7R, respectively. Further insight into the molecular mechanisms controlling rye TCR was gained by the application of the GDDSC (genetically directed differential subtraction chain) method. We have isolated several GDDSC-products that are potentially connected with callus induction and somatic embryo formation (Hromada et al. 2007).
Molecular mechanisms of somatic embryogenesis have also been investigated in somatic embryo induction systems and classical tissue culture systems of immature embryos of carrot and Arabidopsis. The employment of various molecular techniques has led to the identification of several embryogenesis-related genes such as LEA (Late Embryogenesis Abundant), SERK (Somatic Embryogenesis Receptor-like Kinase), AGL15 (Agamous-like15), BBM (Baby Boom), LEC1, FUS3 (Fusca3) and ABI3 (ABA-Insensitive 3) (Ikeda et al. 2006).
In carrot cell suspension cultures, presence of the SERK mRNA was found to indicate the capability of single cells to develop into somatic embryos (Schmidt et al. 1997). Similar results were obtained for Dactylis glomerata (Somleva et al. 2000) and Arabidopsis thaliana (Hecht et al. 2001). Moreover, transgenic A. thaliana overexpressing the AtSERK1 gene exhibited an enhanced capacity for somatic embryogenesis in comparison with non-transformed plants (Hecht et al. 2001).
The results obtained by Gaj et al. (2005) indicate that LEC genes have a key role in somatic embryogenesis of A. thaliana. The authors shown that the embryogenic potential of the lec1, lec2 and fus3 mutants was much worse in comparison with a wild type (0.0-3.9% of explants producing somatic embryos via callus stage vs 65-94% explants forming somatic embryos directly). The double (lec1 lec2, lec1 fus3 and lec2 fus3) and triple (fus3 lec1 lec2) mutants did not formed somatic embryos at all. Ectopic expression of LEC1 (Lotan et al. 1998), LEC2 (Stone et al. 2001) and BBM (Boutilier et al. 2002) in Arabidopsis caused the spontaneous formation of somatic embryos on intact plants or explants. The LEC and BBM genes encode transcription factors. Furthermore, LEC2 shares greatest similarity with the B3 domain of transcription factors VIVIPAROUS1 and FUSCA3 (Stone et al. 2001).
The NiR gene, encoding ferredoxin-nitrite reductase, was found to determine regeneration ability in rice (Nishimura et al. 2005). Molecular analysis revealed that the poor regeneration ability of rice is strictly correlated with TCR, especially with plant regeneration (Taguchi-Shiobara et al. 1997).
The aim of the present study was to identify ortologues of SERK, LEC1, VP1 and NiR in rye and analyze their expression in immature embryos during in vitro culture.
Materials and methods
Tissue culture procedure
The tissue culture protocol used for immature embryos was similar to that described previously (Rakoczy-Trojanowska and Malepszy 1995) except that the auxin added to the induction medium was DICAMBA (dichloro-2-metoxy-3,6-benzoic acid) instead of 2,4-D. Briefly, the immature embryos (19-21 days after pollination) were cultured on MS medium (Murashige and Skoog 1962) supplemented with DICAMBA (3 mg/dm3), sucrose (20 g/dm3) and Difco agar (7.5 g/dm3). The calli were subcultured every four weeks. After three subcultures, the explants were transferred to regeneration medium without growth regulators.
RNA isolation
Total RNA was isolated from immature and mature zygotic embryos, leaves and callus of rye from the highly embryogenic line L318 and the non-responding line L9 by the single-step procedure of Chomczynski and Sacchi (1987). The samples of callus were collected from induction medium after two, four, eight or twelve weeks incubation and after two days, two weeks and four weeks growth on the regeneration medium. Tissue samples were homogenized with TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The quality and quantity of the isolated RNA was verified on the agarose gel.
RT-PCR, cloning and sequence analysis
The SuperScript one-Step RT-PCR with Platinum Taq System (Invitrogen) was used for cDNA synthesis and PCR. All reaction components were mixed in a single tube. To design oligonucleotide primers to amplify rye ortologues (Sc) of embryogenesis-related genes, the nucleotide sequences of the selected genes were obtained from GenBank: SERK – mRNA sequences of wheat SERK (GenBank accession no. BT009426), rice SERK (AY652735) and maize SERK1 (NM_001111662); LEC1 – mRNA sequences of rice LEC1 (AY062184, AU088581) and maize LEC1 (AF410176); VP1 – mRNA sequence of maize VP1 (M60214); NiR – mRNA sequence of rice NiR (D50556). Where multiple sequences were available for a gene, they were aligned using the on-line software Primer3 (http://frodo.wi.mit.edu/primer3/) and the region of highest homology was chosen for the selection of gene-specific primers. The primers used in RT-PCR are listed in Table 1.
Table 1.
Primers used to amplify rye homologues of embryogenesis-related genes
| Gene | Primer | Sequence 5’-3’ | Size of product [bp] |
|---|---|---|---|
| SERK | SERK_z | F: TTGCTGGAGGTGTTGCTG R: TACACCTTTCCAAAGCCAC | 234 |
| LEC1 | AU088 | F: CAAGGAGACGATCCAGGAGT R: GGTAGCGGTGGAGGTAGACG | 180 |
| VP1 | VP | F: AGAAGGTGCTGAAGCAGAGC R: CCCTGTGTTTTCCAGCAGAT | 203 |
| NiR | NiR_zyt | F: GAGAAGAGGATGCCGAACG R: ATGTTCTGCTCCACGGTGA | 235 |
| Control 18 S rRNA | 18 S | F: CATCCCAAAGTCCAACTA R: GCTACCACATCCAAGGAA | 250 |
The amplified RT-PCR products were cloned into the pCRII-TOPO vector, using an Invitrogen TOPO TA Cloning Kit (Invitrogen) according to the manufacturer's instructions and sequenced by the Sequencing Service at the Institute of Biochemistry and Biophysics Polish Academy of Sciences (http://www.ibb.waw.pl/services/). The RT-PCR fragment sequences were analyzed using BLAST (http://www.ncbi.nlm.nih.gov/); alignments and comparisons were performed using ClustalW on Bioedit.
Real Time and in situ RT-PCR
The relative expression of putative ortologues of the genes SERK, LEC1, VP1 and NiR was measured by Real Time RT-PCR analysis using a Roche system (LightCycler with the Software v. 3.5) and reagents (LightCycler-RNA Amplification Kit SYBR Green I, LightCycler-Control Kit RNA). Expression of the rye gene coding for 18 S RNA was used to normalize the expression data for the other genes. RNA concentration in reaction mixture was 10 ng/μl. The three samples were used for q-PCR and each experiment was repeated three times. The sample with RNA isolated from immature embryos of line L318 was used as a calibrator.
In situ RT-PCR analysis of sections prepared from immature zygotic embryos was performed according to the protocol described by Przybecki et al. (2006). Stained sections were visualized and recorded using the Olympus Provis AX70 light microscope with analysis software.
Results
Amplification and sequence analysis of rye ortologues
SERK
Using primers designed to a conserved region of the SERK gene (Table 1), a 234-bp fragment was amplified by RT-PCR (ScSERK, GenBank accession no. EG999307) as presented in Fig. 1. The nucleotide sequence of this fragment showed the highest similarity to the SERK1 mRNAs of rice, Oryza sativa (90%, AY652735) and wheat, Triticum aestivum (100%, AK333001).
Fig. 1.
Sequence alignments of SERK (EG999307 – rye, AK333001 – wheat, AY652735 – rice), LEC1 (ES351494 – rye, AB288036 and AY264284 – rice, NM_001112048 – maize) and VP1 (ES584596 – rye, AY150678 – barley, FJ640559 – wheat, NM_001112070 maize) homologues. The nucleotides differing from the consensus are shaded
LEC1
Primers based on LEC1 sequences of monocotyledonous species (Table 1) were used in RT-PCR to amplify a fragment of 180 bp (ScLEC1, GenBank accession no. ES351494), which showed a high degree of sequence similarity to the Oryza sativa HAP3 mRNA (98%, AB288036), OsLEC1 mRNA (98%, AY264284) and Zea mays LEC1 mRNA (96%, NM_001112048) as shown in Fig. 1.
VP1
The 203-bp fragment amplified by RT-PCR using VP primers (Table 1), (ScVP1, GenBank accession no. ES584596) shared a high degree of sequence similarity with the VP1 mRNAs of Hordeum vulgare (97%, AY150678), Triticum aestivum (97%, FJ640559) and Zea mays (91%, NM_001112070) as demonstrated in Fig. 1.
NiR
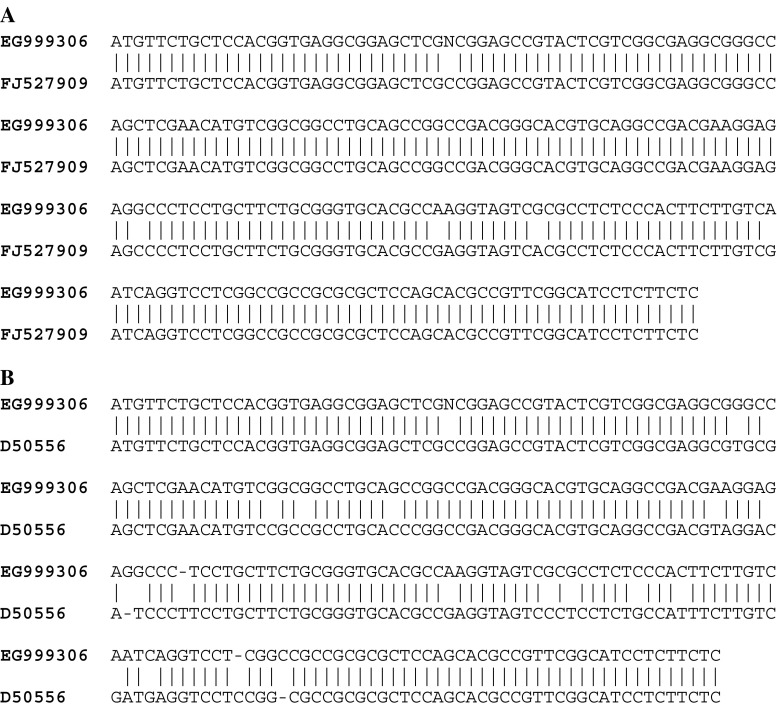
Use of the NiR_zyt primers (Table 1) in an RT-PCR amplified a DNA fragment of 235 bp (ScNiR, GenBank accession no. EG999306) which showed highest sequence similarity to the Triticum aestivum ferredoxin-nitrite reductase mRNA (97%, FJ527909) (Fig. 2a), and to the NiR mRNA of rice (91%, D50556) (Fig. 2b).
Fig. 2.
Nucleotide sequence similarity among NiR homologues. a – NiR of rye (GenBank accession no. EG999306) and wheat (FJ527909), b – ScNiR (EG999306) and rice NiR (D50556)
Expression analysis of ScSERK, ScLEC1, ScVP1 and ScNiR
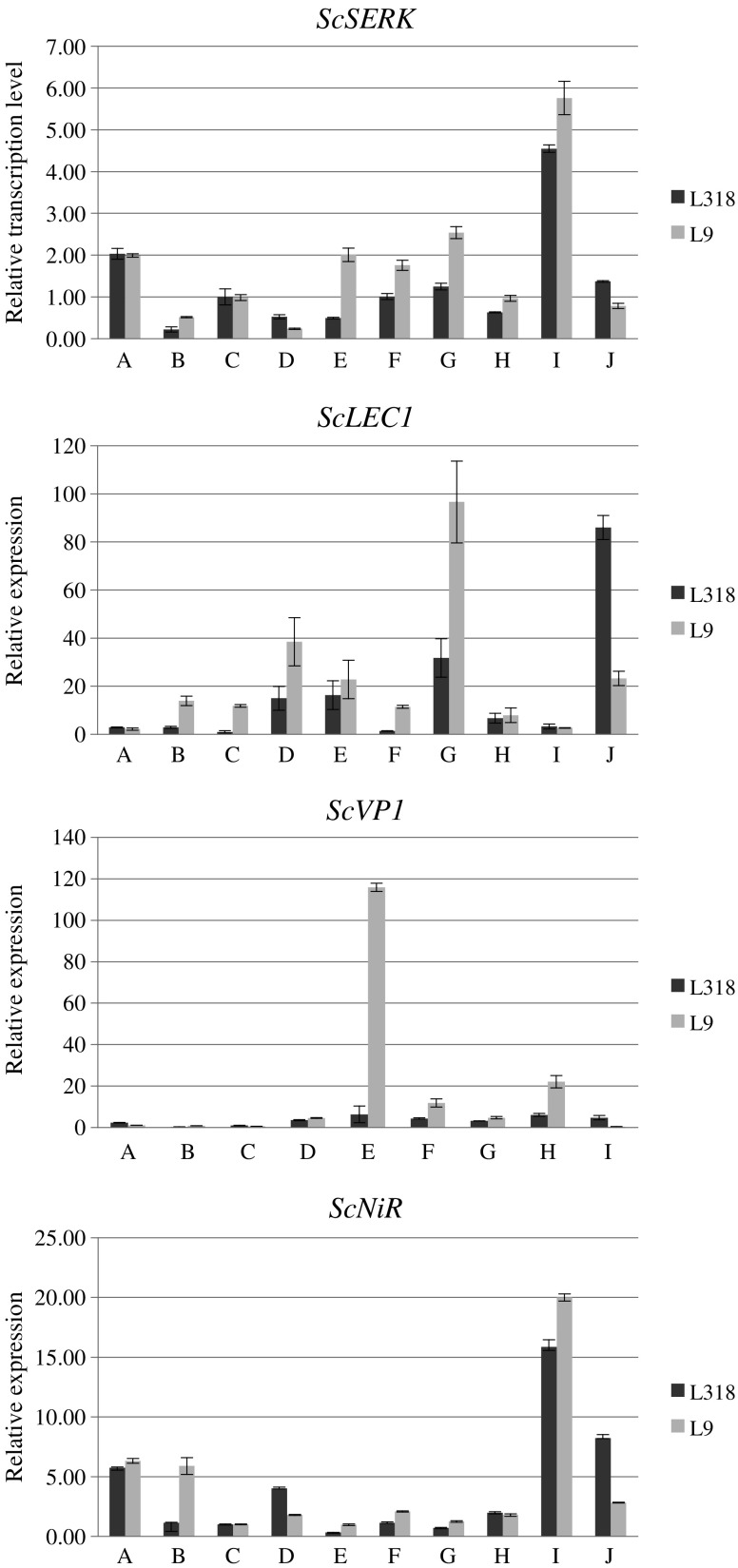
Real Time RT-PCR analysis demonstrated that the rye ortologues of the SERK, LEC1, VP1 and NiR genes were expressed in all investigated tissues in both rye lines (Fig. 3). The expression level of ScSERK and ScNiR was higher in line L9 in most stages of tissue culture except for two: after two weeks on induction medium and four weeks on regeneration medium. After two weeks on regeneration medium, the expression of these genes was increased in both lines. There was no difference between rye lines L318 and L9 in the amount of ScSERK and ScNiR mRNA observed in leaves and immature embryos.
Fig. 3.
Relative expression levels (Y axis) of rye SERK, LEC1, VP1 and NiR ortologues in two lines L318 and L9 with different regeneration ability. a – leaves; b – mature zygotic embryos; c – immature zygotic embryos; d – g (callus derived from induction medium after two, four, eight or twelve weeks incubation, respectively); h – j (callus collected from regeneration medium after two days, two weeks and four weeks, respectively). The expression level of immature embryos of line L318 was used as a calibrator
The level of ScLEC1 expression was relatively low throughout the whole period in in vitro culture and was highest in line L9 after 12 weeks on induction medium, and in line L318 after 4 weeks on regeneration medium. Similarly, the expression level of ScVP1 was low in almost all investigated stages apart from callus of line L9 collected after 4 weeks of culture on induction medium (16-fold higher than in line L318).
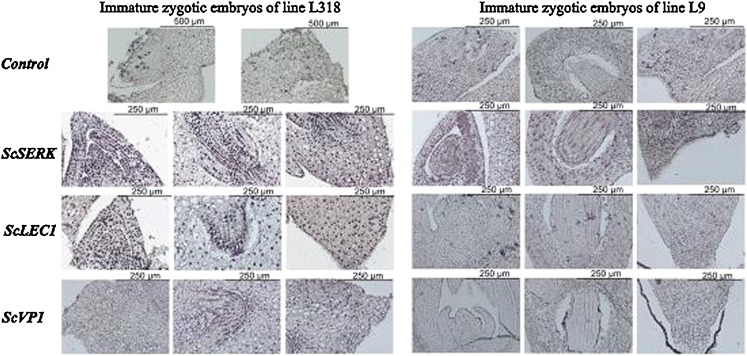
In situ RT-PCR analysis demonstrated that ScSERK mRNA was localized in immature zygotic embryos of L318 and L9 and was present at a similar level in both lines (Fig. 4). The most intense signals came from the shoot and root regions of the embryo. In opposition to the q-PCR data, the strongest ScLEC1 expression was located in the shoot part of immature L318 embryos. No accumulation of the ScLEC1 transcript was detected in embryos of line L9 (Fig. 4). Similarly, the hybridization signal of the ScVP1 transcript was only observed in embryo roots of line L318, whereas it was not detected in the same tissue of L9 embryos (Fig. 4).
Fig. 4.
In situ RT-PCR detection of ScSERK, ScLEC1, ScVP1 mRNA in immature zygotic embryos of rye. The negative control was a specimen without specific primers added to the amplification mixture. The blue-purple staining indicates the location of transcript accumulation
Discussion
Somatic embryogenesis is controlled by several key genes including SERK, LEC1, LEC2, FUS3/ABI3, LEA and BBM (Ikeda et al. 2006), but until now these genes have not been examined in rye. The main aim of this study was to identify rye ortologues of SERK, LEC1, VP1 and NiR and examine their expression in terms of the regeneration processes in cultured immature rye embryos. NiR gene is not connected with somatic embryogenesis directly but it plays a key role in morphogenesis initiation in vitro. Therefore we decided to include this gene in our analysis.
Using specific oligonucleotide primers, four sequences designated ScSERK, ScLEC1, ScVP1 and ScNiR were amplified by RT-PCR from rye RNA. Expression of the rye ScSERK ortologue was detected at similar levels in both the highly regenerative line L318 and the poorly regenerative line L9 during the in vitro culture period. These results differ from those obtained for carrot (Schmidt et al. 1997) and Arabidopsis (Hecht et al. 2001) the expression of the SERK genes in which was detected only in the embryogenic calli and never in non-embryogenic tissues. But they are similar with those reported for the maize ZmSERK1 and ZmSERK2 genes the expression of which was found in the embryogenic and non-embryogenic cultures (Baudino et al. 2001). Similar levels of the MtSERK1 transcript were found in leaf explants of two lines of Medicago truncatula that differ in their regeneration potential (Nolan et al. 2003). Although the expression of ScSERK was detected at all tissue culture stages, its level in line L9 was lightly higher. Comparable levels of ScSERK transcripts in immature embryos of L318 and L9 may indicate their similar embryogenic potential. During the first 2 weeks of culture the expression of ScSERK decreased by half in line L318 and by about 75% in L9, in comparison to the explants. We suppose that at this culture stage the SERK protein is activated in callus of line L318. SERK is a receptor kinase that can participate in signal transduction pathways and in the initiation of processes leading to somatic embryogenesis (Schmidt et al. 1997). However, SERK protein seems to exert a negative influence on plant regeneration as the transcript level was decreased in line L9 after 4 weeks on regeneration medium.
A similar pattern of expression was observed in the ScNiR gene. The expression profile of ScNiR during the in vitro culture of rye confirmed its relationtion in the regeneration process. The 4-fold higher levels of the ScNiR transcript in calli of line L318 collected after two weeks of culturing on medium containing auxins may indicate the positive effect of its gene product on embryogenic callus formation. Nitrate is added as the nitrogen source for in vitro culture, but its metabolite, nitrite, has a toxic effect on the plant cell growth. The rapid metabolism of nitrite is therefore essential for callus growth and so the NiR-encoded ferredoxin-nitrite reductase, which catalyzes the reduction of nitrite to ammonium ions, plays a key role. The increased expression of ScNiR in the embryogenic calli of line L318 subcultured on medium without hormones is probably due to the elevated level of metabolic activity connected with the intensive regeneration process. Metabolic products, including the toxic NO-2, are released into the medium. Presumably ScNiR expression is enhanced to counter the accumulation of nitrite ions.
The LEC1 gene is an important regulator of somatic embryogenesis (Lotan et al. 1998; Yazawa et al. 2004; Zhang et al. 2002). LEC1 encodes a protein related to the heme-activated protein 3 (HAP3) subunit of the CCAAT box-binding factor (CBF), a eukaryotic transcriptional regulator (Lotan et al. 1998). Because LEC1 is a component of a plant CBF, it may regulate embryonic processes by activating the transcription of specific genes (Lotan et al. 1998). Ectopic expression of the LEC1 gene in post-embryonic Arabidopsis plants induces embryonic processes and a set of specific genes required for embryo development (Lotan et al. 1998).
The expression of ScLEC1 was detected in all tissues and at all culture stages, but was always higher in line L9 than in L318, except during the 4 weeks on regeneration medium. It is possible that ScLEC1, as an element of a CBF complex, is connected with the suppression of genes regulating somatic embryogenesis. A significant increase in the expression of LEC1 in 4-week-old callus of line L318 may indicate that this gene is crucial for the induction of plant regeneration. However, further studies are required to precisely characterize its function. In this respect it would be informative to identify the other components of the CBF complex and to examine the expression of the PKL (Pickle) gene, which acts as the repressor of LEC1 (Lotan et al. 1998).
The ScVP1 transcript was barely detectable in zygotic embryos and leaves of both rye lines. Only callus of line L9 collected after 4 weeks of in vitro culture showed definite expression of this gene. Similarly in wheat, expression of the VP1 gene was not detected in leaves, but some of the transcript was present in mature zygotic embryos (Nakamura and Toyama 2001). VP1 has been shown to be a major regulator of late embryo development in wheat and ancestral species (McKibbin et al. 2002). Inactivation of VP1 leads to the vivipary (McKibbin et al. 2002). Due to the good regeneration ability of L318 it might have been expected that ScVP1 would be expressed in this line. However, the very low ScVP1 expression in 4-week-old L318 callus compared to a significant increase in a transcript level in L9, suggests that it functions as a suppressor of somatic embryogenesis and plant regeneration. It is possible that in its role as a transcription factor, VP1 may act both as activator and a repressor for different genes (Suzuki et al. 1997). Among the genes that are regulated by VP1 are two kinases (Suzuki et al. 2003, supplemental data 1, www.plantphysiol.org), implying a potential function for this factor in the suppression of ScSERK during the culture of immature embryos of rye. The findings of the present study are in agreement with those of a previous study employing classical genetic analysis (Rakoczy-Trojanowska and Malepszy 1995). This analysis showed that the lack of response is determined by at least two interacting genes of a suppressive character. Three of the genes examined here – ScSERK, ScLEC1 and ScVP1, may be appropriate candidates.
While further research is essential to gain a greater understanding of the molecular mechanisms controlling the tissue culture response of rye, it is hoped that most of the presented data will find application in plant biotechnology and molecular breeding.
Conclusions
Expression of rye ortologue of the SERK gene is likely to be involved in the induction of somatic embryogenesis initiated from immature embryos and its expression is suppressed in the later stages of in vitro culture.
The expression profiles of the genes ScLEC1 and ScVP1 in line L9 suggest that their complementary interaction negatively influences somatic tissue development.
Enhanced expression of the ScNiR gene in line L318 at the start of tissue culture and in callus in medium without hormones (4th week of culture), may indicate its important double role: in callus induction and in plant regeneration.
Acknowledgements
We would like to thank Grzegorz Koczyk (Institute of Plant Genetics Polish Academy of Sciences, Poznań) for bioinformatics instruction when designing the primers.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Baudino S, Hansen S, Brettschneider R, Hecht VRG, Dresselhaus T, Lörz H, Dumas C, Rogowsky PM. Molecular characterisation of two novel maize LRR receptor-like kinases, which belong to the SERK gene family. Planta. 2001;213:1–10. doi: 10.1007/s004250000471. [DOI] [PubMed] [Google Scholar]
- Bolibok H, Gruszczyńska A, Hromada-Judycka A, Rakoczy-Trojanowska M. The identification of QTLs associated with the in vitro response of rye (Secale cereale L.) Cell Mol Biol Lett. 2007;12:523–535. doi: 10.2478/s11658-007-0023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu C, van Lammeren AAM, Miki BLA, Custers JBM, van Lookeren Campagne MM. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 2002;14:1737–1749. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidiniumthiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Gaj MD, Zhang S, Harada JJ, Lemaux PG. Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta. 2005;222:997–988. doi: 10.1007/s00425-005-0041-y. [DOI] [PubMed] [Google Scholar]
- Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt EDL, Boutilier K, Grossniklaus U, de Vries SC. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 2001;127:803–816. doi: 10.1104/pp.010324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hromada A, Bolibok H, Rakoczy-Trojanowska M. Application of the GDDSC for the isolation of winter rye (Secale cereale L.) genome regions connected with in vitro reaction of immature embryos. Vortr Pflanzenzüchtg. 2007;71:217–224. [Google Scholar]
- Ikeda M, Umehara M, Kamada H. Embryogenesis-related genes; its expression and roles during somatic and zygotic embryogenesis in carrot and Arabidopsis. Plant Biotech. 2006;23:153–161. [Google Scholar]
- Linacero R, Vasquez AM. Somatic embryogenesis from immature inflorescences of rye. Plant Sci. 1990;72:253–258. doi: 10.1016/0168-9452(90)90089-7. [DOI] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/S0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Ma R, Guo Y-D, Pulli S. Somatic embryogenesis and fertile green plant regeneration from suspension cell-derived protoplasts of rye (Secale cereale L.) Plant Cell Rep. 2003;22:320–327. doi: 10.1007/s00299-003-0694-6. [DOI] [PubMed] [Google Scholar]
- McKibbin RS, Wilkinson MD, Bailey PC, Flintham JE, Andrew LM, Lazzeri PA, Gale MD, Lenton JR, Holdsworth MJ. Transcripts of VP1 homeologues are misspliced in modern wheat and ancestral species. Proc Natl Acad Sci USA. 2002;99:10203–10208. doi: 10.1073/pnas.152318599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige F, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Nakamura S, Toyama T. Isolation of a VP1 homologue from wheat and analysis of its expression in embryos of dormant and non-dormant cultivars. J Exp Bot. 2001;52(357):875–876. doi: 10.1093/jexbot/52.357.875. [DOI] [PubMed] [Google Scholar]
- Nishimura A, Ashikari M, Lin S, Takashi T, Angeles ER, Yamamoto T, Matsuoka M. Isolation of rice regeneration quantitative trait loci gene and its application to transformation system. Proc Natl Acad Sci USA. 2005;102:11940–11944. doi: 10.1073/pnas.0504220102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan KE, Irwanto RR, Rose RJ. Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiol. 2003;133:218–230. doi: 10.1104/pp.103.020917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybecki Z, Siedlecka E, Filipecki M, Urbańczyk-Wochniak E. In situ reverse transcription PCR on plant tissues. Meth Mol Biol. 2006;334:181–197. doi: 10.1385/1-59745-068-5:181. [DOI] [PubMed] [Google Scholar]
- Rakoczy-Trojanowska M, Malepszy S. The influence of genetic factors on regeneration ability in plant cultures in vitro. Post Biol Kom. 1990;17(13):247–257. [Google Scholar]
- Rakoczy-Trojanowska M, Malepszy S. Genetic factors influencing regeneration ability in rye (Secale cereale L.). I. Immature inflorescences. Theor Appl Genet. 1993;86:406–410. doi: 10.1007/BF00838554. [DOI] [PubMed] [Google Scholar]
- Rakoczy-Trojanowska M, Malepszy S. Genetic factors influencing regeneration ability in rye (Secale cereale L.). II. Immature embryos. Theor Appl Genet. 1995;83:233–239. doi: 10.1007/BF00838554. [DOI] [PubMed] [Google Scholar]
- Schmidt EDL, Guzzo F, Toonen MAJ, de Vries SC. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development. 1997;124:2049–2062. doi: 10.1242/dev.124.10.2049. [DOI] [PubMed] [Google Scholar]
- Somleva MN, Schmidt EDL, de Vries SC. Embryogenic cells in Dactylis glomerata L. (Poaceae) explants identified by cell cracking and by SERK expression. Plant Cell Rep. 2000;19:718–726. doi: 10.1007/s002999900169. [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fisher RL, Goldberg LB, Harada JJ. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Katterling MG, Li Q, McCarty DR. Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol. 2003;132:1664–1677. doi: 10.1104/pp.103.022475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Lin SY, Tanno K, Komatsuda T, Yano M, Sasaki T, Oka S. Mapping quantitative trait loci associated with regeneration ability of seed callus in rice, Oryza sativa L. Theor Appl Genet. 1997;95:828–833. doi: 10.1007/s001220050632. [DOI] [Google Scholar]
- Ward KA, Jordan MC. Callus formation and plant regeneration from immature and mature embryos of rye (Secale cereale L.) In vitro Cell Dev Biol Plant. 2001;37:361–368. doi: 10.1007/s11627-001-0064-4. [DOI] [Google Scholar]
- Yazawa K, Takahata K, Kamada H. Isolation of the gene encoding Carrot leafy cotyledon1 and expression analysis during somatic and zygotic embryogenesis. Plant Physiol Biochem. 2004;42:215–223. doi: 10.1016/j.plaphy.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Zhang S, Wong L, Meng L, Lemaux PG. Similarity of expression patterns of knotted1 and ZmLEC1 during somatic and zygotic embryogenesis in maize (Zea mays L.) Planta. 2002;215:191–194. doi: 10.1007/s00425-002-0735-3. [DOI] [PubMed] [Google Scholar]