Abstract
Huntington disease is an autosomal dominant neurodegenerative disorder caused by a toxic expansion in the CAG repeat region of the huntingtin gene. Oligonucleotide approaches based on RNAi and antisense oligonucleotides provide promising new therapeutic strategies for direct intervention through reduced production of the causative mutant protein. Allele-specific and simultaneous mutant and wild-type allele–lowering strategies are being pursued with local delivery to the brain, each with relative merits. Delivery remains a key challenge for translational success, especially with chronic therapy. The potential of disease-modifying oligonucleotide approaches for Huntington disease will be revealed as they progress into clinical trials.
Introduction
Huntington disease (HD) has the highest prevalence of all autosomal CAG repeat neurodegenerative diseases. The mutant huntingtin gene, which has an expansion in the CAG repeat region to more than 36 CAGs within exon 1 (1), confers toxicity ascribed to the mutant protein, although mRNA contribution to the disease process has yet to be fully tested. Neuropathology is characterized by striatal and cortical neuronal atrophy (2). Patients with HD usually develop involuntary movements, cognitive dysfunction, and behavioral changes in the fourth decade of life.
Despite the identification of mutant huntingtin as the genetic cause of HD in 1993 (1), there is a high unmet need for disease-modifying therapy. Several therapeutic candidates with the potential to alter the underlying progression of disease have been tested in clinical trials (3–8). These agents include neuroprotective strategies to counter the toxic cellular effects of mutant huntingtin protein and cellular replacement strategies to counter the loss of neurons in the striatum. Such strategies are indirect and function downstream of the effects of mutant huntingtin protein. At best, these approaches repair damage after it has occurred. Moreover, mutant huntingtin protein has multiple deleterious molecular and cellular consequences, each of which could be the basis of a therapeutic approach. However, targeting these individually might be insufficient for significant clinical benefit (Figure 1). With the advent of oligonucleotide approaches to gene suppression such as RNAi and antisense oligonucleotides (ASOs), therapeutic strategies directly targeting the causative gene — mutant huntingtin — may be developed and tested. In theory, by reducing huntingtin mRNA levels, the synthesis of mutant huntingtin protein would be reduced, representing a strategy upstream in the pathogenetic process, potentially preventing cellular damage.
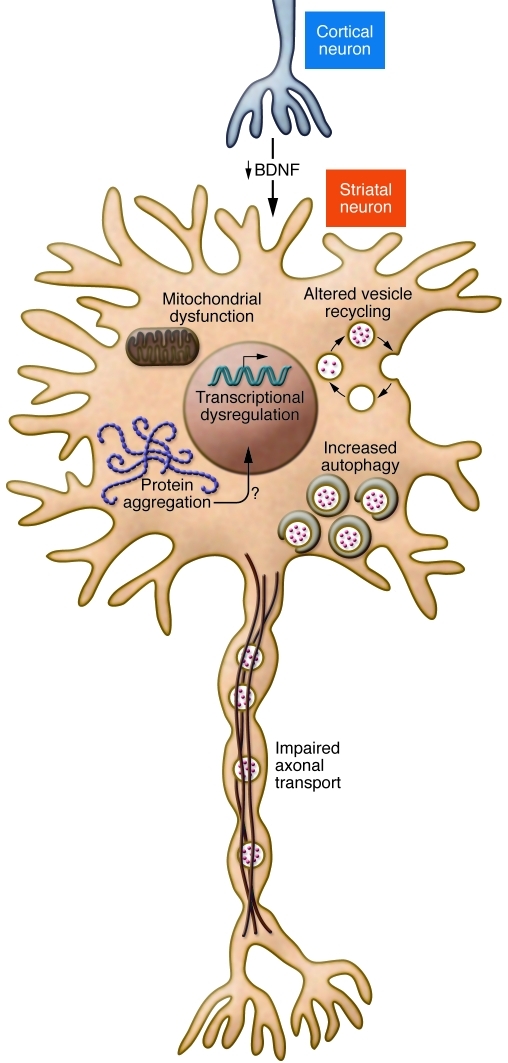
Figure 1. Multiple pathogenic mechanisms of mutant huntingtin include loss of BDNF neurotrophic support for striatal neurons, impaired axonal transport, altered vesicle recycling, mitochondrial dysfunction, increased autophagy, protein aggregation, and transcriptional dysregulation.
No single aberrant effect of mutant huntingtin explains neuronal dysfunction and early death. Mutant huntingtin disrupts the transcriptional activation of BDNF expression, thereby reducing BDNF delivery from cortex to striatum. Striatal neurons depend on BDNF to maintain their health (21, 22). Huntingtin has been known to associate with vesicles (34). Mutant huntingtin impairs endosomal recycling (39), thereby reducing uptake of transferrin receptor and EAAC1, probably among other recycled receptors (39, 40). Loss of cysteine (EAAC1 recycling defect) decreases glutathione and increases reactive oxygen species (40). Mutant huntingtin is associated with impaired axonal trafficking (37) and loss of BDNF delivery from the cortical to striatal neurons (36). Mitochondria defects (107) are well described, but it is unclear whether these are early events. A role in HD pathogenesis of protein aggregation in nuclear inclusions or cytoplasmic aggregates (48) is not established. Cell death can occur without generation of mutant huntingtin aggregation (108), and aggregates might be protective (109). Interference with HDAC efficiency has led to the idea that HDAC inhibition can counter a harmful effect of mutant huntingtin (110). Autophagy clears mutant huntingtin associated with organelles; an increase in autophagy might have therapeutic value (111). A recent review details these and other postulated molecular mechanisms in HD (15).
Gene silencing through RNAi or ASO action should be a viable therapeutic strategy in the treatment of HD. Current oligonucleotide-based approaches have not allowed for the targeting of mutant alleles with CAG repeat expansions, such as mutant huntingtin, with high selectivity or potency in vitro (9, 10). Since wild-type huntingtin has numerous physiological activities in cells that are important for neuronal function, complete suppression of both mutant and wild-type huntingtin may not be desirable (11), and allele-specific silencing of mutant huntingtin by targeting associated SNPs (12–14) represents a promising alternative. Another potential therapeutic strategy comprises simultaneous partial (rather than complete) lowering of wild-type and mutant huntingtin expression, with the aim of reducing mutant huntingtin expression sufficiently for therapeutic benefit while preserving sufficient wild-type huntingtin expression for maintaining normal cellular physiology. Clearly, specific and potent allelic silencing, if achievable, would be ideally suited for therapy in autosomal dominant diseases such as HD.
Allele-specific silencing is predicated on experimentally verified functions of wild-type huntingtin. Huntingtin has multiple effects in development (15) and neuronal physiology (15), including regulation of transcription (16–18), membrane dynamics (19), mitochondrial efficiency (20), BDNF transcription (15, 21, 22), autophagy (23–26), and endosomal recycling (ref. 7 and Figure 1). Huntingtin knockout mice (Hdh–/–) exhibit embryonic lethality by day 8.5, at a time preceding nervous system development (27). Elimination of wild-type huntingtin in adult mice is associated with neuronal loss in several brain regions, especially cortex and striatum, motor deficits, behavioral changes, and shortened lifespan (11). Overexpression of wild-type huntingtin improves survival in cultured neuron-derived cells (28, 29) and neuroblastoma cells expressing mutant huntingtin (30). Loss of wild-type huntingtin increases caspase-3 activity and apoptosis (31). During development, elimination of wild-type huntingtin reduces neuronal survival and produces mice with behavioral changes (32). Overexpression of wild-type huntingtin in the YAC128 mouse model of HD reduces the extent of striatal neuropathology (33).
Wild-type huntingtin is associated with vesicles in neurons (34). Huntingtin in axons moves in anterograde and retrograde directions (35), and reduction of wild-type protein levels is associated with reduced BDNF (36) and mitochondrial trafficking in neurons (37). Post-translational modification of wild-type huntingtin provides further regulation of axonal trafficking (38). Wild-type huntingtin forms a complex with Rab11 and affects recycling of transferrin receptor and EAAC1, a receptor that controls uptake of cysteine in neurons (39, 40). Loss of wild-type huntingtin impairs vesicle recycling and increases production of reactive oxygen species (40). Mutant huntingtin has similar effects in neurons as disrupting the Rab11 complex (39).
To summarize, elimination of wild-type huntingtin has multiple harmful impacts on adult neurons: loss of pro-survival mechanisms, decreased levels of an essential growth factor (BDNF), and impaired axonal trafficking and endosomal recycling, leading to accumulation of toxic reactive oxygen species. Thus, silencing of the huntingtin gene must be done with the recognition that wild-type huntingtin expression should be maintained at safe levels.
Huntingtin is a large protein (~350 kDa) with many potential sites of protein interaction, including the polyglutamine region. In theory, expansion of the polyglutamine series can strengthen protein interactions or permit new interactions (41). Immunoisolation and pull-down experiments have been used to create protein networks of huntingtin (42, 43). Wild-type huntingtin has many sites for proteolytic cleavage and may be targeted by caspases, calpains, and aspartyl proteases (44–47). Mutant huntingtin shares these sites of proteolysis (45, 47). It is unclear if differences in cleavage rates contribute to toxicity of mutant huntingtin fragments through changes in folding, clearance of mutant huntingtin fragments, or the propensity for mutant huntingtin (and fragments) to aggregate or form aberrant protein interactions (48–50). Thus, combinatorial changes by the mutant huntingtin and its fragments might underlie complexities in HD pathogenesis. Thus, directly silencing mutant huntingtin offers a most rational therapeutic strategy.
RNAi and ASOs are the two major approaches to therapeutic gene suppression (Figure 2). They offer powerful solutions for selectively inhibiting disease targets, including those such as huntingtin that are difficult to modulate specifically with traditional pharmaceutical classes such as small molecules or proteins, due to protein size or intracellular location. Although small-molecule screens have been conducted, initially to prevent huntingtin aggregation (51) and more recently to reverse pathogenic processes (52), clinical testing has not yet been initiated with these small molecules. Furthermore, neither the function of the pathogenic protein nor its interacting partners needs to be known to implement either protein lowering approach; the only consideration is that excess protein drives pathology, as is the case with mutant huntingtin protein. A contribution of mutant mRNA to pathology would also be susceptible to gene silencing. Both therapeutic approaches require adequate in vivo delivery of drug; this critical aspect for drug development is discussed in detail later in this review.
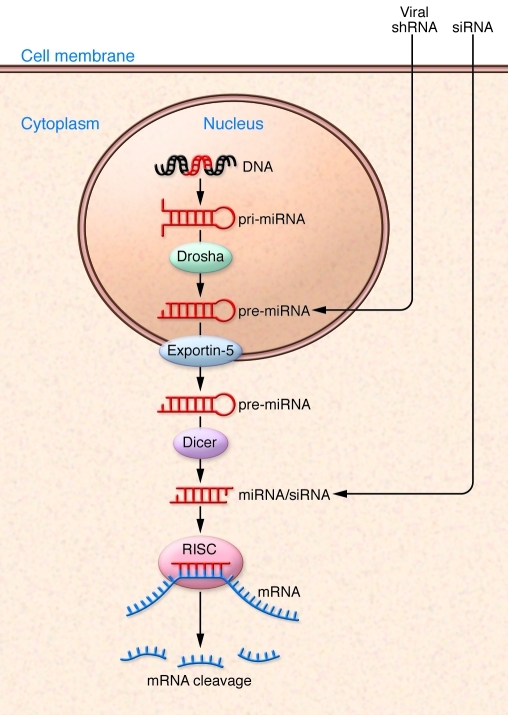
Figure 2. siRNA, miRNA, and shRNA cellular pathways.
With successful delivery, siRNAs enter the cell and gain access to the cytoplasmic compartment, where they are incorporated into RNAi silencing complex (RISC). The RISC complex with the active guide strand (in red) binds the complementary sequence within the target mRNA, resulting in Argonaut 2–mediated cleavage and subsequent mRNA degradation. Endogenous miRNAs are derived from miRNA genes that are transcribed to primary miRNAs (pri-mRNA) that are cleaved by Drosha to hairpin precursors (pre-miRNAs) within the nucleus. Viral delivery of shRNAs requires entry into the nucleus, where promoter-directed expression of pre-miRNA occurs. These pre-miRNAs are exported to the cytoplasm by Exportin-5, and then the hairpin precursor is cleaved by Dicer to form miRNAs that are incorporated into RISC, leading to mRNA binding and cleavage.
After entry into cells, synthetic siRNAs leverage the naturally occurring process of RNAi in a consistent and predictable manner by directing sequence-specific degradation of mRNA. Since siRNAs harness an endogenous catalytic mechanism, potent and selective siRNAs with picomolar EC50s in vitro can usually be identified if the starting pool of siRNAs is sufficiently large. Endogenous miRNA can be derived from a stem-loop structure in which opposite RNA strands have complementarity (Figure 2). miRNA precursor structures can be found in introns and give rise to endogenous miRNAs through a series of enzymatic steps (53). The extent of complementarity between miRNAs and target mRNAs is quite variable. In theory, the lack of exact complementarity could lead to off-target miRNA effects. Recent evidence suggests that the miRNA silencing confers a slow degradation of target mRNA, in contrast to the abrupt cleavage found in RNAi (54). ASOs are single-stranded oligodeoxynucleotides of approximately 15–25 nucleotides that suppress the synthesis of the targeted protein. After entering the cell and binding to the complementary mRNA, ASOs reduce gene expression by enabling the RNase-H–mediated degradation of the target mRNA or physically blocking translation of the target mRNA (55).
With careful oligonucleotide sequence design and selection, these approaches can be highly specific for the target of interest (Table 1), and appropriate chemical modifications can provide stability in vivo (56, 57). A preferred sequence design for potential therapeutics comprises alignment of mRNAs from rodent, monkey, and human and identification of all possible conserved target regions (58). Typically, the starting pool comprises hundreds or thousands of possible sequences. For a large gene such as huntingtin, with a transcript size of ~13 kb, approximately 13,000 sequences can be initially considered as candidates, since the starting pool of sequences is obtained by tiling across the entire huntingtin transcript (Figure 3). If sufficient homology exists across species, hundreds of sequences against conserved regions can be designed and screened in vitro for subsequent testing in preclinical models and potential clinical advancement. The possible target regions within the mRNA of interest are subjected to a BLAST-like analysis to select sequences that are unlikely to result in off-target silencing of genes that have partial homology to the gene of interest (59). These selected sequences are evaluated empirically in vitro to identify the most potent siRNAs from dose-response studies in cell culture systems.
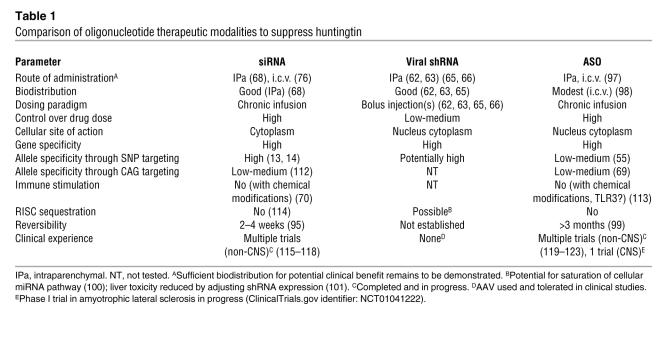
Table 1 .
Comparison of oligonucleotide therapeutic modalities to suppress huntingtin
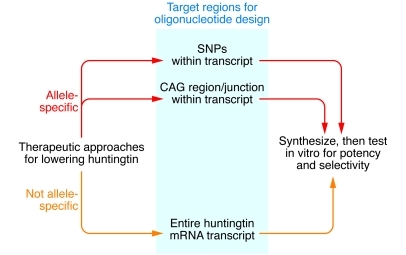
Figure 3. Oligonucleotide therapeutic approaches for lowering huntingtin.
Either a non–allele-specific or an allele-specific approach can be taken. If a non–allele-specific approach is taken, then the entire huntingtin mRNA transcript can be used to design oligonucleotide sequences with bioinformatics algorithms, generating hundreds of sequences for in vitro potency and selectivity screens. If an allele-specific approach is taken, then either SNP targeting or CAG targeting are possible options. With both of these options, tens of sequences can be designed based on current knowledge of targeting SNPs or the expanded CAG region, for testing in vitro for potency and selectivity.
RNAs can potentially stimulate innate immunity (60). With use of appropriate chemically modified nucleotides (e.g., 2′-O-methyl substitutions), immune stimulation can be eliminated (Table 1) without compromising silencing activity (61). These same chemical modifications, together with modifications at the 3′ end such as phosphorothioate linkages, provide the added benefit of stability against nucleases that are present in biological fluids (57). Thus, with appropriate design, selection, and chemical modification, potent and specific oligonucleotides can be identified for in vivo studies and potential clinical testing.
Silencing the gene versus selectively silencing the mutant allele: a comparison
Allele-specific silencing of mutant huntingtin by targeting associated SNPs (12–14) and simultaneous partial lowering of wild-type and mutant huntingtin expression (62, 63) represent alternative potential therapeutic approaches. shRNAs and siRNAs targeting wild-type and/or mutant huntingtin have been evaluated in vivo. Suppression of wild-type huntingtin mRNA in the adult mouse striatum by 70% is tolerated for at least 4 months, based on histological evaluation (64). Partial lowering of mutant huntingtin over months in adult rodent models of HD is effective in reducing neuropathology, improving motor behavior, and prolonging survival (62, 63, 65–68). In the first proof-of-principle studies, the mutant human huntingtin transgene was selectively suppressed by targeting a region of the huntingtin gene unique to the human (versus wild-type mouse) transcript (62, 63, 65–68). These experiments therefore addressed an allele-specific huntingtin-lowering approach and demonstrated significant benefit on neuropathology and motor behavior with approximately 50%–80% lowering of mutant huntingtin mRNA in the presence of unaltered expression of endogenous wild-type gene. However, the effects of nonallele-specific huntingtin reduction had not yet been evaluated. Would partial lowering of both mutant and wild-type huntingtin be tolerated and beneficial in ameliorating the disease phenotype?
Adeno-associated viral delivery of an inhibitory shRNA (62) designed to silence both mutant human and wild-type endogenous mouse huntingtin simultaneously were administered into the striatum of HD-N171-82Q transgenic mice. These mice express a fragment of human huntingtin containing 82 CAG repeats and exhibit neuropathology in the striatum, loss of body weight, and motor dysfunction. In this mouse model, intrastriatal administration of RNAi initiated at 7 weeks of age resulted in suppression of mutant and wild-type huntingtin mRNAs by approximately 60% and 75% at weeks 11 and 20, respectively. Histological evaluation for toxicity with markers of striatal neurons (DARPP32) and microglia (Iba1) showed that after 3 months treatment was well tolerated compared with control groups. Rotarod performance was improved by 10 and 14 weeks of age in the treated group. Body weight loss was not affected by intrastriatal treatment, as expected with local administration into the striatum, and at 18 weeks of age, wild-type mice weighed significantly more than transgenic mice, independent of treatment. Since the treated and control transgenic mouse groups were not significantly different in body weight at 14 or 18 weeks of age, better rotarod performance could not be attributed to lower body weight. In addition, the treated transgenic mice exhibited a trend toward extended survival, with more than 75% of animals surviving to week 20 (when the experiment was terminated) compared with 45% in the control groups. This study demonstrated that, despite significant suppression of wild-type huntingtin, lowering of nonallele-specific huntingtin by up to 75% over 3 months can improve behavioral abnormalities in a transgenic mouse model of HD and is tolerated based on histological markers.
These observations were extended to longer time frames by evaluating the impact of concomitant silencing of mutant human and wild-type mouse huntingtin on neuropathology in one mouse model of HD (63). Using lentiviral delivery of an shRNA that targets a region that the two transcripts have in common, the authors reported a substantial (though not quantified) reduction in huntingtin expression level. Nine months following co-injection of the vectors with human htt171-82Q and shRNA, there was amelioration of striatal neuropathology in the treated group compared with the control group, with almost complete elimination of ubiquitin-stained huntingtin inclusions and a complete restoration of DARPP-32 staining. Thus, this study demonstrated that simultaneous suppression of wild-type and mutant huntingtin over 9 months can essentially normalize two markers of disease pathology in the striatum (63). Taken together, these studies support therapeutic potential of simultaneous partial lowering of wild-type and mutant huntingtin, with the proviso that a sufficient therapeutic index is demonstrated in more extensive preclinical toxicology studies with chronic dosing.
These studies indicate that reduction of huntingtin can be tolerated in mice up to 9 months. The safety of this suppression should be contrasted with data (presented earlier) that huntingtin has significant roles in the adult brain. Patients with HD will likely need to be treated for decades, and longer-term neuronal tolerance for reduction in huntingtin has not yet been determined. Therefore, strategies to silence the mutant huntingtin allele selectively have been studied.
Ideal genetic therapy in autosomal dominant disease, as in HD, comprises specific knock down of the mutant allele. CAG repeat expansion diseases pose a particular complexity to this paradigm. To date, oligonucleotides targeting the CAG repeat region have only been reported to distinguish between mutant and wild-type huntingtin mRNA with modest discrimination (up to ~7-fold selectivity) and/or relatively high (nanomolar) in vitro concentrations (62, 69), in contrast to the picomolar potencies typically achieved with siRNAs in vitro. A significant consideration is the small starting pool of sequences, which greatly reduces the odds of identifying a potent molecule and severely limits the possibility of selecting a molecule that is highly specific for huntingtin over all other genes (Figure 3). Nonetheless, this is clearly an exciting area of emerging research, and with further effort, more potent oligonucleotides targeting the CAG repeat region may be identified, particularly as additional chemical modifications are applied (70). A key step for this approach will be to demonstrate selective in vivo silencing of mutant huntingtin by targeting the CAG repeat region; this has yet to be reported.
An alternative approach to selective silencing of the mutant allele is SNP targeting. Numerous SNPs have been identified in the huntingtin gene and mRNA; several of these SNPs are associated only with the mutant huntingtin allele. The HD mRNA has been associated with three SNPs in 75% to 80% of patients with HD (13, 14). Approximately 50% of patients with HD are associated with a SNP that has linkage disequilibrium with the mutant huntingtin allele and is much less frequent in the general population. The SNP (RS362307) is located in the 3′ UTR, and only 13% of subjects without HD have this SNP heterozygosity. The other sites are found in the open reading frame, exon 39 (RS363125) and exon 57 (RS362237). The latter two SNP heterozygosities are equally present in subjects with and without HD. The presence of identifiable SNP heterozygosities serves as the foundation for allelic mRNA silencing, which is distinguishable from gene silencing (which targets both wild-type and mutant alleles).
Advances in understanding RNAi structure and function make allele-selective gene silencing practicable by targeting these SNPs. Since a particular SNP may not be readily susceptible to RNAi, the presence of SNP heterozygosities expands therapeutic targets in the mutant allele. Deploying intentional nucleotide mismatches between the guide strand and the target mRNA can confer nucleotide specificity in silencing. Purine-purine mismatches provide the best nucleotide discrimination (71). Some mutations or SNP heterozygosities are amenable to siRNA mismatches for providing selectivity. The SNP site RS362307 in mutant huntingtin linkage disequilibrium is particularly troublesome because it is GC rich and because a single mismatch is insufficient to discriminate between the wild-type and mutant huntingtin alleles (14). Ordinarily, mismatch in the seed sequence can abrogate RNAi, but in the circumstance of huntingtin SNP site RS362307, dual mismatches provide RNAi allelic discrimination with more than 10-fold selectivity (14). The selectivity of the siRNA for mutant huntingtin over other transcripts and the demonstration of in vivo silencing and doses required remain to be determined.
Practical considerations for a SNP-based RNAi approach to selectively target mutant huntingtin include sequence specificity and potency, given the small starting pool of possible allele-specific sequences for each SNP (Figure 3) versus the more than 13,000 sequences considered for gene silencing of both alleles. Potency may be improved with chemical modifications (70), but specificity for huntingtin over other genes is inherent to the oligonucleotide sequence. Moreover, with a SNP-based approach, multiple allele-specific drugs would ultimately need to be developed in order to treat most patients with HD. It is likely that each drug would require a separate Investigational New Drug–enabling safety/toxicity package, and separate drug manufacturing and analytical chemistry assays would be needed to support initial clinical testing. Approximately 25% of the HD population do not have a SNP associated with the mutant allele and would not be candidates for SNP-based RNAi therapy (13, 14).
Delivery of RNAi therapeutics and ASO
Securing consistent in vivo delivery of siRNA and ASO therapeutic candidates is critical to treating CNS disorders. These molecules are not suitable for oral delivery due to poor absorption from the gastrointestinal tract, and the blood-brain barrier remains a formidable challenge for accessing cells within the CNS (72). Thus, these drug candidates require direct delivery to the brain via intraparenchymal or intracerebroventricular routes of administration (Table 1). Although transfection reagents have been used to enhance local CNS delivery in animal models (73–78), such reagents are not required for successful neuronal delivery and carry risk of cytotoxicity. The mechanism of siRNA and ASO uptake into CNS neurons with simple saline formulations is unknown, although there have been multiple demonstrations of effective oligonucleotide delivery in vivo (68, 79–82). In vivo efficacy studies with siRNAs in the nervous system have included inhibition of well-known molecular targets such as dopamine and serotonin transporters (79, 80), P2X3 (81), δ-opioid receptor (74), α-synuclein (82), and huntingtin (68, 76). In these studies, only the expected phenotypic effects were reported, which supports the specificity of siRNAs. Silencing huntingtin in mouse models of HD attenuated neuropathology and behavioral deficits. In an acute mouse model generated by intrastriatal injection of adeno-associated virus (AAV) to express a fragment of mutant huntingtin, a single injection of cholesterol-conjugated siRNA targeting huntingtin into the adult striatum lowered huntingtin protein by 66%, resulting in a reduction of neuropil aggregates by approximately 65%, an amelioration of clasping behavior by approximately 30%–40%, and a reduction in foot slips on a balance beam by approximately 70% (68, 76). In the R6/2 transgenic mouse, which expresses exon 1 of huntingtin with 144 CAG repeats, a single injection of siRNA targeting huntingtin into the lateral ventricle at postnatal day 2 led to approximately 55% decrease in huntingtin mRNA levels that recovered to baseline after 14 days and resulted in phenotypic benefit in the adult (68, 76). These studies demonstrate that partial lowering of huntingtin with direct CNS delivery of siRNA can lead to substantial effects in mouse models of HD, suggesting that this approach may provide significant therapeutic benefit.
For chronic treatment of neurodegenerative diseases, continuous or repeated long-term infusion of oligonucleotide directly into the CNS may be required. Silencing in the CNS after intravenous administration of siRNA complexed to the rabies virus glycoprotein peptide, postulated to be facilitated via interaction with the nicotinic acetylcholine receptor, has been reported (77). Successful delivery to intracranial tumors in rats using intravenous administration of a biotinylated siRNA attached via a biotin-streptavidin linker to a transferrin receptor antibody, has also been reported (83). These approaches need to be confirmed and optimized for drug development.
For direct delivery of therapeutics into the CNS, implantable infusion systems have been used widely for chronic infusion (Table 1), e.g., for intrathecal administration of pain medications and for clinical trials of intraparenchymally administered glial cell derived neurotrophic factor (GDNF) for the treatment of Parkinson disease (84–86). Although differing efficacy results were obtained in the GDNF studies, the trials provide precedence for the use of implantable infusion systems for direct intraparenchymal delivery of therapeutics to the CNS, such as oligonucleotides for the treatment of HD.
A key remaining challenge for CNS therapeutics, including RNAi and ASO, is distribution to an area of brain sufficient for a clinically meaningful effect. Local delivery of siRNA has produced silencing in neurons but with limited spatial distribution (56, 68, 74, 77, 79, 81). Convection-enhanced delivery is a method of infusing molecules under positive pressure to actively distribute the molecules throughout the tissue region of interest. This method has been used successfully for delivering small molecules (87, 88), proteins (89–91), viral vectors (92, 93), and nanoparticles (94) throughout brain tissue regions in animals and humans, resulting in far more extensive distribution than possible by passive diffusion. For RNAi and ASO approaches to HD, successful clinical translation of local CNS delivery will require scaling of drug distribution and effect over distances substantially greater than those in preclinical species.
How long does the silencing last? The mRNA lowering effect of siRNAs is durable, gradually returning to baseline over 2 to 4 weeks after dosing in vivo (Table 1 and ref. 95). ASO effects can persist for several months following termination of dosing in animals (96). ASOs targeting huntingtin are being tested in preclinical studies with direct CNS delivery (ref. 97 and Table 1). The half-lives of ASOs in mouse and monkey tissues after direct CNS administration are 2–6 weeks (ref. 98 and Table 1). In the clinic, ASOs are beginning to be evaluated for the treatment of neurodegenerative disease, with a Phase I clinical study initiated recently in patients with amyotrophic lateral sclerosis. In an earlier clinical study of an ASO targeting apolipoprotein B, the reduction of circulating apolipoprotein B was found to persist for at least 3 months after cessation of dosing, likely resulting from the extended half-life of ASOs in tissue (ref. 99 and Table 1). However, for ASO-based therapies, a significant consideration, as with viral delivery of shRNA, is the ability to rapidly terminate drug exposure should there be safety issues, whether due to on-target or non-specific effects.
AAV and lentivirus are promising neurotrophic vehicles for the shRNA precursor to siRNA (Figure 2). AAV shRNA has been demonstrated to successfully silence huntingtin mRNA in transgenic HD animal models, reducing mutant huntingtin protein and delaying onset of disease (65). A complication of the shRNA, neuronal damage, has been recognized (64). The cause of the neuronal damage is not established, but medium-sized spiny neurons in the striatum (the same that are first affected in HD) exhibited neurotoxicity. Overexpression of shRNA can interfere with small RNA pathways (100). A compelling example is overexpression of AAV-shRNA in mouse liver, which led to hepatocyte death (100). The cause is unclear, but excess siRNA has been postulated to consume RNAi silencing complex (RISC) components to a point that normal molecular events are impaired (Table 1). To alleviate this complication but maintain effective mutant huntingtin silencing, the stem-loop has been altered to include miRNA-type structure and the promoter has been changed from U6 based to Pol-II based (64, 101). With these modifications, the AAV shRNAmir (short hairpin RNA expressing microRNA) expression is reduced and the RNAi is well tolerated in the brain (64). It is speculated that the critical changes include the use of the Pol-II–based promoter and the miRNA-related shuttle, which protect against overproduction of shRNA. The authors posit that use of a miRNA shuttle protects against excessive huntingtin knock down, keeping huntingtin expression to more than 50% steady state to maintain neuronal health (102).
AAV delivery is epigenetic, while lentivirus becomes integrated into the cellular genome, with the possibility of disrupting native genes (103). AAV has many serotypes, expanding the neurotropism and expression of cargo. AAV is not passed between subjects and has no known pathology. It has been used in Phase I gene therapy studies of CNS with good safety in Parkinson disease (104). Human complications are attributed to the surgical administration of the virus, rather than toxicity of the virus itself (105). Self-complementary AAV (scAAV) presents an advance in AAV technology. Traditional AAV is single stranded and needs to form double-stranded DNA for expression. This process has limited efficiency and slows AAV expression. scAAV contains an inverted repeat genome that directly forms double-stranded DNA, thereby improving delivery of the scAAV cargo (106). New directions in AAV-based therapy could include regulated expression of the shRNAmir, use of multiple injections into the brain (Table 1) to yield sufficient spread of the AAV package through bilateral striata and cortex, and subsequent administration of AAV with different serotypes.
Safety and efficacy, associated with dosing, biodistribution, and durability, will need to be evaluated further for all forms of gene silencing: siRNA, ASO, and viral delivery of shRNA. For local CNS delivery in particular, distribution to the region of interest needs to be accurate and adequate for potential clinical benefit.
Concluding remarks
RNAi and ASOs provide novel and promising approaches to disease modification that have been enabled by a unique feature of their mechanisms of action — direct intervention at the mRNA level to reduce production of the causative pathogenic protein. Preclinical studies in animal models of HD have confirmed that siRNAs delivered by direct CNS administration in saline or in a viral package silence mutant huntingtin and ameliorate neuropathology as well as abnormal behaviors. Allele-specific silencing of mutant huntingtin versus simultaneous partial lowering of wild-type and mutant huntingtin represent oligonucleotide therapeutic strategies with their own relative merits. Practical considerations for allele-specific approaches that target SNPs associated with the mutant allele or that target the expanded CAG region include degree of discrimination between mutant and wild-type huntingtin, selectivity for huntingtin over other transcripts, doses required for silencing mutant huntingtin in vivo, and the number of different SNP-targeting sequences required to cover the HD population. For all approaches, translational success will depend on delivery that is scalable to the target region of interest in HD patients and will support chronic treatment. Adequate distribution and accurate targeting will need to be shown, and safety with long-term exposure will need to be demonstrated. Successful outcomes in these future studies will support clinical testing of oligonucleotide drug candidates in patients, providing for the first time potential therapies that could substantially modify HD progression. Demonstrating proof-of-concept for oligonucleotide therapeutics in HD patients would not only be important for this patient population but also have far-reaching implications for the treatment of neurodegenerative diseases in general.
Acknowledgments
This work was partially supported by NIH National Institute of Neurological Disorders and Stroke grant 38194 (to N. Aronin) and by CHDI (to N. Aronin). N. Aronin is a member of the University of Massachusetts Medical School Diabetes and Endocrine Research Center (supported by NIH grant DK32520).
Footnotes
Conflict of interest: D.W.Y. Sah owns equity in Alnylam Pharmaceuticals Inc.
Citation for this article: J Clin Invest. 2011;121(2):500–507. doi:10.1172/JCI45130.
References
- 1. [No authors listed]. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 2. Walker FO. Huntington’s disease. Lancet. 2007;369(9557):218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 3. Hauser RA, Sandberg PR, Freeman TB, Stoessl AJ. Bilateral human fetal striatal transplantation in Huntington’s disease. Neurology. 2002;58(11):1704. doi: 10.1212/wnl.58.11.1704. [DOI] [PubMed] [Google Scholar]
- 4. Huntington Study Group. A randomized, placebo-controlled trial of coenzyme Q10 and remacemide in Huntington’s disease. Neurology. 2001;57(3):397–404. doi: 10.1212/wnl.57.3.397. [DOI] [PubMed] [Google Scholar]
- 5. Huntington Study Group. Dosage effects of riluzole in Huntington’s disease: a multicenter placebo-controlled study. Neurology. 2003;61(11):1551–1556. doi: 10.1212/01.wnl.0000096019.71649.2b. [DOI] [PubMed] [Google Scholar]
- 6. Peyser CE, et al. Trial of d-alpha-tocopherol in Huntington’s disease. Am J Psychiatry. 1995;152(12):1771–1775. doi: 10.1176/ajp.152.12.1771. [DOI] [PubMed] [Google Scholar]
- 7. Huntington Study Group TREND-HD Investigators. Randomized controlled trial of ethyl-eicosapentaenoic acid in Huntington disease: the TREND-HD study. Arch Neurol. 2008;65(12):1582–1589. doi: 10.1001/archneur.65.12.1582. [DOI] [PubMed] [Google Scholar]
- 8. Tabrizi SJ, et al. High-dose creatine therapy for Huntington disease: a 2-year clinical and MRS study. Neurology. 2005;64(9):1655–1656. doi: 10.1212/01.WNL.0000160388.96242.77. [DOI] [PubMed] [Google Scholar]
- 9. Caplen NJ, Taylor JP, Statham VS, Tanaka F, Fire A, Morgan RA. Rescue of polyglutamine-mediated cytotoxicity by double-stranded RNA-mediated RNA interference. Hum Mol Genet. 2002;11(2):175–184. doi: 10.1093/hmg/11.2.175. [DOI] [PubMed] [Google Scholar]
- 10. Aronin N. Expanded CAG repeats in the crosshairs. Nat Biotechnol. 2009;27(5):451–452. doi: 10.1038/nbt0509-451. [DOI] [PubMed] [Google Scholar]
- 11. Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26(3):300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 12. van Bilsen PH, Jaspers L, Lombardi MS, Odekerken JC, Burright EN, Kaemmerer WF. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington’s disease patient-derived fibroblasts. Hum Gene Ther. 2008;19(7):710–719. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- 13. Lombardi MS, et al. A majority of Huntington’s disease patients may be treatable by individualized allele-specific RNA interference. Exp Neurol. 2009;217(2):312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 14. Pfister EL, et al. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Curr Biol. 2009;19(9):774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90(3):905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 16. Nucifora FC, Jr, et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291(5512):2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 17. Benn CL, et al. Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. J Neurosci. 2008;28(42):10720–10733. doi: 10.1523/JNEUROSCI.2126-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zuccato C, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35(1):76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 19. Kegel KB, et al. Polyglutamine expansion in huntingtin alters its interaction with phospholipids. J Neurochem. 2009;110(5):1585–1597. doi: 10.1111/j.1471-4159.2009.06255.x. [DOI] [PubMed] [Google Scholar]
- 20. Aoyama K, et al. Neuronal glutathione deficiency and age-dependent neurodegeneration in the EAAC1 deficient mouse. Nat Neurosci. 2006;9(1):119–126. doi: 10.1038/nn1609. [DOI] [PubMed] [Google Scholar]
- 21. Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in Huntington’s disease. Prog Neurobiol. 2007;81(5-6):294–330. doi: 10.1016/j.pneurobio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 22. Shimojo M. Huntingtin regulates RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) nuclear trafficking indirectly through a complex with REST/NRSF-interacting LIM domain protein (RILP) and dynactin p150 Glued. J Biol Chem. 2008;283(50):34880–34886. doi: 10.1074/jbc.M804183200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kegel KB, et al. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20(19):7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qin ZH, et al. Autophagy regulates the processing of amino terminal huntingtin fragments. Hum Mol Genet. 2003;12(24):3231–3244. doi: 10.1093/hmg/ddg346. [DOI] [PubMed] [Google Scholar]
- 25. Jeong H, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137(1):60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 27. Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat Genet. 1995;11(2):155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 28. Rigamonti D, et al. Wild-type huntingtin protects from apoptosis upstream of caspase-3. J Neurosci. 2000;20(10):3705–3713. doi: 10.1523/JNEUROSCI.20-10-03705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rigamonti D, Sipione S, Goffredo D, Zuccato C, Fossale E, Cattaneo E. Huntingtin’s neuroprotective activity occurs via inhibition of procaspase-9 processing. J Biol Chem. 2001;276(18):14545–14548. doi: 10.1074/jbc.C100044200. [DOI] [PubMed] [Google Scholar]
- 30. Ho LW, Brown R, Maxwell M, Wyttenbach A, Rubinsztein DC. Wild type Huntingtin reduces the cellular toxicity of mutant Huntingtin in mammalian cell models of Huntington’s disease. J Med Genet. 2001;38(7):450–452. doi: 10.1136/jmg.38.7.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y, et al. Huntingtin inhibits caspase-3 activation. EMBO J. 2006;25(24):5896–5906. doi: 10.1038/sj.emboj.7601445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nasir J, et al. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81(5):811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 33. Van Raamsdonk JM, Pearson J, Murphy Z, Hayden MR, Leavitt BR. Wild-type huntingtin ameliorates striatal neuronal atrophy but does not prevent other abnormalities in the YAC128 mouse model of Huntington disease. BMC Neurosci. 2006;7:80. doi: 10.1186/1471-2202-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DiFiglia M, et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14(5):1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 35. Block-Galarza J, et al. Fast transport and retrograde movement of huntingtin and HAP 1 in axons. Neuroreport. 1997;8(9-10):2247–2251. doi: 10.1097/00001756-199707070-00031. [DOI] [PubMed] [Google Scholar]
- 36. Gauthier LR, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118(1):127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 37. Trushina E, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24(18):8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Her LS, Goldstein LS. Enhanced sensitivity of striatal neurons to axonal transport defects induced by mutant huntingtin. J Neurosci. 2008;28(50):13662–13672. doi: 10.1523/JNEUROSCI.4144-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X, et al. Mutant huntingtin impairs vesicle formation from recycling endosomes by interfering with Rab11 activity. Mol Cell Biol. 2009;29(22):6106–6116. doi: 10.1128/MCB.00420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li X, et al. Aberrant Rab11-dependent trafficking of the neuronal glutamate transporter EAAC1 causes oxidative stress and cell death in Huntington’s disease. J Neurosci. 2010;30(13):4552–4561. doi: 10.1523/JNEUROSCI.5865-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28(8):425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 42. Goehler H, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington’s disease. Mol Cell. 2004;15(6):853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 43. Kaltenbach LS, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3(5):e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gafni J, Ellerby LM. Calpain activation in Huntington’s disease. J Neurosci. 2002;22(12):4842–4849. doi: 10.1523/JNEUROSCI.22-12-04842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gafni J, Hermel E, Young JE, Wellington CL, Hayden MR, Ellerby LM. Inhibition of calpain cleavage of huntingtin reduces toxicity: accumulation of calpain/caspase fragments in the nucleus. J Biol Chem. 2004;279(19):20211–20220. doi: 10.1074/jbc.M401267200. [DOI] [PubMed] [Google Scholar]
- 46. Goffredo D, et al. Calcium-dependent cleavage of endogenous wild-type huntingtin in primary cortical neurons. J Biol Chem. 2002;277(42):39594–39598. doi: 10.1074/jbc.C200353200. [DOI] [PubMed] [Google Scholar]
- 47. Kim YJ, et al. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington’s disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc Natl Acad Sci U S A. 2001;98(22):12784–12789. doi: 10.1073/pnas.221451398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 49. Wang CE, et al. Accumulation of N-terminal mutant huntingtin in mouse and monkey models implicated as a pathogenic mechanism in Huntington’s disease. Hum Mol Genet. 2008;17(17):2738–2751. doi: 10.1093/hmg/ddn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ellerby LM, Orr HT. Neurodegenerative disease: cut to the chase. Nature. 2006;442(7103):641–642. doi: 10.1038/442641a. [DOI] [PubMed] [Google Scholar]
- 51. Chopra V, et al. A small-molecule therapeutic lead for Huntington’s disease: preclinical pharmacology and efficacy of C2-8 in the R6/2 transgenic mouse. Proc Natl Acad Sci U S A. 2007;104(42):16685–16689. doi: 10.1073/pnas.0707842104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fecke W, Gianfriddo M, Gaviraghi G, Terstappen GC, Heitz F. Small molecule drug discovery for Huntington’s Disease. Drug Discov Today. 2009;14(9–10):453–464. doi: 10.1016/j.drudis.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 53. Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123(4):621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 54. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 56. Andronescu M, Zhang ZC, Condon A. Secondary structure prediction of interacting RNA molecules. J Mol Biol. 2005;345(5):987–1001. doi: 10.1016/j.jmb.2004.10.082. [DOI] [PubMed] [Google Scholar]
- 57. Choung S, Kim YJ, Kim S, Park HO, Choi YC. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem Biophys Res Commun. 2006;342(3):919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 58. de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6(6):443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2(12):711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hornung V, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11(3):263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 61. Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13(3):494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 62. Boudreau RL, et al. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol Ther. 2009;17(6):1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Drouet V, et al. Sustained effects of nonallele-specific Huntingtin silencing. Ann Neurol. 2009;65(3):276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 64. McBride JL, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105(15):5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Harper SQ, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci U S A. 2005;102(16):5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS, Mandel RJ. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington’s disease transgenic mice. Mol Ther. 2005;12(4):618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Machida Y, Okada T, Kurosawa M, Oyama F, Ozawa K, Nukina N. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem Biophys Res Commun. 2006;343(1):190–197. doi: 10.1016/j.bbrc.2006.02.141. [DOI] [PubMed] [Google Scholar]
- 68. DiFiglia M, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci U S A. 2007;104(43):17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gagnon KT, et al. Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry. 2010;49(47):10166–10178. doi: 10.1021/bi101208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug Discov Today. 2008;13(19–20):842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 71. Schwarz DS, et al. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2(9):e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Boudreau RL, Davidson BL. RNAi therapeutics for CNS disorders. Brain Res. 2010;1338:112–121. doi: 10.1016/j.brainres.2010.03.038. [DOI] [PubMed] [Google Scholar]
- 73. Hassani Z, et al. Lipid-mediated siRNA delivery down-regulates exogenous gene expression in the mouse brain at picomolar levels. J Gene Med. 2005;7(2):198–207. doi: 10.1002/jgm.659. [DOI] [PubMed] [Google Scholar]
- 74. Luo MC, et al. An efficient intrathecal delivery of small interfering RNA to the spinal cord and peripheral neurons. Mol Pain. 2005;1:29. doi: 10.1186/1744-8069-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tan PH, Yang LC, Shih HC, Lan KC, Cheng JT. Gene knockdown with intrathecal siRNA of NMDA receptor NR2B subunit reduces formalin-induced nociception in the rat. Gene Ther. 2005;12(1):59–66. doi: 10.1038/sj.gt.3302376. [DOI] [PubMed] [Google Scholar]
- 76. Wang YL, Liu W, Wada E, Murata M, Wada K, Kanazawa I. Clinico-pathological rescue of a model mouse of Huntington’s disease by siRNA. Neurosci Res. 2005;53(3):241–249. doi: 10.1016/j.neures.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 77. Kumar P, et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448(7149):39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 78. Salahpour A, Medvedev IO, Beaulieu JM, Gainetdinov RR, Caron MG. Local knockdown of genes in the brain using small interfering RNA: a phenotypic comparison with knockout animals. Biol Psychiatry. 2007;61(1):65–69. doi: 10.1016/j.biopsych.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 79. Thakker DR, et al. Neurochemical and behavioral consequences of widespread gene knockdown in the adult mouse brain by using nonviral RNA interference. Proc Natl Acad Sci U S A. 2004;101(49):17270–17275. doi: 10.1073/pnas.0406214101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Thakker DR, Hoyer D, Cryan JF. Interfering with the brain: use of RNA interference for understanding the pathophysiology of psychiatric and neurological disorders. Pharmacol Ther. 2006;109(3):413–438. doi: 10.1016/j.pharmthera.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 81. Dorn G, et al. siRNA relieves chronic neuropathic pain. Nucleic Acids Res. 2004;32(5):e49. doi: 10.1093/nar/gnh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. McCormack AL, Mak SK, Henderson JM, Bumcrot D, Farrer MJ, Di Monte DA. Alpha-synuclein suppression by targeted small interfering RNA in the primate substantia nigra. PLoS One. 2010;5:e12122. doi: 10.1371/journal.pone.0012122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Xia CF, Zhang Y, Boado RJ, Pardridge WM. Intravenous siRNA of brain cancer with receptor targeting and avidin-biotin technology. Pharm Res. 2007;24(12):2309–2316. doi: 10.1007/s11095-007-9460-8. [DOI] [PubMed] [Google Scholar]
- 84. Patel NK, Bunnage M, Plaha P, Svendsen CN, Heywood P, Gill SS. Intraputamenal infusion of glial cell line-derived neurotrophic factor in PD: a two-year outcome study. Ann Neurol. 2005;57(2):298–302. doi: 10.1002/ana.20374. [DOI] [PubMed] [Google Scholar]
- 85. Lang AE, et al. Randomized controlled trial of intraputamenal glial cell line-derived neurotrophic factor infusion in Parkinson disease. Ann Neurol. 2006;59(3):459–466. doi: 10.1002/ana.20737. [DOI] [PubMed] [Google Scholar]
- 86. Slevin JT, Gerhardt GA, Smith CD, Gash DM, Kryscio R, Young B. Improvement of bilateral motor functions in patients with Parkinson disease through the unilateral intraputaminal infusion of glial cell line-derived neurotrophic factor. J Neurosurg. 2005;102(2):216–222. doi: 10.3171/jns.2005.102.2.0216. [DOI] [PubMed] [Google Scholar]
- 87. Murad GJ, et al. Image-guided convection-enhanced delivery of gemcitabine to the brainstem. J Neurosurg. 2007;106(2):351–356. doi: 10.3171/jns.2007.106.2.351. [DOI] [PubMed] [Google Scholar]
- 88. Heiss JD, Walbridge S, Asthagiri AR, Lonser RR. Image-guided convection-enhanced delivery of muscimol to the primate brain. J Neurosurg. 2010;112(4):790–795. doi: 10.3171/2009.7.JNS09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lonser RR, et al. Image-guided, direct convective delivery of glucocerebrosidase for neuronopathic Gaucher disease. Neurology. 2007;68(4):254–261. doi: 10.1212/01.wnl.0000247744.10990.e6. [DOI] [PubMed] [Google Scholar]
- 90. Murad GJ, et al. Real-time, image-guided, convection-enhanced delivery of interleukin 13 bound to pseudomonas exotoxin. Clin Cancer Res. 2006;12(10):3145–3151. doi: 10.1158/1078-0432.CCR-05-2583. [DOI] [PubMed] [Google Scholar]
- 91. Song DK, Lonser RR. Convection-enhanced delivery for the treatment of pediatric neurologic disorders. J Child Neurol. 2008;23(10):1231–1237. doi: 10.1177/0883073808321064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bankiewicz KS, et al. Convection-enhanced delivery of AAV vector in parkinsonian monkeys; in vivo detection of gene expression and restoration of dopaminergic function using pro-drug approach. Exp Neurol. 2000;164(1):2–14. doi: 10.1006/exnr.2000.7408. [DOI] [PubMed] [Google Scholar]
- 93. Hadaczek P, et al. Convection-enhanced delivery of adeno-associated virus type 2 (AAV2) into the striatum and transport of AAV2 within monkey brain. Hum Gene Ther. 2006;17(3):291–302. doi: 10.1089/hum.2006.17.291. [DOI] [PubMed] [Google Scholar]
- 94. Voges J, et al. Imaging-guided convection-enhanced delivery and gene therapy of glioblastoma. Ann Neurol. 2003;54(4):479–487. doi: 10.1002/ana.10688. [DOI] [PubMed] [Google Scholar]
- 95. Zimmermann TS, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441(7089):111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 96. Crooke RM, Graham MJ, Lemonidis KM, Whipple CP, Koo S, Perera RJ. An apolipoprotein B antisense oligonucleotide lowers LDL cholesterol in hyperlipidemic mice without causing hepatic steatosis. J Lipid Res. 2005;46(5):872–884. doi: 10.1194/jlr.M400492-JLR200. [DOI] [PubMed] [Google Scholar]
- 97. Cleveland DW. RNaseH-dependent Antisense Approaches for Huntington Disease. In:Oligonucleotide Therapeutics Society . 6th Annual Meeting. October 22, 2010. [Google Scholar]
- 98. Mazur C, et al. Pharmacokinetics and pharmacodynamics of antisense oligonucleotides in the central nervous system of mice, dogs and monkeys after intrathecal, intracerebroventricular and intrastriatal administration, abstract. In:Oligonucleotide Therapeutics Society . 6th Annual Meeting. October 22, 2010. [Google Scholar]
- 99. Kastelein JJ, et al. Potent reduction of apolipoprotein B and low-density lipoprotein cholesterol by short-term administration of an antisense inhibitor of apolipoprotein B. Circulation. 2006;114(16):1729–1735. doi: 10.1161/CIRCULATIONAHA.105.606442. [DOI] [PubMed] [Google Scholar]
- 100. Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 101. Giering JC, Grimm D, Storm TA, Kay MA. Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol Ther. 2008;16(9):1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- 102. White JK, et al. Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nat Genet. 1997;17(4):404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- 103. Ciuffi A. Mechanisms governing lentivirus integration site selection. Curr Gene Ther. 2008;8(6):419–429. doi: 10.2174/156652308786848021. [DOI] [PubMed] [Google Scholar]
- 104. Eberling JL, et al. Results from a phase I safety trial of hAADC gene therapy for Parkinson disease. Neurology. 2008;70(21):1980–1983. doi: 10.1212/01.wnl.0000312381.29287.ff. [DOI] [PubMed] [Google Scholar]
- 105. Christine CW, et al. Safety and tolerability of putaminal AADC gene therapy for Parkinson disease. Neurology. 2009;73(20):1662–1669. doi: 10.1212/WNL.0b013e3181c29356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. McCarty DM. Self-complementary AAV vectors; advances and applications. Mol Ther. 2008;16(10):1648–1656. doi: 10.1038/mt.2008.171. [DOI] [PubMed] [Google Scholar]
- 107. Panov AV, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5(8):731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 108. Kim M, et al. Mutant huntingtin expression in clonal striatal cells: dissociation of inclusion formation and neuronal survival by caspase inhibition. J Neurosci. 1999;19(3):964–973. doi: 10.1523/JNEUROSCI.19-03-00964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 110. Hockly E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci U S A. 2003;100(4):2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 112. Hu J, Liu J, Corey DR. Allele-Selective Inhibition of Huntingtin Expression by Switching to an miRNA-like RNAi Mechanism. Chem Biol. 2010;17(11):1183–1188. doi: 10.1016/j.chembiol.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Henry S, Stecker K, Brooks D, Monteith D, Conklin B, Bennett CF. Chemically modified oligonucleotides exhibit decreased immune stimulation in mice. J Pharmacol Exp Ther. 2000;292(2):468–479. [PubMed] [Google Scholar]
- 114. John M, et al. Effective RNAi-mediated gene silencing without interruption of the endogenous microRNA pathway. Nature. 2007;449(7163):745–747. doi: 10.1038/nature06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kaiser PK, et al. RNAi-based treatment for neovascular age-related macular degeneration by Sirna-027. Am J Ophthalmol. 2010;150(1):33–39 e32. doi: 10.1016/j.ajo.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 116. DeVincenzo J, et al. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc Natl Acad Sci U S A. 2010;107(19):8800–8805. doi: 10.1073/pnas.0912186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Davis ME, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wyszko E, et al. A multivariate analysis of patients with brain tumors treated with ATN-RNA. Acta Pol Pharm. 2008;65(6):677–684. [PubMed] [Google Scholar]
- 119. Akdim F, et al. Effect of mipomersen, an apolipoprotein B synthesis inhibitor, on low-density lipoprotein cholesterol in patients with familial hypercholesterolemia. Am J Cardiol. 2010;105(10):1413–1419. doi: 10.1016/j.amjcard.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 120. Hau P, Jachimczak P, Bogdahn U. Treatment of malignant gliomas with TGF-beta2 antisense oligonucleotides. Expert Rev Anticancer Ther. 2009;9(11):1663–1674. doi: 10.1586/era.09.138. [DOI] [PubMed] [Google Scholar]
- 121. Leighl NB, et al. A phase I/II study of GTI-2040 plus docetaxel as second-line treatment in advanced non-small cell lung cancer: a study of the PMH phase II consortium. J Thorac Oncol. 2009;4(9):1163–1169. doi: 10.1097/JTO.0b013e3181a949b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Fidias P, et al. Phase I study of the c-raf-1 antisense oligonucleotide ISIS 5132 in combination with carboplatin and paclitaxel in patients with previously untreated, advanced non-small cell lung cancer. J Thorac Oncol. 2009;4(9):1156–1162. doi: 10.1097/JTO.0b013e3181b2793f. [DOI] [PubMed] [Google Scholar]
- 123. Schimmer AD, et al. Phase I/II trial of AEG35156 X-linked inhibitor of apoptosis protein antisense oligonucleotide combined with idarubicin and cytarabine in patients with relapsed or primary refractory acute myeloid leukemia. J Clin Oncol. 2009;27(28):4741–4746. doi: 10.1200/JCO.2009.21.8172. [DOI] [PMC free article] [PubMed] [Google Scholar]