Abstract
The role of the Notch signaling pathway in tumor development is complex, with Notch1 functioning either as an oncogene or as a tumor suppressor in a context-dependent manner. To further define the role of Notch1 in tumor development, we systematically surveyed for tumor suppressor activity of Notch1 in vivo. We combined the previously described Notch1 intramembrane proteolysis–Cre (Nip1::Cre) allele with a floxed Notch1 allele to create a mouse model for sporadic, low-frequency loss of Notch1 heterozygosity. Through this approach, we determined the cell types most affected by Notch1 loss. We report that the loss of Notch1 caused widespread vascular tumors and organism lethality secondary to massive hemorrhage. These findings reflected a cell-autonomous role for Notch1 in suppressing neoplasia in the vascular system and provide a model by which to explore the mechanism of neoplastic transformation of endothelial cells. Importantly, these results raise concerns regarding the safety of chronic application of drugs targeting the Notch pathway, specifically those targeting Notch1, because of mechanism-based toxicity in the endothelium. Our strategy also can be broadly applied to induce sporadic in vivo loss of heterozygosity of any conditional alleles in progenitors that experience Notch1 activation.
Introduction
The Notch pathway is an evolutionarily conserved signaling mechanism that plays essential roles both during development and in the maintenance of adult tissue homeostasis in metazoan (1). Activation of Notch receptors involves the sequential cleavage by ADAM10 and γ-secretase, which results in the release of the Notch intracellular domain (NICD). NICD translocates into the nucleus, where it binds RBPJK and recruits Mastermind and coactivators to turn on target gene expression (Figure 1A and ref. 1). The physiological outcome is highly context dependent: Notch1 can promote differentiation in some tissues while acting to maintain stem cell proliferation in others. Consequently, Notch1 can act either as a tumor suppresser or as an oncogene (2). In contrast to gain-of-function mutations in Notch1 and their oncogenic role in T-ALL (3, 4), and perhaps in solid tumors, Notch1 loss-of-function mutations have only been implicated in a limited number of tumors, such as skin (2), where Notch1 loss promotes tumor formation non–cell-autonomously by creating a wound-like environment and autonomously by enhancing progression to cancer (5, 6).
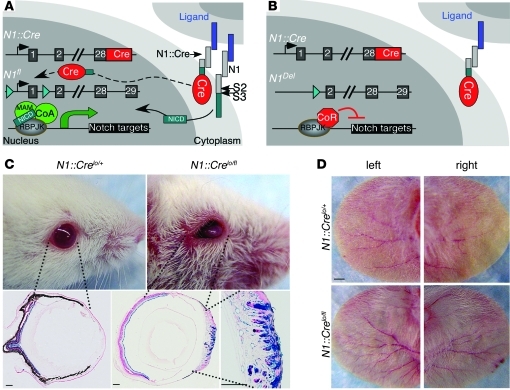
Figure 1. N1::Cre excises floxed Notch1 after activation.
(A and B) Loss of functional Notch1 is a low-frequency event, since only a small number of Cre6MT molecules reach the nucleus. C-terminal 6MT is omitted from the diagram for clarity. CoA, coactivator; CoR, corepressor; MAM, Mastermind. (C) All N1::Crelo/fl mice developed corneal plaques after 2–3 months, and this phenotype became more severe as animals aged. LacZ staining revealed that the transparent corneal was transformed into a skin-like structure with neoangiogenesis. Not all ECs and keratinocytes were labeled with lacZ. (D) N1::Crelo/fl mice showed increased angiogenesis in the ear. Scale bars: 150 μm (C); 10 mm (D).
To determine whether Notch1 acts as a tumor suppressor in additional tissues, we combined a previously reported knockin line, NIP1::Cre allele (N1::Crelo; ref. 7), with a floxed Notch1 allele (8) to perform an unbiased survey of cell types that will gain a growth advantage when Notch1 is lost. We observed that mice with the genotype of N1::Crelo/fl had a shorter median life expectancy and developed vascular tumors in various organs, but most frequently in the liver. Our findings are in agreement with reports suggesting that blockade of the Notch signaling pathway could enhance EC proliferation (9, 10) and suggest a link between loss of Notch1 signaling and formation of vascular tumors. This finding raises concerns about Notch-targeting therapies in humans, in which vascular tumors may arise after chronic exposure to inhibitory Notch1 antibodies (11, 12), decoy ligands (such as Dll4; ref. 9), anti-Dll4 antibodies (9, 10, 13), dominant-negative Mastermind-like 1 (MAML1) mimetics (14), or γ-secretase inhibitors, which are being considered as therapeutic interventions for cancer. Moreover, our model can facilitate biomarker identification and help uncover molecular mechanisms of vascular tumors.
Results
A model of sporadic Notch1 loss of heterozygosity.
Ligand-mediated activation of Notch involves 2 sequential cleavage events that release the NICD from the cell membrane (Figure 1A). This unique activation mechanism allowed us to generate N1::Cre knockin mice (7), in which exons 28–31 of a Notch1 allele (N1) are replaced with cDNA encoding the λ-phage–derived site-specific recombinase Cre and a C-terminal 6Xmyc tag (6MT). Consequently, this is a null Notch1 allele producing a Notch1-Cre6MT fusion protein that releases Cre6MT from the cell membrane following ligand binding and subsequent proteolysis (Figure 1A and ref. 7). We have previously reported that when N1::Cre is combined with the RosaLacZ reporter, the excision of the floxed stop cassette by ligand-dependent release of Cre indelibly labels cells that have experienced Notch1 activation. An active LacZ allele is then inherited by all their descendents, providing a powerful tool for lineage mapping (7).
We reasoned that a combination of N1::Cre expressed from one allele with a floxed Notch1 expressed from the other will produce mice in which the remaining functional (floxed) Notch1 allele would be inactivated at some frequency after every round of Notch1 activation by the N1::Cre protein. This will create progressive, age-dependent Notch1 loss of heterozygosity (LOH; Figure 1B). Because of the low expression level (Supplemental Figure 1), the fusion with 6MT, and the inherently inefficient, ligand-dependent release of Cre from the membrane, very few cells will lose Notch1 after each round of Notch1 activation in this line (referred to herein as N1::Crelo). Cells that experience multiple rounds of Notch1 activation have a higher probability of becoming Notch1 null. Whereas loss of Notch1 may have no consequences on cells that only need 1 round of Notch1 activation and/or in which other Notch receptors can compensate for Notch1 loss, cells depending on Notch1 to maintain tissue homeostasis or promote the exit from cell cycle may begin to accumulate after Notch1 LOH. Their expansion could be detected by 1 of 3 reporter alleles knocked into the Rosa locus downstream of a floxed stop cassette: first, an allele containing click-beetle red (CBR) luciferase that permits periodic noninvasive in vivo luciferase imaging of tumor progression (RosaCBR; Supplemental Figure 2); second, an allele harboring LacZ that allows single-cell resolution postmortem analysis (RosaLacZ; ref. 15); third, an allele bearing enhanced yellow fluorescent protein (EYFP) that facilitates fluorescence-activated cell sorting to purify cells that have experienced Cre activity (RosaEYFP; ref. 16). To control for the possible contribution from the Cre protein (17) or from Notch1 heterozygosity (18, 19), we compared mutant N1::Crelo/fl mice with control N1::Crelo/+ littermates throughout this study.
N1::Crelo/fl mice develop corneal plaques and display increased angiogenesis.
N1::Crelo/fl pups were born at the expected Mendelian ratio and were indistinguishable from their littermate controls at weaning. In contrast, when the floxed Notch1 allele was combined with a knockin allele with improved expression of untagged Cre (i.e., N1::Crehi), a higher LOH frequency was observed, resulting in embryonic lethality at E10.5 due to defective angiogenic remodeling within the yolk sac (Supplemental Figure 1 and refs. 20, 21). Thus, the low frequency of Notch1 LOH in N1::Crelo/fl animals better mimics Notch1 LOH in human patients.
LOH in N1::Crelo/fl mice became evident when all animals developed corneal hyperplasia after 2–3 months (Figure 1C), as previously reported for mice with corneal Notch1 deletion (22). X-gal staining showed that the normal transparent, avascular cornea was replaced by a vascularized epidermis-like structure. Not all keratinocytes and ECs expressed lacZ (Figure 1C), which indicates that Notch1 loss in a fraction of corneal epithelial cells was sufficient to disrupt corneal differentiation.
N1::Crelo/fl mice develop vascular tumors, predominantly in the liver.
The design of our experiment permitted both control and mutant animals to activate the CBR luciferase reporter gene by removal of a floxed stop cassette in a Notch1 activation–dependent manner. In addition to the corneal phenotype and to increased angiogenesis in ears (Figure 1D), no other phenotypes were detected during monthly scans for elevated luciferase activity (see below). However, Kaplan-Meier analysis of 45 control and 41 mutant animals demonstrated that the mutants had significantly reduced life expectancy compared with their littermate controls (~420 versus ~600 days; P < 10–4, log-rank test; Figure 2A). To investigate the underlying pathology, we performed comprehensive necropsy analysis on 13 pairs of control and mutant animals. Postmortem analysis of mutant animals revealed reactive splenomegaly (Figure 2C), characterized by marked expansion of the interfollicular compartments with extramedullary hematopoiesis of granulocytic, erythrocytic, and megakaryotic lineages and no bias in B/T cell lineages (Supplemental Figure 3), and consistent with the observed inflammation in the liver (Supplemental Figure 4). The presence of blood in the abdominal cavity (hemoperitoneum) was noted in 6 of 7 naturally deceased animals (Figure 2B). Different from control liver (Figure 2D), the mutant liver from naturally deceased mutant mice was often pale, with many irregular reddened foci (Figure 2E). Some patches were almost white. Hemoperitoneum, in association with the irregular contour of the liver lobes, indicated that these patches might represent the collapse of blood-filled vascular spaces. Indeed, inspection of the liver in euthanized nonmoribund mutant animals of similar age revealed no blood in the abdominal cavity. Instead, the liver often contained numerous reddened, large blood-filled spaces, consistent with a diagnosis of a vascular tumor (Figure 2F).
Figure 2. N1::Crelo/fl mice developed vascular tumors.
(A) Kaplan-Meier survival analysis of 41 mutant and 45 control animals. Animals euthanized or alive at the time of analysis were censored (44 control and 26 mutant). (B) Summary of the major pathological findings from a comprehensive analysis of 13 pairs of control and mutant animals. Some animals developed hemangiomas in multiple tissues, making the tumor count greater than 13. The distribution of hemangiomas in different organs is further dissected (see Supplemental Table 1). (C–F) Representative images of spleen (C) and liver from N1::Crelo/+ (D) and N1::Crelo/fl animals (E and F). (E) Liver from a naturally deceased animal. (F) Liver from a euthanized N1::Crelo/fl animal, in which the tumors were still intact and filled with blood. (G–I) H&E liver sections from N1::Crelo/+ (G) and N1::Crelo/fl animals (H and I). See Results for details. (J–L) Immunostaining of liver tumor for PECAM (J), vWF (K), and PECAM and Ki67 (L). Inset in L shows the enlarged view of boxed area. Representative vascular tumors in other organs are shown in M (skin; H&E) and N (ovary; vWF). Scale bars: 50 μm; 10 μm (L, inset).
Examination of H&E sections of mutant liver revealed various abnormalities (Supplemental Figure 4). In 11 of 13 animals, the normal homogenous and compact hepatic parenchyma seen in controls (Figure 2G) was replaced either with solid, hypercellular tissue composed of spindle and epithelioid cells interspaced with numerous vascular channels (Figure 2H) or with irregular vascular channels lined by cells with a scant-to-moderate eosinophilic cytoplasm and round-to-oval nuclei (Figure 2I). The nuclei in cells lining vascular cavities either were hyperchromatic or contained coarse chromatin (Figure 2I and Supplemental Figure 5). The endothelial origin of these cells was confirmed by strong staining with EC markers PECAM-1 (CD-31) and vWF (Figure 2, J and K). Many cells were also positive for the cell proliferation marker Ki67 (Figure 2L). These findings were consistent with a diagnosis of vascular tumor/hemangioma in most animals, with incipient hemangiosarcoma in a few.
Although comprehensive analysis on all 13 pairs of animals exposed a plethora of minor anatomic phenotypes and symptoms in addition to those described above (Figure 2B), the statistically significant finding (P = 10–5, 1-tailed Fisher exact test) was that 11 of 13 mutant animals (84.6%) had hemangioma/vascular tumors, some with multiple involved organs. These included the ovary, testis, skin, lymph node, uterus, and colon (Figure 2, B, M, N, and Supplemental Table 1). Nonetheless, the liver was the primary site affected in 81.8% of mutant animals (9 of 11; Figure 2B and Supplemental Table 1), within which the tumor progression could be assessed with MRI (Figure 3A and Supplemental Videos 1–3). In contrast to mutant animals, none of the control animals developed vascular tumors (Figure 2B), even late in life (~2 years).
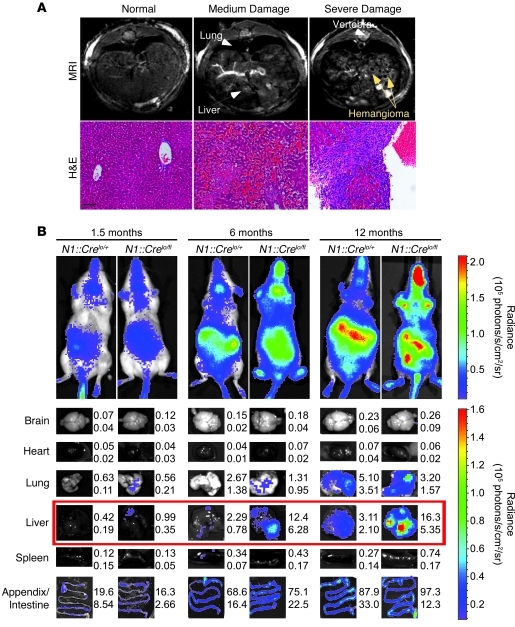
Figure 3. Imaging tumor progression.
(A) MRI was used to assess tumor progression in the liver. H&E images of the liver from respective animals are also shown. Scale bar: 100 μm. (B) Bioluminescence imaging of live animals and freshly dissected organs at the specified ages. Shown are mean and SD of organ luciferase signal (105 photons/s) from at least 4 different individuals of the same genotype. Organs showing statistically significant differences in signal intensity are boxed in red. See Supplemental Figure 6 for images of more organs.
Incidence of vascular tumors in various organs correlates with efficiency of EC labeling.
The high incidence rate of hemangioma in the liver prompted us to ask whether Notch1 activation is similar in all vascular (i.e., red) organs. Flow cytometry analysis of EYFP+ ECs from control N1::Crelo/+ animals revealed that the liver ECs had significantly higher EYFP labeling efficiency (Figure 4), perhaps reflecting higher remodeling rates. Interestingly, these differences mirrored tumor incidence rates (Figure 2B).
Figure 4. Comparison of the RosaEYFP reporter labeling efficiency in ECs of different organs in N1::Crelo/+RosaEYFP/LacZ mice.
(A) ECs from liver, kidney, spleen, heart, and lung were enriched, stained with CD45 and CD31, and subjected to flow cytometry analysis. Percent CD31+EYFP– and CD31+EYFP+ cells among CD45– cells is indicated. (B) Statistical analysis on 4 N1::Crelo/+RosaEYFP/LacZ mice showed that the liver had the highest percentage of EYFP+ ECs (about 20%, compared with less than 5% in other organs). *P < 0.05.
Bioluminescence imaging of tumor progression.
Our attempt to monitor tumor growth in mutant animals by in vivo imaging with CBR luciferase failed, because of the overwhelmingly strong signal from LOH of Notch1 in the intestine (Figure 3B and ref. 7). However, no lesions were found in the intestinal system, consistent with the redundant role for Notch2 in this niche (23). Notch2 activation was not detected in any EC (data not shown). To confirm that vascular tumor cells experienced Cre activity, we performed ex vivo bioluminescence imaging on freshly dissected organs and observed statistically significant differences between control and mutant liver starting at as early as 1 month of age, long before animal survival was compromised (Figure 3B). Notably, bioluminescence increased with tumor progression (Figure 3B and Supplemental Figure 6). The failure to identify any other foci with significantly increased bioluminescence among 12 N1::Crelo/fl animals indicates that other tumors were unlikely to be present.
EC proliferation is a direct consequence of Notch1 loss.
Histological analysis identified multiple LacZ-positive ECs within the mutant liver parenchyma; costaining of PECAM/Ki67, PECAM/EYFP, and EYFP/BrdU confirmed that proliferating (Ki67+ or BrdU+) cells were of endothelial origin, expressed the reporter, and may have lost functional Notch1 (Figure 2L and Figure 5A).
Figure 5. Characterization of vascular tumor cells.
(A) LacZ-, PECAM/EYFP-, and BrdU/EYFP-stained liver sections. A single, 2-hour-long pulse of BrdU labeled 5%–10% of ECs. Inset in right panel shows the enlarged view of boxed area. Scale bars: 50 μm; 10 μm (inset). (B–E) Immunostaining of phospho-Rb. Phospho-Rb signal was absent in areas without tumor (B and C); the presence of the signal (brown) is shown in D and E. Sections were counterstained with hematoxylin. The tumor identity in D and E was confirmed with PECAM and Ki67 costaining in Figure 2L (adjacent sections). Boxed regions in B and D are shown at higher magnification in C and E, respectively. Scale bars: 100 μm.
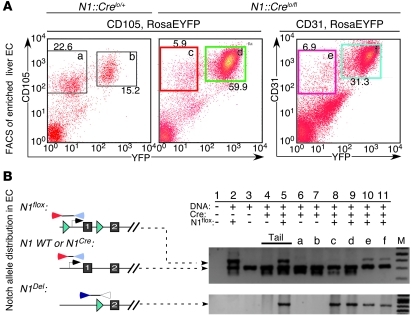
The analyses performed thus far suggest that Notch1 deletion occurred in many ECs in the N1::Crelo/fl animal. To evaluate the status of the Notch1 locus in liver ECs, we enriched for these cells by a Percoll gradient isolation (24, 25) followed by fluorescence-activated cell sorting (FACS). CD105+CD45– proliferating liver ECs from N1::Crelo/flRosaEYFP/LacZ and N1::Crelo/+RosaEYFP/LacZ mice were collected (Figure 6A), and the DNA was purified and subjected to PCR amplification in order to detect the 3 Notch1 alleles: a normal promoter in the N1::Crelo allele, a floxed promoter in N1fl allele, and a rearranged N1del allele lacking the proximal promoter and exon1 (Figure 6B). In all tissues isolated from N1::Crelo/fl animals, N1del was readily detected (Figure 6B, bottom). In addition, both the N1fl and the wild-type N1::Crelo promoters were detected in multiple tissues and in unsorted cells (Figure 6B, top). In contrast, only the intact N1::Crelo allele, but not the N1fl allele, was detected in purified, proliferating ECs (CD45–CD105+), regardless of their EYFP expression status (Figure 6A, gates c and d). This indicates that the vast majority (>90%; Supplemental Figure 7) of proliferating liver ECs have lost the functional (N1fl) allele. Analysis of DNA from cells sorted with the pan-EC marker CD31 detected the N1fl allele in both CD31+EYFP+ and CD31+EYFP– ECs (Figure 6A, gates e and f). The presence of a functional Notch1 in pan-ECs but not CD105+ proliferating ECs, the exclusive BrdU labeling of EYFP+ ECs (but not any other linage in the liver; Figure 5A), and the presence of hyperphosphorylated retinoblastoma protein (Rb; Figure 5, B–E) strongly argue that Notch1 suppresses proliferation and vascular tumor formation cell autonomously, as previously reported in cultured ECs (26, 27).
Figure 6. Loss of Notch1 in ECs is associated with their rapid expansion.
(A) Liver ECs were sorted to remove CD45+ cells and then separated based on the expression of EYFP and either CD105 (proliferating EC marker) or CD31 (pan-EC marker). (B) PCR analysis confirmed the deletion of floxed Notch1 allele in the majority of CD45–CD105+EYFP+ and CD45–CD105+EYFP– cells; in contrast, a substantial amount of floxed alleles was still detected in CD45–CD31+EYFP+ and CD45–CD31+EYFP– cells. The primers bind at positions illustrated in the diagram. Lanes a–f correspond with gates in A. M, molecular marker.
Discussion
The Notch signaling pathway is an essential component in a complex network that controls proper development and maintains tissue homeostasis in the adult. The embryonic lethality associated with loss of Notch1 (28, 29) prevents a comprehensive evaluation of its role in various organs in later embryonic stages or in the adult. To circumvent this problem, most genetic approaches adopt transgenic or Cre-LoxP–based conditional loss-of-function strategies (30) to express or remove a gene of interest in most or all cells within a population. An obvious limitation of these methods as models for human cancer is that they do not mimic sporadic occurrence of disease-causing mutations in a few tumor-initiating cells, which later evolve into a tumor. Moreover, transgenic approaches could overwhelm the immune system or disrupt homeostatic mechanisms that might otherwise play significant roles in determining disease progression. To overcome these limitations, hormone-controlled CreER — a fusion protein of Cre recombinase and the binding domain of estrogen receptor — is often used to randomly induce sporadic overexpression or deletion of a gene of interest. Such strategy has been recently used to induce the sporadic expression of a SYT-SSX2 fusion gene in multiple tissues, resulting in exclusively synovial sarcoma-like tumors (31).
Here we devised an alternative strategy to hormone-induced CreER by taking advantage of the low-frequency, activation-dependent trans-excision of a floxed Notch1 allele in N1::Crelo/fl animals to investigate its role in the adult stage. We identified the EC population as dependent on Notch1 activity in the adult and defined the consequence of Notch1 loss: the development of vascular tumors. Unlike the skin, in which tumor promotion is elicited by the non–cell-autonomous consequences of a wound-like environment (5), the tumor suppressor function of Notch1 ECs appears to be cell autonomous. Nevertheless, although these tumors could be found in various organs, they were most prevalent in the liver. Consistent with this observation, we found that ECs from liver have the highest EYFP labeling efficiency, suggestive of higher levels of angiogenic activity and thus higher probability of Notch1 LOH. Alternatively, some unique features of liver ECs (32), as well as the specific anatomical or environmental factor(s), may predispose the liver to vascular tumor formation.
Our study demonstrates the utility of NIP::Cre mice in screening for tumor suppressor functions in vivo and indicates that NIP::Cre mice can serve as a general model in which to initiate sporadic cancer. When compared with transgenic or regular Cre-floxed–based strategies (30), the advantages of NIP::Cre mice are as follows: first, the loss or gain of mutations is sporadic, and their probability can be manipulated by selecting either the N1::Crelo or the N1::Crehi alleles to better mimic cancer initiation in human patients; second, in gain-of-function analysis, tissues that normally do not express Notch1 will not be forced to express it; third, in loss-of-function paradigms, most cells will have an active Notch1 allele during development, and a normal physiological niche could therefore be maintained; and fourth, unlike a tamoxifen-dependent pulse of gene loss, cells that continuously depend on Notch1 have a higher probability of LOH than do cells that activate Notch1 only once in their history. Considering that Notch1 is continuously required in multiple tissues during development and in the adult (33), especially in the gut (7, 34) and mammary gland (35) stem cell populations, a NIP::Cre allele coupled with floxed alleles of tumor suppressors offers a better model to mimic LOH disease development in human patients.
The Notch pathway has emerged as a target to block angiogenesis in solid tumors (9, 10, 36) and eliminate tumor-initiating cells (13), either by the administration of anti-Dll4 antibody (9, 10, 13) or by the overexpression and/or application of a soluble Dll4 protein (9). However, here we showed that chronic reduction in Notch1 signaling could promote vascular tumor formation, and may do so in humans. Consistent with our observation, Yan and colleagues recently showed that chronic injection of Dll4 antibodies into adult mice, rats, and monkeys induces vascular neoplasm only in rats (37). We reproduced this observation in the mouse using a Notch1 LOH genetic model, which indicates that the vascular tumors found in rats treated with Dll4 antibodies could be caused by a mechanism-based toxicity due solely to the loss of Notch1 signaling in ECs. Concomitant loss of signals from other Notch receptors, such as Notch4 in ECs (38) and Notch2/3 in pericytes (39–41), may have accelerated the tumor formation in their model (37). Although no other types of tumors appear sensitive to anti-Notch1 therapy, the therapeutic window of anti-Notch therapies in general, and anti-Notch1 therapies in particular (11), needs to be carefully reevaluated.
Methods
Mice.
N1::Crelo (NIP-Cre; Jax Mice strain name Notch1tm3(cre)Rko; ref. 7), Notch1 floxed conditional mice (Jax Mice strain name Notch1tm2Rko; ref. 8), and the RosaLacZ (15) and RosaEYFP (16) reporter animals have been described previously. N1::Crehi, an enhanced version of N1::Crelo with an increased amount of N1::Cre fusion protein, is described previously (21, 20) and in Supplemental Figure 1. RosaCBR, a Cre reporter line in which the gene encoding CBR luciferase is under the control of the ubiquitous Rosa26 promoter and preceded by a floxed stop cassette, is constructed in a similar way as the RosaLacZ (15) and RosaEYFP (16) knockin animals (Supplemental Figure 2). All animals were housed in the mouse facility of Washington University, and all protocols were approved by the Animal Studies Committee of Washington University.
Immunohistochemistry.
Paraffin sections were prepared with standard procedures from postmortem or euthanized animals. Antigens were retrieved by boiling at 121°C for 20 minutes in 10 mM sodium citrate buffer (pH 6.0) after deparaffinization. Endogenous peroxidase activity was inactivated in 3% H2O2 for 15 minutes before the sections were blocked with blocking buffer (3% BSA, 0.1% Tween in 1× PBS) for 1 hour at room temperature. This was then followed by overnight incubation with different primary antibodies at 4°C. After 3 5-minute washes with 1× PBS, sections were incubated at room temperature for 1 hour with HRP-conjugated anti-rabbit or anti-goat secondary antibodies (Vector Labs, 1:100), depending on the host species of the primary antibodies. DAB reactions were performed following 3 5-minute washes in 1× PBS with substrate from Vector Labs. Sections were counterstained with hematoxylin for 30 seconds, dehydrated, and mounted with VectaMount (Vector Labs). For immunofluorescence staining, 3% H2O2 treatment was omitted, and DyLight488- or DyLight549-conjugated secondary antibodies (Jackson ImmunoResearch, 1:500) were used. For frozen sections, tissues were fixed in 4% PFA, equilibrated with 30% sucrose in 1× PBS overnight, embedded in OCT compound, and cut at 8 μm. For BrdU staining, 10 μl 10 mg/ml 5-bromo-2-deoxyuridine solution (Sigma-Aldrich) per gram body weight was injected intraperitoneally into animals 2 hours before sacrifice. Frozen sections were first stained with chick anti-EYFP primary antibody and Alexa Fluor 488–conjugated donkey anti-chick secondary antibody, followed by treatment with 2N HCl at 37°C for 30 minutes and BrdU staining. The following primary antibodies were used: rabbit anti-Ki67 (Novocastra, 1:300, paraffin sections), goat anti–PECAM-1 (M-20) (Santa Cruz, 1:50, paraffin and frozen sections), rabbit anti-vWF (AbCam, 1:800, paraffin sections), chick anti-GFP (Aves Lab, 1:1,000, frozen sections), rat anti-BrdU (clone BU1/75, Accurate Chemical, 1:200, frozen sections), and rabbit anti–phospho-Rb (Ser780) (Cell Signaling, 1:100, paraffin sections).
X-gal staining.
Animals were anesthetized and perfused with 2% paraformaldehyde plus 0.25% glutaraldehyde in 1× PBS with 2 mM MgCl2. Tissues were then dissected and postfixed in 4% paraformaldehyde in 1× PBS with 2 mM MgCl2 on ice for 1 hour, washed, equilibrated with 30% sucrose in 1× PBS with 2 mM MgCl2 overnight, and embedded in OCT compound. 8-mm frozen sections were cut and stained with X-gal staining solution as described previously (7). Tissues from postmortem animals were processed similarly, except for perfusion.
Purification of liver ECs and flow cytometry.
Freshly dissected liver from control and mutant animals was mechanically disrupted between 2 glass slides and digested with collagenase IV (Sigma-Aldrich). ECs were enriched by Percoll gradient centrifugation (25) and stained with PerCP-Cy5.5–conjugated anti-CD45 (BD Biosciences) and PE-conjugated anti-CD105 or -CD31 (BioLegend) antibodies. FACS sorting was done on a MoFlo instrument (Beckman Coulter). For the comparison of EYFP labeling efficiency in the liver, kidney, spleen, heart, and lung, tissues were first mechanically disrupted and then digested with collagenase IV (Sigma-Aldrich, liver or kidney, 5 mg per animal), collagenase D (Roche, lung or spleen, 5 mg per animal), or collagenase II (Worthington, 3,000 U) supplemented with 200 U DNase I (Roche). Single-cell suspensions from peripheral blood, thymus, spleen, and bone marrow were prepared and analyzed with flow cytometry as described previously (42). Flow cytometry analyses were performed on FACScan (BD Biosciences), and data were analyzed with FlowJo (Treestar). Fluorescence-conjugated antibodies were purchased from Biolegend.
PCR.
Primers 1c (5′-TGGAAGCTACTGACTTAGTAGGGGGAAAAC-3′) and 2d (5′-GCAAGCATGAAGTGGTCCAGGGTGTGAGTG-3′) were used for the simultaneous detection of floxed and wild-type Notch1 allele and produced a 317-bp band for wild-type Notch1 locus and a 367-bp band for floxed Notch1 locus. Primers 1b (5′-TGGCCTGCCTGTCTGGAACAACAGTTCAGG-3′) and 2e (5′-ACCCTTGCCTCAGTTCAAACACAAGATACG-3′) amplified the deleted Notch1 allele with a product size about 532 bp.
Bioluminescence imaging.
For bioluminescence imaging of living animals, mice were anesthetized with 2.5% isoflurane, injected intraperitoneally with 150 μg/g d-luciferin (Biosynth) in PBS, and imaged with a charge-coupled device (CCD) camera–based bioluminescence imaging system (IVIS 50, Caliper; exposure time 1–60 seconds, binning 8, field of view 12, f stop 1, open filter, anterior and posterior views). Signal was displayed as photons/s/cm2/sr (43). Regions of interest (ROIs) were defined manually around the abdomen, throat, and whole body using Living Image and Igor Pro Software (version 2.50), and results were analyzed as photon flux (photons/s). Ex vivo images were obtained immediately after live imaging. Animals were euthanized, and organs were quickly removed and imaged for 60 seconds.
MRI.
All MRI experiments were performed in Washington University’s Biomedical MR Laboratory. Respiratory-gated, spin-echo MR images of mice were collected with a small-animal MR scanner based on an Oxford Instruments (Oxford) 4.7-tesla, 40-cm bore magnet. The magnet is equipped with Magnex Scientific (Oxford) actively shielded, high-performance (21 cm ID, ~30 G/cm, ~400 ms rise time) gradient coils and Oy International Electric Company power supplies and is interfaced with a Varian NMR Systems DirectDrive console. All data were collected using a Stark Contrast 2.5-cm birdcage-style radio frequency coil. Prior to the imaging experiments, mice were anesthetized with isoflurane and were maintained on isoflurane/O2 (1%–1.25% v/v) throughout data collection. Animal core body temperature was maintained at 37°C ± 1°C by circulation of warm air through the bore of the magnet. During the imaging experiments, the respiration rates for all mice were regular and approximately 2 s–1. Synchronization of MR data collection with animal respiration was achieved with a home-built respiratory-gating unit (44), and all images were collected during postexpiratory periods. 24–30 contiguous coronal slices, ventral to dorsal, were collected for each mouse. Imaging parameters were as follows: excitation pulse repetition time, approximately 3 s; time to echo, 30 ms; field of view, 2.5 cm2; slice thickness, 1.0 mm; data matrix, 128 × 128; 4 averages.
Statistics.
Kaplan-Meier survival analysis was done with SPSS. Unpaired 2-tailed Student’s t test was performed using Excel 2008 software. 1-tailed Fisher’s exact test was computed with Statistics Calculators ( http://www.danielsoper.com/statcalc/calc29.aspx). A P value of 0.05 or less was considered significant.
Supplementary Material
Acknowledgments
We thank Sullen Greco for necropsy analysis; Edi Brogi, Shadmehr Demehri, and Omar Jassim for advice; Andrew Zhang for technical assistance; and Kopan lab members for helpful discussion. This work was supported by NIH/NCI Small Animal Imaging Resource Program (SAIRP) grant U24CA83060, NIH/NCI In vivo Cellular and Molecular Imaging Center (ICMIC) grant P50CA094056, and the Alvin J. Siteman Cancer Center at Washington University, an NCI Comprehensive Cancer Center (NIH grant P30CA91842). Z. Liu was supported in part by NIH grant R01DK066408 awarded to R. Kopan.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(2):800–808. doi:10.1172/JCI43114.
See the related Commentary beginning on page 508.
References
- 1. Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64(21):2746–2762. doi: 10.1007/s00018-007-7164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 4. Jeannet R, et al. Oncogenic activation of the Notch1 gene by deletion of its promoter in Ikaros-deficient T-ALL [published online ahead of print September 9, 2010].Blood . doi: 10.1182/blood-2010-05-286658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Demehri S, Turkoz A, Kopan R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell. 2009;16(1):55–66. doi: 10.1016/j.ccr.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nicolas M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33(3):416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 7. Vooijs M, et al. Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development. 2007;134(3):535–544. doi: 10.1242/dev.02733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang X, et al. Notch1 signaling influences V2 interneuron and motor neuron development in the spinal cord. Dev Neurosci. 2006;28(1–2):102–117. doi: 10.1159/000090757. [DOI] [PubMed] [Google Scholar]
- 9. Noguera-Troise I, et al. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444(7122):1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- 10. Ridgway J, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444(7122):1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 11. Wu Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464(7291):1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 12. Aste-Amezaga M, et al. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS ONE. 2010;5(2):e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoey T, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009;5(2):168–177. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 14. Moellering RE, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 16. Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Loonstra A, et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci U S A. 2001;98(16):9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Costa RM, Honjo T, Silva AJ. Learning and memory deficits in Notch mutant mice. Curr Biol. 2003;13(15):1348–1354. doi: 10.1016/S0960-9822(03)00492-5. [DOI] [PubMed] [Google Scholar]
- 19. Givogri MI, et al. Notch signaling in astrocytes and neuroblasts of the adult subventricular zone in health and after cortical injury. Dev Neurosci. 2006;28(1–2):81–91. doi: 10.1159/000090755. [DOI] [PubMed] [Google Scholar]
- 20. Liu Z, Obenauf AC, Speicher MR, Kopan R. Rapid identification of homologous recombinants and determination of gene copy number with reference/query pyrosequencing (RQPS). Genome Res. 2009;19(11):2081–2089. doi: 10.1101/gr.093856.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morimoto M, Liu Z, Cheng HT, Winters N, Bader D, Kopan R. Canonical Notch signaling in the developing lung is required for determination of arterial smooth muscle cells and selection of Clara versus ciliated cell fate. J Cell Sci. 2010;123(pt 2):213–224. doi: 10.1242/jcs.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vauclair S, Majo F, Durham AD, Ghyselinck NB, Barrandon Y, Radtke F. Corneal epithelial cell fate is maintained during repair by Notch1 signaling via the regulation of vitamin A metabolism. Dev Cell. 2007;13(2):242–253. doi: 10.1016/j.devcel.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 23. Riccio O, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27(Kip1) and p57(Kip2). EMBO Rep. 2008;9(4):377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Beijnum, JR, Rousch M, Castermans K, van der Linden E, Griffioen AW. Isolation of endothelial cells from fresh tissues. Nat Protoc. 2008;3(6):1085–1091. doi: 10.1038/nprot.2008.71. [DOI] [PubMed] [Google Scholar]
- 25. Gomez DE, Thorgeirsson UP. Lectins as tools for the purification of liver endothelial cells. In: Rhodes JM, Milton JD, eds.Lectin Methods and Protocols . Totowa, New Jersey, USA: Humana Press; 1998:319–328. [DOI] [PubMed] [Google Scholar]
- 26. Noseda M, et al. Notch activation induces endothelial cell cycle arrest and participates in contact inhibition: role of p21Cip1 repression. Mol Cell Biol. 2004;24(20):8813–8822. doi: 10.1128/MCB.24.20.8813-8822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sainson RC, et al. Cell-autonomous notch signaling regulates endothelial cell branching and proliferation during vascular tubulogenesis. FASEB J. 2005;19(8):1027–1029. doi: 10.1096/fj.04-3172fje. [DOI] [PubMed] [Google Scholar]
- 28. Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8(6):707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 29. Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121(5):1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 30. Keller C, Capecchi MR. New genetic tactics to model alveolar rhabdomyosarcoma in the mouse. Cancer Res. 2005;65(17):7530–7532. doi: 10.1158/0008-5472.CAN-05-0477. [DOI] [PubMed] [Google Scholar]
- 31. Haldar M, Hedberg ML, Hockin MF, Capecchi MR. A CreER-based random induction strategy for modeling translocation-associated sarcomas in mice. Cancer Res. 2009;69(8):3657–3664. doi: 10.1158/0008-5472.CAN-08-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chi JT, et al. Endothelial cell diversity revealed by global expression profiling. Proc Natl Acad Sci U S A. 2003;100(19):10623–10628. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dou GR, et al. RBP-J, the transcription factor downstream of Notch receptors, is essential for the maintenance of vascular homeostasis in adult mice. FASEB J. 2008;22(5):1606–1617. doi: 10.1096/fj.07-9998com. [DOI] [PubMed] [Google Scholar]
- 34. van Es JH, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435(7044):959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 35. Bouras T, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3(4):429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 36. Thurston G, Noguera-Troise I, Yancopoulos GD. The Delta paradox: DLL4 blockade leads to more tumour vessels but less tumour growth. Nat Rev Cancer. 2007;7(5):327–331. doi: 10.1038/nrc2130. [DOI] [PubMed] [Google Scholar]
- 37. Yan M, et al. Chronic DLL4 blockade induces vascular neoplasms. Nature. 2010;463(7282):E6–E7. doi: 10.1038/nature08751. [DOI] [PubMed] [Google Scholar]
- 38. Krebs LT, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14(11):1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 39. Domenga V, et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004;18(22):2730–2735. doi: 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Campos AH, Wang W, Pollman MJ, Gibbons GH. Determinants of Notch-3 receptor expression and signaling in vascular smooth muscle cells: implications in cell-cycle regulation. Circ Res. 2002;91(11):999–1006. doi: 10.1161/01.RES.0000044944.99984.25. [DOI] [PubMed] [Google Scholar]
- 41. Varadkar P, Kraman M, Despres D, Ma G, Lozier J, McCright B. Notch2 is required for the proliferation of cardiac neural crest-derived smooth muscle cells. Dev Dyn. 2008;237(4):1144–1152. doi: 10.1002/dvdy.21502. [DOI] [PubMed] [Google Scholar]
- 42. Demehri S, et al. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6(5):e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gross S, Piwnica-Worms D. Real-time imaging of ligand-induced IKK activation in intact cells and in living mice. Nat Methods. 2005;2(8):607–614. doi: 10.1038/nmeth779. [DOI] [PubMed] [Google Scholar]
- 44. Garbow J, Dugas J, Song SK, Conradi M. A simple, robust hardware device for passive or active respiratory gating in MRI and MRS experiments. Concepts Magn Reson. 2004;21B(1):40–48. doi: 10.1002/cmr.b.20010. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.