Abstract
Recent studies have focused on understanding the neural mechanisms underlying the emergence of clinical signs and symptoms in early stage Huntington disease (HD). Although cell-based assays have focused on cell autonomous effects of mutant huntingtin, animal HD models have revealed alterations in the function of neuronal networks, particularly those linking the cerebral cortex and striatum. These findings are complemented by metabolic imaging studies of disease progression in premanifest subjects. Quantifying metabolic progression at the systems level may identify network biomarkers to aid in the objective assessment of new disease-modifying therapies and identify new regions that merit mechanistic study in HD models.
Neuropathology in Huntington disease
Huntington disease (HD) is caused by expansion of a CAG repeat in the huntingtin (Htt) gene. Expansion of the repeat length beyond 35 CAGs significantly elevates disease risk (1). The Htt gene is ubiquitously expressed in the brain with relative enrichment in cortical pyramidal neurons projecting to the striatum but relatively low levels in striatal projection neurons (2, 3).
Although there are clear effects in the cerebral cortex and hippocampus, the most pronounced neuropathology in HD is found in the striatum (4, 5). The striatum is a key component of the basal ganglia, an interconnected set of subcortical nuclei involved in the regulation of how to act (and not act) in particular contexts (6). The striatum is composed primarily of GABAergic projection neurons with dendrites that are heavily studded with spines. These spiny projection neurons (SPNs) can be subdivided into two major classes on the basis of their axonal projection, dendritic size, and physiology as well as their expression of dopamine receptors and releasable peptides (7, 8). SPNs that project to the external segment of the globus pallidus (GPe) form what is called the indirect SPN (iSPN) pathway because they indirectly control the nuclei that interface between the basal ganglia and the rest of the brain. SPNs that project to the internal segment of the globus pallidus (GPi) and substantia nigra pars reticulata form the direct SPN (dSPN) pathway. iSPNs are smaller and more excitable than dSPNs (Figure 1 and ref. 9); this difference is very likely to reflect a tonic negative modulation of iSPNs by dopamine acting at D2 receptors. In contrast, dSPNs are positively modulated by D1 receptors that appear to only be activated by transient elevations in dopamine release produced by bursts of action potentials in dopaminergic neurons (10).
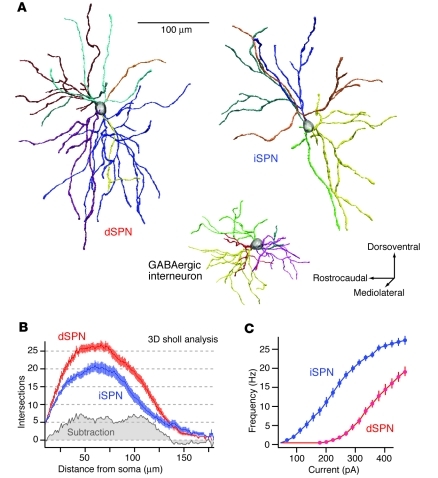
Figure 1. Dichotomous anatomy and physiology of dSPNs and iSPNs.
(A) Reconstructions of biocytin-filled dSPNs and iSPNs from P35–P45 BAC transgenic mice; a GABAergic interneuron is shown for comparison. (B) Plot of dendritic intersections in dSPNs and iSPNs as a function of distance from the soma that was derived from 3-dimensional reconstructions. (C) Summary of the responses of dSPNs and iSPNs to a range of intrasomatic current steps; iSPNs were more excitable over a broad range of injected currents. Adapted with permission from Journal of Neuroscience (9).
iSPNs and dSPNs are generally thought to play complementary roles in movement control and action selection. Both SPN populations have a rich, highly convergent synaptic connectivity with glutamatergic neurons distributed throughout much of the cerebral cortex (11). Both populations also have rich connectivity with glutamatergic neurons in the thalamus (12). A broad array of experimental evidence supports the proposition that iSPNs help to suppress cortical selection of unwanted movements or actions, whereas dSPNs help to promote cortical selection of particular actions (6, 13). These roles are particularly well characterized in situations in which the action leads to reward or punishment.
In the early stages of HD, chorea (hyperkinesia) is one of the most prominent symptoms. This feature is strongly correlated with iSPN dysfunction and phenotypic decline (4, 14, 15). What is less clear is why there is loss of function in this projection system. The ability of D2 receptor antagonists and an inhibitor of the type 2 vesicular monoamine transporter (tetrabenazine) — which decreases striatal dopamine (DA) release — to ameliorate chorea (16–18) suggests that the loss of function is a consequence of a deficit in the ability of cortical neurons to drive activity in iSPNs. D2 DA receptors are important negative modulators of excitability and glutamatergic synaptic transmission in iSPNs (19) (see below for more discussion of the potential role of DA in HD).
A reduction in iSPN GABA release will disinhibit autonomously active GPe neurons, resulting in diminished basal ganglia inhibition of motor thalamus, hyperkinesia, and altered cortical activity. In the later stages of HD, the pathology has spread to dSPNs, resulting in difficulty in initiating movements (5, 14).
Although less severe than the pathology in the striatum, there is clear pathology elsewhere in the brain. Neuropathological changes have been found in the cerebral cortex, globus pallidus, thalamus, subthalamic nucleus (STN), substantia nigra, hypothalamus, and cerebellum in later stages of the disease (5, 20–24). Imaging studies also have shown widespread volumetric changes in HD patients, particularly in the cerebral cortex (25).
HD pathogenesis is rooted in network dysfunction
The mechanisms underlying the neurodegeneration seen in HD are actively being pursued on several fronts. As detailed in recent reviews, much of the study in the HD field has focused on cell autonomous mechanisms, revealing that mutant Htt (mHtt) can alter a panoply of critical processes including gene transcription, protein trafficking, autophagy, Ca2+ homeostasis, and mitochondrial respiration (1, 26).
In the simplest scenario, in which cell autonomous effects dominate pathogenesis, the consequences of mHtt expression would be directly related to expression level. That is, the more mHtt, the more severe the pathophysiology and degeneration. Although there is a correlation between global expression of mHtt and pathology, this clearly is not the case when comparing different regions within an individual brain (1). Even within the striatum, neuron vulnerability and mHtt expression level are not correlated (3). The alternative possibility is that the impact of mHtt is modulated by cellular phenotype or interactions between cells (non-autonomous deficits). As discussed below, most of the available evidence point to some combination of these two possibilities.
One of the most widely discussed “interaction” models of HD casts it as a neurotrophic disorder (27). The evidence for this model comes from several lines of study. Of the four neurotrophins released in the brain, brain derived neurotrophic factor (BDNF) is the one implicated in HD. BDNF expression is decreased in the cortex, and its receptor (TrkB) is decreased in the striatum of human HD patients, suggesting a deficit in cortical neurotrophic support of the striatum (28). In animal models of HD, cortical BDNF expression is reduced (29, 30). Moreover, downregulating BDNF in striatum in mice worsens the HD phenotype, whereas elevating BDNF expression in the forebrain alleviates the HD phenotype (31–34). Studies demonstrating a role for Htt in axonal transport of BDNF strengthen this argument by providing a mechanism by which BDNF delivery to the striatum might be impaired in HD (35), but this point is somewhat controversial (36).
As appealing as it is, there are significant gaps in this story line. One gap is the tissue specificity of the defect. Alterations in BDNF expression appear to be regionally selective (1), suggesting that expression defects are shaped by cellular phenotype or the network in which neurons are embedded. This opens the door to the possibility that the changes in BDNF expression in HD are secondary to some other effect of mHtt, in spite of the data linking it to the expression of Htt. Along these lines, an elegant series of experiments using regionally selective expression of mHtt has shown that recapitulation of the key features of the HD phenotype demand simultaneous expression in both cortex and striatum (37). Identifying which circuits are most affected in HD, and when, could be very instructive. Metabolic mapping (as described below) in HD patients is one way to obtain this information.
Another gap in the neurotrophic theory is that it does not clearly define the role BDNF plays in the pathophysiology of the adult striatum. Loss of cortical BDNF delivery has little impact on the development and maturation of the striatum (38), perhaps because both the mesencephalon and thalamus also provide BDNF to the striatum. In other brain regions, BDNF and TrkB signaling shape synaptic signaling in adult neurons by either increasing basal AMPA receptor signaling at glutamatergic synapses (39) or by facilitating the late phase of long-term potentiation (LTP) of glutamatergic synapses (40). Much less is known about the functions of BDNF and TrkB signaling in the striatum. A recent study has shown that although both classes of SPN express TrkB receptors, expression was dramatically higher in iSPNs of wild-type mice (41). The implications of this difference have yet to be fully determined. What the authors of this study did show was that deleting TrkB receptors significantly increased the somatic excitability of iSPNs, but not dSPNs. This shift in iSPNs could reflect a homeostatic adaptation to a reduction in the strength or number of glutamatergic synapses following TrkB deletion. Consistent with this conjecture, BDNF and TrkB signaling enhanced LTP induction at glutamatergic synapses on striatal SPNs (42) and, thus, their downregulation in HD would be expected to impair LTP (43, 44), as described in other regions (45, 46). This could prevent cortical neurons from effectively activating iSPNs to suppress unwanted activity.
This impairment could be exacerbated by elevated DA release and increased D2 receptor signaling in HD. D2 receptor signaling promotes long-term depression (LTD) at corticostriatal synapses (19). Consistent with this view, impairing DA uptake exacerbated the HD phenotype and reducing DA release with tetrabenazine improved motor behavior in a mouse model (47, 48). The drop in CB1 receptor expression in HD patients (49) and HD models (50–52) could be a compensation in response to elevated D2 receptor signaling in HD: LTD in iSPN is mediated by presynaptic CB1 receptors activated by postsynaptically generated endocannabinoids (53). Deficits in striatal glutamatergic synaptic transmission have been seen in HD models, particularly at ages at which symptoms are evident (54). However, much remains to be done to tie these deficits to mechanisms that could be therapeutically manipulated. In particular, it remains to be determined whether these deficits are specific to iSPNs, dSPNs, or interneurons (44), and whether they are presynaptic or postsynaptic in origin. It is also unclear whether they are present at cortical synapses or thalamic synapses (or both).
Lastly, the BDNF hypothesis does not account for the neurodegeneration seen in HD. Indeed, there is cell loss in mice in which the Bdnf gene has been deleted from corticostriatal projection neurons (31), but this loss is modest and is only evident at the end of the mouse’s life. Thus, it is likely that there is another factor mediating neuronal loss in HD. Two new studies have provided a potential clue. In SPNs from HD mice, there has long been the suggestion that NMDA receptor (NMDAR) function is altered (1). In a recent study of SPNs in a presymptomatic mouse HD model that expresses full-length human mHtt with a 128-glutamine tract (YAC128), it was found that extrasynaptic NMDARs enriched in the NR2B subunit were elevated (55). These receptors were activated only by intense synaptic stimulation that led to glutamate spillover out of the synapse. In contrast, at this early point in time, synaptic AMPA receptors and NMDARs appeared normal, suggesting that BDNF signaling was not altered. In older symptomatic mice, partial blockade of these extrasynaptic NMDARs with memantine alleviated motor symptoms. This is obviously of potential clinical importance, but the role of these extrasynaptic NMDARs was unclear. An earlier study had shown that, unlike synaptic NMDARs that promoted aggregation and sequestration of mHtt, extrasynaptic NMDARs inhibited CREB signaling, leading to disaggregation of toxic mHtt (56). These studies suggest that activation of extrasynaptic NMDARs in SPNs could be an early event in HD pathogenesis and SPN loss, notwithstanding previous work suggesting that in the R6/1 and R6/2 mouse models, SPNs are resistant to NMDAR toxicity (57, 58).
These studies do not address why extrasynaptic NMDARs rise in HD models (or whether the change was peculiar to SPNs). There is no reason to believe that alterations in BDNF signaling underlie this change. Milnerwood and Raymond suggested that activation of the Ca2+-dependent protease calpain could cleave a synaptic targeting sequence of NR2B subunits, causing their “misplacement” (55, 59–61). There is evidence of elevated calpain activity in the striatum of YAC128 mice, perhaps due to a greater propensity for SPNs in HD mice to elevate dendritic Ca2+ concentration in response to synaptic activity. This could happen in a number of ways; for example, there could be greater synchrony in cortical or thalamic activity, leading to stronger engagement of NMDARs (54). A relevant and poorly understood feature of SPNs in this regard is their cycling between down-states and up-states (62). In each of these states, the membrane potential of SPNs is relatively stereotyped. In the down-state, the membrane potential is hyperpolarized (approximately –85 mV), about 30–40 mV from the action potential threshold. In the up-state, the membrane potential is more depolarized (approximately –60 mV), close to the threshold for action potential generation. During up-states, dendritic Ca2+ concentration surely rises as NMDARs lose their Mg2+ block and voltage-dependent Ca2+ channels open, potentially into the range needed to activate calpain. However, it isn’t clear whether this process is altered in HD. Modest mHtt-induced alterations in the targeting of voltage-dependent ion channels controlling the duration of these up-states could result in elevations in dendritic Ca2+ concentration, increasing calpain activation. Another possibility is that mHtt alters mobilization of intracellular Ca2+ stores in response to normal glutamatergic synaptic input. Some time ago, it was shown that mHtt is capable of interacting with inositol trisphosphate receptors (IP3Rs) in the endoplasmic reticulum, enhancing their open probability and elevating intracellular Ca2+ release (63). Increased activation of D1 DA receptors, activation of protein kinase A, and phosphorylation of IP3Rs could exacerbate this dysregulation of intracellular Ca2+ (64). Whether one or more of these mechanisms is altered in HD models and contributes to misplacement of NMDARs remains to be determined.
It is also worth noting in this context that the excitotoxicity hypothesis of HD (65), once thought to be dead, has been resurrected by new insights into kynurenine metabolism (66). This new work raises the possibility that extrasynaptic NMDARs might be targeted by quinolinic acid released by microglia, promoting neurodegeneration in the later stages of the disease.
Homeostatic alterations in network activity
Even though the pathophysiology in HD is likely to originate in the corticostriatal connection, with time it is likely to spread. Neurons are homeostatic entities, preferring to spike within a particular range of frequencies set by their developmentally determined phenotype (67). Sustained deviations from their set point typically trigger a cascade of adaptations ranging from synaptic scaling (adjusting synaptic strength up or down to achieve a desired level of activity) to alterations in the expression of intrinsic ionic mechanisms governing spiking — all aimed at enforcing homeostasis (67). In models of Parkinson disease (PD), for example, the loss of DA and inhibitory D2 receptor signaling triggers a rapid and profound pruning of corticostriatal synapses in iSPNs, ostensibly in an attempt to reduce spiking (68). This adaptation is not entirely successful, and there are a host of secondary changes in intrinsic excitability and dendritic morphology that evolve over the weeks following lesioning dopaminergic fibers (69). Even though the principal target of the dopaminergic innervation lost in PD is the striatum, the response to its loss spreads to other parts of the basal ganglia with time. A recently described example involves the GPe. Soon after near complete dopaminergic denervation, STN and GPe neurons begin to spike in frequent, rhythmic bursts (70). This pathological activity pattern is widely thought to underlie motor symptoms in PD. In response to this shift in activity, GPe neurons adapt by downregulating the expression of hyperpolarization and cyclic nucleotide–gated channels that help support autonomous pacemaking (71). Adaptations of this kind are likely to occur throughout the basal ganglia and the networks it influences (e.g., cerebral cortex) (72).
As in PD, there is compelling evidence from HD patients that the activity of the GPe-STN network is altered and that the pathophysiology is linked to motor symptoms (73, 74). Electrical stimulation of basal ganglia output neurons in the GPi ameliorates HD symptoms (75). What is less clear is the origin of this pathological activity; it could be the cerebral cortex, the striatum, or the GPe-STN network itself. Brain imaging in premanifest HD patients offers a potential method of tracking the emergence of pathophysiology as well as delineating a “functional natural history” of the disorder. Knowledge of the specific time line of the changes in brain structure and function that occur before and during phenoconversion can help direct translational studies aimed at the development of new therapeutics for HD.
Functional imaging in HD
Among neurodegenerative disorders, HD is unique in that individuals destined to develop symptoms can be identified through genetic testing before clinical signs of the disease begin. This raises the possibility of developing therapies to prevent or delay the onset of clinical manifestations in HD gene carriers. Therefore, substantial effort has been dedicated to the characterization of quantitative descriptors of disease progression in premanifest individuals. In particular, imaging tools such as PET and MRI have been employed to assess changes in regional brain structure and function in early-stage disease (see refs. 76 and 77 for recent reviews). As we shall see, these approaches have relied mainly on the measurements of structure (local tissue volume) and function (cerebral metabolism/blood flow, D2 neuroreceptor binding) in single brain regions, particularly the striatum. The rationale behind the “striato-centric” approach of most human imaging studies in HD is rooted in the experimental literature. While striatal cell death and consequent volume loss are consistent features of HD mouse models, a period of sustained (and potentially reversible) metabolic dysfunction has been found to precede the observed histopathological changes (78). Indeed, these functional changes correlated significantly with CAG repeat length, gene expression levels, and mHtt content. Of note, the R6/2 model exhibited substantial metabolic changes in the cerebral cortex in addition to the striatum (78). These observations in experimental models, and the demonstration of widespread cortical changes in the human HD brain (20, 79), have set the stage for further in vivo studies of cerebral function in this disorder. These efforts may prove to have particular value in their use of novel analytic strategies to capture the broader spatial topography of the disease process.
A variety of functional imaging strategies have been applied to study the natural history of HD, particularly in premanifest subjects for whom clinical ratings, even if quantitative, are not suitable for use as indicators of disease progression. The earliest approaches involved direct measurements of regional cerebral blood flow and metabolism in the resting state, employing mainly radionuclide imaging with PET and single photon emission computed tomography (76, 77, 80). The potential utility of such measures as disease biomarkers was highlighted by the consistent presence of striatal metabolic abnormalities in HD mutation carriers and by the association of these changes with progressive deterioration in functional capacity (81). Moreover, the changes in frontoparietal and temporo-occipital cortex that were observed in these subjects correlated with specific cognitive deficits (80).
The description of early reductions in D2 receptors on striatal projection neurons in HD mouse models, which were found to precede local cell loss (82), led to the development of [11C] raclopride (RAC) PET as the basis for an alternative imaging biomarker of disease progression (83, 84). In fact, in a later cross-sectional RAC PET study, striatal D2 receptor binding was found to be more sensitive to early HD pathology than corresponding assessments of regional metabolism in the same subjects (85). Nonetheless, efforts again shifted when it was observed that ongoing volume loss in the striatum and in other regions can be assessed simply and non-invasively using MRI volumetrics (86, 87). Irrespective of the relative strengths and weaknesses of these imaging approaches, the measurement of progression in single brain regions (whether of local metabolic activity, D2 receptor binding, or tissue volume) does not take into account the systems-level changes that are likely to be present even at the earliest disease stages. Recent computational advances have indeed allowed for the characterization of network-based functional imaging biomarkers to quantify disease progression.
Metabolic network activity in premanifest HD
Network analysis of metabolic imaging data acquired in the rest state has recently been performed to identify and validate HD-related spatial covariance patterns (HDRPs) in premanifest subjects (88). The mathematical-statistical model that was employed had been used extensively in prior studies of the network changes associated with the progression and treatment of PD as well as other movement disorders (89–91). It is based on the notion that small and biologically relevant signals can be detected in functional brain images after minimizing the substantive variability that exists across subjects and brain regions. This approach, which is based on principal components analysis, generates spatial covariance patterns associated with the clinically manifest disease or with the preclinical prodromal state. This method of pattern detection is performed in the entire sample, blind to subject class (patient or healthy subject) or genotype (mutation carriers or control). Moreover, spatial covariance analysis utilizes metabolic data from the entire brain without a priori assumptions regarding the contributions of specific regions to the resulting patterns. Importantly, once a significant pattern has been identified, its expression in a given subject can be computed prospectively from that individual’s scan. The resulting network values (subject scores) can discriminate between groups and/or be correlated with independent clinical, behavioral, neurophysiologic, or biochemical disease descriptors. These patterns typically exhibit increased signal-to-noise with disease progression, which together with the objective, data-driven features of this algorithm make it an optimal tool to evaluate early abnormalities in functional brain organization in neurodegenerative disorders such as HD.
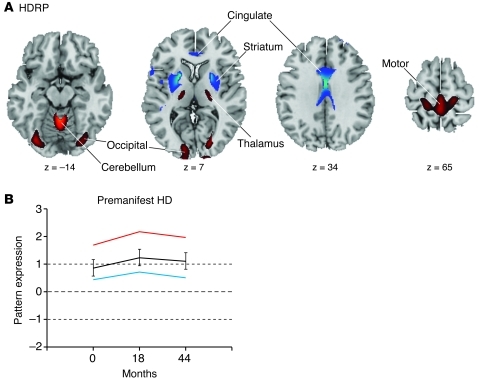
The utility of this approach is evident in the longitudinal study published by our group (88), in which baseline metabolic imaging data from 12 premanifest gene carriers and 12 healthy control subjects were analyzed as a combined group. The resulting HDRP was characterized by reductions in striatal and anterior cingulate metabolic activity, with associated metabolic increases in the ventrolateral (VL) thalamus, cerebellum/dentate nucleus, and in the primary motor and visual regions of the cerebral cortex (Figure 2A). Subject scores for this pattern were elevated in the premanifest cohort relative to healthy controls. Longitudinal analysis (Figure 2B) revealed a significant increase in HDRP expression from baseline to 18 months and a subsequent decline from 18 to 44 months. Interestingly, the changes in HDRP activity over time did not correlate with simultaneous measurements of striatal D2 receptor binding, suggesting that the two processes evolve independently at the early stages of the disease. Further, HDRP expression was found to correlate with predicted years to clinical onset, a function of subject CAG repeat length and age (92). Indeed, gene carriers with elevated HDRP scores at baseline were more likely to phenoconvert (i.e., develop overt clinical manifestations of HD) over the ensuing 44 months than those with baseline HDRP expression in the normal range (60% for scores ≥1.5; 14% for scores in the normal range) (88). These observations suggest that this disease-related functional measure is a relevant descriptor of the underlying biological process.
Figure 2. HDRP.
(A) This pattern was identified by spatial covariance analysis of 18F-Fluorodeoxyglucose (FDG) PET scans at baseline from 12 premanifest HD gene carriers and 12 age-matched healthy control subjects (115). This pattern is characterized by metabolic decreases in the striatum and cingulate cortex, with relative increases in the ventral thalamus, primary motor cortex (Brodmann areas [BA] 4), and occipital cortex (BA 17, 18). The covariance map was overlaid on T1-weighted magnetic resonance–template images. Voxel weights for the pattern were thresholded at P = 0.005. Voxels with positive region weights (increases) are shown in red, and voxels with negative region weights (decreases) are shown in blue. (B) Time course of HDRP expression in the 12 premanifest gene carriers scanned at baseline and at 18 and 44 months. Mean subject scores are displayed for the 8 premanifest subjects who subsequently phenoconverted (red), the 4 subjects who did not phenoconvert during the period of follow-up (blue), and all 12 subjects (black). Subject scores, reflecting the expression of this pattern, separate the premanifest from the control groups (P < 0.01). (The mean value [± 1 SD] for the healthy control group is represented by dotted lines. Error bars represent 1 SEM at each time point). Reprinted with permission from Brain (88).
The HDRP topography likely represents a combination of regional cell loss, metabolic effects occurring distally at remote nodes of the network, and compensatory changes in brain function. Widespread changes in tissue volume occur in premanifest HD subjects at both subcortical and cortical sites (20, 93–95), which can contribute to the changes seen on imaging. Nonetheless, in a recent study combining network quantification with MRI voxel-based morphometry, it was found that volume loss in the longitudinal scan data did not affect estimated rates of network progression in premanifest subjects (96). This suggests that the functional changes captured by metabolic imaging in premanifest subjects are generally independent of ongoing loss of tissue volume. Moreover, the non-linear trajectory of HDRP progression in the preclinical period, with a decline (rather than an increase) in network activity between 18 and 44 months, suggests the presence of adaptive and/or compensatory processes that slow as symptom onset nears.
Changes in regional metabolic activity with disease progression
Insights into the potential compensatory function of the HDRP have been provided by the analysis of the time course of regional metabolic activity at key nodes of the HDRP network (88). Measures of striatal metabolic activity (Figure 3A) were found to be significantly below normal throughout the duration of the study, with relatively greater reductions in the premanifest HD subjects who eventually phenoconverted during the 44-month follow-up period. Interestingly, significant elevations in thalamic metabolism, localized to the VL and mediodorsal (MD) nuclei (Figure 3B), were evident in the premanifesting baseline scans of these subjects. Whereas increased levels of thalamic metabolic activity were sustained in the premanifesting subjects who did not subsequently phenoconvert, a progressive loss of this compensatory response was observed in those who became symptomatic during the ensuing follow-up period. Of note, increased activation in the MD thalamus was observed in the same subjects during the performance of a motor learning task (97). The precise mechanism mediating the thalamic response to progressive dysfunction and/or loss of striatal projection neurons is not understood. Nonetheless, it is likely that positive feedback from the thalamus, particularly the intralaminar nuclei, to the striatum is involved (98). As the disease progresses, this compensatory response appears to attenuate, perhaps as a consequence of inherent thalamic cell loss (95, 99–102). Moreover, the observed metabolic decline in the VL thalamus may also reflect changes in afferent synaptic activity from basal ganglia output structures. In any event, the regional data are consistent with the notion that symptom onset in HD is associated with a cortical degree of attrition in inhibitory pallidothalamic output with concomitant increases in the metabolic activity of cortical motor regions.
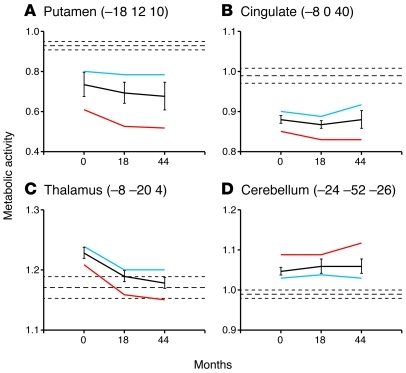
Figure 3. Metabolic activity at key network nodes.
(A) In the putamen, metabolic activity declined over time in premanifest subjects (P < 0.005). Relatively greater reductions were observed in the subgroup of premanifest gene carriers who subsequently phenoconverted (red) as compared to those who remained asymptomatic (blue). Mean metabolic activity is also displayed for the whole group (black). (B) Regional metabolic activity was also reduced in the cingulate cortex at all 3 time points, without significant longitudinal change. (C) In the thalamus, metabolic activity declined over time (P < 0.01). In the premanifest subjects who did not phenoconvert, thalamic metabolism was elevated at baseline and remained above normal at the subsequent two longitudinal time points. By contrast, baseline thalamic metabolic activity was also elevated in the premanifest subjects who subsequently phenoconverted, but then declined to normal levels as symptoms emerged. (D) Cerebellar metabolism was also elevated at baseline in premanifest subjects, but no subsequent changes were discerned. Mean globally normalized metabolic activity values (± 1 SEM) for the healthy control subjects are represented by horizontal dashed lines. Error bars represent 1 SEM at each time point. Reprinted with permission from Brain (88).
Abnormally reduced metabolic activity was also noted in the cingulate cortex of premanifest subjects, as well as elevated metabolism in the cerebellum (88). The cingulate region (Figure 3C) has been found to be involved in HD both functionally (103, 104) and as a locus of primary cell loss (20, 105). Indeed, significant reductions in cingulate metabolism were present at the earliest preclinical time point, suggesting that this functional change is central to the disease process. This observation in premanifest individuals accords with the notion that corticostriatal neurons are critical to the development of the earliest behavioral features of the disease (see, for example, ref. 37). By contrast, metabolic increases in the cerebellum (Figure 3D) were greater in the premanifest subjects who ultimately phenoconverted. However, unlike thalamic hypermetabolism, metabolic activity in the cerebellum continued to increase as subjects phenoconverted (88). This suggests that the metabolic changes in the region are linked to the appearance of abnormal involuntary movements.
That the HDRP metabolic network and its nodes exhibit distinctive temporal profiles (Figure 4 and ref. 88) is consistent with the substantial regional variability that has been described in relation to the disease process (79). That said, several lines of evidence suggest that abnormal cellular metabolism may be a feature of the neuropathological process underlying HD. Non-neuronal tissues demonstrate metabolic impairments in HD patients (106, 107), and in brain, increases in lactate levels have been demonstrated utilizing magnetic resonance spectroscopy (108). Multiple biochemical studies have identified abnormalities in mitochondrial electron transport in HD (109–111). Mitochondrial toxins can induce selective striatal neurodegeneration in animal models that mimic HD neuropathology (112). Although the data suggest that the hypermetabolism observed in VL thalamus and cerebellum is compensatory in nature (88), it is also possible that this phenomenon reflects impending metabolic failure and cell death. Further studies will be needed to evaluate whether metabolic increases precede the onset of neurodegeneration in other brain regions, including the striatum.
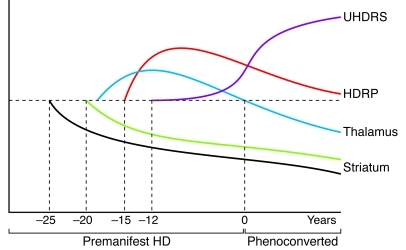
Figure 4. Hypothesized time course of changes in striatal D2 binding, pattern expression, and regional metabolism in premanifest HD.
The individual growth curve model (114) was applied to the longitudinal imaging data (88) to estimate the time course of these imaging measures in the period prior to clinical diagnosis. The decline in striatal D2 receptor binding (black) begins approximately 25 years before diagnosis (year 0). About 5 years later, striatal metabolism (green) begins to decline. Thalamic metabolic activity (blue) is likely elevated early on (exactly when is not known), but declines to normal levels as clinical signs (Unified HD Rating Scale [UHDRS] score, purple) become evident. HDRP activity is estimated to increase beginning approximately 15 years before diagnosis. This network measure likely increases for a number of years, then declines prior to symptom onset (see text). The horizontal dashed line represents the normal mean. Reprinted with permission from Brain (88).
Future directions
Recent theoretical advances in the multivariate modeling of time series data have allowed for substantial improvements in the statistical power and accuracy of large-scale brain networks in neurodegenerative disorders. In this regard, within-subject spatial covariance algorithms such as ordinal trends canonical variates analysis (113) are finding currency in the network analysis of longitudinal brain imaging data. In simple terms, this approach identifies metabolic patterns in which subject expression changes over time in most, if not all, members of the cohort. This rigorous computational approach is likely to be particularly useful in the analysis of longitudinal data from premanifest HD subjects, in which progressive network-level changes are likely to occur consistently in individual mutation carriers.
The combination of powerful network computational algorithms with subject level regression modeling techniques (114) is likely to generate a new class of progression biomarkers for preclinical HD. In addition to generating important translational data regarding the systems-level changes that underlie preclinical disease progression and phenoconversion, HD-related functional brain networks may prove valuable as biomarkers for the objective assessment of potential disease-modifying therapies.
Acknowledgments
This work was supported by a grant from the Cure Huntington’s Disease Initiative to D.J. Surmeier, NIH NINDS R01 NS 37564 to D. Eidelberg, the General Clinical Research Center (GCRC) of The Feinstein Institute for Medical Research (NCRR M01 RR 018535), and a generous grant from CHDI/High Q Foundation to D. Eidelberg.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(2):484–492. doi:10.1172/JCI45646.
References
- 1. Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90(3):905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 2. Sapp E, et al. Huntingtin localization in brains of normal and Huntington’s disease patients. Ann Neurol. 1997;42(4):604–612. doi: 10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- 3. Fusco FR, et al. Cellular localization of huntingtin in striatal and cortical neurons in rats: lack of correlation with neuronal vulnerability in Huntington’s disease. J Neurosci. 1999;19(4):1189–1202. doi: 10.1523/JNEUROSCI.19-04-01189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reiner A, Albin RL, Anderson KD, D’Amato CJ, Penney JB, Young AB. Differential loss of striatal projection neurons in Huntington disease. Proc Natl Acad Sci U S A. 1988;85(15):5733–5737. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57(5):369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 6. Balleine BW, Liljeholm M, Ostlund SB. The integrative function of the basal ganglia in instrumental conditioning. Behav Brain Res. 2009;199(1):43–52. doi: 10.1016/j.bbr.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 7. Gerfen CR, et al. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science. 1990;250(4986):1429–1432. doi: 10.1126/science.2147780. [DOI] [PubMed] [Google Scholar]
- 8. Surmeier DJ, Song WJ, Yan Z. Coordinated expression of dopamine receptors in neostriatal medium spiny neurons. J Neurosci. 1996;16(20):6579–6591. doi: 10.1523/JNEUROSCI.16-20-06579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gertler TS, Chan CS, Surmeier DJ. Dichotomous anatomical properties of adult striatal medium spiny neurons. J Neurosci. 2008;28(43):10814–10824. doi: 10.1523/JNEUROSCI.2660-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Surmeier DJ, Ding J, Day M, Wang Z, Shen W. D1 and D2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons. Trends Neurosci. 2007;30(5):228–235. doi: 10.1016/j.tins.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 11. Bolam JP, Hanley JJ, Booth PA, Bevan MD. Synaptic organisation of the basal ganglia. J Anat. 2000;196(pt 4):527–542. doi: 10.1046/j.1469-7580.2000.19640527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doig NM, Moss J, Bolam JP. Cortical and thalamic innervation of direct and indirect pathway medium-sized spiny neurons in mouse striatum. J Neurosci. 2010;30(44):14610–14618. doi: 10.1523/JNEUROSCI.1623-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kravitz AV, et al. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466(7306):622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Albin RL, et al. Abnormalities of striatal projection neurons and N-methyl-D-aspartate receptors in presymptomatic Huntington’s disease. N Engl J Med. 1990;322(18):1293–1298. doi: 10.1056/NEJM199005033221807. [DOI] [PubMed] [Google Scholar]
- 15. Sapp E, et al. Evidence for a preferential loss of enkephalin immunoreactivity in the external globus pallidus in low grade Huntington’s disease using high resolution image analysis. Neuroscience. 1995;64(2):397–404. doi: 10.1016/0306-4522(94)00427-7. [DOI] [PubMed] [Google Scholar]
- 16. Leavitt BR, Hayden MR. Is tetrabenazine safe and effective for suppressing chorea in Huntington’s disease? Nat Clin Pract Neurol. 2006;2(10):536–537. doi: 10.1038/ncpneuro0299. [DOI] [PubMed] [Google Scholar]
- 17. Guay DR. Tetrabenazine, a monoamine-depleting drug used in the treatment of hyperkinetic movement disorders. Am J Geriatr Pharmacother. 2010;8(4):331–373. doi: 10.1016/j.amjopharm.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 18. Grimbergen YA, Roos RA. Therapeutic options for Huntington’s disease. Curr Opin Investig Drugs. 2003;4(1):51–54. [PubMed] [Google Scholar]
- 19. Surmeier DJ, et al. The role of dopamine in modulating the structure and function of striatal circuits. Prog Brain Res. 2010;183:149–167. doi: 10.1016/S0079-6123(10)83008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thu DC, et al. Cell loss in the motor and cingulate cortex correlates with symptomatology in Huntington’s disease. Brain. 2010;133(pt 4):1094–1110. doi: 10.1093/brain/awq047. [DOI] [PubMed] [Google Scholar]
- 21. Heinsen H, et al. Nerve cell loss in the thalamic centromedian-parafascicular complex in patients with Huntington’s disease. Acta Neuropathol. 1996;91(2):161–168. doi: 10.1007/s004010050408. [DOI] [PubMed] [Google Scholar]
- 22. Heinsen H, et al. Nerve cell loss in the thalamic mediodorsal nucleus in Huntington’s disease. Acta Neuropathol. 1999;97(6):613–622. doi: 10.1007/s004010051037. [DOI] [PubMed] [Google Scholar]
- 23. Petersen A, Bjorkqvist M. Hypothalamic-endocrine aspects in Huntington’s disease. Eur J Neurosci. 2006;24(4):961–967. doi: 10.1111/j.1460-9568.2006.04985.x. [DOI] [PubMed] [Google Scholar]
- 24. Gabery S, et al. Changes in key hypothalamic neuropeptide populations in Huntington disease revealed by neuropathological analyses. Acta Neuropathol. 2010;120(6):777–788. doi: 10.1007/s00401-010-0742-6. [DOI] [PubMed] [Google Scholar]
- 25. Rosas HD, et al. Evidence for more widespread cerebral pathology in early HD: an MRI-based morphometric analysis. Neurology. 2003;60(10):1615–1620. doi: 10.1212/01.wnl.0000065888.88988.6e. [DOI] [PubMed] [Google Scholar]
- 26. Milnerwood AJ, Raymond LA. Early synaptic pathophysiology in neurodegeneration: insights from Huntington’s disease. Trends Neurosci. 2010;33(11):513–523. doi: 10.1016/j.tins.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 27. Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol. 2009;5(6):311–322. doi: 10.1038/nrneurol.2009.54. [DOI] [PubMed] [Google Scholar]
- 28. Zuccato C, Marullo M, Conforti P, MacDonald ME, Tartari M, Cattaneo E. Systematic assessment of BDNF and its receptor levels in human cortices affected by Huntington’s disease. Brain Pathol. 2008;18(2):225–238. doi: 10.1111/j.1750-3639.2007.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zuccato C, et al. Progressive loss of BDNF in a mouse model of Huntington’s disease and rescue by BDNF delivery. Pharmacol Res. 2005;52(2):133–139. doi: 10.1016/j.phrs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 30. Zuccato C, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001;293(5529):493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 31. Strand AD, et al. Expression profiling of Huntington’s disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J Neurosci. 2007;27(43):11758–11768. doi: 10.1523/JNEUROSCI.2461-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Canals JM, et al. Brain-derived neurotrophic factor regulates the onset and severity of motor dysfunction associated with enkephalinergic neuronal degeneration in Huntington’s disease. J Neurosci. 2004;24(35):7727–7739. doi: 10.1523/JNEUROSCI.1197-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gharami K, Xie Y, An JJ, Tonegawa S, Xu B. Brain-derived neurotrophic factor over-expression in the forebrain ameliorates Huntington’s disease phenotypes in mice. J Neurochem. 2008;105(2):369–379. doi: 10.1111/j.1471-4159.2007.05137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie H, et al. Brain-derived neurotrophic factor rescues and prevents chronic intermittent hypoxia-induced impairment of hippocampal long-term synaptic plasticity. Neurobiol Dis. 2010;40(1):155–162. doi: 10.1016/j.nbd.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 35. Gauthier LR, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118(1):127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 36. Her LS, Goldstein LS. Enhanced sensitivity of striatal neurons to axonal transport defects induced by mutant huntingtin. J Neurosci. 2008;28(50):13662–13672. doi: 10.1523/JNEUROSCI.4144-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gu X, et al. Pathological cell-cell interactions are necessary for striatal pathogenesis in a conditional mouse model of Huntington’s disease. Mol Neurodegener. 2007;2:8. doi: 10.1186/1750-1326-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J Neurosci. 2004;24(17):4250–4258. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kang H, Schuman EM. Long-lasting neurotrophin-induced enhancement of synaptic transmission in the adult hippocampus. Science. 1995;267(5204):1658–1662. doi: 10.1126/science.7886457. [DOI] [PubMed] [Google Scholar]
- 40. Lu Y, Christian K, Lu B. BDNF: a key regulator for protein synthesis-dependent LTP and long-term memory? Neurobiol Learn Mem. 2008;89(3):312–323. doi: 10.1016/j.nlm.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lobo MK, et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330(6002):385–390. doi: 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jia Y, Gall CM, Lynch G. Presynaptic BDNF promotes postsynaptic long-term potentiation in the dorsal striatum. J Neurosci. 2010;30(43):14440–14445. doi: 10.1523/JNEUROSCI.3310-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kung VW, Hassam R, Morton AJ, Jones S. Dopamine-dependent long term potentiation in the dorsal striatum is reduced in the R6/2 mouse model of Huntington’s disease. Neuroscience. 2007;146(4):1571–1580. doi: 10.1016/j.neuroscience.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 44. Picconi B, et al. Plastic and behavioral abnormalities in experimental Huntington’s disease: a crucial role for cholinergic interneurons. Neurobiol Dis. 2006;22(1):143–152. doi: 10.1016/j.nbd.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 45. Usdin MT, Shelbourne PF, Myers RM, Madison DV. Impaired synaptic plasticity in mice carrying the Huntington’s disease mutation. Hum Mol Genet. 1999;8(5):839–846. doi: 10.1093/hmg/8.5.839. [DOI] [PubMed] [Google Scholar]
- 46. Lynch G, et al. Brain-derived neurotrophic factor restores synaptic plasticity in a knock-in mouse model of Huntington’s disease. J Neurosci. 2007;27(16):4424–4434. doi: 10.1523/JNEUROSCI.5113-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang H, Chen X, Li Y, Tang TS, Bezprozvanny I. Tetrabenazine is neuroprotective in Huntington’s disease mice. Mol Neurodegener. 2010;5:18. doi: 10.1186/1750-1326-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cyr M, Sotnikova TD, Gainetdinov RR, Caron MG. Dopamine enhances motor and neuropathological consequences of polyglutamine expanded huntingtin. FASEB J. 2006;20(14):2541–2543. doi: 10.1096/fj.06-6533fje. [DOI] [PubMed] [Google Scholar]
- 49. Glass M, Dragunow M, Faull RL. The pattern of neurodegeneration in Huntington’s disease: a comparative study of cannabinoid, dopamine, adenosine and GABA(A) receptor alterations in the human basal ganglia in Huntington’s disease. Neuroscience. 2000;97(3):505–519. doi: 10.1016/S0306-4522(00)00008-7. [DOI] [PubMed] [Google Scholar]
- 50. Denovan-Wright EM, Robertson HA. Cannabinoid receptor messenger RNA levels decrease in a subset of neurons of the lateral striatum, cortex and hippocampus of transgenic Huntington’s disease mice. Neuroscience. 2000;98(4):705–713. doi: 10.1016/S0306-4522(00)00157-3. [DOI] [PubMed] [Google Scholar]
- 51. McCaw EA, Hu H, Gomez GT, Hebb AL, Kelly ME, Denovan–Wright EM. Structure, expression and regulation of the cannabinoid receptor gene (CB1) in Huntington’s disease transgenic mice. Eur J Biochem. 2004;271(23–24):4909–4920. doi: 10.1111/j.1432-1033.2004.04460.x. [DOI] [PubMed] [Google Scholar]
- 52. Lastres–Becker I, Gomez M, De Miguel R, Ramos JA, Fernandez–Ruiz J. Loss of cannabinoid CB(1) receptors in the basal ganglia in the late akinetic phase of rats with experimental Huntington’s disease. Neurotox Res. 2002;4(7–8):601–608. doi: 10.1080/10298420290030514. [DOI] [PubMed] [Google Scholar]
- 53. Lovinger DM. Neurotransmitter roles in synaptic modulation, plasticity and learning in the dorsal striatum. Neuropharmacology. 2010;58(7):951–961. doi: 10.1016/j.neuropharm.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cepeda C, Cummings DM, Andre VM, Holley SM, Levine MS. Genetic mouse models of Huntington’s disease: focus on electrophysiological mechanisms. ASN Neuro. 2010;2(2):e00033. doi: 10.1042/AN20090058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Milnerwood AJ, et al. Early increase in extrasynaptic NMDA receptor signaling and expression contributes to phenotype onset in Huntington’s disease mice. Neuron. 2010;65(2):178–190. doi: 10.1016/j.neuron.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 56. Okamoto S, et al. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat Med. 2009;15(12):1407–1413. doi: 10.1038/nm.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hansson O, et al. Resistance to NMDA toxicity correlates with appearance of nuclear inclusions, behavioural deficits and changes in calcium homeostasis in mice transgenic for exon 1 of the huntington gene. Eur J Neurosci. 2001;14(9):1492–1504. doi: 10.1046/j.0953-816x.2001.01767.x. [DOI] [PubMed] [Google Scholar]
- 58. Petersen A, Chase K, Puschban Z, DiFiglia M, Brundin P, Aronin N. Maintenance of susceptibility to neurodegeneration following intrastriatal injections of quinolinic acid in a new transgenic mouse model of Huntington’s disease. Exp Neurol. 2002;175(1):297–300. doi: 10.1006/exnr.2002.7885. [DOI] [PubMed] [Google Scholar]
- 59. Cowan CM, et al. Polyglutamine-modulated striatal calpain activity in YAC transgenic huntington disease mouse model: impact on NMDA receptor function and toxicity. J Neurosci. 2008;28(48):12725–12735. doi: 10.1523/JNEUROSCI.4619-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Prybylowski K, Chang K, Sans N, Kan L, Vicini S, Wenthold RJ. The synaptic localization of NR2B-containing NMDA receptors is controlled by interactions with PDZ proteins and AP-2. Neuron. 2005;47(6):845–857. doi: 10.1016/j.neuron.2005.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu HY, Hsu FC, Gleichman AJ, Baconguis I, Coulter DA, Lynch DR. Fyn-mediated phosphorylation of NR2B Tyr-1336 controls calpain-mediated NR2B cleavage in neurons and heterologous systems. J Biol Chem. 2007;282(28):20075–20087. doi: 10.1074/jbc.M700624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wilson CJ, Kawaguchi Y. The origins of two-state spontaneous membrane potential fluctuations of neostriatal spiny neurons. J Neurosci. 1996;16(7):2397–2410. doi: 10.1523/JNEUROSCI.16-07-02397.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tang TS, et al. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington’s disease. Proc Natl Acad Sci U S A. 2005;102(7):2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tang TS, Chen X, Liu J, Bezprozvanny I. Dopaminergic signaling and striatal neurodegeneration in Huntington’s disease. J Neurosci. 2007;27(30):7899–7910. doi: 10.1523/JNEUROSCI.1396-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Beal MF. Huntington’s disease, energy, and excitotoxicity. Neurobiol Aging. 1994;15(2):275–276. doi: 10.1016/0197-4580(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 66. Schwarcz R, Guidetti P, Sathyasaikumar KV, Muchowski PJ. Of mice, rats and men: Revisiting the quinolinic acid hypothesis of Huntington’s disease. Prog Neurobiol. 2010;90(2):230–245. doi: 10.1016/j.pneurobio.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Turrigiano G. Homeostatic signaling: the positive side of negative feedback. Curr Opin Neurobiol. 2007;17(3):318–324. doi: 10.1016/j.conb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 68. Day M, et al. Selective elimination of glutamatergic synapses on striatopallidal neurons in Parkinson disease models. Nat Neurosci. 2006;9(2):251–259. doi: 10.1038/nn1632. [DOI] [PubMed] [Google Scholar]
- 69. Warre R, et al. Altered function of glutamatergic cortico-striatal synapses causes output pathway abnormalities in a chronic model of parkinsonism. Neurobiol Dis. doi: 10.1016/j.nbd.2010.10.013. [published online ahead of print October 21, 2010]. doi:10.1016/j.nbd.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 70. Bevan MD, Magill PJ, Terman D, Bolam JP, Wilson CJ. Move to the rhythm: oscillations in the subthalamic nucleus-external globus pallidus network. Trends Neurosci. 2002;25(10):525–531. doi: 10.1016/S0166-2236(02)02235-X. [DOI] [PubMed] [Google Scholar]
- 71. Chan CS, et al. HCN channelopathy in external globus pallidus neurons in models of Parkinson’s disease. Nat Neurosci. 2011;14(1):85–92. doi: 10.1038/nn.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mallet N, et al. Disrupted dopamine transmission and the emergence of exaggerated beta oscillations in subthalamic nucleus and cerebral cortex. J Neurosci. 2008;28(18):4795–4806. doi: 10.1523/JNEUROSCI.0123-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tang JK, et al. Firing rates of pallidal neurons are similar in Huntington’s and Parkinson’s disease patients. Exp Brain Res. 2005;166(2):230–236. doi: 10.1007/s00221-005-2359-x. [DOI] [PubMed] [Google Scholar]
- 74. Starr PA, Kang GA, Heath S, Shimamoto S, Turner RS. Pallidal neuronal discharge in Huntington’s disease: support for selective loss of striatal cells originating the indirect pathway. Exp Neurol. 2008;211(1):227–233. doi: 10.1016/j.expneurol.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fasano A, Mazzone P, Piano C, Quaranta D, Soleti F, Bentivoglio AR. GPi-DBS in Huntington’s disease: results on motor function and cognition in a 72-year-old case. Mov Disord. 2008;23(9):1289–1292. doi: 10.1002/mds.22116. [DOI] [PubMed] [Google Scholar]
- 76. Paulsen JS. Functional imaging in Huntington’s disease. Exp Neurol. 2009;216(2):272–277. doi: 10.1016/j.expneurol.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Esmaeilzadeh M, Ciarmiello A, Squitieri F. Seeking brain biomarkers for preventive therapy in Huntington disease. CNS Neurosci Ther. doi: 10.1111/j.1755-5949.2010.00157.x. [published online ahead of print June 11, 2010]. doi:10.1111/j.1755-5949.2010.00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jenkins BG, et al. Effects of CAG repeat length, HTT protein length and protein context on cerebral metabolism measured using magnetic resonance spectroscopy in transgenic mouse models of Huntington’s disease. J Neurochem. 2005;95(2):553–562. doi: 10.1111/j.1471-4159.2005.03411.x. [DOI] [PubMed] [Google Scholar]
- 79. Rosas HD, Salat DH, Lee SY, Zaleta AK, Hevelone N, Hersch SM. Complexity and heterogeneity: what drives the ever-changing brain in Huntington’s disease? Ann N Y Acad Sci. 2008;1147:196–205. doi: 10.1196/annals.1427.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Montoya A, Price BH, Menear M, Lepage M. Brain imaging and cognitive dysfunctions in Huntington’s disease. J Psychiatry Neurosci. 2006;31(1):21–29. [PMC free article] [PubMed] [Google Scholar]
- 81. Kuwert T, Lange HW, Langen KJ, Herzog H, Aulich A, Feinendegen LE. Cortical and subcortical glucose consumption measured by PET in patients with Huntington’s disease. Brain. 1990;113(pt 5):1405–1423. doi: 10.1093/brain/113.5.1405. [DOI] [PubMed] [Google Scholar]
- 82. Cha JH, et al. Altered neurotransmitter receptor expression in transgenic mouse models of Huntington’s disease. Philos Trans R Soc Lond B Biol Sci. 1999;354(1386):981–989. doi: 10.1098/rstb.1999.0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Antonini A, et al. Striatal glucose metabolism and dopamine D2 receptor binding in asymptomatic gene carriers and patients with Huntington’s disease. Brain. 1996;119(pt 6):2085–2095. doi: 10.1093/brain/119.6.2085. [DOI] [PubMed] [Google Scholar]
- 84. Antonini A, Leenders KL, Eidelberg D. [11C]raclopride-PET studies of the Huntington’s disease rate of progression: relevance of the trinucleotide repeat length. Ann Neurol. 1998;43(2):253–255. doi: 10.1002/ana.410430216. [DOI] [PubMed] [Google Scholar]
- 85. van Oostrom JC, et al. Striatal dopamine D2 receptors, metabolism, and volume in preclinical Huntington disease. Neurology. 2005;65(6):941–943. doi: 10.1212/01.wnl.0000176071.08694.cc. [DOI] [PubMed] [Google Scholar]
- 86. Rosas HD, et al. Striatal volume loss in HD as measured by MRI and the influence of CAG repeat. Neurology. 2001;57(6):1025–1028. doi: 10.1212/wnl.57.6.1025. [DOI] [PubMed] [Google Scholar]
- 87. Aylward EH. Change in MRI striatal volumes as a biomarker in preclinical Huntington’s disease. Brain Res Bull. 2007;72(2–3):152–158. doi: 10.1016/j.brainresbull.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 88. Feigin A, et al. Thalamic metabolism and symptom onset in preclinical Huntington’s disease. Brain. 2007;130(pt 11):2858–2867. doi: 10.1093/brain/awm217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Eidelberg D. Metabolic brain networks in neurodegenerative disorders: a functional imaging approach. Trends Neurosci. 2009;32(10):548–557. doi: 10.1016/j.tins.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Spetsieris PG, Eidelberg D. Scaled subprofile modeling of resting state imaging data in Parkinson’s disease: methodological issues. NeuroImage. doi: 10.1016/j.neuroimage.2010.10.025. [published online ahead of print October 20, 2010]. doi:10.1016/j.neuroimage.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tang CC, Poston KL, Dhawan V, Eidelberg D. Abnormalities in metabolic network activity precede the onset of motor symptoms in Parkinson’s disease. J Neurosci. 2010;30(3):1049–1056. doi: 10.1523/JNEUROSCI.4188-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR. A new model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin Genet. 2004;65(4):267–277. doi: 10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- 93. Rosas HD, et al. Regional and progressive thinning of the cortical ribbon in Huntington’s disease. Neurology. 2002;58(5):695–701. doi: 10.1212/wnl.58.5.695. [DOI] [PubMed] [Google Scholar]
- 94. Rosas HD, Feigin AS, Hersch SM. Using advances in neuroimaging to detect, understand, and monitor disease progression in Huntington’s disease. NeuroRx. 2004;1(2):263–272. doi: 10.1602/neurorx.1.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Paulsen JS, et al. Brain structure in preclinical Huntington’s disease. Biol Psychiatry. 2006;59(1):57–63. doi: 10.1016/j.biopsych.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 96. Tang C, et al. A metabolic network associated with the progression of preclinical Huntington’s disease: application of the ordinal trends analysis to a longitudinal PET study. Neurotherapeutics. 2009;6(1):207–208. [Google Scholar]
- 97. Feigin A, et al. Preclinical Huntington’s disease: compensatory brain responses during learning. Ann Neurol. 2006;59(1):53–59. doi: 10.1002/ana.20684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Smith Y, Raju DV, Pare JF, Sidibe M. The thalamostriatal system: a highly specific network of the basal ganglia circuitry. Trends Neurosci. 2004;27(9):520–527. doi: 10.1016/j.tins.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 99. Sotrel A, Paskevich PA, Kiely DK, Bird ED, Williams RS, Myers RH. Morphometric analysis of the prefrontal cortex in Huntington’s disease. Neurology. 1991;41(7):1117–1123. doi: 10.1212/wnl.41.7.1117. [DOI] [PubMed] [Google Scholar]
- 100. Heinsen H, et al. Nerve cell loss in the thalamic centromedian-parafascicular complex in patients with Huntington’s disease. Acta Neuropathol. 1996;91(2):161–168. doi: 10.1007/s004010050408. [DOI] [PubMed] [Google Scholar]
- 101. Heinsen H, et al. Nerve cell loss in the thalamic mediodorsal nucleus in Huntington’s disease. Acta Neuropathol. 1999;97(6):613–622. doi: 10.1007/s004010051037. [DOI] [PubMed] [Google Scholar]
- 102. Kassubek J, Juengling FD, Ecker D, Landwehrmeyer GB. Thalamic atrophy in Huntington’s disease co-varies with cognitive performance: a morphometric MRI analysis. Cereb Cortex. 2005;15(6):846–853. doi: 10.1093/cercor/bhh185. [DOI] [PubMed] [Google Scholar]
- 103. Mayberg HS, Starkstein SE, Peyser CE, Brandt J, Dannals RF, Folstein SE. Paralimbic frontal lobe hypometabolism in depression associated with Huntington’s disease. Neurology. 1992;42(9):1791–1797. doi: 10.1212/wnl.42.9.1791. [DOI] [PubMed] [Google Scholar]
- 104. Reading SA, et al. Functional brain changes in presymptomatic Huntington’s disease. Ann Neurol. 2004;55(6):879–883. doi: 10.1002/ana.20121. [DOI] [PubMed] [Google Scholar]
- 105. Pavese N, et al. Microglial activation correlates with severity in Huntington disease: a clinical and PET study. Neurology. 2006;66(11):1638–1643. doi: 10.1212/01.wnl.0000222734.56412.17. [DOI] [PubMed] [Google Scholar]
- 106. Lodi R, et al. Abnormal in vivo skeletal muscle energy metabolism in Huntington’s disease and dentatorubropallidoluysian atrophy. Ann Neurol. 2000;48(1):72–76. doi: 10.1002/1531-8249(200007)48:1<72::AID-ANA11>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 107. Saft C, et al. Mitochondrial impairment in patients and asymptomatic mutation carriers of Huntington’s disease. Mov Disord. 2005;20(6):674–679. doi: 10.1002/mds.20373. [DOI] [PubMed] [Google Scholar]
- 108. Koroshetz WJ, Jenkins BG, Rosen BR, Beal MF. Energy metabolism defects in Huntington’s disease and effects of coenzyme Q10. Ann Neurol. 1997;41(2):160–165. doi: 10.1002/ana.410410206. [DOI] [PubMed] [Google Scholar]
- 109. Gu M, Gash MT, Mann VM, Javoy-Agid F, Cooper JM, Schapira AH. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann Neurol. 1996;39(3):385–389. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- 110. Browne SE, et al. Oxidative damage and metabolic dysfunction in Huntington’s disease: selective vulnerability of the basal ganglia. Ann Neurol. 1997;41(5):646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 111. Tabrizi SJ, Cleeter MW, Xuereb J, Taanman JW, Cooper JM, Schapira AH. Biochemical abnormalities and excitotoxicity in Huntington’s disease brain. Ann Neurol. 1999;45(1):25–32. doi: 10.1002/1531-8249(199901)45:1<25::AID-ART6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 112. Brouillet E, Jacquard C, Bizat N, Blum D. 3-Nitropropionic acid: a mitochondrial toxin to uncover physiopathological mechanisms underlying striatal degeneration in Huntington’s disease. J Neurochem. 2005;95(6):1521–1540. doi: 10.1111/j.1471-4159.2005.03515.x. [DOI] [PubMed] [Google Scholar]
- 113. Moeller JR, Habeck CG. Reciprocal benefits of mass-univariate and bilinear modeling in brain mapping: applications to event-related functional MRI, H215O- and FDG-PET. . Int J Biomed Imag. 2006;2006:1–13. doi: 10.1155/IJBI/2006/79862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Singer JD, Willett JB. Applied Longitudinal Data Analysis: Modeling Change And Event Occurrence. New York, New York, USA: Oxford University Press; 2003. [Google Scholar]
- 115. Feigin A, et al. Metabolic network abnormalities in early Huntington’s disease: an [(18)F]FDG PET study. J Nucl Med. 2001;42(11):1591–1595. [PubMed] [Google Scholar]