Abstract
Repair of cartilage injury with hyaline cartilage continues to be a challenging clinical problem. Because of the limited number of chondrocytes in vivo, coupled with in vitro de-differentiation of chondrocytes into fibrochondrocytes, which secrete type I collagen and have an altered matrix architecture and mechanical function, there is a need for a novel cell source that produces hyaline cartilage. The generation of induced pluripotent stem (iPS) cells has provided a tool for reprogramming dermal fibroblasts to an undifferentiated state by ectopic expression of reprogramming factors. Here, we show that retroviral expression of two reprogramming factors (c-Myc and Klf4) and one chondrogenic factor (SOX9) induces polygonal chondrogenic cells directly from adult dermal fibroblast cultures. Induced cells expressed marker genes for chondrocytes but not fibroblasts, i.e., the promoters of type I collagen genes were extensively methylated. Although some induced cell lines formed tumors when subcutaneously injected into nude mice, other induced cell lines generated stable homogenous hyaline cartilage–like tissue. Further, the doxycycline-inducible induction system demonstrated that induced cells are able to respond to chondrogenic medium by expressing endogenous Sox9 and maintain chondrogenic potential after substantial reduction of transgene expression. Thus, this approach could lead to the preparation of hyaline cartilage directly from skin, without generating iPS cells.
Introduction
Hyaline cartilage sustains body growth as primordial cartilage and growth plate cartilage, and provides shock absorption and lubrication in diarthrodial joints as articular cartilage. Hyaline cartilage is a highly organized tissue with complex biomechanical properties and substantial durability, which are sustained by extracellular matrix. Hyaline cartilage extracellular matrix is produced by chondrocytes and consists of collagen fibrils composed of types II, IX, and XI collagen molecules, proteoglycans, and other matrix proteins. Because hyaline cartilage has a poor intrinsic capacity for healing, the loss of cartilage due to trauma or degeneration with age can result in growth impairment, debilitating conditions, and osteoarthritis. Cartilage damage sometimes heals with fibrocartilage, which differs from hyaline cartilage. Fibrocartilage is a type of scar tissue that expresses types I and II collagen; hyaline cartilage, in contrast, does not express type I collagen (1, 2). As the presence of type I collagen impairs cartilage-specific matrix architecture and mechanical function, repair of cartilage damage by fibrocartilage results in morbidity and functional impairment. Thus, the goal for repair of cartilage injury is the regeneration of organized hyaline cartilage (3). However, because chondrocytes are limited in number, and because they de-differentiate into fibrochondrocytes during monolayer expansion in culture (4), there is a significant need for cell sources that produce hyaline cartilage in regenerative medicine. Healing of cartilage damage with hyaline cartilage rather than fibrocartilage remains a challenging clinical problem (3).
In primordial cartilage and growth plate cartilage, chondrocytes undergo hypertrophy and hypertrophic cartilage is degraded and eventually replaced by bone. In articular cartilage, chondrocytes seldom undergo hypertrophy and cartilage remains throughout life. Thus, it is also critical to avoid the generation of hypertrophic cartilage, particularly in applications involving repair of articular cartilage. Prevention of hypertrophy of hyaline cartilage to calcified cartilage is a barrier to cartilage regeneration.
Sox9, Sox5, and Sox6 play important roles in the commitment of mesenchymal cells to the chondrocyte lineage. In mouse chimeras, Sox9–/– cells are excluded from cartilage primordia throughout embryonic development (5). Cartilage is almost absent in the limbs of mice after conditional inactivation of the Sox9 gene in early mesenchymal limb bud cells (6) and in mice lacking Sox5 and Sox6 (7). Sox9, Sox5, and Sox6 activate transcription of chondrocyte marker genes by binding their enhancers (8–11).
In the search for alternative cell sources, cell type conversion to chondrocytic cells from dermal fibroblasts has been studied. Ectopic expression of chondrogenic transcriptional factors Sox5, Sox6, and Sox9 in dermal fibroblasts causes expression of their target genes, which are chondrocyte markers such as type II collagen (12). Treatment with chondrogenic supplementation including TGF-β results in expression of chondrocyte markers in dermal fibroblasts (13). However, the histology of pellet cultures of those cells appears fibrocartilaginous (12, 13), strongly suggesting that type I collagen expression persists even after these treatments. Although dermal fibroblasts represent a readily accessible cell source, the tendency for high expression of type I collagen is a large obstacle to production of hyaline cartilage. To eliminate fibroblastic characters, cell reprogramming may be necessary.
Myocytes, adipocytes, and chondrocytes are induced in cultures of the mouse embryonic 10T1/2 cell line by 5-aza-2′-deoxycytidine (14), which probably converts murine embryonic cells to various cell types by reducing DNA methylation (15). These findings support the notion that cell reprogramming facilitates the conversion of cells from one lineage to another. The generation of induced pluripotent stem (iPS) cells has provided tools for reprogramming somatic cells to an undifferentiated pluripotent stem cell state. The expression of a defined set of factors (Oct3/4, Sox2, c-Myc, and Klf4, as well as Nanog and human LIN28) can fully reprogram dermal fibroblasts into iPS cells (16–26). It may therefore be possible that these cell reprogramming tools would contribute to erasing the fibroblastic nature during induction of hyaline cartilage from dermal fibroblast culture.
In the present study, we examined whether hyaline chondrogenic cells could be directly induced from mouse adult dermal fibroblast cultures by the combined expression of reprogramming factors and chondrogenic factors. We induced chondrogenic cell lines directly from dermal fibroblast culture by the transduction of c-Myc, Klf4, and SOX9. Induced chondrogenic cells produced homogeneous hyaline cartilage–like tissue without type I collagen expression after subcutaneous injection into nude mice. Hyaline cartilage–like tissue remained for at least 16 weeks without tumor formation. This approach may therefore be a step toward the generation of patient-specific hyaline cartilage from skin without going through the process of generating iPS cells.
Results
Generation of induced chondrogenic cell lines from mouse dermal fibroblast culture.
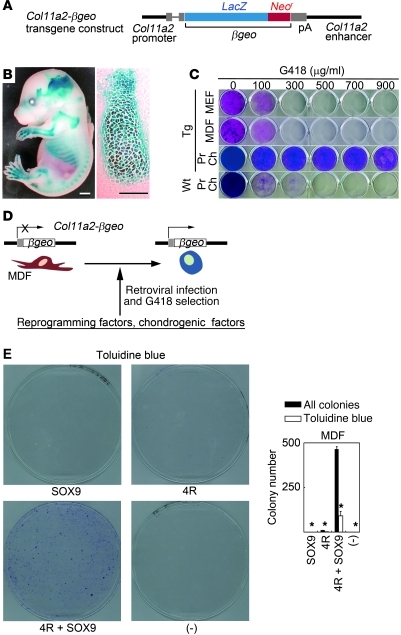
In order to select cells that acquired the chondrogenic phenotype by ectopic expression of factors, we utilized the promoter/enhancer activities of the type XI collagen α2 chain gene (Col11a2), as types II, IX, and XI collagen molecules constitute hyaline cartilage–specific collagen fibrils and sustain cartilage tissue structure. Thus, activation of transcription of Col11a2 was considered to be an indicator of the acquisition of chondrocytic phenotype. We previously identified the Col11a2 promoter/enhancer sequences responsible for cartilage-specific expression (27). The Col11a2 promoter exhibits insulator activities (28) that contribute to stable transgene expression in transgenic mice. Here, we generated transgenic mice expressing βgeo (fusion gene of the β-galactosidase gene [LacZ] and the neomycin resistance gene) under the control of the Col11a2 promoter/enhancer sequences (Figure 1A). Col11a2-βgeo transgenic mice showed LacZ activities specifically in the primordial cartilage of limbs and ribs, but not in other tissues including skin (Figure 1B). Mouse embryonic fibroblasts (MEFs) prepared from transgenic embryos at 13.5 dpc and mouse dermal fibroblasts (MDFs) prepared from adult transgenic mice died in the presence of 300 μg/ml G418 (Figure 1C). On the other hand, primary chondrocytes prepared from transgenic mice survived in the presence of 900 μg/ml G418. Most primary chondrocytes prepared from wild-type littermates died in the presence of 300 μg/ml of G418.
Figure 1. Generation of the Col11a2-βgeo transgenic reporter mouse and selection of cells that acquired chondrogenic characteristics by transduction of reprogramming factors and SOX9.
(A) Schematic representation of the transgene. (B) X-gal staining of the whole body of the Col11a2-βgeo transgenic mouse at 15.5 dpc (left panel). Histological sections of primordial cartilage of the pharynx showed chondrocyte-specific LacZ activity (right panel). Scale bars: 200 μm in left panel, 100 μm in right panel. (C) MEFs prepared from Col11a2-βgeo transgenic embryos at 13.5 dpc, MDFs prepared from 6-month-old Col11a2-βgeo transgenic mice, and primary chondrocytes prepared from Col11a2-βgeo transgenic neonates and wild-type mice were cultured in the presence of various concentrations of G418. Crystal violet staining is shown. Pr Ch, primary chondrocytes. (D) Strategy to induce chondrogenic cells from MDF culture. (E) Effects of 4 reprogramming factors (4R) and/or SOX9 on the colony formation from Col11a2-βgeo reporter MDFs. Black bars indicate the number of all colonies. White bars indicate the number of colonies showing metachromatic staining with toluidine blue (*P < 0.005, compared with 4R + SOX9). Error bars indicate mean ± SD (n = 3). Col11a2-βgeo MDFs were transduced with factors indicated below the graph and selected with G418 for 2 weeks. The MOI for each vector was: pMXs-EGFP, 41; pMXs-c-Myc, 5; pMXs-Klf4, 11; pMXs-Oct3/4, 24; pMXs-Sox2, 16; pMXs-SOX9, 7.
We assessed the ability of a combination of 4 reprogramming factors (Oct3/4, Sox2, c-Myc, and Klf4) and chondrogenic factor SOX9 to induce G418-resistant colony formation from Col11a2-βgeo reporter MDFs. G418 resistance indicated that chondrogenic characteristics had been induced (Figure 1D). We prepared MDFs from Col11a2-βgeo transgenic mice at 3 months of age. We introduced reprogramming factors and SOX9 using the retroviral pMXs/Plat-E vector system (29). Transduction of SOX9 alone into MDFs produced no colonies in the presence of G418 (Figure 1E). Transduction of the 4 reprogramming factors into MDFs resulted in the formation of few colonies. On the other hand, transduction of SOX9 and the 4 reprogramming factors into MDFs induced the formation of approximately 464 G418-resistant colonies from 1.7 × 105 MDFs. To detect glycosaminoglycan production, we stained dishes with toluidine blue. Of 464 colonies, 91 (25%) showed metachromatic purple staining with toluidine blue. On the other hand, very few colonies that were transduced with only the reprogramming factors (but without SOX9) showed metachromatic purple staining with toluidine blue.
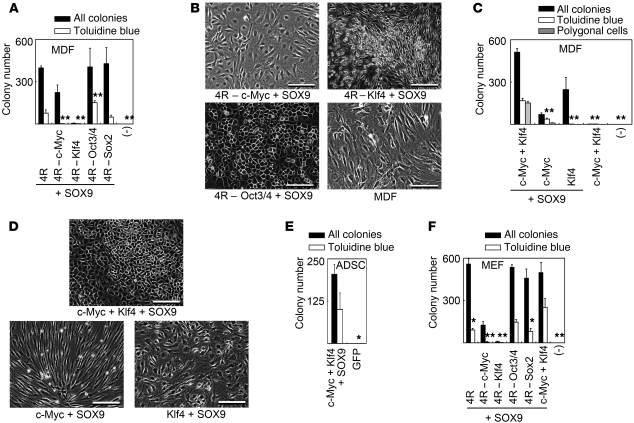
In order to identify the reprogramming factors that are important for colony formation, we removed 1 reprogramming factor and assessed the ability of the remaining 3 factors to induce colony formation from the Col11a2-βgeo reporter MDFs in the presence of SOX9. Removal of either c-Myc or Klf4 from the 4 reprogramming factors in the presence of SOX9 resulted in reduced colony formation (Figure 2A). Very few colonies showed metachromatic staining with toluidine blue. G418-resistant colonies that lacked c-Myc consisted of flatter-shaped cells, and G418-resistant colonies that lacked Klf4 consisted of spindle-shaped cells and small round cells that appeared to be dead (Figure 2B). On the other hand, removal of either Oct3/4 or Sox2 resulted in the formation of substantial numbers of colonies that showed metachromatic staining with toluidine blue (Figure 2A). Most of the colonies with metachromatic toluidine blue staining consisted of polygon-shaped cells, which are typical of chondrocyte morphology (Figure 2B, bottom left). Thus, in this experimental system, c-Myc and Klf4 were needed for the transformation of cells in MDF populations into G418-resistant colonies of cells with metachromatic toluidine blue staining and polygonal chondrocyte-like morphology. The combination of SOX9 plus c-Myc and Klf4 generated 510 G418-resistant colonies from 1.7 × 105 βgeo reporter MDFs (Figure 2C). A total of 168 colonies (33%) showed metachromatic toluidine blue staining, and 152 colonies (30%) consisted of polygon-shaped cells (Figure 2D). The cell morphologies of each colony are shown in Supplemental Figure 1 (supplemental material available online with this article; doi: 10.1172/JCI44605DS1). Of the colonies with metachromatic toluidine blue staining, 84% consisted of polygon-shaped cells, and 92% of the colonies that consisted of polygon-shaped cells showed metachromatic toluidine blue staining. Transduction of c-Myc and SOX9 resulted in the formation of 39 colonies with metachromatic toluidine blue staining, but most of these colonies consisted of spindle-shaped cells with a non-polygonal morphology (Figure 2, C and D). Transduction of Klf4 and SOX9 resulted in the formation of 248 G418-resistant colonies, but none of the colonies showed metachromatic toluidine blue staining. They consisted of cells with flatter, non-polygonal morphology (Figure 2, C and D). In summary, the combination of c-Myc, Klf4, and SOX9 generated 152 G418-resistant colonies of cells with polygonal morphology from 1.7 × 105 βgeo reporter MDFs, and 92% of colonies composed of cells with polygonal morphology showed metachromatic staining with toluidine blue. Removal of any one of these three factors decreased the number of G418-resistant colonies of cells with polygonal morphology.
Figure 2. Effects of combinations of reprogramming factors and SOX9 on formation of G418-resistant colonies from 1.7 × 105 Col11a2-βgeo transgenic reporter cells.
(A) Effects of removal of 1 reprogramming factor from 4 factors plus SOX9 on the formation of G418-resistant colonies from Col11a2-βgeo transgenic reporter MDFs. For metachromatic toluidine blue staining, **P < 0.01 compared with 4R + SOX9. (B) Morphology of cells induced by removal of c-Myc, Klf4, or Oct3/4 from 4 reprogramming factors plus SOX9 from βgeo MDFs and morphology of MDFs. Scale bars: 100 μm. (C) Effects of c-Myc and/or Klf4 plus SOX9 on the formation of G418-resistant colonies from βgeo MDFs. For metachromatic toluidine blue staining, **P < 0.01 compared with c-Myc + Klf4 + SOX9. The number of colonies composed of polygon-shaped cells is indicated by gray bars. (D) Morphology of cells induced by c-Myc, Klf4, and SOX9, c-Myc and SOX9, and Klf4 and SOX9 from βgeo MDFs. Scale bars: 100 μm. (E) Effects of c-Myc, Klf4, and SOX9 on the formation of G418-resistant colonies from ADSCs prepared from Col11a2-βgeo transgenic mice. For metachromatic toluidine blue staining, *P < 0.05. (F) Effects of various combinations of reprogramming factors and SOX9 on the formation of G418-resistant colonies from Col11a2-βgeo transgenic MEFs. For metachromatic toluidine blue staining, *P < 0.05 and **P < 0.01 compared with c-Myc + Klf4 + SOX9. Black bars indicate the number of all colonies, and white bars indicate the number of colonies stained with toluidine blue. Error bars indicate mean ± SD (n = 3). MOI for each vector in A and F was: pMXs-EGFP, 41; pMXs-c-Myc, 5; pMXs-Klf4, 11; pMXs-Oct3/4, 24; pMXs-Sox2, 16; pMXs-SOX9, 7. MOI for each vector in C and D was: pMXs-EGFP, 61; pMXs-c-Myc, 7; pMXs-Klf4, 16; pMXs-SOX9, 11.
Transduction of c-Myc, Klf4, and SOX9 into Col11a2-βgeo reporter adipose tissue–derived stromal cells (ADSCs) prepared from adult mice also generated G418-resistant colonies of cells with metachromatic staining with toluidine blue at a slightly reduced efficiency when compared with that for MDFs (Figure 2E). Transduction of various combinations of reprogramming factors and SOX9 into MEFs showed that c-Myc and Klf4 are necessary and that c-Myc, Klf4, and SOX9 are sufficient for the formation of G418-resistant colonies exhibiting metachromatic staining with toluidine blue (Figure 2F). We examined the effects of Sox5 and Sox6 by transducing c-Myc, Klf4, Sox5, and Sox6 into βgeo reporter MDFs. No G418-resistant colonies were formed by this combination of factors, indicating that Sox5 and Sox6 cannot be substituted for Sox9 in this selection system.
From MDF cultures transduced with c-Myc, Klf4, and SOX9, we selected colonies composed of cells with polygonal morphology and continued their cultivation. We established 27 cell lines (referred to as MK-1 and MK-3 to -28) from individual colonies. However, we lost the MK-2 cell line due to technical errors during expansion of the cell population.
Induced cells were chondrogenic in vitro.
In an effort to determine whether induced cells selected with G418 have chondrogenic potential, we cultured the MK-1 and MK-3 to -28 cell lines in three-dimensional, type I collagen gels for 3 weeks. Histological analysis of the collagen gels showed that all MK cell lines examined produced cartilage-like structures, while MDFs did not (Supplemental Figure 2A).
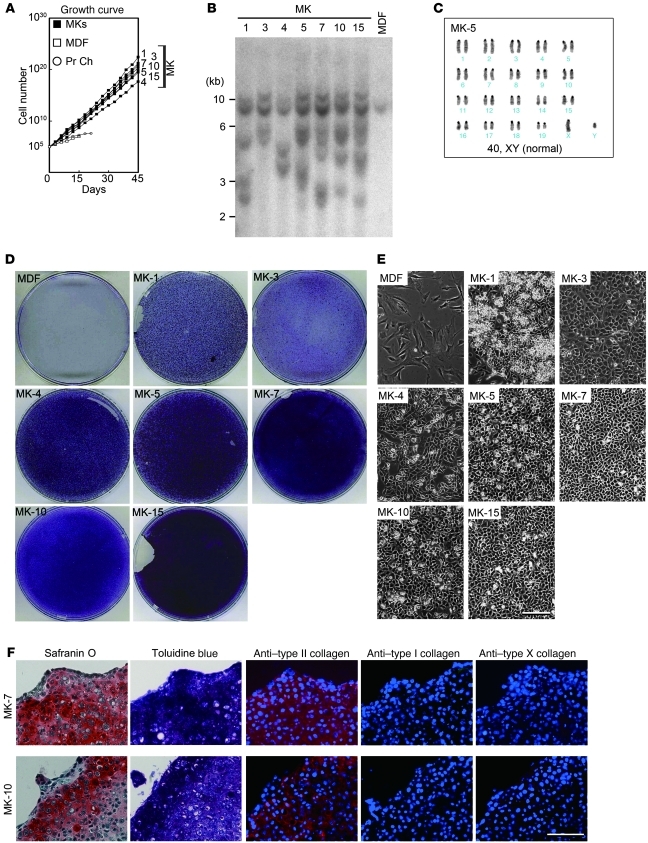
We then randomly selected the MK-1, -3, -4, -5, -7, -10, and -15 cell lines for closer examination. We initiated growth curve analyses of induced cells at passage 6. Under the assumption that each induced cell line arises from a single cell, we estimated that induced cells went through 22 population doublings before they were used in this experiment. Growth curve analysis showed that the MK cell lines grew exponentially for more than 45 days (Figure 3A), while MDFs and primary chondrocytes stopped growing at 15 and 21 days, respectively. Calculated doubling time for each cell line, MDFs, and primary chondrocytes were as follows: MK-1, 38 hours; MK-3, 40 hours; MK-4, 72 hours; MK-5, 49 hours; MK-7, 42 hours; MK-10, 43 hours; MK-15, 49 hours; MDFs, 163 hours; and primary chondrocytes, 100 hours. Southern hybridization analysis of genomic DNAs showed that each MK cell line had a unique retroviral transgene integration pattern (Figure 3B). Karyotyping analyses of induced cell lines showed high frequencies of cells with abnormal karyotypes (Figure 3C and Supplemental Figure 2B). In abnormal karyotypes, chromosome 1, 4, or 19 was duplicated. Induced cells were supposedly clonal cells, indicating that cells became karyotypically heterogeneous with time in culture.
Figure 3. Characterization of induced cell lines.
(A) Growth curves of induced cell lines and MDFs. A total of 1 × 105 cells were passaged every 3 days into each well of 6-well plates. Averages of 4 separate experiments are shown. (B) Southern blot analysis of genomic DNA extracted from MK lines and MDFs. Genomic DNA was digested with EcoRI and BamHI, separated on an agarose gel, transferred to a nylon membrane, and hybridized with Klf4 cDNA probe. (C) Karyotype of the MK-5 cell line. (D) When the calculated cell numbers in the growth curve exceeded 1010, the cells were replated on 10-cm dishes. MDF culture (passage 3) was used for a control. After reaching confluence, induced chondrogenic cells and MDFs were cultured for an additional 14 days and stained with alcian blue. (E) Cell morphologies of induced cell lines. Phase contrast images are shown. Scale bar: 100 μm. (F) Pellet culture of MK-7 and MK-10 cells. After 3 weeks of culture, pellets were recovered and processed for histological sections. Semiserial sections were stained with safranin O-fast green-iron hematoxylin and toluidine blue, and immunostained with anti–type II collagen antibodies, anti–type I collagen antibodies, and anti–type X collagen antibodies. Scale bar: 50 μm.
When the calculated cell numbers in the growth curve exceeded 1010, the cells were split and replated onto 10-cm dishes, cultured for an additional 14 days after reaching confluence, and stained with toluidine blue. These cell line cultures showed metachromatic toluidine blue staining, indicating the presence of glycosaminoglycan, whereas MDFs did not (Figure 3D). The MK-3, -5, -7, -10, and -15 cell lines showed a polygonal cell morphology. The MK-1 and -4 cell lines showed a slightly spindle-shaped morphology (Figure 3E). To assess in vitro cartilaginous tissue formation more strictly, we performed pellet culture. MK-7 and MK-10 cells produced pellets, whereas MDF did not form pellet. Histological analysis of pellet culture showed that the MK-7 and MK-10 cell lines produced cartilage-like structures with cells surrounded by substances that showed metachromatic staining with toluidine blue, indicating a cartilaginous matrix (Figure 3F). Immunohistochemical analysis confirmed that MK-7 and -10 cells expressed type II collagen proteins but not type I and type X collagen proteins in pellet culture. Thus, the induced cell lines had chondrogenic differentiation potential in vitro.
Expression and DNA methylation analyses of induced chondrogenic cells.
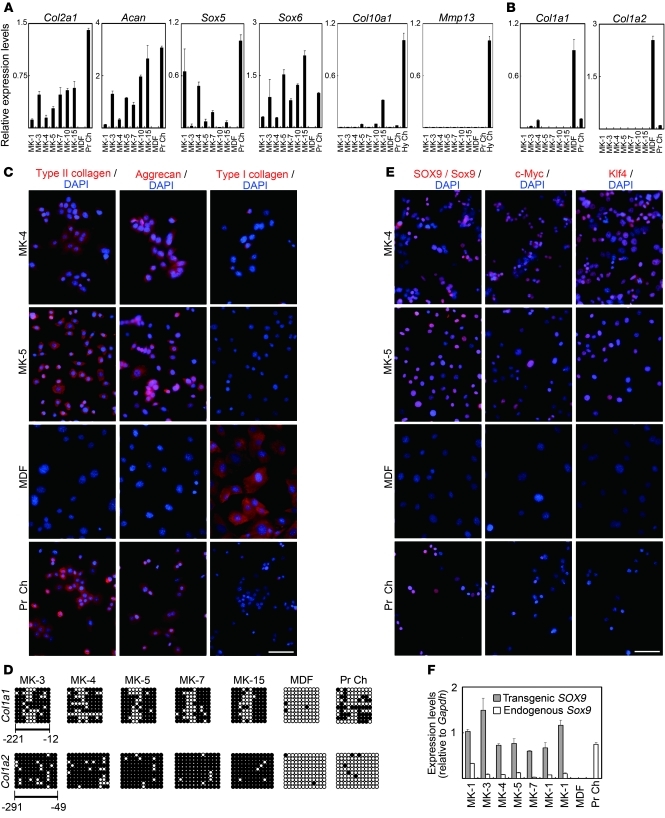
RT-PCR and real-time RT-PCR analysis showed that the MK cell lines expressed chondrocyte marker genes (Figure 4A and Supplemental Figure 3A). Relative expression levels of type II collagen gene (Col2a1) and aggrecan gene (Acan) in each MK cell line tended to be associated with that of Sox6. Parental MDFs did not express these genes. To analyze whether hypertrophy occurs in the induced cells, we examined the expression of the type X collagen gene (Col10a1) and Mmp13 in induced cells. For positive controls, we dissected hypertrophic zones from the femoral and tibial epiphyseal cartilage of neonatal mice and extracted RNAs. Col10a1 expression was higher in MK-5, MK-10, and MK-15 cells and lower in the other MK cell lines than in primary chondrocytes prepared from ribs. However, Col10a1 expression in MK cells was markedly lower compared with that in hypertrophic zones of epiphyseal cartilage. Mmp13 was not expressed in the induced cell lines. Most MK cell lines did not express the fibroblast-associated type I collagen α1 chain gene (Col1a1) and type I collagen α2 chain gene (Col1a2), although the MK-3 and MK-4 cells expressed a small amount of Col1a1 gene expression (Figure 4B). MDFs expressed these genes at high levels. Primary chondrocytes expressed a small amount of Col1a1 and Col1a2, probably due to minor contamination of fibroblasts during the harvesting procedure or de-differentiation of chondrocytes. Immunofluorescence staining showed that almost all MK-4 and MK-5 cells expressed type II collagen and aggrecan (Figure 4C). MK-4 cells weakly expressed type I collagen, whereas none of the MK-5 cells expressed type I collagen (Figure 4C), which is consistent with the results from real-time RT-PCR (Figure 4B). In primary chondrocyte samples, most cells expressed type II collagen and aggrecan, and a few cells (fewer than 10%) expressed type I collagen, probably due to minor contamination of fibroblasts or de-differentiation of chondrocytes.
Figure 4. Chondrocyte marker gene expression analyses of induced cell lines.
(A) Real-time RT-PCR analysis of chondrocyte marker gene expression in MK induced cells, MDFs, and primary chondrocytes. Error bars indicate mean ± SD (n = 3). (B) Real-time RT-PCR analysis of fibroblast marker gene expression in MK induced cells, MDFs, and primary chondrocytes. MK-4 cells expresseda small amount of Col1a1. Error bars indicate mean ± SD (n = 3). (C) Immunofluorescence staining showing expression of marker proteins (red) in MK-4 and -5 cells, MDFs, and primary chondrocytes. Blue nuclear staining is DAPI. Scale bar: 100 μm. (D) Bisulfite genomic sequencing of the promoter regions of Col1a1 and Col1a2. Each horizontal row of circles represents an individual sequencing result from 1 amplicon. Open circles indicate unmethylated CpG dinucleotides, while closed circles indicate methylated CpGs. (E) Immunofluorescence analysis showing protein expression (red) of SOX9/Sox9, Klf4, and c-Myc in MK-4 and MK-5 cells and expression of Sox9 in primary chondrocytes. Blue nuclear staining is DAPI. Scale bar: 100 μm. (F) Transgenic SOX9 mRNA levels and endogenous Sox9 mRNA levels were determined by real-time RT-PCR analysis with primers specific for transgenic SOX9 transcripts and for endogenous Sox9 transcripts, respectively. Shown are individual RNA expression levels normalized to respective Gapdh expression levels. Error bars indicate mean ± SD (n = 3). Hy Ch, hypertrophic chondrocyte zone of epiphyseal cartilage from neonatal femur and tibia.
The cytosine guanine (CpG) dinucleotides in the promoters of the fibroblast-associated genes Col1a1 and Col1a2 were highly methylated in MK cell lines, but were unmethylated in parental MDFs (Figure 4D). This suggests that expression of c-Myc, Klf4, and SOX9 induced silencing of the Col1a1 and Col1a2 genes in MDF culture. The Col1a1 promoter was moderately methylated in primary chondrocytes. Despite the low expression levels of Col1a2, its promoter was not methylated in primary chondrocytes prepared from neonatal rib cartilage. These results suggest that fibroblast marker gene promoters were excessively methylated in induced cells as compared with those in primary chondrocytes.
Expression of c-Myc, Klf4, and Sox9/SOX9 proteins was detected in MK cells by Western blotting (Supplemental Figure 3B) and immunofluorescence staining (Figure 4E). Western blot analysis showed that c-Myc and Klf4 expression were lower in the MK-3 cell line and SOX9 expression was lower in the MK-4 cell line when compared with other MK cell lines. On immunofluorescence staining, immunoreactivity against SOX9, c-Myc, and Klf4 was below detectable levels in some cells from each induced cell line. It is possible that these cells underwent silencing of transgenes. Real-time RT-PCR analysis showed that transgenes cMyc and Klf4 were expressed (Supplemental Figure 3, C and D). Transgene SOX9 expression was obvious in induced cells. Expression of the endogenous Sox9 gene was detected in induced cells but not in parental MDFs (Figure 4F), suggesting that endogenous Sox9 gene transcription was activated in induced cells.
In summary, the induced cell lines, with the exception of MK-3 and MK-4, expressed chondrocyte markers but not fibroblast markers, suggesting their commitment to the chondrogenic lineage. On the other hand, MK-4 cells showed type I collagen expression, which suggests incomplete or only partial chondrogenic commitment.
Induced chondrogenic cells formed homogenous cartilage in vivo.
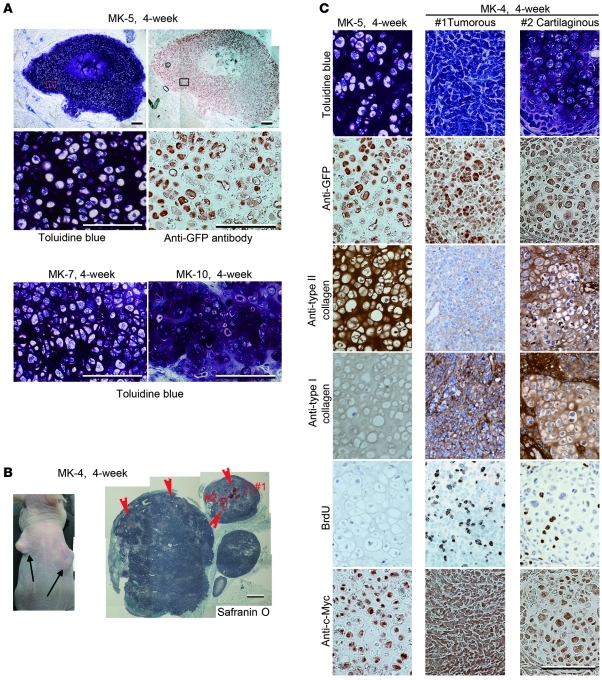
We examined the ability of induced chondrogenic cells to generate ectopic cartilage in vivo by subcutaneously injecting induced cells and parental MDFs into the dorsal flanks of nude mice (Supplemental Table 1). We injected induced cells alone without carrier. Four weeks after injection, we sacrificed the mice and histologically examined the injected sites. Among the 7 cell lines that were examined, MK-5, -7, and -10 produced substantial amounts of homogenous cartilage tissue, as indicated by metachromatic staining with toluidine blue (Figure 5A). We also observed lacuna formation, which is typical of cartilage. The distribution of GFP-expressing cells almost exactly corresponded to regions with metachromatic staining, suggesting that most of the injected MK-5 (Figure 5A), MK-7, and MK-10 cells (Supplemental Figure 4) differentiated into chondrocytic cells. The central region of the cartilage generated by MK-5 cell injection appeared necrotic, probably due to the lack of vasculature (Figure 5A, top). Immunohistochemical analysis showed that the cartilage generated by MK-5 cell injection into nude mice expressed no type I collagen (Supplemental Figure 5, right), similar to embryonic primordial cartilage (Supplemental Figure 5, left). Type I collagen was abundantly expressed in the surrounding host subcutaneous tissue of nude mice (Supplemental Figure 5, right) and most of the surrounding connective tissues, including muscle, dermis, tendon, vessels, and fat tissue in the embryonic limb buds (Supplemental Figure 5, left). These results suggest that the cartilaginous tissue generated by MK-5 cell injection in vivo corresponds to hyaline cartilage rather than fibrocartilage.
Figure 5. In vivo cartilage formation by induced cell lines.
MK-5, MK-7, MK-10, and MK-4 cells were injected into subcutaneous spaces of nude mice. (A) Histology of tissues 4 weeks after injection of MK-5, MK-7, and MK-10 cells. Sections were stained with toluidine blue. Semiserial sections of tissues generated by MK-5 cell injection were immunostained with anti-GFP antibodies. Magnifications of boxed regions in the top panels are shown in the respective second-row panels. Scale bars: 200 μm in top panels, 100 μm in middle and bottom panels. Each top panel was assembled from 2 images. (B) Injection of MK-4 cells produced tumors (arrows) at injected sites (4 weeks after injection; left panel). Histology of tumors generated by MK-4 cell injection 4 weeks after injection (right panel). Arrowheads indicate small cartilaginous portions. Scale bar: 1 mm. Right panel was assembled from 11 images. (C) Semiserial sections of cartilage generated by MK-5 cell injection (left column) and tumorous portion (middle) and cartilaginous portion (right) in tumors generated by MK-4 cell injection at 4 weeks after injection were stained with toluidine blue and immunostained with anti-GFP antibodies, anti–type II collagen antibodies, anti–type I collagen antibodies, anti-BrdU antibodies, and anti–c-Myc antibodies. Scale bar: 100 μm. Tumorous portion (middle column) and cartilaginous portion (right) correspond to the boxed regions #1 and #2 in the right in B, respectively.
On the other hand, MK-4 cells produced tumors within 4 weeks of injection (Figure 5B). Histological examination showed that the tumors consisted of a small cartilaginous portion (Figure 5B, arrowheads) embedded in a large tumorous portion. Low-power views revealed that the tumor was characterized by a solidly packed, round to ovoid cell pattern that was rather uniform and by small foci of immature cartilage. Under high-power observation, the main tumor cells possessed plump nuclei with 1 or 2 prominent nucleoli, showing primitive features without differentiation (Figure 5C, top center). Thus, the tumor did not correspond to teratoma. The cartilaginous islands consisted of immature chondrocytes with large nuclei without any sarcomatous change and chondroid matrix positively stained with toluidine blue (Figure 5C, top right). Tissues generated 4 weeks after MK-5 cell injection expressed type II collagen but not type I collagen, confirming that the generated tissue was cartilage (Figure 5C, left column). However, the tumorous portion generated from MK-4 cells strongly expressed type I collagen and weakly expressed type II collagen (Figure 5C, middle column), which suggests that MK-4 cells are chondrogenic/fibroblastic intermediate cells. The cartilaginous region generated by MK-4 cells showed moderate expression of type II collagen and weak expression of type I collagen (Figure 5C, right column). BrdU labeling revealed a very low amount of proliferation in cartilage generated from MK-5 cells (Figure 5C, left, fifth row) and large amounts of proliferation in tumors generated from MK-4 cells (Figure 5C, middle, fifth row). Mice were intraperitoneally injected with BrdU labeling reagent 2 hours before sacrifice. Immunoreactivity against c-Myc was similar among cartilaginous tissue generated by MK-5 cell injection, the tumorous portion, and the cartilaginous portion in tumors generated by MK-4 cell injection (Figure 5C, bottom row).
To examine the course of cartilage production and MK cell proliferation in vivo, we collected tissues at 1 and 3 weeks after the injection of MK-5 cells (Supplemental Figure 5B). The MK-5 cells gradually produced cartilage extracellular matrix, as indicated by intense safranin O staining (Supplemental Figure 5B, top row), and their rate of proliferation decreased with time, as indicated by a gradual decrease in the number of BrdU-positive cells (Supplemental Figure 5B, bottom row). The proliferating activities of tissues generated 1–4 weeks after MK-5 cell injection were between those of embryonic primordial cartilage and articular cartilage after birth (Supplemental Figure 5B).
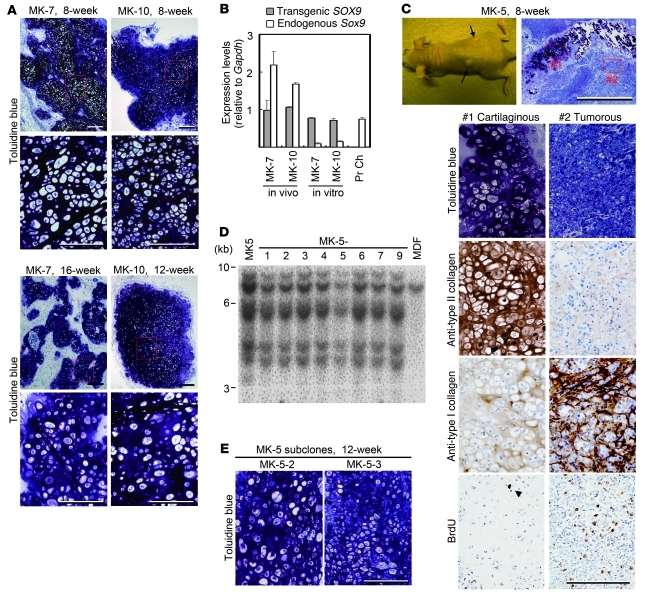
We then followed the mice for 8–16 weeks after MK cell injection (Supplemental Table 1). MK-7 cell–injected sites continued to show homogenous cartilage formation without tumor formation at 8 and 16 weeks (Figure 6A, left panels). MK-10 cell–injected sites showed homogenous cartilage without tumor formation at 8 and 12 weeks (Figure 6A, right panels). To analyze the stability of the chondrogenic phenotype, we extracted RNA from the cartilage tissues generated from MK-7 cells (16 weeks after injection) and MK-10 cells (8 weeks after injection). Transgenic SOX9 mRNA levels and endogenous Sox9 mRNA levels were determined with real-time RT-PCR analysis with primers specific for transgenic SOX9 transcripts and primers specific for endogenous Sox9 transcripts, respectively. Expression of endogenous Sox9 was potently activated after implantation into nude mice as compared with the expression levels in monolayer culture or primary chondrocytes (Figure 6B). The expression of Col2a1 and Acan were also increased as compared with their expression level in monolayer culture (Supplemental Figure 6). The surrounding cartilage matrix produced in vivo may have stimulated MK-7 or MK-10 cells to express endogenous Sox9, which in turn caused further production of cartilage matrix, leading to the stable maintenance of a differentiated chondrocytic phenotype without tumor formation. These results suggest that MK-7 and MK-10 cells are chondrogenic cells.
Figure 6. Prolonged of in vivo cartilage formation.
Semiserial sections were stained with toluidine blue and immunostained with antibodies, as indicated on the left. (A) Histology of tissues where MK-7 cells (8 and 16 weeks after injection) and MK-10 cells (8 and 12 weeks after injection) were injected. For each tissue, lower-magnification images are shown above and higher magnification images of boxed regions are shown below. Scale bars: 200 μm in top panel and 100 μm in bottom panel for each tissue. (B) RNA samples were extracted from cartilage tissue (in vivo) generated in nude mice by subcutaneous injection of MK-7 (16 weeks after injection) and MK-10 (8 weeks after injection) and from monolayer culture (in vitro) of MK-7 and MK-10 cells. Individual RNA expression levels were normalized to respective Gapdh expression levels. Error bars indicate mean ± SD (n = 3). (C) Injection of MK-5 cells produced tumors (arrows) 8 weeks after injection (top left panel). Histological sections of tumors stained with toluidine blue (top right panel). Magnifications of cartilaginous remnant (boxed region #1 in the top right panel) and tumor (boxed region #2 in the top right panel) are shown below. Scale bars: 500 μm in top right panel, 100 μm in bottom panels. (D) Southern blot analysis of genomic DNA extracted from MK-5 subclones, MK-5 cell line, and MDFs with Klf4 probe. (E) Histology of tissues where subclones of MK-5 cells were injected. Tissues were collected 12 weeks after injection. Scale bar: 100 μm.
In contrast to the successful application of the MK-7 and MK-10 cells, the MK-5 cells produced small cartilaginous remnants surrounded by large tumors at 8 weeks after injection (Figure 6C, top row). The cartilaginous remnants strongly expressed type II collagen and weakly expressed type I collagen (Figure 6C, left, third and fourth rows) and contained only one BrdU-positive cell in a representative field (Figure 6C, bottom left, arrowhead). The tumor cells, which were highly proliferative as indicated by the numerous BrdU-positive cells (Figure 6C, bottom right), had strong expression of type I collagen and weak expression of type II collagen (Figure 6C, third and fourth rows, right). We speculated that the MK-5 cell line was contaminated with chondrogenic/fibroblastic intermediate cells that produced the tumors in nude mice and that the number of these contaminating cells was too small to detect the expression of type I collagen in monolayer cultures of MK-5 cells. To examine this possibility, we derived subclones from single MK-5 cells using the limiting dilution technique. Southern hybridization revealed that each MK-5 subclone had a transgene integration pattern that was identical to that of the original MK-5 cells (Figure 6D). The injection of MK-5 subclones produced cartilage but not tumors at 8 and 12 weeks after injection (Figure 6E and Supplemental Table 1). These results suggest that the transduction of c-Myc, Klf4, and SOX9 into dermal fibroblast culture produced hyaline chondrogenic cells, which do not express type I collagen, along with chondrogenic/fibroblastic intermediate cells, which do express type I collagen (Supplemental Figure 7). The induced cell lines, such as MK-7, MK-10, and subclones of MK-5, that formed homogenous hyaline cartilage with high expression levels of endogenous Sox9 and without tumor formation might be hyaline chondrogenic cells.
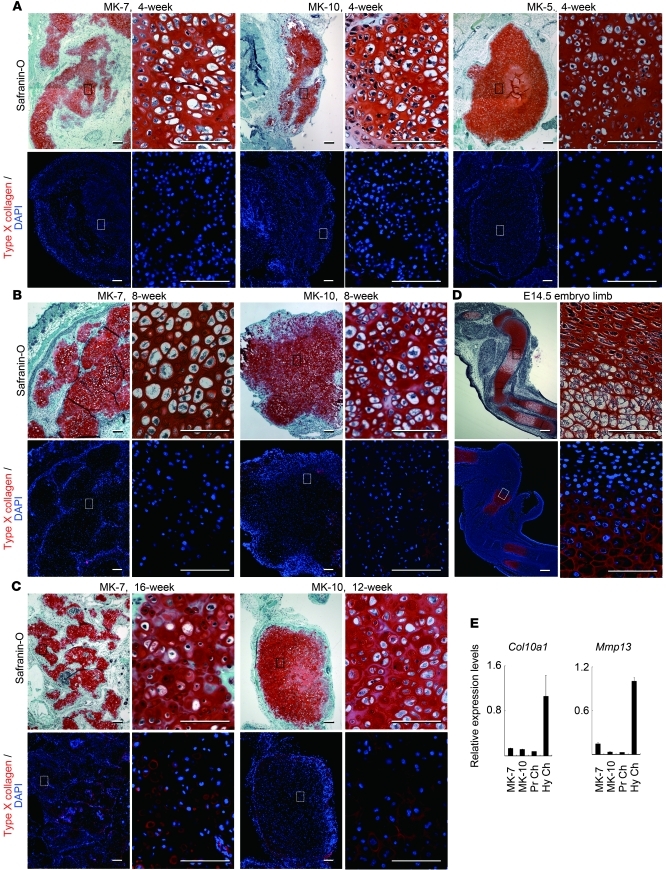
In order to determine whether hypertrophy occurs in the cartilaginous tissues formed in nude mice, we examined type X collagen expression by immunohistochemistry. Type X collagen was not expressed in cartilaginous tissue at 4 weeks after injection of MK-5, -7, and -10 cells (Figure 7A). At 8 weeks after injection, type X collagen was not expressed in cartilaginous tissue by MK-7 cell injection and was expressed in a limited part of the cartilaginous tissue by MK-10 cells (Figure 7B). Type X collagen expression was detected in the cartilaginous tissue formed at 16 weeks after injection of MK-7 cells (Figure 7C). Real-time RT-PCR analysis showed that expression levels of Col10a1 and Mmp13 remained lower in cartilaginous tissues generated by MK-7 (16 weeks after injection) and MK-10 (8 weeks after injection), as compared with the expression levels in the samples collected from the hypertrophic zones of epiphyseal cartilage in neonatal mice (Figure 7E). These results suggest that the induced cells slowly undergo hypertrophy in transplants in vivo.
Figure 7. Slow progression of hypertrophy in cartilaginous tissue by induced cells in vivo.
Semiserial sections were stained with safranin O, fast green, and iron hematoxylin (safranin O) and immunostained with anti-type X collagen antibodies (red). (A) Histology of cartilaginous tissues generated by MK-7, MK-10, and MK5 cells 4 weeks after injection. (B) Histology of cartilaginous tissues generated by MK-7 and MK-10 cells 8 weeks after injection. (C) Histology of cartilaginous tissues generated by MK-7 cells 16 weeks after injection and MK-10 cells 12 weeks after injection. (D) The control, humeral primordial hyaline cartilage at 14.5 dpc expresses type X collagen in hypertrophic chondrocytes. For A–C, lower-magnification images are shown on the left and higher magnification images of boxed regions on the right for each tissue. Scale bars: 100 μm in left panel and 200 μm in right panel for each tissue. (E) Real-time RT-PCR analysis of Col10a1 and Mmp13 expression in cartilage tissue generated in nude mice by subcutaneous injection of MK-7 (16 weeks after injection) and MK-10 (8 weeks after injection). Individual RNA expression levels were normalized to respective Gapdh expression levels. Error bars indicate mean ± SD (n = 3).
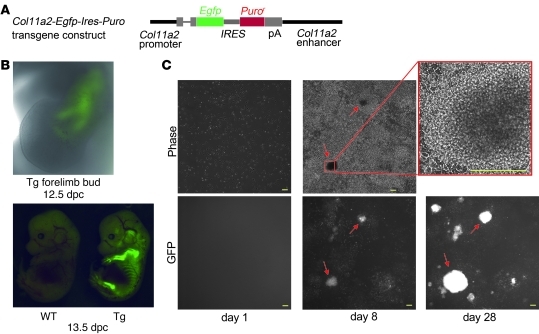
Induction of chondrogenic cells from Col11a2-Egfp-Ires-Puro transgenic MDFs.
To examine the process of induction of chondrogenic cells from MDFs, we generated transgenic mice bearing the Col11a2-Egfp-Ires-Puro transgene (Figure 8A). GFP fluorescence was initially observed during mesenchymal condensation at 12.5 dpc (Figure 8B, top). The mice showed GFP fluorescence specifically in primordial cartilage at 13.5 dpc (Figure 8B, bottom). MDF prepared from the Col11a2-Egfp-Ires-Puro transgenic mice died in the presence of 1 μg/ml puromycin, while primary chondrocytes from the transgenic mice survived in 2.5 μg/ml puromycin (data not shown). We transduced c-Myc, Klf4, and SOX9 retroviral vectors into MDFs prepared from neonatal Col11a2-Egfp-Ires-Puro transgenic mice. On the day of completion of overnight retroviral transduction (day 1), no MDFs expressed GFP in the wells of a 6-well plates (Figure 8C, left column, and Supplemental Figure 8A, left panel), suggesting that the MDF culture does not contain chondrogenic cells, which corresponds to the condensed mesenchymal cell state. One day after retroviral transduction, cells began to proliferate vigorously. Two days after transduction, some cells began to show GFP fluorescence. Three days after the transduction, we began to add a minimal amount of puromycin (1 μg/ml), which kills fibroblasts. A few nodules showing intense GFP fluorescence appeared by 7 days after transduction (Figure 8C, middle column) per 4 mm × 4 mm field. Nodules were composed of polygon-shaped cells that formed multiple layers (Figure 8C, upper right panel), which is characteristic of cultured primary chondrocytes (30). Nodules were stained with alcian blue (Supplemental Figure 8B), which is suggestive of chondrogenic cells. The number of nodules gradually increased with time (Figure 8C, lower right panel). In addition, immunofluorescence staining with anti-Sox9 antibodies showed that very few parental MDFs in wells of a 6-well plate exhibited fluorescence (Supplemental Figure 8, C and D). In fact, most of the cells showing fluorescence in MDF culture appeared to be false-positive. Sox9 is expressed in chondroprogenitor cells (31). The number of possible prechondrogenic cells indicated by Sox9 expression in MDF culture was much lower than the number of G418-resistant colonies (Figure 2C) or chondrogenic nodules (Supplemental Figure 8A) generated by transduction of c-Myc, Klf4, and SOX9.
Figure 8. Course of chondrogenic cell induction and effects of continued expression of the transgenes.
(A) Schematic representation of the Col11a2-Egfp-Ires-Puro transgene. (B) GFP expression in Col11a2-Egfp-Ires-Puro transgenic embryos. Forelimbs of 12.5-dpc transgenic embryo and whole wild-type and transgenic embryos are shown. (C) Induction of nodules in MDF culture by retroviral transduction of c-Myc, Klf4, and SOX9. After an overnight incubation in the retrovirus-containing medium, Col11a2-Egfp-Ires-Puro transgenic MDFs were replated into the 6-well plates (day 1) and subjected to time-lapse observation by Biostation CT (Nikon). Phase and GFP images were taken once a day. On day 1, MDFs did not express GFP. On day 8, nodules (arrows) appeared and expressed GFP. The magnification of the boxed region in the phase image on day 8 (upper middle panel) showed that the nodules formed multiple layers of cells and consisted of polygon-shaped cells (upper right panel). Scale bars: 200 μm.
Generation of induced chondrogenic cells with a doxycycline-inducible system.
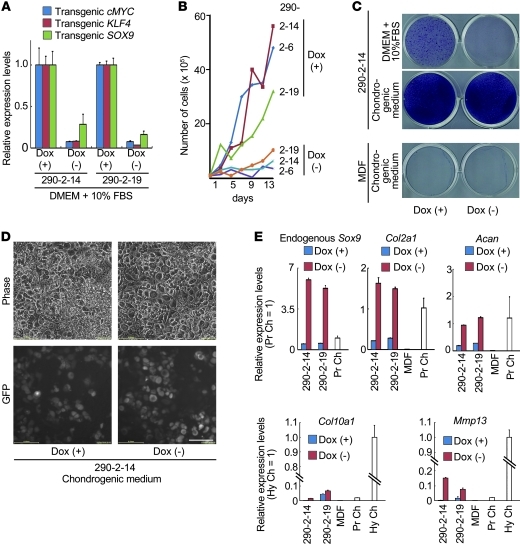
To examine how the continued expression of the transgenes affected characteristics of induced cells, we misexpressed human c-MYC, human KLF4, and human SOX9 from doxycycline-inducible (dox-inducible) lentiviral vectors. We transduced these inducible constructs into MDFs prepared from neonatal Col11a2-Egfp-Ires-Puro transgenic mice and cultured them in the presence of 1 μg/ml dox and 1–2 μg/ml puromycin. We derived pleural-induced cell lines from individual puromycin-resistant colonies. Induced cell lines cultured in DMEM plus 10% FBS in the presence of dox expressed chondrocyte marker genes, but not fibroblast marker genes (Supplemental Figure 8E). The levels of chondrocyte marker genes in these cells in the presence of dox were lower than those in MK cells induced by conventional retroviral vectors (Figure 4A). It is possible that this difference was due to reduced transgene expression in the dox-inducible system. Withdrawal of dox for 2 days resulted in a dramatic reduction of transgene expression (Figure 9A) and decreased proliferation rates in the DMEM plus 10% FBS culture media (Figure 9B). This result suggests that fast proliferation of induced cells in vitro (Figure 3D) depended on continued expression of the transgenes.
Figure 9. Analysis of induced cells from Col11a2-Egfp-Ires-Puro transgenic MDFs by dox-inducible lentiviral c-MYC, KLF4, and SOX9 vectors.
(A) Real-time RT-PCR analysis of the dox-inducible transgenes in induced cell lines (nos. 290-2-14 and 290-2-19) cultured in DMEM + 10% FBS in the presence or absence of dox. Relative expression levels in cells cultured in the presence of dox are set as 1. (B) Growth analysis of induced cell lines (nos. 290-2-6, 290-2-14, and 290-2-19) cultured in DMEM plus 10% FBS in the presence or absence of dox. A total of 1 × 105 induced cells were seeded in 6-well plates, and cell numbers were determined every other day. (C) Toluidine blue staining of induced cells and MDFs. Induced cells were cultured in DMEM PLUS 10% FBS in the presence (top left) or absence (top right) of dox for 14 days. The culture medium of induced cells was changed to chondrogenic medium with dox (middle left panel) or chondrogenic medium without dox (middle right panel), and cells were cultured for 7 days. (D) Phase and GFP images of induced cells cultured in chondrogenic medium in the presence or absence of dox for 7 days. Scale bar: 100 μm. (E) Real-time RT-PCR analysis of induced cells. Culture media of induced cells was changed from DMEM plus 10% FBS and dox to the chondrogenic medium with dox (+) or chondrogenic medium without dox (–). After 3 days, total RNAs were extracted. Error bars indicate mean ± SD (n = 3).
Next, we examined whether the chondrogenic phenotype of induced cells is maintained after withdrawal of dox in vitro. Induced cells were moderately stained with toluidine blue in the presence of dox, whereas they were not stained in the absence of dox in the DMEM plus 10% FBS culture media (Figure 9C, top). These results suggest that induced cells lose the chondrogenic phenotype after withdrawal of dox in the DMEM plus 10% FBS culture media; this result is consistent with the unique properties of primary chondrocytes, which rapidly lose their differentiated phenotype upon culturing (32). On the other hand, induced cells were intensely stained with toluidine blue in the chondrogenic medium (DMEM supplemented with 1% FBS, 500 ng/ml GDF5, 10 ng/ml TGF-β1, 10–7 M dexamethasone, 50 μg/ml ascorbate-2-phosphate, 1 mM pyruvate, 10 μg/ml insulin, 10 μg/ml transferrin, and 0.2 μg/ml sodium selenite), regardless of the presence or absence of dox (Figure 9C, middle). Parental MDFs cultured in chondrogenic medium in the presence or absence of dox were only slightly stained with alcian blue (Figure 9C, bottom). Induced cells were polygon-shaped and expressed GFP in chondrogenic medium in the absence of dox (Figure 9D), suggesting Col11a2 transcription. Real-time RT-PCR analysis showed that the withdrawal of dox greatly activated the expression of endogenous Sox9 in induced cells cultured in the chondrogenic medium; the levels of endogenous Sox9 expression were even higher than that in primary chondrocytes (Figure 9E). The expression levels of the chondrocyte marker genes Col2a1 and Acan were also increased by the withdrawal of dox when induced cells were cultured in chondrogenic medium (Figure 9E). It is possible that continued expression of the transgenes disrupts chondrocytic differentiation in this system. These results suggest that induced cells in which continued expression of the transgenes are reduced can respond well to chondrogenic medium and maintain a chondrogenic potential. Expression levels of Col10a1 and Mmp13 in dox-inducible induced cells were elevated by the withdrawal of dox when cells were cultured in chondrogenic medium, although they were moderately lower compared with those in hypertrophic zones of epiphyseal cartilage from neonatal femur and tibia (Figure 9E). These results suggest that induced cells in which continued expression of the transgenes is reduced undergo hypertrophy.
We then injected dox-inducible induced cells into the subcutaneous spaces of nude mice. When water containing dox was administered to the nude mice continuously, we observed small cartilaginous tissue formation in only 1 of the 8 injection sites at 4 weeks after injection (data not shown). The other 7 injection sites showed no tissue formation. Thus, the dox-inducible induced cells may not be as chondrogenic as MK cell lines, or the dox concentration may not have been sufficiently high to induce transgene expression in vivo. When dox was not administered to the mice, we found no tissue formation in the 16 injected sites in nude mice at 4 weeks after injection (Supplemental Table 1). Based on these results, long-term heterologous expression of transgenes appears to be necessary to induce and maintain the hyaline cartilage–like phenotype at ectopic sites.
Discussion
Inducing hyaline cartilage from dermal cells has been very difficult, because the dermis expresses type I collagen most abundantly, and the absence of type I collagen is prerequisite for proper function of hyaline cartilage. In addition to gaining the phenotype of the destination cell types, elimination of characteristics of the original fibroblastic cell type is another critical issue in converting fibroblasts into different cell types, particularly in the case of forming hyaline cartilage in which the presence of type I collagen critically affects its extracellular matrix architecture and function.
In the present study, we obtained chondrogenic cells expressing no type I collagen through the transduction of c-Myc and Klf4 in addition to the chondrogenic factor SOX9. Induced chondrogenic cells showed extensive methylation of the promoters of the type I collagen genes. We speculate that ectopic expression of c-Myc and Klf4 partially reprograms the fibroblasts, making the cells susceptible to chondrogenic programming, which is initiated by ectopic expression of Sox9. As a result of efficient chondrogenic programming, the type I collagen genes may be silenced. The c-Myc and Klf4 transgenes appear to contribute to the initial expansion of induced chondrogenic cell population. Some induced cell lines formed tumors, while other induced cell lines did not. The latter induced cell lines produced stable hyaline cartilage–like tissue without tumor formation for a sustained period of time in the subcutaneous spaces of nude mice; these results represent advancements in cartilage tissue engineering. We have not determined the origin of the induced chondrogenic cells in MDF culture. Our results suggest that cells other than prechondrogenic cells in MDFs can give rise to induced chondrogenic cells. A small number of multipotent stem cells coexist with fibroblasts in primary dermal fibroblast culture (33), and it is possible that c-Myc, Klf4, and SOX9 modified these stem cells or fibroblasts.
The cartilaginous tissues formed by injection of induced chondrogenic cells in nude mice slowly underwent hypertrophy. The expression levels of Col10a1 in dox-inducible induced cells were increased by the withdrawal of dox when cells were cultured in chondrogenic medium. As Sox9 overexpression in chondrocytes inhibits hypertrophy (34), we speculate that continued expression of SOX9 in these induced cells contributed to delaying the onset of hypertrophy. The results showing that continued expression of Sox9 is needed to maintain hyaline-like cartilage obviously limit the therapeutic application of the technology to articular cartilage regeneration, but alternatively, many should find this strategy useful for generating cartilage from dermal fibroblasts for the study of human congenital skeletal defects.
Along with chondrogenic cells, chondrogenic/fibroblastic intermediate cells were also produced. These cells express type I collagen and form tumors when transplanted into nude mice. The tumor-producing MK-4 cell line expressed lower SOX9 protein and lower Sox6, Col2a1, and Acan mRNA levels in vitro when compared with other induced chondrogenic cell lines. A possible explanation is that chondrogenic/fibroblastic intermediate cells arose from fibroblasts that were not properly transduced with transgenes and were insufficiently reprogrammed to the chondrogenic lineage, although this is only speculation.
One goal in cartilage tissue engineering is to obtain expandable chondrogenic cells that do not lose their chondrogenic properties. However, chondrocytes do not rapidly proliferate for a sustained period of time. Chondrogenic progenitor cells collected from human osteoarthritic cartilage reach senescence after expansion (35). In the present study, it appears that the c-Myc and Klf4 transgene products, in cooperation with SOX9, may reprogram cells in fibroblast culture to become chondrogenic cells and also subsequently contribute to the rapid proliferation of induced cells. After being subcutaneously implanted into nude mice, some induced cell lines formed tumors, whereas other induced cell lines did not. The latter induced cells were gradually embedded in cartilage extracellular matrix and lost their proliferating activity, producing stable cartilage without tumor formation for a prolonged period of time, despite continued c-Myc expression. We have not identified the molecular mechanisms responsible for the controlled proliferation of induced chondrocytic cells in vivo. The control of proliferation rates of induced cells in vivo appears to be associated with the activation of endogenous Sox9 gene expression and chondrocytic differentiation. Reduced proliferation and greatly increased activation of the endogenous Sox9 gene was also seen in the induced cells produced by dox-inducible lentiviral vectors when they were cultured in the absence of dox in chondrogenic medium. However, as long as the c-Myc and Klf4 transgenes are present in the induced cells, it is possible that these cells may eventually become tumorigenic. To reduce the risk of tumorigenicity, c-Myc and Klf4 constructs should be transiently expressed to induce chondrogenic cells and then eliminated after a sufficient number of induced cells are produced.
“Kick-starting” the chondrocytic program by transient expression of the three factors would be ideal, but the existing challenges associated with maintaining hyaline cartilage will emerge. Preparation of a chondrogenic environment, such as chondrogenic supplementation and scaffold, may be a prerequisite for maintaining a chondrogenic phenotype, as dox-inducible induced cells without dox respond to chondrogenic medium and cannot form ectopic cartilage in nude mice by subcutaneous injection of cells alone. The increased Col10a1 expression in dox-inducible induced cells without dox indicates that it is necessary to develop chondrogenic supplementation that inhibits chondrocyte hypertrophy. Finally, we should note that induced cell lines showed karyotypic instability during culture. Karyotypic instability is a serious problem with this technology. It remains to be confirmed whether shortening the culture period or modifying the culture condition improves the karyotypic instability of induced cells.
Although significant challenges remain, the present approach may be a step toward the generation of patient-specific cartilage from skin without going through the process of generating iPS cells.
Methods
Generation of Col11a2-βgeo transgenic mice.
The α2(XI) collagen gene–based expression vector 742LacZInt contains the mouse Col11a2 promoter (–742 to +380), an SV40 RNA splice site, the LacZ reporter gene, the SV40 polyadenylation signal, and a 2.3-kb segment of the first intron sequence of Col11a2 as an enhancer (27). To create the βgeo transgene, a 0.8-kb neomycin resistance gene fragment was linked to the 3ι-end of a 3.1-kb cDNA fragment encoding LacZ. The βgeo fragment was cloned into the NotI sites of the 742lacZInt expression vector by replacing the LacZ gene to create Col11a2-βgeo. The plasmid Col11a2-βgeo was digested with EcoRI and PstI to release the inserts. Transgenic mice were produced by microinjection of the inserts into the pronuclei of fertilized eggs from F1 hybrid mice (C57BL/6 × DBA), as described previously (27). Transgenic mice were identified by PCR assays of genomic DNA extracted from the tail. Genomic DNA was amplified by transgene-specific PCR by using primers derived from the LacZ gene (CGCTACCATTACCAGTTG) and from the neomycin resistance gene (CCAGTCATAGCCGAATAG) to amplify a 135-bp product from βgeo transgenic mice. Mice were backcrossed to the C57BL/6 strain for at least 4 generations. X-gal staining of mouse bodies and sections was performed as previously described (36).
Cell culture.
For MDF isolation, skin was prepared from 3- to 6-month-old mice. After hair was shaved off, skin was minced and trypsinized at 37°C for 4 hours. Dissociated cells were filtered through a nylon mesh (pore size, 40 μm; Tokyo Screen Co.) to generate a single-cell suspension and then seeded onto 100-mm dishes and cultured in DMEM supplemented with 10% FBS (passage 1). Epidermal cells did not appear to survive under these culture conditions, as immunofluorescence staining showed that most cells expressed type I collagen (Figure 4C).
ADSCs were isolated as previously described (37). Subcutaneous fat pads were prepared from 3- to 6-month-old mice, minced, and digested in 0.2% collagenase medium at 37°C for 2–4 hours. The dissociated cells were filtered through nylon mesh (pore size, 70 μm; Tokyo Screen Co.). Released cells were collected by centrifugation (630 g for 10 minutes at 4°C). Cells were resuspended in a fresh medium, filtered through nylon mesh (pore size, 40 μm; Tokyo Screen Co.), and collected by centrifugation (630 g for 10 minutes at 4°C). Cells were resuspended in a fresh medium and seeded onto 100-mm dishes and cultured in DMEM supplemented with 5% FBS (passage 1).
MEFs were isolated as previously described (16). Briefly, the head and visceral tissues were removed from 13.5-dpc embryos. The remaining bodies were minced, trypsinized, and transferred into a tube. Cells were collected by centrifugation and then resuspended in DMEM containing 10% FBS. A total of 1 × 106 cells were cultured on each 100-mm dish (passage 1). MDFs, ADSCs, and MEFs at passage 1 were trypsinized and stored in liquid nitrogen until use.
Primary chondrocytes were isolated as previously described (38, 39). The ventral sections of rib cages or the epiphyseal sections of humerus and femur were dissected from newborn mice in PBS. Adherent soft tissues were removed from the cartilage after digestion with 2 mg/ml collagenase type II (Sigma-Aldrich) in DMEM at 37°C for 30 minutes. Chondrocytes were released from the cartilage by soaking the tissue in fresh collagenase medium for 4–5 hours. Released cells were collected by centrifugation (200 g for 10 minutes at 4°C) and resuspended in fresh medium. Cells were seeded into 60-mm or 100-mm dishes and cultured in DMEM supplemented with 10% FBS (passage 1).
Retroviral vectors and transduction.
pMXs-c-Myc (Addgene plasmid 13375), pMXs-Klf4 (Addgene plasmid 13370), pMXs-Oct3/4 (Addgene plasmid 13366), pMXs-Sox2 (Addgene plasmid 13367), pMXs-EGFP, and pMXs-gw were gifts from S. Yamanaka (Center for iPS Cell Research and Application [CiRA], Kyoto University, Kyoto, Japan) (16). The Gateway cassette (Invitrogen) was inserted into a pMXs retroviral vector (40) in order to prepare pMXs-gw. Human SOX9 cDNA, a gift from H. Akiyama (Graduate School of Medicine, Kyoto University, Kyoto, Japan), was cloned into pENTR-1A (Invitrogen) to prepare pENTR1A-hSOX9 and recombined with pMXs-gw by LR reaction (Invitrogen) to prepare pMXs-hSOX9.
Retroviral infection was performed as described previously (16). Plat-E cells were gifts from T. Kitamura (The Institute of Medical Science, The University of Tokyo, Tokyo, Japan). One day after seeding Plat-E cells (29) at 8 × 106 cells per 10-cm dish, we transfected the cells with pMXs-based retroviral vectors by using FuGENE 6 transfection reagent (Roche). Twenty-four hours after transfection, the medium was replaced. Twenty-four hours after medium replacement, the medium was collected as the virus-containing supernatant from Plat-E cultures.
Frozen stored MEFs, MDFs, or ADSCs were thawed, seeded onto 100-mm dishes, and cultured in DMEM supplemented with 10% FBS. One day before transduction, cells were trypsinized and replated at 1.7 × 105 cells per 6-cm dish (passage 3).
The virus-containing supernatants were filtered through a 0.45-μm cellulose acetate filter (Schleicher & Schuell) and supplemented with 4 mg/ml Polybrene (Nacalai Tesque). To determine the titer of each retroviral vector, we transduced MDFs with each retrovirus or lentivirus vector, extracted genomic DNAs from MDFs 2 days after infection, and quantified copy numbers of retroviral transgenes that were integrated into MDF genome per cell by real-time PCR. Titers for each vector were as follows: pMXs-EGFP, 1.0 × 107 IU/ml; pMXs-c-Myc, 1.2 × 106 IU/ml; pMXs-Klf4, 2.7 × 106 IU/ml; pMXs-Oct3/4, 6.1 × 106 IU/ml; pMXs-Sox2, 4.0 × 106 IU/ml; pMXs-SOX9, 1.8 × 106 IU/ml. MOI for each vector was estimated using these titers.
Equal amounts of supernatants containing each of the retroviruses were mixed and added to the MEF or MDF cultures. After a 16-hour incubation in the virus-containing medium, each fibroblast culture in the 6-cm dish was trypsinized and replated into a 10-cm dishes in fresh medium. After 2 days, the medium was removed and replaced with fresh medium that contained 500 μg/ml G418. The medium was changed every other day for 2 weeks for selection. Dishes were subjected to toluidine blue staining followed by crystal violet staining to determine colony numbers. Colony numbers were determined using NIS Element software (Nikon). We defined a colony as a cell cluster that was more than 0.5 mm in diameter. Other dishes were used for picking up colonies to generate induced chondrogenic cells.
Generation of induced chondrogenic cell lines.
After setting up a sterile cylinder surrounding each colony, we harvested the cells by trypsinization and replated them in 96-well dishes. After 6–10 days, cells were replated in 24-well dishes and cultured for 24–31 days. Then, cells were replated in 6-well dishes. After 18–31 days, cells were replated in 10-cm dishes. We defined this stage as passage 4. Induced chondrogenic cells were cultured in DMEM containing 10% FBS in the presence of 500 μg/ml G418. Induced chondrogenic cells were passaged every 6 days. We initiated growth curve analyses of induced chondrogenic cells and MDFs at passage 6.
Cell culture in collagen gel.
Collagen gel culture was performed with a collagen gel culture kit (Nitta Gelatin Inc.), according to the manufacturer’s instructions. MK-1 and MK-3 to -28 induced chondrogenic cells and MDFs were digested with trypsin/EDTA. Cells were then adjusted to 2 × 107 cells/ml in 0.25% type I acid–soluble collagen prepared at 4°C. Cell suspensions (500 μl) were placed in the center of each well of 6-well plates and gelated at 37°C. The gel-cell composite was overlaid with 3 ml of DMEM containing 10% FBS. The medium was replaced with fresh medium every other day. After 3 weeks of culture, gel-cell composites were fixed in 4% paraformaldehyde, processed, and embedded in paraffin.
Pellet culture.
MDFs or induced cells were suspended at 5 × 105 cells/ml in DMEM containing 10% FBS, transferred into a 15-ml tube (Falcon), and centrifuged at 190 g for 5 minutes. The resulting cell pellet was incubated for 3 weeks.
In vivo cartilaginous tissue formation.
MDFs or induced cells were suspended at 1 × 107 cells/ml in DMEM containing 10% FBS. Then, 100 μl of the cell suspension was injected subcutaneously into the dorsal flank of 6-week-old female nude mice (BALB/cA Jcl-nu/nu). No carrier was used. Mice were sacrificed after 1–16 weeks, and the injected sites were dissected from the mice. The majority of samples were fixed in 4% paraformaldehyde, processed, and embedded in paraffin. Some samples were used for extraction of the total RNA for real-time RT-PCR analysis.
Toluidine blue staining.
Cells were fixed with 10% neutral buffered formalin at 25°C for 10 minutes, washed with water, incubated with 0.05% toluidine blue solution (Wako 206-14555) for 30 minutes at 25°C, and washed 3 times with water.
Alcian blue staining.
Cells were fixed with methanol at –20°C for 2 minutes, incubated with 0.1% alcian blue (Sigma-Aldrich) in 0.1N HCl for 2 hours at 25°C, and washed 3 times with distilled water.
Immunohistochemical staining.
Semi-serial histological sections were immunostained with the primary and secondary antibodies listed in Supplemental Table 4. For goat polyclonal anti–type II collagen antibody, immune complexes were detected by using a Histofine SAB-PO kit (Nichirei) with 3-amino-9-ethyl carbazole (AEC) as a chromogen. For anti–c-Myc antibody and anti-GFP antibody, immune complexes were detected by EnVision+ System HRP–labeled polymer anti-rabbit kit (Dako, K4002) and AEC as a chromogen.
Southern blot analysis.
Genomic DNA isolated from induced chondrogenic cells and MDFs was digested with EcoRI and BamHI, separated on an agarose gel, transferred to a nylon membrane, and hybridized with Klf4 cDNA probe.
Determination of karyotypes.
Karyotypes of induced chondrogenic cells were determined with quinacrine-Hoechst staining at the International Council for Laboratory Animal Science (ICLAS) Monitoring Center (Japan).
Real-time RT-PCR and RT-PCR analyses.
Induced chondrogenic cells (passage 6), MDFs (passage 3), and primary chondrocytes (passage 1) were cultured in 60-mm dishes. After the cells reached confluence, the total RNA was extracted by using RNeasy Mini Kits (QIAGEN). The total RNA was also extracted from tissues generated by subcutaneous injection of induced cells into nude mice. In addition, we dissected hypertrophic zones from femoral and tibial epiphyseal cartilage of neonatal mice and extracted total RNAs. Total RNAs were digested with DNase to eliminate any contaminating genomic DNA. For real-time quantitative RT-PCR, 1 μg of total RNA was reverse transcribed into first-strand cDNA by using SuperScript III (Invitrogen) and an oligo(dT)20 primer. The PCR amplification occurred in a reaction volume of 20 μl containing 2 μl cDNA, 10 μl SYBER PremixExTaq (Takara), and 7900HT (Applied Biosystems). The RNA expression levels were normalized to the level of Gapdh expression. For RT-PCR analysis, 1 μg of total RNA was reverse transcribed into first-strand cDNA by using QuantiTect Reverse Transcription (QIAGEN). PCR was performed with ExTaq (Takara). The primers used are listed in Supplemental Tables 2 and 3.
Western blot analysis.
Induced chondrogenic cells (passage 6), MDFs (passage 3), and primary chondrocytes (passage 1) were cultured in 100-mm dishes. After reaching confluence, cells were lysed. Cell lysates were subjected to SDS-PAGE. The separated proteins were then electroblotted and immunostained with the antibodies listed in Supplemental Table 4.
Immunofluorescence staining.
Cells were cultured on culture slides, fixed in 4% paraformaldehyde, and permeabilized with 0.5% Tween 20. Cells were incubated with the primary antibodies listed in Supplemental Table 4. Immune complexes were detected by using the appropriate secondary antibodies conjugated to Alexa Fluor (Supplemental Table 4).
Bisulfite genomic sequencing.
MK induced cells, MDFs, and primary chondrocytes were examined. Bisulfite treatment was performed by using the EpiTect Bisulfite kit (QIAGEN) according to the manufacturer’s instructions. PCR primers are listed in Supplemental Table 3. Amplified products were cloned into pMD20-T vector by using a Mighty TA-cloning Kit (Takara). Ten randomly selected clones were sequenced with M13 primer RV and M13 primer M4 for each gene. Nucleotide numbers for Col1a1 and Col1a2 are indicated below the panels in the left column of Figure 4D. The 5ι boundaries of first exons are set as +1 for Col1a1 (GenBank NC 000077, nt 94797584) and Col1a2 (GenBank NC 000072, nt 4455697).
BrdU staining.
Mice were intraperitoneally injected with BrdU labeling reagent (10 μl/g body weight) (Zymed, Invitrogen). Two hours later, the mice were sacrificed. Tissues were dissected and sectioned. Incorporated BrdU was detected by using a BrdU staining kit (Zymed, Invitrogen) to distinguish actively proliferating cells. The average number of BrdU-positive cells/total cells ± SD was calculated.
Generation of Col11a2-Egfp-Ires-Puro transgenic mice.
To create the Egfp-Ires-Puro transgene, Egfp, Ires, and puromycin-resistant gene sequences were cloned into pENTR-1A (Invitrogen) to prepare pENTR1A-mcs/Egfp-Ires-Puro and recombined with p742-gw-Int by LR reaction (Invitrogen) to prepare Col11a2-Egfp-Ires-Puro. The Col11a2-Egfp-Ires-Puro insert was released from the vector backbone with EcoRI and PstI. Transgenic mice were produced by microinjecting the inserts into the pronuclei of fertilized eggs. To identify transgenic mice, genomic DNA was amplified by transgene-specific PCR by using primers derived from the Gfp gene (ATGGACGAGCTGTACAAGTAAA) and from the IRES sequence (TTGCCAAAAGACGGCAATATG) to amplify a 155-bp product from Egfp-Ires-Puro transgenic mice.
Dox-inducible lentiviral vectors and transduction.
The Tet-responsive Ptight promoter was PCR amplified from pTRE-Tight plasmid (BD Biosciences — Clontech) and cloned into the pENTR5′ plasmid (Invitrogen) to prepare pENTER5′-XEmcs/(Ptight). Human c-MYC and human KLF4 cDNA from pMXs-hc-MYC (Addgene plasmid 17220) and pMXs-hKLF4 (Addgene plasmid 17219) were cloned into pDONR221 (Invitrogen) by BP clonase (Invitrogen) to prepare pDONR221-hc-MYC and pDONR221-h-KLF4, respectively. Human SOX9 cDNA was PCR amplified by using primers NT566 (ATTCAGTCGACATGGGATCCGAGGACTCCGCGGGC) and NT567 (GTGTTTCTAGATCAAGGTCGAGTGAGCTGTGTGTA) and cloned into pENTR-1A (Invitrogen) to prepare pENTR1A-F(–)hSOX9. Lentiviral vector pLe6Δ was prepared by deleting the PGKpromoter-EM7-blastcidin sequence at KpnI sites from pLenti6.4/R4R2/V5-DEST (Invitrogen). pDONR221-hc-MYC, pDONR221-h-KLF4, and pENTR1A-F(–)hSOX9 were recombined with pENTER5′-XEmcs/(Ptight) and pLe6Δ by LR clonase II Plus reaction (Invitrogen) to prepare pLe6Δ-Ptight-hc-MYC, pLe6Δ-Ptight-hKLF4, and pLe6Δ-Ptight-F(–)hSOX9, respectively. HiPerform lentivirus was prepared according to the manufacturer’s instructions (Invitrogen), concentrated by using PEG-it (System Bioscience), and stored at –80°C.
The rtTA2S-M2 sequence was PCR amplified from pTet-On Advanced (BD Biosciences — Clontech), cloned into pENTR-1A (Invitrogen), and recombined with pMXs-gw by LR reaction (Invitrogen) to prepare pMXs-rtTA2-M2.
MDFs were prepared from neonatal Col11a2-Egfp-Ires-Puro transgenic mice. A total of 5 × 105 MDFs in a 10-cm dish were transduced with rtTA2-M2 retrovirus overnight. On the next day, the MDFs were transduced with inducible c-MYC, KLF4, and SOX9 lentivirus for 48 hours. The MOI of each virus vector was as follows: pLe6Δ-Ptight-hc-MYC, 2.1; pLe6Δ-Ptight-hKLF4, 2.1; pLe6Δ-Ptight-F(–)hSOX9, 4.2. Medium was replaced with new virus-containing medium every 12 hours. After incubation with virus, MDFs were trypsinized and replated into five 10-cm dishes in fresh DMEM plus 10% FBS medium in the presence of 1 μg/ml dox. Medium was changed every other day.
Statistics.
Data are shown as means and standard deviations. The 2-tailed Student’s t test was used to compare data. P values less than 0.05 were considered to be statistically significant.
All experiments were approved by the IACUC of Osaka University Graduate School of Medicine and the Osaka University Living Modified Organism [LMO] Experiments Safety Committee.
Supplementary Material
Acknowledgments
We thank Shinya Yamanaka for providing advice and vectors and Kazutoshi Takahashi for providing instructions for retroviral gene transduction. We also thank Toshio Kitamura for the Plat-E cells and pMX retroviral vectors. We thank Daisuke Ikegami, Mina Okamoto, and Mari Shinkawa for their assistance and discussions. We thank Toru Nakano and Tomoatsu Kimura for their advice and Masahito Ikawa for providing instructions for lentiviral gene transduction. This study was supported in part by Scientific Research Grants 18390415, 19659378, and 21390421 from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (MEXT) and the Japan Science and Technology Agency (JST), CREST.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(2):640–657. doi:10.1172/JCI44605.
References
- 1. Frisbie DD, et al. Arthroscopic subchondral bone plate microfracture technique augments healing of large chondral defects in the radial carpal bone and medial femoral condyle of horses. Vet Surg. 1999;28(4):242–255. doi: 10.1053/jvet.1999.0242. [DOI] [PubMed] [Google Scholar]
- 2. Bae DK, Yoon KH, Song SJ. Cartilage healing after microfracture in osteoarthritic knees. Arthroscopy. 2006;22(4):367–374. doi: 10.1016/j.arthro.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 3. Bedi A, Feeley BT, Williams RJ., 3rd Management of articular cartilage defects of the knee. J Bone Joint Surg Am. 2010;92(4):994–1009. doi: 10.2106/JBJS.I.00895. [DOI] [PubMed] [Google Scholar]
- 4. Goessler UR, et al. Expression of collagen and fiber-associated proteins in human septal cartilage during in vitro dedifferentiation. Int J Mol Med. 2004;14(6):1015–1022. [PubMed] [Google Scholar]
- 5. Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22(1):85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 6. Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16(21):2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smits P, et al. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1(2):277–290. doi: 10.1016/S1534-5807(01)00003-X. [DOI] [PubMed] [Google Scholar]
- 8. Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17(4):2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17(19):5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Y, Li H, Tanaka K, Tsumaki N, Yamada Y. Identification of an enhancer sequence within the first intron required for cartilage-specific transcription of the alpha2(XI) collagen gene. J Biol Chem. 2000;275(17):12712–12718. doi: 10.1074/jbc.275.17.12712. [DOI] [PubMed] [Google Scholar]
- 11. Han Y, Lefebvre V. L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol Cell Biol. 2008;28(16):4999–5013. doi: 10.1128/MCB.00695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ikeda T, et al. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 2004;50(11):3561–3573. doi: 10.1002/art.20611. [DOI] [PubMed] [Google Scholar]
- 13. Sudo K, et al. Mesenchymal progenitors able to differentiate into osteogenic, chondrogenic, and/or adipogenic cells in vitro are present in most primary fibroblast-like cell populations. Stem Cells. 2007;25(7):1610–1617. doi: 10.1634/stemcells.2006-0504. [DOI] [PubMed] [Google Scholar]
- 14. Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17(4):771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 15. Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 16. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 17. Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1(1):55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 18. Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 19. Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448(7151):318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 20. Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 21. Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 22. Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26(1):101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 23. Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2(1):10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 24. Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451(7175):141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 25. Kim JB, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454(7204):646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 26. Aoi T, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321(5889):699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 27. Tsumaki N, Kimura T, Matsui Y, Nakata K, Ochi T. Separable cis-regulatory elements that contribute to tissue- and site-specific alpha 2(XI) collagen gene expression in the embryonic mouse cartilage. J Cell Biol. 1996;134(6):1573–1582. doi: 10.1083/jcb.134.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murai J, Ikegami D, Okamoto M, Yoshikawa H, Tsumaki N. Insulation of the ubiquitous RXRB promoter from the cartilage–specific adjacent gene, COL11A2. J Biol Chem. 2008;283(41):27677–27687. doi: 10.1074/jbc.M803657200. [DOI] [PubMed] [Google Scholar]
- 29. Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7(12):1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 30. Argentin G, Cicchetti R, Nicoletti B. Mouse chondrocytes in monolayer culture. In Vitro Cell Dev Biol Anim. 1993;29A(8):603–606. doi: 10.1007/BF02634541. [DOI] [PubMed] [Google Scholar]
- 31. Akiyama H, et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci U S A. 2005;102(41):14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gosset M, Berenbaum F, Thirion S, Jacques C. Primary culture and phenotyping of murine chondrocytes. Nat Protoc. 2008;3(8):1253–1260. doi: 10.1038/nprot.2008.95. [DOI] [PubMed] [Google Scholar]
- 33. Toma JG, et al. Isolation of multipotent adult stem cells from the dermis of mammalian skin. Nat Cell Biol. 2001;3(9):778–784. doi: 10.1038/ncb0901-778. [DOI] [PubMed] [Google Scholar]
- 34. Akiyama H, et al. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18(9):1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koelling S, et al. Migratory chondrogenic progenitor cells from repair tissue during the later stages of human osteoarthritis. Cell Stem Cell. 2009;4(4):324–335. doi: 10.1016/j.stem.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 36. Nagy A, Gertsenstein M, Vintersten K, Behringer R.Manipulating the Mouse Embryo . New York, New York, USA: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 37. Bjorntorp P, Karlsson M, Pertoft H, Pettersson P, Sjostrom L, Smith U. Isolation and characterization of cells from rat adipose tissue developing into adipocytes. J Lipid Res. 1978;19(3):316–324. [PubMed] [Google Scholar]
- 38. Lefebvre V, Garofalo S, Zhou G, Metsaranta M, Vuorio E, De Crombrugghe B. Characterization of primary cultures of chondrocytes from type II collagen/beta-galactosidase transgenic mice. Matrix Biol. 1994;14(4):329–335. doi: 10.1016/0945-053X(94)90199-6. [DOI] [PubMed] [Google Scholar]
- 39. Aszodi A, Hunziker EB, Brakebusch C, Fassler R. Beta1 integrins regulate chondrocyte rotation, G1 progression, and cytokinesis. Genes Dev. 2003;17(19):2465–2479. doi: 10.1101/gad.277003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kitamura T, et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31(11):1007–1014. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.