Abstract
Huntington disease (HD) is a dominantly inherited neurodegenerative disorder that results from expansion of the polyglutamine repeat in the huntingtin (HTT) gene. There are currently no effective treatments for this devastating disease. Given its monogenic nature, disease modification therapies for HD should be theoretically feasible. Currently, pharmacological therapies aimed at disease modification by altering levels of HTT protein are in late-stage preclinical development. Here, we review current efforts to develop new treatments for HD based on our current understanding of HTT function and the main pathological mechanisms. We emphasize the need to enhance translational efforts and highlight the importance of aligning the clinical and basic research communities to validate existing hypotheses in clinical studies. Human and animal therapeutic trials are presented with an emphasis on cellular and molecular mechanisms relevant to disease progression.
Introduction
The broad spectrum of neurodegenerative diseases (NDDs) is characterized by the selective death of specific neuronal populations. Identification of the genes that cause the inherited forms of these diseases has led to a greater understanding of pathogenic mechanisms. Among the most common NDDs, the inherited forms are only a small subset of all cases, notable exceptions being spinal muscular atrophy, the spinocerebellar ataxias, and Huntington disease (HD). For instance, mutations in superoxide dismutase-1 in amyotrophic lateral sclerosis, of α-synuclein or the leucine-rich repeat kinase-2 in Parkinson disease (PD), or of the amyloid precursor protein in Alzheimer disease (AD) account for just 1%–5% of all cases in the general population (1–3). By contrast, although the prevalence of HD (5–10 per 100,000) (2) is much lower than for PD or AD, the complete penetrance of the HD mutation makes this one of the most common inherited NDDs. HD is unique in that allele carriers can be identified prior to the development of clinically meaningful symptoms, making it a model for the development of disease-modifying therapies with the potential to influence similar strategies — from scientific and regulatory perspectives — for other NDDs with more heterogeneous etiologies. Given the monogenic nature of HD, its prevalence and penetrance, and the existence of worldwide clinical networks ( http://www.euro-hd.net), we stress that HD is a disease for which this ambitious goal might be achieved.
HD is an autosomal dominant disease exclusively caused by the expansion of a CAG repeat in the huntingtin (HTT) gene, which encodes a stretch of polyglutamines at the amino terminus (4). Expansion length (>35 CAGs) is negatively correlated with age of onset of clinical symptoms and accounts for 60%–70% of the variation (5). Clinically, HD is characterized by motor, cognitive, and psychiatric disturbances. These include deficits in movement control (chorea, dyskinesias), impairments in executive function, working memory, attention, impulsivity, loss of motivation and self care, emotional lability, and a high incidence of depressive disorders (6–8).
Traditionally, therapeutic approaches to HD have included compounds developed for psychiatric indications based on the affected neuronal circuitry: the frontal and motor corticostriatal circuits (9, 10). None of these were initially developed for the treatment of HD. In this review we focus on the cellular and biological pathways affected by mutant HTT (mHTT) and the current status of associated drug discovery efforts (Figure 1). We also emphasize the need for further clinical research to validate existing hypotheses, which are mostly derived from animal studies and postmortem human tissues. It is generally accepted that most candidate therapeutics fail due to lack of efficacy in pivotal clinical studies. Leaving aside issues arising from inadequate clinical rating scales or trial design flaws, a simple explanation for this failure is that the pathogenic mechanistic hypotheses developed for a given indication, or the chosen intervention points within those mechanisms, the “targets”, are incorrect. The critical question for both the basic and clinical research communities is how we can work together more effectively to better define targets to maximize success. In this context, success is defined as developing therapies to slow the progression of HD, leading to significantly improved quality of life and extended functional lifespan. Although an ambitious goal, a disease such as HD represents a unique opportunity in which true disease modification should be attainable.
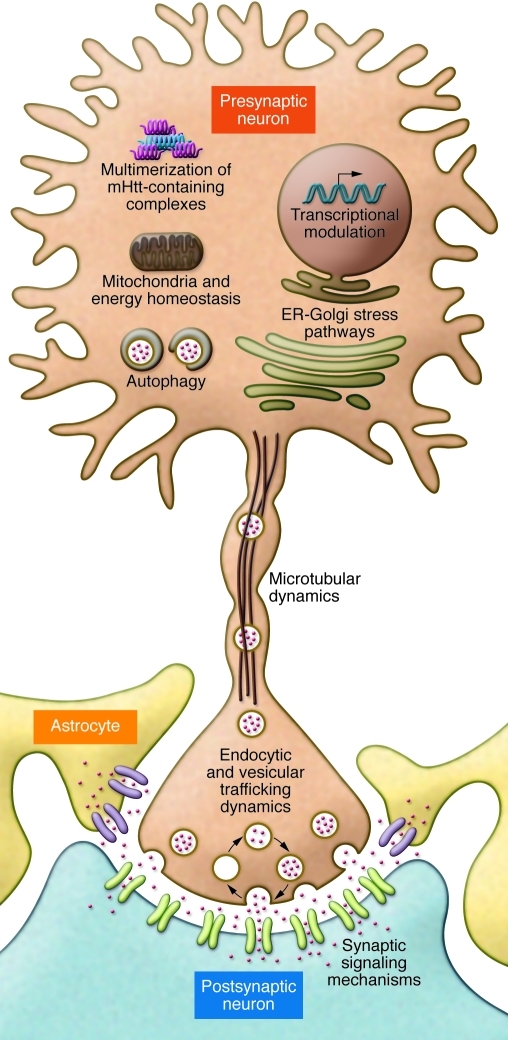
Figure 1. Cellular mechanisms implicated in HD pathogenesis.
The major mechanisms associated with HD pathogenesis are depicted here. The schematic shows a presynaptic neuron and a postsynaptic neuron flanked by two astrocytes. HTT itself is depicted as a “solenoid,” based on the presumed folding due to its HEAT repeats. The mechanisms depicted are multimerization of mHtt-containing complexes, transcriptional modulation, ER-Golgi stress pathways, mitochondria and energy homeostasis, microtubular dynamics, endocytic and vesicular trafficking dynamics, autophagy, and synaptic signaling mechanisms. mHTT, mutant HTT protein.
HD is characterized by the progressive degeneration of a subset of neurons in the corpus striatum, populations of cortical pyramidal neurons in the motor, frontal, and occipital cortices (8, 11–14), as well as neurons in other brain regions such as the hypothalamus (15). Current clinical diagnosis usually occurs in mid-life and is generally defined by the onset of motor symptoms. Intracellular inclusions of nuclear or neuropil HTT also contain other ubiquitinated proteins (4, 16). Many of these neurological and neuropathological features are, perhaps surprisingly, associated with other NDDs of different molecular etiology. Traditionally, NDDs have been defined by the cardinal symptoms that arise from the affected circuitry; for instance, the executive, attention, and planning deficits manifest in HD can be linked to dysfunction of frontostriatal circuits (8). Since all HD patients share the same mutation, a treatment aimed at a mechanism proximal to HTT might benefit all patients; this is in contrast to AD or PD, in which only a minority of cases arise from well-known molecular alterations.
Identifying mutations causative for a given disease enables the development of genetic animal models; there are now many rodent models of HD, and sheep and primate models have been engineered more recently (17–20). The R6/2 mouse is the most widely used and expresses an N-terminal fragment of the HTT gene under the control of the human HTT promoter (21). The finding that a fragment of HTT was sufficient to cause HD-like symptoms, and that the progression was faster than in mice expressing full-length mHTT, supported the toxic fragment hypothesis (22). This theory postulates that the cleavage of HTT into N-terminal fragments is an early causative event in HD pathogenesis. Disease progression in the R6/2 mouse is rapid and recapitulates some of the pathological findings in postmortem HD tissues, including inclusion formation, some striatal and cortical neuronal death, ventricular enlargement, widespread white matter atrophy, and similar patterns of transcriptional dysregulation (21, 23–28). Other full-length models of HD include knockin mouse models (19, 26) and transgenic YAC and BAC mice and rats (20, 29, 30). These differ in mHTT expression levels, length of the CAG repeat, age of phenotype onset, rate of disease progression, extent of neuronal death, and the robustness of behavioral (cognitive, psychiatric, and motor) disturbances. Most of the mechanistic hypotheses driving the field have been identified or explored within these rodent models.
Although molecular changes observed in HD seem to be well conserved (26, 31–33), relatively minimal neuronal death occurs in rodents. Also, because frontal cortex anatomy is vastly different from rodents to primates, these models will likely only recapitulate some aspects of HD (34). Ideally, the clinical relevance of a particular intervention would be ascertained as rapidly as possible. In this regard, the main challenge in designing observational or exploratory interventional clinical studies is to gain insight into the exact nature of the deficits within complex biological mechanisms (in humans), which would support specific targets amenable to pharmacological intervention. It might only be possible to achieve this by “stressing” the system in a clinical context in order to uncover a statistically significant effect. For instance, an evaluation of energetic homeostatic responses (through direct measurements in muscle tissue) after an exercise stress paradigm might be necessary to uncover robust changes in energetic endpoints. To identify selective deficits that can be targeted therapeutically, an analysis of specific molecular alterations might only be possible through the use of peripheral tissues also affected in HD (35). Finally, to understand functional alterations in synaptic networks, or the involvement of specific neurotransmitter pathways, stressors might be applied to uncover these deficits prior to overt clinical symptoms (36, 37). Clinically available drugs such as sub-anesthetic doses of ketamine to probe the NMDA receptor system might be used to investigate the effects in cognition in HD patients. These specific approaches, coupled with imaging technologies, can be informative of specific alterations in HD.
In developing disease-modifying strategies, it is important to understand the link between initial pathogenesis related to mHTT function and compensatory mechanisms that develop over the extended disease course. For this reason, the importance of conducting longitudinal studies in pre-manifest individuals cannot be overemphasized. Most published clinical studies involve manifest HD patients (who may be on multiple psychiatric medications), are cross-sectional, and typically have a sample population that is too small to draw significant conclusions (see Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI45364DS1). The continued support of physicians and individuals at risk is required to better understand the emergence of early HD-related changes and their correlation with onset and progression of clinically relevant symptoms. To achieve this, two studies — PREDICT-HD and TRACK-HD (6, 7, 38, 39) — are evaluating disease symptom progression in important clinical domains, as well as circuitry changes at and prior to clinical diagnosis. Similarly, developing optimal symptomatic therapies will also require an understanding of the heterogeneity in the manifestation and timing of symptoms.
Existing animal and clinical studies with an emphasis on mechanisms
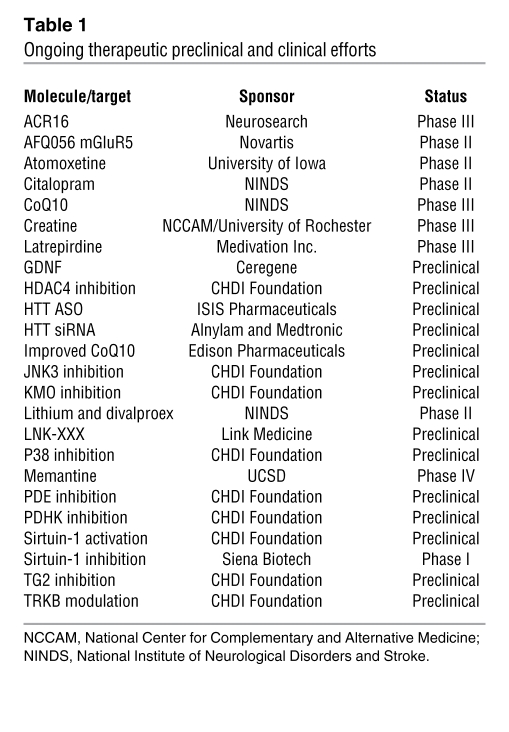
Ongoing and completed HD therapeutic clinical trials (Table 1, Supplemental Table 1, and refs. 9, 10, 40) have largely focused on the mechanistic areas of synaptic transmission and energy homeostasis. A gene delivery tolerability study has been conducted with ciliary neurotrophic factor (41), minocycline was used to inhibit caspase-1 and modafinil was studied for its potential effects in cognition and alertness (42). The Cochrane Collaboration has systematically reviewed therapeutic intervention trials for both symptomatic treatments (10) and disease progression (40) in HD. Many of the symptomatic treatment trials included few patients, and the primary outcome measure was total functional capacity and/or motor performance. Tetrabenazine is the only symptomatic treatment that has shown efficacy in reducing chorea in ambulatory HD patients (10) and has since been approved for clinical use. Most HD symptoms are currently treated ineffectively or not at all, and therefore this is an important area of clinical research. Ongoing symptomatic trials include a metabotropic glutamate receptor-5 (mGluR5) inhibitor (Table 1 and ref. 43) and latrepirdine (44). However, the development of disease-modifying treatments is the primary focus of HD therapeutic research and of this review. To date, no disease-modifying clinical efficacy trials have demonstrated treatment efficacy (40). A major limitation is that clinical assessment tools used as outcome measures lack sensitivity, meaning that the statistical power to detect improvement is poor even when hundreds of patients are tracked over two or three years. The generation and validation of improved assessment measures is a major focus of the European Huntington’s Disease Network and of the PREDICT-HD and TRACK-HD studies.
Table 1 .
Ongoing therapeutic preclinical and clinical efforts
The discovery of the disease-causing mutation in HTT and the development of rodent models facilitated the investigation into potential pathogenic mechanisms through genetic manipulation as well as pharmacologic or molecular intervention (Supplemental Tables 2 and 3). Some notable recent pharmacological and molecular approaches include modulation of adenosine signaling (45), histone deacetylase (HDAC) inhibition (refs. 46 and 47 and Supplemental Table 3), phosphodiesterases (PDEs; refs. 48 and 49), kinase modulation (50, 51), CB2 cannabinoid receptor (52), and the growth factor pathways modulated by glial cell derived neurotrophic factor (GDNF), neurturin, and brain derived neurotrophic factor (BDNF) (53–57). Targets that have proved efficacious in genetic crosses notably include the involvement of caspases (22), HDAC4 (58), sirtuins (59), BDNF (54), p53 (60–62), and transglutaminase 2 (63, 64). In addition, deleterious effects associated with the genetic reduction of specific targets has highlighted the potential involvement of some mechanisms in the pathology of HD, notably CB2 (52), CREB1 (65, 66), mGluR-2 and -5 (43), Hsp70 (67), and PGC1α (68).
Pathogenic mechanisms in HD and current approaches to intervention
HTT as a therapeutic target.
The most expeditious way to modify the course of HD would be to prevent the expression or function of mHTT itself. Currently, mHTT is not a target for traditional pharmacologic modulation, since it has complex functions that remain incompletely understood. However, approaches to decrease HTT expression are in late preclinical development (Table 1 and ref. 58). One approach uses antisense oligonucleotides (ASOs) that function in vivo through an RNase-H mechanism to degrade the mRNAs of both alleles of HTT and is delivered through infusion into the ventricular or intrathecal spaces. The other uses siRNA therapeutics targeting both alleles via intraparenchymal administration. Both strategies aim to decrease the WT and mutant alleles of HTT and therefore share similar on-target mediated toxicity challenges. Significant loss of WT HTT expression is known to be detrimental both in developmental and adult contexts (69, 70). It is noteworthy that allele-specific therapies are being developed to mitigate on-target effects due to excessive lowering of WT HTT protein.
Currently, rodents are being used to determine the therapeutic window between efficacy due to decreased mHTT and adverse effects that might be triggered by insufficient WT HTT. There might be significant challenges associated with both approaches, and given their different mechanisms of action, modality-dependent side effects might differ. Perhaps the most significant challenge is identifying markers sensitive to the reduction of HTT in the brain that could assess whether an adequate dose has been achieved within the predicted therapeutic window. The propensity of mHTT to oligomerize could possibly be used to assess dosage effects on the formation of these aggregates in vivo. In AD, imaging tools can now visualize plaque load and various species of TAU protein and Aβ peptides can be detected in rodent and human CSF (71), an approach that would be invaluable for the clinical development of HTT-lowering therapies. While HTT is an intracellular protein (like TAU protein), it might accumulate in CSF due to neuronal loss, a possibility that will be investigated once sensitive assays are optimized.
Transcriptional dysregulation is robustly correlated between HD animal models and human postmortem samples, suggesting a central role of mutant HTT in these molecular changes. Genes encoding neurotransmitter receptors are downregulated early in disease, including dopamine (D1 and D2; refs. 72 and 73), adenosine (A2a; ref. 74), and cannabinoid (CB1; ref. 75) receptors for which existing imaging tracers could potentially act as indirect markers of mHTT function in clinical studies. Energetic alterations in patients and animal models of HD (76–78) may be of relevance since energetic endpoints can be monitored non-invasively in vivo through imaging or MRS techniques (79, 80). Finally, it will be vital to identify degeneration-relevant markers, such as MSN or cortically expressed proteins found in CSF, which could be used to track degeneration longitudinally. Animal models will be invaluable in determining which measures are sensitive to decreased HTT levels (and which are reversible after loss of mHTT), that might guide clinical development.
The PREDICT-HD and TRACK-HD studies are evaluating longitudinal changes in premanifest HD and individuals with early-stage disease. Previous studies by Tabrizi et al. have identified robust cross-sectional changes (39), and currently this group is examining parameters that may be sensitive enough to track disease progression over a short time span (1–3 years). Such markers sufficiently sensitive for Phase II studies would warrant further investment for disease-modification trials. Given the widespread degeneration observed in the basal ganglia and cortical areas in manifest and advanced HD (12, 14, 38, 39, 79, 81–83), it is plausible that non-invasive techniques such as quantitative EEG (qEEG) could gauge progression. However, validating these approaches together with assessing sensitive tasks in functions important for quality of life of HD patients remains an important area of investigation for disease modification therapies.
HTT aggregation and protein homeostasis.
In all NDDs, seemingly soluble proteins are mutated and form a multitude of oligomeric species and intracellular inclusions (16). The processes governing oligomerization and the mechanisms by which they cause cellular dysfunction are fundamental areas of investigation. Any strategy to rebalance the equilibrium of this process is a potential therapeutic approach. One possibility is to manipulate the cellular mechanisms that ensure correct protein folding (16, 84) or eliminate misfolded proteins: the ubiquitin proteasome system (85) and autophagy (84, 86–88). Autophagy induction can decrease aggregate load in various neurodegeneration models, including HD (88). To date, the main pharmacologic approach in clinical development to directly enhance autophagy is the inhibition of farnesyl transferase, a protein responsible for the farnesylation (a lipid modification) of a number of substrate proteins and implicated in autophagy regulation (Table 1 and ref. 89). However, recent evidence from a knockin mouse HD model suggests that HD-specific alterations in autophagy might lead to a block in the trafficking or degradation of HTT (86). This could have implications for the exact therapeutic approach (that is, for which step in the autophagy cascade to target) and the disease stage at which a therapeutic intervention might be effective. For instance, as this block in the degradation of HTT exists in HD-derived lymphoblasts, we could use these cells in the development of autophagy-directed therapeutics. The main challenge for this area is to understand whether peripheral autophagy mechanisms are predictive of central modulation of autophagy, and to bypass the known adverse effects associated with chronic peripheral inhibition of mTOR signaling, such as ulcerative mucositis, anemia, and neutropenia, among others (90).
Energetics.
Mitochondrial dysfunction is implicated in most CNS disorders, and energetic disturbances in HD are well documented (27, 68, 76, 78, 91–93). The absence of mutations in the mitochondrial genome suggests indirect effects of mHTT on mitochondrial integrity (assuming nuclearly-encoded mitochondrial genes are not affected specifically in HD). Abnormalities in the electron transport chain and the glycolytic machinery have been reported, but few define the precise lesion(s) that would suggest therapeutic strategies (93, 94). Many clinical trials have attempted to alleviate mitochondrial dysfunction (Supplemental Tables 2 and 3) using a variety of anti-oxidants and energetic supplements such as ethyl-EPA, idebenone (coenzyme Q10 [CoQ10]), or creatine without much success. These compounds suffer from poor pharmacokinetic properties or unclear correlation between brain exposure levels and their biological effects. A more potent CoQ10 analog with improved tissue distribution is being developed to treat mitochondrial myopathies and HD (Table 1 and ref. 58). A comprehensive longitudinal investigation of central and peripheral mitochondrial and glycolytic function in HD patients is required to define the relationship between peripheral energetic changes and central and peripheral mechanisms (35, 78). Recently, modulation of sirtuin and its downstream targets — the transcription factors PGC1α and PPR1γ (76, 95–98, and Supplemental Tables 1 and 2) — has been shown to modulate the expression of genes important in mitochondrial function; the relevance for HD is supported by the association of PGC1α polymorphisms with age of onset (99). Both SIRT1 and PPR1γ appear tractable as therapeutic targets and, therefore, as validation of this mechanistic hypothesis.
Transcriptional changes.
Transcriptional dysregulation has been extensively documented as a pathogenic mechanism in HD. The transcriptional changes that occur are robust and highly conserved between rodent models and HD postmortem brain (33). However, dysregulated transcriptional signatures have not been studied longitudinally in humans, and whether these can track disease progression (at least peripherally) is unclear. Altered expression of specific neurotransmitter receptors can be tracked in human imaging studies (73–75, 100) and likely influences the excitability of vulnerable neurons, rendering them susceptible to deregulated calcium signaling, leading to cell death. The role of mHTT protein in transcriptional processes modulated by Sp1, p53, REST/NSRF, and CREB is well documented (31, 32, 61, 62, 65, 66, 101–105). A feasible strategy to modulate CREB signaling in the brain through the modulation of PDEs is in place. Rolipram, a PDE4 inhibitor, has been shown to modify some of the symptoms in HD models (striatal death, survival, motor deficits; Supplemental Table 2 and refs. 48, 49), and other selective PDE inhibitors such as PDE10 are being investigated (106). Similarly, the beneficial effects of non-selective HDAC inhibitors such as SAHA (46, 47, 107–109) prompted the genetic investigation of individual HDACs in the R6/2 mouse (Supplemental Table 3). Based on these findings, class II HDAC-selective inhibitors are in preclinical development. The Sirt proteins regulate many pathways that are significant in HD pathogenesis, and both activation and inhibition of SIRT1 has been reported to be beneficial in HD models. Resveratrol improved peripheral glucose levels but did not affect survival or striatal pathology in HD mice (Supplemental Table 3; refs. 58, 59). This target requires further investigation with selective brain-penetrant compounds.
Synaptic biology.
Circuitry changes (neuronal death, white matter alterations, retraction of processes, and synaptic dysfunction) directly underlie alterations in symptomatic functional domains. The release of GABA by MSNs and their vulnerability in HD led to the initial investigation of GABAergic agents to treat HD, although these therapies proved ineffective (see references in Supplemental Table 1). Other neurotransmitters that have been investigated include glutamate (the major afferent transmitter modulating the firing of the MSNs), acetylcholine, and dopamine (the basal ganglia being the major target of substantia nigra projection neurons). Dopamine agonism has been shown in animal models to be detrimental to HD rodent models, whereas D2 antagonism is associated with improved motor performance in patients (9, 72, 110–113). The only approved drug for HD is tetrabenazine, a vesicular monoamine transporter-2 (VMAT2) inhibitor that lowers extracellular dopamine and norepinephrine (9, 10, 113). However, despite beneficial effects on chorea and motor subscores, tetrabenazine fails to improve the cognitive and psychiatric deficits, or to slow disease progression. Cholinergic modulation with galantamine has been shown to have potential beneficial effects (114) but a larger clinical trial to demonstrate efficacy has not been conducted.
The major hypothesis driving HD synaptic research is that of the excitability of MSNs. In this regard, approaches to reduce extrasynaptic glutamate signaling have been explored and include modulation of NR2B signaling (115), lowering glutamate receptor activation with NMDA receptor antagonists (ketamine and memantine; Supplemental Table 1), modulating the interplay of glutamate and dopamine on MSNs (72), and recently with mGluR5 antagonists (43). Currently there is conflicting evidence to support the inhibition of glutamate receptors as a disease-modifying strategy in HD (10, 21, 43, 96, 110, 116–118). Despite potential elevated glutamate signaling early in the disease course in rodent models, extensive deafferentation occurs at later stages, decreasing cortical and thalamic input to the basal ganglia (80, 119) and affecting regulated MSN firing. Biphasic changes in glutamate and dopamine transmission may explain why decreasing extrasynaptic signaling via NR2B appears effective in the YAC128 (a mouse model expressing human mHTT with 128 CAGs; refs. 29, 110), but not in the faster-developing R6/2 HD model (120, 121). The re-uptake of synaptically released glutamate by astrocytes occurs through the EAAT2 transporter, a target that is downregulated in HD (refs. 122, 123, and Figure 1). This downregulation leads to enhanced extracellular glutamate; the pharmacologic upregulation of EAAT2 is currently being explored preclinically. As in PD, electrical modulation of the output nuclei (in PD, the subthalamic nucleus; in HD, the globus pallidus) might confer significant motor relief, a hypothesis currently being tested clinically (124, 125).
Insights into neurotransmitter alterations in HD individuals have been gained through imaging studies that provide a static and nonfunctional window into disease pathophysiology. Other techniques — such as qEEG, electrical stimulation, and fMRI monitoring of activity changes during functional tasks compromised in HD — have only recently been explored and, so far, only cross-sectionally (36, 80, 119). The most challenging goal is to understand system-wide changes in neural connectivity and the responsiveness of the affected circuitry to specific stimulations. The evaluation of selective agents aimed at neurotransmitter signaling components which control the excitability of affected neuronal populations in HD is needed to assess their potential effectiveness as symptomatic treatments. For instance, the identification of the earliest molecular mechanisms which contribute to the enhanced excitability of indirect pathway neurons will be critical to define novel intervention strategies. This should involve an understanding of firing properties of cholinergic and fast-spiking interneurons in an HD context, as well as a detailed investigation of membrane conductance alterations during disease progression. A greater emphasis on pallidal and subthalamic activity will be an important area to explore pharmacologically, as the loss of the MSNs has a significant effect in the activity of these output nuclei. Whether symptomatic agents will also modify disease progression is hard to predict (and therefore should be pursued) in the absence of a better understanding how mHTT regulates the synaptic properties of vulnerable neurons.
Concluding remarks
Fundamental change is required in the clinical exploration of HD biology in humans. Rather than cross-sectional alterations, an understanding of changes over time — from pre-manifest to early manifest disease — that includes investigation of disease-specific molecular alterations is essential. In order to uncover early changes, experimental medicine and interventional trials that stress a given cellular mechanism might be useful. Most pathogenic mechanism hypotheses are developed from animal models that are amenable to experimental or genetic manipulation; clinical researchers will have to devise experimental, non-invasive approaches that can query specific mechanisms and targets in humans to either validate or invalidate these hypotheses. This must not involve multi-year trials that recruit hundreds of patients, the most precious asset in a rare disorder.
Perhaps the most critical component of observational studies will be the standardization of best practice to ensure that small-sample studies can be meaningfully compared. This is particularly true of biofluid analyses (e.g., plasma and CSF) for which collection, shipment, and storage practices must be standardized to ensure high-quality data. In addition, an understanding of the longitudinal change in particular parameters will be critical to validate the role of specific mechanisms in disease progression, and possibly in patient selection for therapeutic trials. The reviews in this series explore in greater detail the recent advances in understanding of the synaptic changes and energetic dysfunction characteristic of HD (126, 127), as well as the development of oligonucleotide strategies for HTT reduction (128).
To conclude, the wider medical community should know that significant advances have been made in understanding the etiology of HD and in approaches to its treatment. Current efforts toward disease modification are at least as advanced as for any other neurological indication. The hope that effective treatments will be developed is realistic, and this message needs to be communicated to the patient community to encourage enrollment in clinical studies.
Supplementary Material
Acknowledgments
The authors would like to thank Robi Blumenstein and Simon Noble for comments on the manuscript and Michael Palazzolo and James Wang for help in compiling Supplemental Tables 1–3.
Footnotes
Conflict of interest: I. Munoz-Sanjuan is an employee of CHDI, which funds research into HD therapies.
Citation for this article: J Clin Invest. 2011;121(2):476–483. doi:10.1172/JCI45364.
References
- 1. Bertram L, Tanzi RE. The genetic epidemiology of neurodegenerative disease. J Clin Invest. 2005;115(6):1449–1457. doi: 10.1172/JCI24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Spinney L. Uncovering the true prevalence of Huntington’s disease. Lancet Neurol. 2010;9(8):760–761. doi: 10.1016/S1474-4422(10)70160-5. [DOI] [PubMed] [Google Scholar]
- 3. Farrer MJ. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat Rev Genet. 2006;7(4):306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 4. Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90(3):905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 5. Langbehn DR, Hayden MR, Paulsen JS, PREDICT-HD Investigators of the Huntington Study Group CAG-repeat length and the age of onset in Huntington disease (HD): a review and validation study of statistical approaches. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):397–408. doi: 10.1002/ajmg.b.30992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Biglan KM, et al. Motor abnormalities in premanifest persons with Huntington’s disease: the PREDICT-HD study. Mov Disord. 2009;24(12):1763–1772. doi: 10.1002/mds.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duff K, Paulsen JS, Beglinger LJ, Langbehn DR, Stout JC. Psychiatric symptoms in Huntington’s disease before diagnosis: the predict-HD study. Biol Psychiatry. 2007;62(12):1341–1346. doi: 10.1016/j.biopsych.2006.11.034. [DOI] [PubMed] [Google Scholar]
- 8. Duff K, et al. “Frontal” behaviors before the diagnosis of Huntington’s disease and their relationship to markers of disease progression: evidence of early lack of awareness. J Neuropsychiatry Clin Neurosci. 2010;22(2):196–207. doi: 10.1176/appi.neuropsych.22.2.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frank S, Jankovic J. Advances in the pharmacological management of Huntington’s disease. Drugs. 2010;70(5):561–571. doi: 10.2165/11534430-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 10. Mestre T, Ferreira J, Coelho MM, Rosa M, Sampaio C. Therapeutic interventions for symptomatic treatment in Huntington’s disease. Cochrane Database Syst Rev. 2009;2009(3):CD006456. doi: 10.1002/14651858.CD006456.pub2. [DOI] [PubMed] [Google Scholar]
- 11. Paulsen JS, et al. Striatal and white matter predictors of estimated diagnosis for Huntington disease. Brain Res Bull. 2010;82(3–4):201–207. doi: 10.1016/j.brainresbull.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pavese N, et al. Cortical dopamine dysfunction in symptomatic and premanifest Huntington’s disease gene carriers. Neurobiol Dis. 2010;37(2):356–361. doi: 10.1016/j.nbd.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 13. Schippling S, et al. Abnormal motor cortex excitability in preclinical and very early Huntington’s disease. Biol Psychiatry. 2009;65(11):959–965. doi: 10.1016/j.biopsych.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thu DC, et al. Cell loss in the motor and cingulate cortex correlates with symptomatology in Huntington’s disease. Brain. 2010;133(pt 4):1094–1110. doi: 10.1093/brain/awq047. [DOI] [PubMed] [Google Scholar]
- 15. Hult S, Schultz K, Soylu R, Petersen A. Hypothalamic and neuroendocrine changes in Huntington’s disease. Curr Drug Targets. 2010;11(10):1237–1249. doi: 10.2174/1389450111007011237. [DOI] [PubMed] [Google Scholar]
- 16. Truant R, Atwal RS, Desmond C, Munsie L, Tran T. Huntington’s disease: revisiting the aggregation hypothesis in polyglutamine neurodegenerative diseases. FEBS J. 2008;275(17):4252–4262. doi: 10.1111/j.1742-4658.2008.06561.x. [DOI] [PubMed] [Google Scholar]
- 17. Jacobsen JC, et al. An ovine transgenic Huntington’s disease model. Hum Mol Genet. 2010;19(10):1873–1882. doi: 10.1093/hmg/ddq063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Graham RK, et al. Levels of mutant huntingtin influence the phenotypic severity of Huntington disease in YAC128 mouse models. Neurobiol Dis. 2006;21(2):444–455. doi: 10.1016/j.nbd.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 19. Heng MY, Detloff PJ, Albin RL. Rodent genetic models of Huntington disease. Neurobiol Dis. 2008;32(1):1–9. doi: 10.1016/j.nbd.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 20. von Horsten S, et al. Transgenic rat model of Huntington’s disease. Hum Mol Genet. 2003;12(6):617–624. doi: 10.1093/hmg/12.6.617. [DOI] [PubMed] [Google Scholar]
- 21. Cha JH, et al. Altered brain neurotransmitter receptors in transgenic mice expressing a portion of an abnormal human huntington disease gene. Proc Natl Acad Sci U S A. 1998;95(11):6480–6485. doi: 10.1073/pnas.95.11.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Graham RK, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125(6):1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 23. Bjorkqvist M, et al. Progressive alterations in the hypothalamic-pituitary-adrenal axis in the R6/2 transgenic mouse model of Huntington’s disease. Hum Mol Genet. 2006;15(10):1713–1721. doi: 10.1093/hmg/ddl094. [DOI] [PubMed] [Google Scholar]
- 24. Johnson MA, Rajan V, Miller CE, Wightman RM. Dopamine release is severely compromised in the R6/2 mouse model of Huntington’s disease. J Neurochem. 2006;97(3):737–746. doi: 10.1111/j.1471-4159.2006.03762.x. [DOI] [PubMed] [Google Scholar]
- 25. Luthi-Carter R, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington’s disease. Hum Mol Genet. 2000;9(9):1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 26. Strand AD, et al. Expression profiling of Huntington’s disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J Neurosci. 2007;27(43):11758–11768. doi: 10.1523/JNEUROSCI.2461-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tabrizi SJ, et al. Mitochondrial dysfunction and free radical damage in the Huntington R6/2 transgenic mouse. Ann Neurol. 2000;47(1):80–86. doi: 10.1002/1531-8249(200001)47:1<80::AID-ANA13>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 28. Tsang TM, et al. Metabolic characterization of the R6/2 transgenic mouse model of Huntington’s disease by high-resolution MAS 1H NMR spectroscopy. J Proteome Res. 2006;5(3):483–492. doi: 10.1021/pr050244o. [DOI] [PubMed] [Google Scholar]
- 29. Slow EJ, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12(13):1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 30. Gray M, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28(24):6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buckley NJ, Johnson R, Zuccato C, Bithell A, Cattaneo E. The role of REST in transcriptional and epigenetic dysregulation in Huntington’s disease. Neurobiol Dis. 2010;39(1):28–39. doi: 10.1016/j.nbd.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 32. Zuccato C, et al. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington’s disease. J Neurosci. 2007;27(26):6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Runne H, et al. Dysregulation of gene expression in primary neuron models of Huntington’s disease shows that polyglutamine-related effects on the striatal transcriptome may not be dependent on brain circuitry. J Neurosci. 2008;28(39):9723–9731. doi: 10.1523/JNEUROSCI.3044-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yin D, et al. Striatal volume differences between non-human and human primates. J Neurosci Methods. 2009;176(2):200–205. doi: 10.1016/j.jneumeth.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sassone J, Colciago C, Cislaghi G, Silani V, Ciammola A. Huntington’s disease: the current state of research with peripheral tissues. Exp Neurol. 2009;219(2):385–397. doi: 10.1016/j.expneurol.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 36. Komssi S, Kahkonen S. The novelty value of the combined use of electroencephalography and transcranial magnetic stimulation for neuroscience research. Brain Res Rev. 2006;52(1):183–192. doi: 10.1016/j.brainresrev.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 37. Norra C. Challenge tests of monoaminergic systems: neurophysiological aspects. Clin EEG Neurosci. 2007;38(2):66–73. doi: 10.1177/155005940703800207. [DOI] [PubMed] [Google Scholar]
- 38. Paulsen JS, et al. Detection of Huntington’s disease decades before diagnosis: the Predict-HD study. . J Neurol Neurosurg Psychiatry. 2008;79(8):874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tabrizi SJ, et al. Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol. 2009;8(9):791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mestre T, Ferreira J, Coelho MM, Rosa M, Sampaio C. Therapeutic interventions for disease progression in Huntington’s disease. Cochrane Database Syst Rev. 2009;2009(3):CD006455. doi: 10.1002/14651858.CD006455.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bloch J, et al. Neuroprotective gene therapy for Huntington’s disease, using polymer-encapsulated cells engineered to secrete human ciliary neurotrophic factor: results of a phase I study. Hum Gene Ther. 2004;15(10):968–975. doi: 10.1089/hum.2004.15.968. [DOI] [PubMed] [Google Scholar]
- 42. Blackwell AD, Paterson NS, Barker RA, Robbins TW, Sahakian BJ. The effects of modafinil on mood and cognition in Huntington’s disease. Psychopharmacology (Berl). 2008. 199 1 29 36 . 10.1007/s00213-008-1068-0 [DOI] [PubMed] [Google Scholar]
- 43. Schiefer J, et al. The metabotropic glutamate receptor 5 antagonist MPEP and the mGluR2 agonist LY379268 modify disease progression in a transgenic mouse model of Huntington’s disease. Brain Res. 2004;1019(1–2):246–254. doi: 10.1016/j.brainres.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 44. Kieburtz K, et al. A randomized, placebo-controlled trial of latrepirdine in Huntington disease. Arch Neurol. 2010;67(2):154–160. doi: 10.1001/archneurol.2009.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chou SY, et al. CGS21680 attenuates symptoms of Huntington’s disease in a transgenic mouse model. J Neurochem. 2005;93(2):310–320. doi: 10.1111/j.1471-4159.2005.03029.x. [DOI] [PubMed] [Google Scholar]
- 46. Hockly E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci U S A. 2003;100(4):2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thomas EA, et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington’s disease transgenic mice. Proc Natl Acad Sci U S A. 2008;105(40):15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Giampa C, et al. Phosphodiesterase type IV inhibition prevents sequestration of CREB binding protein, protects striatal parvalbumin interneurons and rescues motor deficits in the R6/2 mouse model of Huntington’s disease. Eur J Neurosci. 2009;29(5):902–910. doi: 10.1111/j.1460-9568.2009.06649.x. [DOI] [PubMed] [Google Scholar]
- 49. DeMarch Z, Giampa C, Patassini S, Bernardi G, Fusco FR. Beneficial effects of rolipram in the R6/2 mouse model of Huntington’s disease. Neurobiol Dis. 2008;30(3):375–387. doi: 10.1016/j.nbd.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 50. Li M, Huang Y, Ma AA, Lin E, Diamond MI. Y-27632 improves rotarod performance and reduces huntingtin levels in R6/2 mice. Neurobiol Dis. 2009;36(3):413–420. doi: 10.1016/j.nbd.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 51. Apostol BL, et al. CEP-1347 reduces mutant huntingtin-associated neurotoxicity and restores BDNF levels in R6/2 mice. Mol Cell Neurosci. 2008;39(1):8–20. doi: 10.1016/j.mcn.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 52. Palazuelos J, et al. Microglial CB2 cannabinoid receptors are neuroprotective in Huntington’s disease excitotoxicity. Brain. 2009;132(pt 11):3152–3164. doi: 10.1093/brain/awp239. [DOI] [PubMed] [Google Scholar]
- 53. Canals JM, et al. Brain-derived neurotrophic factor regulates the onset and severity of motor dysfunction associated with enkephalinergic neuronal degeneration in Huntington’s disease. J Neurosci. 2004;24(35):7727–7739. doi: 10.1523/JNEUROSCI.1197-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gharami K, Xie Y, An JJ, Tonegawa S, Xu B. Brain-derived neurotrophic factor over-expression in the forebrain ameliorates Huntington’s disease phenotypes in mice. J Neurochem. 2008;105(2):369–379. doi: 10.1111/j.1471-4159.2007.05137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zuccato C, et al. Progressive loss of BDNF in a mouse model of Huntington’s disease and rescue by BDNF delivery. Pharmacol Res. 2005;52(2):133–139. doi: 10.1016/j.phrs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 56. Ebert AD, Barber AE, Heins BM, Svendsen CN. Ex vivo delivery of GDNF maintains motor function and prevents neuronal loss in a transgenic mouse model of Huntington’s disease. Exp Neurol. 2010;224(1):155–162. doi: 10.1016/j.expneurol.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 57. McBride JL, et al. Viral delivery of glial cell line-derived neurotrophic factor improves behavior and protects striatal neurons in a mouse model of Huntington’s disease. Proc Natl Acad Sci U S A. 2006;103(24):9345–9350. doi: 10.1073/pnas.0508875103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gagnon KT. HD Therapeutics - CHDI Fifth Annual Conference. IDrugs. 2010;13(4):219–223. [PubMed] [Google Scholar]
- 59. Ho DJ, Calingasan NY, Wille E, Dumont M, Beal MF. Resveratrol protects against peripheral deficits in a mouse model of Huntington’s disease. Exp Neurol. 2010;225(1):74–84. doi: 10.1016/j.expneurol.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 60. Ryan AB, Zeitlin SO, Scrable H. Genetic interaction between expanded murine Hdh alleles and p53 reveal deleterious effects of p53 on Huntington’s disease pathogenesis. Neurobiol Dis. 2006;24(2):419–427. doi: 10.1016/j.nbd.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Steffan JS, et al. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci U S A. 2000;97(12):6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bae BI, et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron. 2005;47(1):29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 63. McConoughey SJ, et al. Inhibition of transglutaminase 2 mitigates transcriptional dysregulation in models of Huntington disease. EMBO Mol Med. 2010;2(9):349–370. doi: 10.1002/emmm.201000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mastroberardino PG, et al. ‘Tissue’ transglutaminase ablation reduces neuronal death and prolongs survival in a mouse model of Huntington’s disease. Cell Death Differ. 2002;9(9):873–880. doi: 10.1038/sj.cdd.4401093. [DOI] [PubMed] [Google Scholar]
- 65. Jiang H, et al. Depletion of CBP is directly linked with cellular toxicity caused by mutant huntingtin. Neurobiol Dis. 2006;23(3):543–551. doi: 10.1016/j.nbd.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 66. Klevytska AM, Tebbenkamp AT, Savonenko AV, Borchelt DR. Partial depletion of CREB-binding protein reduces life expectancy in a mouse model of Huntington disease. J Neuropathol Exp Neurol. 2010;69(4):396–404. doi: 10.1097/NEN.0b013e3181d6c436. [DOI] [PubMed] [Google Scholar]
- 67. Hay DG, et al. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet. 2004;13(13):1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 68. Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127(1):59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 69. Auerbach W, et al. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum Mol Genet. 2001;10(22):2515–2523. doi: 10.1093/hmg/10.22.2515. [DOI] [PubMed] [Google Scholar]
- 70. Reiner A, Dragatsis I, Zeitlin S, Goldowitz D. Wild-type huntingtin plays a role in brain development and neuronal survival. Mol Neurobiol. 2003;28(3):259–276. doi: 10.1385/MN:28:3:259. [DOI] [PubMed] [Google Scholar]
- 71. Hampel H, et al. Biomarkers for Alzheimer’s disease: academic, industry and regulatory perspectives. Nat Rev Drug Discov. 2010;9(7):560–574. doi: 10.1038/nrd3115. [DOI] [PubMed] [Google Scholar]
- 72. Andre VM, Cepeda C, Levine MS. Dopamine and glutamate in Huntington’s disease: A balancing act. CNS Neurosci Ther. 2010;16(3):163–178. doi: 10.1111/j.1755-5949.2010.00134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Brandt J, et al. D2 receptors in Huntington’s disease: positron emission tomography findings and clinical correlates. J Neuropsychiatry Clin Neurosci. 1990;2(1):20–27. doi: 10.1176/jnp.2.1.20. [DOI] [PubMed] [Google Scholar]
- 74. Brooks DJ, et al. Positron emission tomography analysis of [11C]KW-6002 binding to human and rat adenosine A2A receptors in the brain. Synapse. 2008;62(9):671–681. doi: 10.1002/syn.20539. [DOI] [PubMed] [Google Scholar]
- 75. Van Laere K, et al. Widespread decrease of type 1 cannabinoid receptor availability in Huntington disease in vivo. J Nucl Med. 2010;51(9):1413–1417. doi: 10.2967/jnumed.110.077156. [DOI] [PubMed] [Google Scholar]
- 76. Chaturvedi RK, et al. Impaired PGC-1alpha function in muscle in Huntington’s disease. Hum Mol Genet. 2009;18(16):3048–3065. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Napolitano M, et al. Inhibition of mitochondrial complex II alters striatal expression of genes involved in glutamatergic and dopaminergic signaling: possible implications for Huntington’s disease. Neurobiol Dis. 2004;15(2):407–414. doi: 10.1016/j.nbd.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 78. Rosenstock TR, Duarte AI, Rego AC. Mitochondrial–associated metabolic changes and neurodegeneration in Huntington’s disease – from clinical features to the bench. Curr Drug Targets. 2010;11(10):1218–1236. doi: 10.2174/1389450111007011218. [DOI] [PubMed] [Google Scholar]
- 79. Reynolds NC, Jr, Prost RW, Mark LP. Heterogeneity in 1H-MRS profiles of presymptomatic and early manifest Huntington’s disease. Brain Res. 2005;1031(1):82–89. doi: 10.1016/j.brainres.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 80. Taylor-Robinson SD, et al. Proton magnetic resonance spectroscopy in Huntington’s disease: evidence in favour of the glutamate excitotoxic theory. Mov Disord. 1996;11(2):167–173. doi: 10.1002/mds.870110209. [DOI] [PubMed] [Google Scholar]
- 81. Hicks SL, Robert MP, Golding CV, Tabrizi SJ, Kennard C. Oculomotor deficits indicate the progression of Huntington’s disease. Prog Brain Res. 2008;171:555–558. doi: 10.1016/S0079-6123(08)00678-X. [DOI] [PubMed] [Google Scholar]
- 82. Ruocco HH, Bonilha L, Li LM, Lopes-Cendes I, Cendes F. Longitudinal analysis of regional grey matter loss in Huntington disease: effects of the length of the expanded CAG repeat. J Neurol Neurosurg Psychiatry. 2008;79(2):130–135. doi: 10.1136/jnnp.2007.116244. [DOI] [PubMed] [Google Scholar]
- 83. Sanchez-Pernaute R, Garcia-Segura JM, del Barrio Alba A, Viano J, de Yebenes JG. Clinical correlation of striatal 1H MRS changes in Huntington’s disease. Neurology. 1999;53(4):806–812. doi: 10.1212/wnl.53.4.806. [DOI] [PubMed] [Google Scholar]
- 84. Krainc D. Clearance of mutant proteins as a therapeutic target in neurodegenerative diseases. Arch Neurol. 2010;67(4):388–392. doi: 10.1001/archneurol.2010.40. [DOI] [PubMed] [Google Scholar]
- 85. Davies JE, Sarkar S, Rubinsztein DC. The ubiquitin proteasome system in Huntington’s disease and the spinocerebellar ataxias. BMC Biochem. 2007;8(suppl 1):S2. doi: 10.1186/1471-2091-8-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Martinez-Vicente M, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13(5):567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Williams A, et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4(5):295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13(7):805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pan J, Song E, Cheng C, Lee MH, Yeung SC. Farnesyltransferase inhibitors-induced autophagy: alternative mechanisms? Autophagy. 2009;5(1):129–131. doi: 10.4161/auto.5.1.7329. [DOI] [PubMed] [Google Scholar]
- 90. Mesa RA. Tipifarnib: farnesyl transferase inhibition at a crossroads. Expert Rev Anticancer Ther. 2006;6(3):313–319. doi: 10.1586/14737140.6.3.313. [DOI] [PubMed] [Google Scholar]
- 91. Wu IC, Ohsawa I, Fuku N, Tanaka M. Metabolic analysis of 13C-labeled pyruvate for noninvasive assessment of mitochondrial function. Ann N Y Acad Sci. 2010;1201:111–120. doi: 10.1111/j.1749-6632.2010.05636.x. [DOI] [PubMed] [Google Scholar]
- 92. Yang L, et al. Combination therapy with coenzyme Q10 and creatine produces additive neuroprotective effects in models of Parkinson’s and Huntington’s diseases. J Neurochem. 2009;109(5):1427–1439. doi: 10.1111/j.1471-4159.2009.06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Oliveira JM. Nature and cause of mitochondrial dysfunction in Huntington’s disease: focusing on huntingtin and the striatum. J Neurochem. 2010;114(1):1–12. doi: 10.1111/j.1471-4159.2010.06741.x. [DOI] [PubMed] [Google Scholar]
- 94. Pandey M, Mohanakumar KP, Usha R. Mitochondrial functional alterations in relation to pathophysiology of Huntington’s disease. J Bioenerg Biomembr. 2010;42(3):217–226. doi: 10.1007/s10863-010-9288-5. [DOI] [PubMed] [Google Scholar]
- 95. Quintanilla RA, Jin YN, Fuenzalida K, Bronfman M, Johnson GV. Rosiglitazone treatment prevents mitochondrial dysfunction in mutant huntingtin-expressing cells: possible role of peroxisome proliferator-activated receptor-gamma (PPARgamma) in the pathogenesis of Huntington disease. J Biol Chem. 2008;283(37):25628–25637. doi: 10.1074/jbc.M804291200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Okamoto S, et al. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat Med. 2009;15(12):1407–1413. doi: 10.1038/nm.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yu J, Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann N Y Acad Sci. 2009;1173(suppl 1):E10–E19. doi: 10.1111/j.1749-6632.2009.04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chiang MC, et al. Modulation of energy deficiency in Huntington’s disease via activation of the peroxisome proliferator-activated receptor gamma. Hum Mol Genet. 2010;19(20):4043–4058. doi: 10.1093/hmg/ddq322. [DOI] [PubMed] [Google Scholar]
- 99. Weydt P, et al. The gene coding for PGC-1alpha modifies age at onset in Huntington’s Disease. Mol Neurodegener. 2009;4:3. doi: 10.1186/1750-1326-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. van Oostrom JC, Dekker M, Willemsen AT, de Jong BM, Roos RA, Leenders KL. Changes in striatal dopamine D2 receptor binding in pre-clinical Huntington’s disease. Eur J Neurol. 2009;16(2):226–231. doi: 10.1111/j.1468-1331.2008.02390.x. [DOI] [PubMed] [Google Scholar]
- 101. Chen-Plotkin AS, et al. Decreased association of the transcription factor Sp1 with genes downregulated in Huntington’s disease. Neurobiol Dis. 2006;22(2):233–241. doi: 10.1016/j.nbd.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 102. Cornett J, Smith L, Friedman M, Shin JY, Li XJ, Li SH. Context-dependent dysregulation of transcription by mutant huntingtin. J Biol Chem. 2006;281(47):36198–36204. doi: 10.1074/jbc.M607839200. [DOI] [PubMed] [Google Scholar]
- 103. Jiang H, Nucifora FC, Jr, Ross CA, DeFranco DB. Cell death triggered by polyglutamine-expanded huntingtin in a neuronal cell line is associated with degradation of CREB-binding protein. Hum Mol Genet. 2003;12(1):1–12. doi: 10.1093/hmg/ddg002. [DOI] [PubMed] [Google Scholar]
- 104. Zuccato C, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001;293(5529):493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 105. Zuccato C, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35(1):76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 106. Kleiman RJ, et al. Chronic suppression of PDE10A alters striatal expression of genes responsible for neurotransmitter synthesis, neurotransmission and signaling pathways implicated in Huntington’s Disease. J Pharmacol Exp Ther. 2011;336:64–76. doi: 10.1124/jpet.110.173294. [DOI] [PubMed] [Google Scholar]
- 107. Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413(6857):739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 108. Zadori D, Geisz A, Vamos E, Vecsei L, Klivenyi P. Valproate ameliorates the survival and the motor performance in a transgenic mouse model of Huntington’s disease. Pharmacol Biochem Behav. 2009;94(1):148–153. doi: 10.1016/j.pbb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 109. Ferrante RJ, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J Neurosci. 2003;23(28):9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Joshi PR, et al. Age-dependent alterations of corticostriatal activity in the YAC128 mouse model of Huntington disease. J Neurosci. 2009;29(8):2414–2427. doi: 10.1523/JNEUROSCI.5687-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. . J Comp Neurol. 2003;465(1):11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 112. Ortiz AN, Kurth BJ, Osterhaus GL, Johnson MA. Dysregulation of intracellular dopamine stores revealed in the R6/2 mouse striatum. J Neurochem. 2010;112(3):755–761. doi: 10.1111/j.1471-4159.2009.06501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang H, Chen X, Li Y, Tang TS, Bezprozvanny I. Tetrabenazine is neuroprotective in Huntington’s disease mice. Mol Neurodegener. 2010;5:18. doi: 10.1186/1750-1326-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Petrikis P, Andreou C, Piachas A, Bozikas VP, Karavatos A. Treatment of Huntington’s disease with galantamine. Int Clin Psychopharmacol. 2004;19(1):49–50. doi: 10.1097/00004850-200401000-00010. [DOI] [PubMed] [Google Scholar]
- 115. Heng MY, Detloff PJ, Wang PL, Tsien JZ, Albin RL. In vivo evidence for NMDA receptor-mediated excitotoxicity in a murine genetic model of Huntington disease. J Neurosci. 2009;29(10):3200–3205. doi: 10.1523/JNEUROSCI.5599-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Huang K, et al. Palmitoylation and function of glial glutamate transporter-1 is reduced in the YAC128 mouse model of Huntington disease. Neurobiol Dis. 2010;40(1):207–215. doi: 10.1016/j.nbd.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 117. Cummings DM, et al. Alterations in cortical excitation and inhibition in genetic mouse models of Huntington’s disease. J Neurosci. 2009;29(33):10371–10386. doi: 10.1523/JNEUROSCI.1592-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Cepeda C, Wu N, Andre VM, Cummings DM, Levine MS. The corticostriatal pathway in Huntington’s disease. Prog Neurobiol. 2007;81(5–6):253–271. doi: 10.1016/j.pneurobio.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kloppel S, et al. Magnetic resonance imaging of Huntington’s disease: preparing for clinical trials. Neuroscience. 2009;164(1):205–219. doi: 10.1016/j.neuroscience.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Milnerwood AJ, et al. Early increase in extrasynaptic NMDA receptor signaling and expression contributes to phenotype onset in Huntington’s disease mice. Neuron. 2010;65(2):178–190. doi: 10.1016/j.neuron.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 121. Tallaksen-Greene SJ, Janiszewska A, Benton K, Ruprecht L, Albin RL. Lack of efficacy of NMDA receptor-NR2B selective antagonists in the R6/2 model of Huntington disease. Exp Neurol. 2010;225(2):402–407. doi: 10.1016/j.expneurol.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Miller BR, et al. Up-regulation of GLT1 expression increases glutamate uptake and attenuates the Huntington’s disease phenotype in the R6/2 mouse. Neuroscience. 2008;153(1):329–337. doi: 10.1016/j.neuroscience.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Sari Y, Prieto AL, Barton SJ, Miller BR, Rebec GV. Ceftriaxone-induced up-regulation of cortical and striatal GLT1 in the R6/2 model of Huntington’s disease. J Biomed Sci. 2010;17:62. doi: 10.1186/1423-0127-17-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Moro E, et al. Bilateral globus pallidus stimulation for Huntington’s disease. Ann Neurol. 2004;56(2):290–294. doi: 10.1002/ana.20183. [DOI] [PubMed] [Google Scholar]
- 125. Fawcett AP, Moro E, Lang AE, Lozano AM, Hutchison WD. Pallidal deep brain stimulation influences both reflexive and voluntary saccades in Huntington’s disease. Mov Disord. 2005;20(3):371–377. doi: 10.1002/mds.20356. [DOI] [PubMed] [Google Scholar]
- 126. Eidelberg D, Surmeier DJ. Brain networks in Huntington disease. J Clin Invest. 2011;121(2):484–492. doi: 10.1172/JCI45646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Mochel F, Haller RG. Energy deficit in Huntington disease: why it matters. J Clin Invest. 2011;121(2):493–499. doi: 10.1172/JCI45691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Sah DWY, Aronin N. Oligonucleotide therapeutic approaches for Huntington disease. J Clin Invest. 2011;121(2):500–507. doi: 10.1172/JCI45130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.