Abstract
Huntington disease (HD) is an autosomal dominant neurodegenerative disease with complete penetrance. Although the understanding of the cellular mechanisms that drive neurodegeneration in HD and account for the characteristic pattern of neuronal vulnerability is incomplete, defects in energy metabolism, particularly mitochondrial function, represent a common thread in studies of HD pathogenesis in humans and animal models. Here we review the clinical, biochemical, and molecular evidence of an energy deficit in HD and discuss the mechanisms underlying mitochondrial and related alterations.
Introduction
Huntington disease (HD) is an autosomal dominant neurodegenerative disease with age-dependent complete penetrance. HD is caused by a CAG repeat expansion in the first exon of the HTT gene that encodes huntingtin (Htt) (1, 2). Individuals who have 36 CAG repeats or more may develop the clinical symptoms and signs of HD, including neuropsychiatric, motor, and cognitive abnormalities that cause a progressive loss of functional capacity and shortened life span. The clinical features of HD typically emerge in adulthood between 30 and 50 years of age, after which the disease progresses relentlessly over the next 15–20 years (3). Presymptomatic testing — preceded by genetic counseling according to internationally accepted guidelines — allows an at-risk person to assess their genetic status and thus predict whether a carrier will develop HD before he or she shows clinical symptoms and signs. Of importance, the presymptomatic phase in HD provides a unique window for therapeutic intervention and neuroprotection.
γ-Aminobutyric acid (GABAergic) medium spiny neurons of the striatum that contain enkephalin or substance P and project to the globus pallidus and substantia nigra are particularly vulnerable in HD and are key to the characteristic involuntary movements in this disease (4). The basis of the selective vulnerability of these neurons in HD remains elusive but may involve the reduced phosphorylation of Htt at serine 421 in the striatum (5), the specific localization of Rhes (Ras homolog enriched in striatum; a small guanine nucleotide–binding protein partner of Htt) to the striatum (6), and/or the preferential vulnerability of the striatum to mitochondrial dysfunction (7). However, the neuropathology is ultimately more widespread, affecting cortical neurons as well as the globus pallidus, substantia nigra, and brainstem. Understanding of the cellular mechanisms that drive neurodegeneration in HD and account for the characteristic pattern of neuronal vulnerability remains incomplete, but defects in energy metabolism, particularly mitochondrial function, represent a common thread in studies of HD pathogenesis in humans and animal models (7, 8). Here we provide an overview of clinical, biochemical, and molecular evidence of an energy deficit in HD, in both the brain and the peripheral organs. We also discuss the mechanisms underlying mitochondrial and related alterations in the context of both a toxic gain-of-function from mutant Htt and the loss of function of normal Htt (Figure 1). The emerging availability of surrogate markers for clinical trials in HD, i.e., neuroimaging, neurophysiological, and neuropsychiatric changes that may be detectable as early as the presymptomatic stages of the disease (9), should open new avenues for testing promising therapeutic approaches to ameliorate the energy deficit while providing favorable safety profiles.
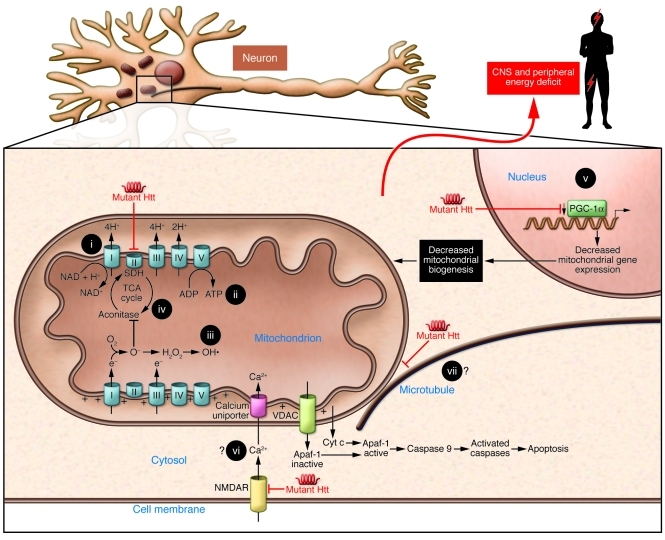
Figure 1. Summary figure indicating the main putative mechanisms by which mutated Htt causes mitochondrial dysfunction.
Such mechanisms would result in decreased mitochondrial biogenesis, oxidative stress, ATP deficit, increased apoptosis, and, ultimately, a central and peripheral energy deficit. Energy-related therapeutic approaches that have been used in preclinical models and/or HD patients include (i) coenzyme Q10, (ii) creatine, (iii) antioxidant therapies, (iv) anaplerotic therapies, and (v) PPAR agonists. Potential therapeutic targets are also shown, i.e., (vi) calcium homeostasis and (vii) mitochondrial transport. Apaf-1, apoptotic protease activating factor 1; NMDAR, N-Methyl-D-aspartic acid receptor; VDAC, voltage-dependent anion channel.
Normal function of Htt
Despite its identification almost two decades ago, understanding of the normal function of Htt protein is still incomplete. Although mainly found in the cytoplasm, Htt is also present in the nucleus (10, 11). Htt contains a series of up to 36 HEAT, α helix–loop–α helix, domains that mediate its interaction with numerous proteins involved in gene expression, endocytosis, vesicle trafficking, intracellular signaling, and metabolism (12–14). In particular, the role of Htt in transcription and intracellular transport is well established (15, 16). In the cytoplasm, Htt interacts with the dynein/dynactin complex and kinesin in microtubule-dependent transport of organelles in neurons, including mitochondria (17). For example, WT Htt enhances the vesicular transport of brain derived neurotrophic factor along microtubules (18). Htt may regulate transcription by shuttling transcription factors between the nucleus and the cytoplasm and by interacting with spliceosome-related proteins (19). Studies in Htt knockout mice have shown that Htt is required for normal embryonic development and neurogenesis (20). Interestingly, expression of Htt is also essential for normal development in zebrafish because knock-down by antisense morpholino oligonucleotides leads to multiple developmental abnormalities, including defects in cellular iron trafficking (21), which supports previous in vitro findings that Htt may be regulated by iron levels (22). Recently, it was shown that Htt also plays a role in dividing cells through the regulation of spindle orientation during mitosis together with dynein/dynactin (23).
Pathophysiology of HD
Knockout of Htt is lethal in the mouse early in embryogenesis (24). Reduced Htt levels in knockout embryonic stem cells induce an abnormal distribution and morphology of organelles such as the endoplasmic reticulum, Golgi, and mitochondria (22). In addition, conditional Htt deletion in forebrain neurons at late embryonic or early neonatal stages leads to a rapidly progressive neurodegenerative phenotype (25). Reduced Htt levels have been shown to sensitize neurons to the toxic effect of mutant Htt (26). Moreover, increased expression of WT Htt in transgenic mice protected neurons from cell death (27). Similarly, increased expression of WT Htt reduced toxicity in peripheral cells overexpressing mutant Htt (28). Thus, these data indicate that a loss-of-function mechanism likely contributes to HD pathogenesis (29).
Nonetheless, cell and animal model systems as well as the direct correlation between length of CAG repeat and onset of HD strongly support the view that mutant HTT also possesses a new toxic gain of function (30). Like many neurodegenerative diseases, HD is characterized by a cascade of events leading to neuronal cell death. While the sequence of these events remains unclear, several mechanisms implicate energy defects. These include transcriptional dysregulation, changes in axonal transport, excitotoxicity, signaling dysfunction, and neuronal aggregates (31). Here we focus on the role of mitochondrial dysfunction and altered energy metabolism secondary to mechanisms such as impaired trafficking and transcriptional interference (12, 15).
Evidence of energy deficit in HD
Brain energy deficit in HD.
Brain energy metabolism has been a focus of HD research for many years due to several observations in both patients and models of the disease (32). In HD patients, there is strong evidence for reduced glucose consumption in the brain, especially in the basal ganglia, even in presymptomatic mutation carriers (33–35). A brain energy deficit has also been suspected based on increased lactate concentrations in the basal ganglia and the occipital cortex of HD patients (36, 37) as well as an elevated lactate-to-pyruvate ratio in the CSF (38). Interestingly, reduced ATP synthesis was found in immortalized HD striatal neuronal cell lines (39). However, ATP depletion was directly demonstrated in HD brain tissues only recently by employing a microwave fixation system that instantly inactivates brain enzymes in rodents to preserve in vivo levels of metabolites such as ATP and phosphocreatine (40). Various mechanisms that underlie the energy deficit in HD brain have been proposed, including impaired oxidative phosphorylation (41), oxidative stress (42), impaired mitochondrial calcium handling (43), abnormal mitochondria trafficking (44), deregulation of key factors of mitochondrial biogenesis, such as the transcriptional coactivator PPARγ coactivator-1α (PGC-1α) (45), and decreased glycolysis (46). Furthermore, experimental evidence supports the view that excitatory glutaminergic inputs, which activate NMDA receptors in vulnerable medium spiny neurons, promote cell death by increasing energy demand in the setting of impaired energy capacity (47).
Energy deficit in HD: not only in the brain.
The existence of a peripheral energy deficit in HD is an attractive hypothesis because it suggests that clues to the disease pathogenesis may be detectable outside the brain (48). A peripheral mitochondrial defect is suggested by the observation that the ATP/ADP ratio in HD patient–derived lymphoblastoid cell lines is low, which mirrors the findings in striatal cells that contain a full-length mutant Htt (49). Furthermore, the ATP/ADP ratio in lymphoblasts is inversely correlated with the length of the mutant polyglutamine tract, implying an increasingly severe limitation in mitochondrial ATP production with the polyglutamine expansion (49). Skeletal muscle, another differentiated excitable cell type with high metabolic demand, shows energetic disturbances in HD patients similar to those evident in HD striatal neurons (50, 51). Studies utilizing 31P-nuclear magnetic resonance spectroscopy (NMRS) have also revealed a reduced phosphocreatine/inorganic phosphate ratio and low ATP levels in resting muscle of symptomatic HD patients and presymptomatic individuals (52, 53). These findings raise the possibility that the ubiquitous expression of mutant Htt may place other cell types at risk, particularly those with high metabolic demand. Notably, it was found that R6/2 mice, the most commonly used model of HD that contains a microinjected fragment from the 5′ end of the human HTT gene isolated from a phage genomic clone derived from an HD patient, develop cardiac dysfunction by 8 weeks of age, progressing to severe failure at 12 weeks (54). Moreover, weight loss has long been recognized in symptomatic HD patients (55) and is not explained by decreased food intake (56, 57), higher sedentary energy expenditure (58), or involuntary movements (59). Instead, there is evidence for an early hypermetabolic state in HD (60), in which weight loss is associated with a systemic metabolic defect at the presymptomatic stages of the disease. These findings of increased metabolic rate in HD were confirmed in several studies in patients (61, 62) and in mouse models (63, 64). Hypothalamic dysfunction has been suggested to contribute to the negative energy balance in HD through the deregulation of the ghrelin/leptin/orexin system (65, 66). However, increased ghrelin levels and decreased leptin levels in HD, both of which promote increased appetite and food intake, may reflect an adaptation to the hypercatabolism in HD, such that increased calorie intake is necessary to compensate for the decreased body weight (60).
Mutated Htt may exert direct deleterious effects on the metabolism of other peripheral organs. A deregulation of adipose tissue metabolism in HD models involves transcriptional interference with PGC-1α (67). In addition, the brain is entirely dependent on energy substrates provided by peripheral organs, so it is conceivable that early weight loss reflects the activation of compensatory mechanisms in peripheral organs in order to provide energetic substrates for a chronic brain energy deficit in HD (60). The slower disease progression in patients exhibiting a higher BMI at presentation supports this hypothesis (68).
Biochemical evidence of energy dysfunction in HD
Is oxidative phosphorylation deficient in HD?
Deficiency of respiratory chain complex II, i.e., succinate dehydrogenase (SDH), has been a focus of attention in HD due to the fact that the inhibition of SDH by 3-nitropropionate (3-NP) or malonate has been shown to closely mimic the neuropathology and clinical features of HD in humans, non-human primates (69), and rodents (70). The possible relevance of respiratory chain deficiency to the human disease is supported by postmortem studies of symptomatic patients showing marked deficiency of complexes II and III and lesser deficiency of complex IV in the caudate or putamen, with normal levels in the frontal cortex or cerebellum (71–73). Benchoua and colleagues identified reductions in the 70-kD FAD subunit (SDH A) and the 30-kD iron-sulfur cluster subunit (SDH B) of complex II in HD caudate and putamen that were disproportionate to decreases in the levels of calbindin, a marker of medium spiny neurons, thus implying that complex II subunit depletion preceded neuronal death (74). Furthermore, cultured striatal neurons transfected with a pathogenic N-terminus Htt fragment showed decreased complex II enzymatic activity associated with selective depletion of SDH A and B, and cells were protected from death upon overexpression of complex II/SDH subunits in this model (74).
In a yeast model, a mutant Htt fragment suppressed mitochondrial respiration by suppressing the function of complexes II and III (75). Other cell models of HD have implicated selective deficiency of complex III in cellular pathophysiology (76). However, no significant deficiency of respiratory chain complexes has been demonstrated in presymptomatic patients (77) or in mice expressing full-length mutant Htt (77), suggesting that respiratory chain defects are a secondary feature in the pathogenesis of HD (78). Nevertheless, selective reduction of respiratory chain complexes implies that certain pathophysiological mechanisms target them. One possible mechanism is oxidative stress, to which iron-sulfur cluster–containing proteins, including the SDH B subunit of complex II and the Rieske protein of complex III, are particularly vulnerable (79, 80).
Mitochondria, a source and a target of oxidative stress in HD.
Oxidative stress is mediated by increases in ROS including superoxide (O2–), hydrogen peroxide (H2O2), hydroxyl radical (OH–), and reactive nitrogen species such as peroxynitrite (ONOO–), and ROS impairs cellular function by degrading proteins, lipids, and nucleic acids. Evidence of enhanced oxidative stress in HD brains includes an increase in accumulation of lipofuscin, a product of unsaturated fatty acid peroxidation, that is most prominent in vulnerable striatal neurons (81). Other markers of ROS include strand breaks in DNA and the accumulation of 8-hydroxy-2′-deoxyguanosine (OH8dG) in HD brain and blood (82). Oxidative modification of proteins (protein carbonyls) and lipids (malondialdehyde and 4-hydroxynonenal) are also increased in HD brain and in animal models (83), and increased malondialdehyde is found in blood (84). Another indicator of increased oxidative stress is the finding that oxidative defense mechanisms including mitochondrial and cytoplasmic superoxide dismutase (MnSOD and Cu-Zn SOD, respectively) are induced in HD brains (85) and transgenic animals (86). While the origin of increased ROS in HD and HD models has not been identified conclusively, the main suspects are mitochondria, where it is estimated that 2 percent of electron flow in the respiratory chain escapes as superoxide, predominantly via complexes I and III. Respiratory chain defects increase ROS production, and complex II inhibition models of HD display evidence of oxidative stress (87).
Among the most vulnerable targets of oxidative stress is mitochondrial aconitase. Aconitase contains an iron-sulfur cluster that is readily inactivated due to superoxide-mediated loss of iron at its catalytic site (88). In addition, the iron-sulfur cluster makes this protein highly susceptible to oxidative modification and ultimate degradation (89). In keeping with other evidence of increased oxidative stress in HD is the finding of a profound (more than 90%) decrease in aconitase activity in HD caudate, with lesser decreases in putamen (~70%) and cortex (~50%) (42). Decreased striatal aconitase with normal levels in cortex has also been found in the R6/2 transgenic mouse model (90). α-Ketoglutarate dehydrogenase (KGDH) has also been implicated as a target of oxidative stress, though data for altered activity of this enzyme in HD is meager (91). Aconitase, KGDH, and SDH are enzymes of the TCA cycle. Defects in these enzymes in HD as well as the susceptibility of medium spiny neurons to SDH inhibition by 3-NP and malonate suggest that these neurons are particularly vulnerable to TCA cycle inhibition and that known and suspected targets of oxidative stress may restrict mitochondrial energy production in these cells by impairing TCA cycle function.
Cellular mechanisms of altered bioenergetics in HD
Impaired mitochondrial calcium handling.
Mitochondria avidly accumulate calcium and play an important role in buffering changing cytoplasmic calcium levels in response to neuronal activity (92). Mitochondrial calcium transport is powered by the mitochondrial proton gradient, and increased neuronal calcium modifies mitochondrial ATP production by uncoupling oxidative phosphorylation. Calcium overload may result in discharge of the mitochondrial membrane potential, opening of the mitochondrial permeability transition (MPT) pore, release of cytochrome c, and activation of cell death pathways (92). Increased calcium influx through NMDA receptors, increased mitochondrial susceptibility to calcium transients, or both may contribute to neuronal degeneration in HD. Mitochondria isolated from lymphoblasts of HD patients and the brains of transgenic mice have a reduced membrane potential and depolarize at lower calcium concentrations than control mitochondria (93). Immunocytochemical identification of N-terminal mutant Htt on neuronal mitochondrial membranes from transgenic mice and reproduction of the mitochondrial calcium defect by incubation of normal human lymphoblast mitochondria with a fusion protein containing a long polyglutamine repeat suggest that the mitochondrial calcium defect is a direct effect of mutant Htt on mitochondria (93). The reduced mitochondrial ATP levels and decreased ATP/ADP ratio found in mutant Htt-containing striatal cells is linked to increased calcium influx through NMDA receptors, and cell ATP/ADP ratio is normalized by blocking calcium influx (49). Choo et al. (94) identified binding of both WT and mutant Htt to the outer mitochondrial membrane in mitochondria from human neuroblastoma cells and from cultured striatal cells from WT and transgenic mice. They demonstrated increased sensitivity to calcium-induced opening of the MPT and release of cytochrome c in normal liver mitochondria incubated with mutant but not WT Htt, consistent with a direct effect of mutant Htt upon mitochondrial calcium handling (94).
A possible link between impaired intracellular transport and energy deficit.
There are several possible ways in which altered intracellular transport might affect energy metabolism in HD. Full-length mutant Htt may impair mitochondrial motility in mammalian neurons through both a toxic gain of function from the polyglutamine tract and a loss of function of WT Htt (95). Altered mitochondrial trafficking precedes mitochondrial and neuronal abnormalities in both in vitro and in vivo murine models of HD (95). Although aggregates were suspected to impede mitochondrial trafficking (96), N-terminal fragments of mutant Htt alone were later shown to interfere with the association of microtubule-based transport proteins with mitochondria in vitro (97). Mutant Htt altered the distribution and reduced the transport rate of mitochondria together with lowered ATP levels in the synaptosomal fraction isolated from knockin mouse brain (97). However, the mechanisms by which mutant Htt affects intracellular organelle trafficking such as mitochondria are not fully understood (44). A recent study of HD brains identified a reduced number and altered distribution of mitochondria within vulnerable, calbindin-positive striatal neurons that was more pronounced with disease progression (98). Reductions were seen preferentially in large- and medium-size mitochondria in conjunction with an increase in levels of Drp1 protein, a mediator of mitochondrial fission and, with disease progression, a decrease in levels of Mfn1, a mediator of mitochondrial fusion (98). These results support the view that altered mitochondrial dynamics represent an important mechanism of mitochondrial dysfunction contributing to the mismatch between energy supply and demand that is a recurring theme in HD and HD models.
Transcriptional deregulation contributes to altered bioenergetics.
Several lines of evidence suggest that transcriptional dysregulation underlies the toxicity of mutant Htt. Whether it is mediated by aggregates, soluble fractions of Htt, or by Htt itself, many transcription factors have been shown to be downregulated in HD. These include CREB-binding protein, TATA-binding protein, and Sp1/TAFII130 (99). Mutant Htt binds the tumor suppression gene p53 more avidly than does WT Htt and has been reported to increase p53 protein levels, nuclear localization, and transcriptional activity in neuronal cultures and transgenic mice (100). Augmented p53 activity was shown to mediate mitochondrial membrane depolarization and a decrease in complex IV activity, as indicated by the fact that p53 inhibition or genetic deletion ameliorated these changes in a cell culture model (100). A specific effect of mutant Htt upon p53 activity is suggested by the fact that p53 activity was not altered by another polyQ-containing protein, ataxin-1 (100). Mutant Htt has also been shown to directly decrease the expression of PGC-1α in the striatum of HD mice and patients through CREB-dependent transcriptional inhibition (45). PGC-1α is a transcriptional coactivator that regulates key energetic metabolic pathways in both brain and peripheral tissues (101). The possible role of PGC-1α in HD pathogenesis was initially suspected from the observation of selective striatal lesions in the PGC-1α knockout mouse (102). Downregulation of PGC-1α in HD striatum was then shown to affect mitochondrial energy metabolism, possibly by impairing oxidative phosphorylation (45). Conversely, HD striatal neurons expressing exogenous PGC-1α were resistant to 3-NP treatment (103). Decreasing levels of PGC-1α were shown to parallel markers of mitochondrial dysfunction with disease progression in HD brains (98). The PPARγ agonist rosiglitazone has been shown to increase mitochondrial mass in the striatal cell model of HD and to prevent the decrease in mitochondrial membrane potential and the increase in oxidative stress that otherwise occurs when these cells are challenged with pathogenic increases in intracellular calcium levels (104). Of note, PGC-1α polymorphisms in HD patients may account for some variability in symptoms onset (105). In addition, adipose-derived stem cells slowed striatal degeneration and behavioral deterioration of HD models while increasing PGC-1α expression (106). The implication that the PGC-1α dysfunction contributes to HD disease progression is not restricted to the brain (67). Thermoregulatory defects were found in the R6/2 mouse model, associated with impaired activation of brown fat (103). The expression of PGC-1α and its target genes were also decreased in muscle of HD transgenic mice as well as in muscle of HD patients (107).
Although PGC-1α function provides an important link between transcriptional dysregulation and mitochondrial dysfunction in HD, it does not readily explain a selective decrease in respiratory chain proteins such as subunits of SDH/complex II in the pathophysiology of the disease (74). Moreover, mutant Htt may impair cellular bioenergetics by affecting extra-mitochondrial pathways. This was suggested by the dissimilarity of gene expression profiles in the 3-NP models and the HdhQ111/Q111 knockin mouse, in which a gene-targeted replacement of exon 1 of the mouse Hdh with a chimeric mouse/human exon 1 codes for approximately 111 CAG repeats. Whereas both 3-NP and the HD mutation led to impaired energy homeostasis, only 3-NP altered nuclear-encoded mitochondrial gene expression (108).
Therapeutic approaches targeting the energy deficit in HD
As in other neurodegenerative disorders, proven disease-modifying drugs are not yet available in HD, and most therapeutic approaches to improving energetics have aimed at providing substrates designed to slow progression or otherwise compensate for the energy deficit in HD. One such therapy is creatine, which encompasses several potential neuroprotective effects such as buffering intracellular energy pools and stabilizing intracellular calcium concentrations (109). The identification of decreased creatine levels in brain tissues from R6/2 mice (110) encouraged several trials of creatine supplementation in HD mouse models (110–113) despite a lack of overt benefit in HD patients so far (114, 115). Instead, more recent studies in which in vivo concentrations of brain metabolites were preserved by using high-resolution 1H-NMRS (116) or enzymatic microwave fixation (40) found increased brain levels of creatine and phosphocreatine in the same mouse model of HD. Moreover, clinical trials of antioxidants designed to slow the progression of the disease have been disappointing. One large-scale study assessing the potential neuroprotective effects of coenzyme Q10, a potential antioxidant and promoter of respiratory chain function, demonstrated a trend toward slowing the decline of the total functional capacity score over 30 months (117) but did not achieve statistical significance. Trials with higher doses are currently ongoing. Other antioxidant approaches were initiated in preclinical models (118, 119) but have not translated into clinical trials. Based on the hypothesis that there is a deficit of substrates for the TCA cycle in HD (60), a short-term therapeutic clinical trial was performed using an anaplerotic compound, triheptanoin, a triglyceride containing seven carbon fatty acids that is metabolized to acetyl-CoA and propionyl-CoA (a precursor of the TCA cycle intermediate, succinate). This study displayed encouraging results for the peripheral metabolism in HD patients, and in particular oxidative phosphorylation in skeletal muscle (120). However, the benefit of anaplerotic approaches for brain energy metabolism remains to be established.
The identification of PGC-1α as a key component of HD pathophysiology supports the use of therapies targeting mitochondrial biogenesis. Activation of the SIRT1 gene family by the polyphenol resveratrol potently induces mitochondrial activity through activating PGC-1α, as evidenced by increased oxidative-type muscle fibers, enhanced resistance to muscle fatigue, and increased tolerance to cold, all PGC-1α–dependent effects (121). Likewise, resveratrol was shown to partially suppress the deleterious effects of mutant polyglutamine in nematode, mouse, and mouse neuronal culture models (122, 123) and significantly reversed 3-NP–induced motor and cognitive impairment (124). In addition, treatments interfering with transcriptional deregulation, such as histone deacetylase inhibitors, were shown to improve mitochondrial dysfunction in striatal neurons from HD mice through the regulation of calcium homeostasis (125). Overall, the multiple functions of Htt in cell metabolism argue for combined therapeutic approaches in order to significantly alter the course of HD and improve energy metabolism, including restoring the normal function of Htt in intracellular transport.
Concluding remarks
Energy deficit is a prominent feature of HD. Energy deficit contributes to the progression of the disease in the brain and can also be traced to the periphery, as reflected in weight loss in HD patients. Importantly, the onset of energy-related manifestations at the presymptomatic stages of the disease, i.e., alterations in brain and muscle metabolism as well as weight loss, underscore the fact that energy deficit is likely to be an early phenomenon in the cascade of events leading to HD pathogenesis. Studies of energy metabolism in HD are therefore worth pursuing to identify potential biomarkers as well as therapeutic targets. Although the data are not entirely concordant, biochemical studies support the view that mitochondrial dysfunction — impaired oxidative phosphorylation, TCA cycle dysfunction, and oxidative stress — is an important determinant of altered energy metabolism in HD. Impaired bioenergetics in HD likely represent downstream effects of both a mutant Htt toxic gain-of-function and a loss-of-function of the WT protein, which, nonetheless, may extend beyond mitochondrial dysfunction. Better understanding of the bioenergetics in HD will more fully illuminate the role of impaired intracellular transport and transcriptional dysregulation in the disease. While treatments designed to improve energy metabolism in the brain and peripheral organs of patients and presymptomatic individuals may not reverse the primary defect in HD on their own, they are likely to significantly alter the course of the disease, delaying disease onset and the pace of disease progression.
Acknowledgments
The authors are grateful to Alexandra Dürr, Sandrine Humbert, and Frédéric Saudou for fruitful discussions.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(2):493–499. doi:10.1172/JCI45691
References
- 1. [No authors listed]. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 2. Gusella JF, et al. A polymorphic DNA marker genetically linked to Huntington’s disease. Nature. 1983;306(5940):234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- 3. Riley BE, Lang AE. Movement Disorders: Huntington’s Disease. In: Bradley WG, Daroff RB, Fenichel GM, Jankovic J, eds.Neurology in Clinical Practice , 5th ed. Philadelphia, Pennsylvania, USA: Elsevier Publishing; 2008:1585–1586. [Google Scholar]
- 4. Mitchell IJ, Cooper AJ, Griffiths MR. The selective vulnerability of striatopallidal neurons. Prog Neurobiol. 1999;59(6):691–719. doi: 10.1016/S0301-0082(99)00019-2. [DOI] [PubMed] [Google Scholar]
- 5. Warby SC, et al. Huntingtin phosphorylation on serine 421 is significantly reduced in the striatum and by polyglutamine expansion in vivo. Hum Mol Genet. 2005;14(11):1569–1577. doi: 10.1093/hmg/ddi165. [DOI] [PubMed] [Google Scholar]
- 6. Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324(5932):1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Damiano M, Galvan L, Deglon N, Brouillet E. Mitochondria in Huntington’s disease. Biochim Biophys Acta. 2010;1802(1):52–61. doi: 10.1016/j.bbadis.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 8. Oliveira JM. Nature and cause of mitochondrial dysfunction in Huntington’s disease: focusing on huntingtin and the striatum. J Neurochem. 2010;114(1):1–12. doi: 10.1111/j.1471-4159.2010.06741.x. [DOI] [PubMed] [Google Scholar]
- 9. Tabrizi SJ, et al. Biological and clinical changes in premanifest and early stage Huntington’s disease in the TRACK-HD study: the 12-month longitudinal analysis. Lancet Neurol. 2011;10(1):31–42. doi: 10.1016/S1474-4422(10)70276-3. [DOI] [PubMed] [Google Scholar]
- 10. Hoogeveen AT, et al. Characterization and localization of the Huntington disease gene product. Hum Mol Genet. 1993;2(12):2069–2073. doi: 10.1093/hmg/2.12.2069. [DOI] [PubMed] [Google Scholar]
- 11. Kegel KB, et al. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. J Biol Chem. 2002;277(9):7466–7476. doi: 10.1074/jbc.M103946200. [DOI] [PubMed] [Google Scholar]
- 12. Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28(8):425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 13. Li SH, Li XJ. Huntingtin-protein interactions and the pathogenesis of Huntington’s disease. Trends Genet. 2004;20(3):146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 14. Caviston JP, Holzbaur EL. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19(4):147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19(5):233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- 16. Li XJ, Li SH. HAP1 and intracellular trafficking. Trends Pharmacol Sci. 2005;26(1):1–3. doi: 10.1016/j.tips.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 17. Caviston JP, Ross JL, Antony SM, Tokito M, Holzbaur EL. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc Natl Acad Sci U S A. 2007;104(24):10045–10050. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gauthier LR, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118(1):127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 19. Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7(9):1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 20. White JK, et al. Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nat Genet. 1997;17(4):404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- 21. Lumsden AL, Henshall TL, Dayan S, Lardelli MT, Richards RI. Huntingtin-deficient zebrafish exhibit defects in iron utilization and development. Hum Mol Genet. 2007;16(16):1905–1920. doi: 10.1093/hmg/ddm138. [DOI] [PubMed] [Google Scholar]
- 22. Hilditch-Maguire P, Trettel F, Passani LA, Auerbach A, Persichetti F, MacDonald ME. Huntingtin: an iron-regulated protein essential for normal nuclear and perinuclear organelles. Hum Mol Genet. 2000;9(19):2789–2797. doi: 10.1093/hmg/9.19.2789. [DOI] [PubMed] [Google Scholar]
- 23. Godin JD, et al. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67(3):392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 24. Nasir J, et al. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81(5):811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 25. Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26(3):300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 26. Auerbach W, et al. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum Mol Genet. 2001;10(22):2515–2523. doi: 10.1093/hmg/10.22.2515. [DOI] [PubMed] [Google Scholar]
- 27. Leavitt BR, et al. Wild-type huntingtin reduces the cellular toxicity of mutant huntingtin in vivo. Am J Hum Genet. 2001;68(2):313–324. doi: 10.1086/318207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ho LW, Brown R, Maxwell M, Wyttenbach A, Rubinsztein DC. Wild type Huntingtin reduces the cellular toxicity of mutant Huntingtin in mammalian cell models of Huntington’s disease. J Med Genet. 2001;38(7):450–452. doi: 10.1136/jmg.38.7.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington’s disease. Nat Rev Neurosci. 2005;6(12):919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 30. Imarisio S, et al. Huntington’s disease: from pathology and genetics to potential therapies. Biochem J. 2008;412(2):191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 31. Roze E, Saudou F, Caboche J. Pathophysiology of Huntington’s disease: from huntingtin functions to potential treatments. Curr Opin Neurol. 2008;21(4):497–503. doi: 10.1097/WCO.0b013e328304b692. [DOI] [PubMed] [Google Scholar]
- 32. Browne SE, Beal MF. The energetics of Huntington’s disease. Neurochem Res. 2004;29(3):531–546. doi: 10.1023/B:NERE.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- 33. Grafton ST, et al. Serial changes of cerebral glucose metabolism and caudate size in persons at risk for Huntington’s disease. Arch Neurol. 1992;49(11):1161–1167. doi: 10.1001/archneur.1992.00530350075022. [DOI] [PubMed] [Google Scholar]
- 34. Kuwert T, et al. Striatal glucose consumption in chorea-free subjects at risk of Huntington’s disease. J Neurol. 1993;241(1):31–36. doi: 10.1007/BF00870669. [DOI] [PubMed] [Google Scholar]
- 35. Antonini A, et al. Striatal glucose metabolism and dopamine D2 receptor binding in asymptomatic gene carriers and patients with Huntington’s disease. Brain. 119(pt 6):2085–2095. doi: 10.1093/brain/119.6.2085. [DOI] [PubMed] [Google Scholar]
- 36. Jenkins BG, Koroshetz WJ, Beal MF, Rosen BR. Evidence for impairment of energy metabolism in vivo in Huntington’s disease using localized 1H NMR spectroscopy. Neurology. 1993;43(12):2689–2695. doi: 10.1212/wnl.43.12.2689. [DOI] [PubMed] [Google Scholar]
- 37. Jenkins BG, et al. 1H NMR spectroscopy studies of Huntington’s disease: correlations with CAG repeat numbers. Neurology. 1998;50(5):1357–1365. doi: 10.1212/wnl.50.5.1357. [DOI] [PubMed] [Google Scholar]
- 38. Koroshetz WJ, Jenkins BG, Rosen BR, Beal MF. Energy metabolism defects in Huntington’s disease and effects of coenzyme Q10. Ann Neurol. 1997;41(2):160–165. doi: 10.1002/ana.410410206. [DOI] [PubMed] [Google Scholar]
- 39. Gines S, et al. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington’s disease knock-in mice. Hum Mol Genet. 2003;12(5):497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- 40. Mochel F, Durant B, Schiffmann R, Durr A. Characterization of the locoregional brain energy profile in wild-type mice and identification of an energy deficit in a neurodegenerative model. J Inherit Metab Dis. 2010;33(1):S181. [Google Scholar]
- 41. Milakovic T, Johnson GV. Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant huntingtin. J Biol Chem. 2005;280(35):30773–30782. doi: 10.1074/jbc.M504749200. [DOI] [PubMed] [Google Scholar]
- 42. Tabrizi SJ, Cleeter MW, Xuereb J, Taanman JW, Cooper JM, Schapira AH. Biochemical abnormalities and excitotoxicity in Huntington’s disease brain. Ann Neurol. 1999;45(1):25–32. doi: 10.1002/1531-8249(199901)45:1<25::AID-ART6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 43. Lim D, et al. Calcium homeostasis and mitochondrial dysfunction in striatal neurons of Huntington disease. J Biol Chem. 2008;283(9):5780–5789. doi: 10.1074/jbc.M704704200. [DOI] [PubMed] [Google Scholar]
- 44. Li XJ, Orr AL, Li S. Impaired mitochondrial trafficking in Huntington’s disease. Biochim Biophys Acta. 2010;1802(1):62–65. doi: 10.1016/j.bbadis.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127(1):59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 46. Powers WJ, et al. Selective defect of in vivo glycolysis in early Huntington’s disease striatum. Proc Natl Acad Sci U S A. 2007;104(8):2945–2949. doi: 10.1073/pnas.0609833104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Milnerwood AJ, et al. Early increase in extrasynaptic NMDA receptor signaling and expression contributes to phenotype onset in Huntington’s disease mice. Neuron. 2010;65(2):178–190. doi: 10.1016/j.neuron.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 48. van der Burg JM, Bjorkqvist M, Brundin P. Beyond the brain: widespread pathology in Huntington’s disease. Lancet Neurol. 2009;8(8):765–774. doi: 10.1016/S1474-4422(09)70178-4. [DOI] [PubMed] [Google Scholar]
- 49. Seong IS, et al. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum Mol Genet. 2005;14(19):2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 50. Strand AD, et al. Gene expression in Huntington’s disease skeletal muscle: a potential biomarker. Hum Mol Genet. 2005;14(13):1863–1876. doi: 10.1093/hmg/ddi192. [DOI] [PubMed] [Google Scholar]
- 51. Gizatullina ZZ, et al. Low stability of Huntington muscle mitochondria against Ca2+ in R6/2 mice. Ann Neurol. 2006;59(2):407–411. doi: 10.1002/ana.20754. [DOI] [PubMed] [Google Scholar]
- 52. Lodi R, et al. Abnormal in vivo skeletal muscle energy metabolism in Huntington’s disease and dentatorubropallidoluysian atrophy. Ann Neurol. 2000;48(1):72–76. doi: 10.1002/1531-8249(200007)48:1<72::AID-ANA11>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 53. Saft C, et al. Mitochondrial impairment in patients and asymptomatic mutation carriers of Huntington’s disease. Mov Disord. 2005;20(6):674–679. doi: 10.1002/mds.20373. [DOI] [PubMed] [Google Scholar]
- 54. Mihm MJ, Amann DM, Schanbacher BL, Altschuld RA, Bauer JA, Hoyt KR. Cardiac dysfunction in the R6/2 mouse model of Huntington’s disease. Neurobiol Dis. 2007;25(2):297–308. doi: 10.1016/j.nbd.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sanberg PR, Fibiger HC. Body weight, feeding, and drinking behaviors in rats with kainic acid-induced lesions of striatal neurons--with a note on body weight symptomatology in Huntington’s disease. Exp Neurol. 1979;66(3):444–466. doi: 10.1016/0014-4886(79)90194-8. [DOI] [PubMed] [Google Scholar]
- 56. Sanberg PR, Fibiger HC, Mark RF. Body weight and dietary factors in Huntington’s disease patients compared with matched controls. Med J Aust. 1981;1(8):407–409. doi: 10.5694/j.1326-5377.1981.tb135681.x. [DOI] [PubMed] [Google Scholar]
- 57. Trejo A, Tarrats RM, Alonso ME, Boll MC, Ochoa A, Velasquez L. Assessment of the nutrition status of patients with Huntington’s disease. Nutrition. 2004;20(2):192–196. doi: 10.1016/j.nut.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 58. Pratley RE, Salbe AD, Ravussin E, Caviness JN. Higher sedentary energy expenditure in patients with Huntington’s disease. Ann Neurol. 2000;47(1):64–70. [PubMed] [Google Scholar]
- 59. Djousse L, Knowlton B, Cupples LA, Marder K, Shoulson I, Myers RH. Weight loss in early stage of Huntington’s disease. Neurology. 2002;59(9):1325–1330. doi: 10.1212/01.wnl.0000031791.10922.cf. [DOI] [PubMed] [Google Scholar]
- 60. Mochel F, et al. Early energy deficit in Huntington disease: identification of a plasma biomarker traceable during disease progression. PLoS One. 2007;2(7):e647. doi: 10.1371/journal.pone.0000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Goodman AO, et al. The metabolic profile of early Huntington’s disease--a combined human and transgenic mouse study. Exp Neurol. 2008;210(2):691–698. doi: 10.1016/j.expneurol.2007.12.026. [DOI] [PubMed] [Google Scholar]
- 62. Aziz NA, van der Marck MA, Pijl H, Olde Rikkert MG, Bloem BR, Roos RA. Weight loss in neurodegenerative disorders. J Neurol. 2008;255(12):1872–1880. doi: 10.1007/s00415-009-0062-8. [DOI] [PubMed] [Google Scholar]
- 63. van der Burg JM, et al. Increased metabolism in the R6/2 mouse model of Huntington’s disease. Neurobiol Dis. 2008;29(1):41–51. doi: 10.1016/j.nbd.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 64. Zabel C, et al. A large number of protein expression changes occur early in life and precede phenotype onset in a mouse model for huntington disease. Mol Cell Proteomics. 2009;8(4):720–734. doi: 10.1074/mcp.M800277-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Popovic V, et al. Circulating and cerebrospinal fluid ghrelin and leptin: potential role in altered body weight in Huntington’s disease. Eur J Endocrinol. 2004;151(4):451–455. doi: 10.1530/eje.0.1510451. [DOI] [PubMed] [Google Scholar]
- 66. Petersen A, Bjorkqvist M. Hypothalamic-endocrine aspects in Huntington’s disease. Eur J Neurosci. 2006;24(4):961–967. doi: 10.1111/j.1460-9568.2006.04985.x. [DOI] [PubMed] [Google Scholar]
- 67. Phan J, Hickey MA, Zhang P, Chesselet MF, Reue K. Adipose tissue dysfunction tracks disease progression in two Huntington’s disease mouse models. Hum Mol Genet. 2009;18(6):1006–1016. doi: 10.1093/hmg/ddn428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Myers RH, et al. Factors associated with slow progression in Huntington’s disease. Arch Neurol. 1991;48(8):800–804. doi: 10.1001/archneur.1991.00530200036015. [DOI] [PubMed] [Google Scholar]
- 69. Palfi S, et al. Chronic 3-nitropropionic acid treatment in baboons replicates the cognitive and motor deficits of Huntington’s disease. J Neurosci. 1996;16(9):3019–3025. doi: 10.1523/JNEUROSCI.16-09-03019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Brouillet E, et al. Partial inhibition of brain succinate dehydrogenase by 3-nitropropionic acid is sufficient to initiate striatal degeneration in rat. . J Neurochem. 1998;70(2):794–805. doi: 10.1046/j.1471-4159.1998.70020794.x. [DOI] [PubMed] [Google Scholar]
- 71. Stahl WL, Swanson PD. Biochemical abnormalities in Huntington’s chorea brains. Neurology. 1974;24(9):813–819. doi: 10.1212/wnl.24.9.813. [DOI] [PubMed] [Google Scholar]
- 72. Gu M, Gash MT, Mann VM, Javoy-Agid F, Cooper JM, Schapira AH. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann Neurol. 1996;39(3):385–389. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- 73. Browne SE, et al. Oxidative damage and metabolic dysfunction in Huntington’s disease: selective vulnerability of the basal ganglia. Ann Neurol. 1997;41(5):646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 74. Benchoua A, et al. Involvement of mitochondrial complex II defects in neuronal death produced by N-terminus fragment of mutated huntingtin. Mol Biol Cell. 2006;17(4):1652–1663. doi: 10.1091/mbc.E05-07-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Solans A, Zambrano A, Rodriguez M, Barrientos A. Cytotoxicity of a mutant huntingtin fragment in yeast involves early alterations in mitochondrial OXPHOS complexes II and III. Hum Mol Genet. 2006;15(20):3063–3081. doi: 10.1093/hmg/ddl248. [DOI] [PubMed] [Google Scholar]
- 76. Fukui H, Moraes CT. Extended polyglutamine repeats trigger a feedback loop involving the mitochondrial complex III, the proteasome and huntingtin aggregates. Hum Mol Genet. 2007;16(7):783–797. doi: 10.1093/hmg/ddm023. [DOI] [PubMed] [Google Scholar]
- 77. Guidetti P, et al. Early degenerative changes in transgenic mice expressing mutant huntingtin involve dendritic abnormalities but no impairment of mitochondrial energy production. Exp Neurol. 2001;169(2):340–350. doi: 10.1006/exnr.2000.7626. [DOI] [PubMed] [Google Scholar]
- 78. Browne SE. Mitochondria and Huntington’s disease pathogenesis: insight from genetic and chemical models. Ann N Y Acad Sci. 2008;1147:358–382. doi: 10.1196/annals.1427.018. [DOI] [PubMed] [Google Scholar]
- 79. Li Y, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11(4):376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 80. Wallace DC. A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found Symp. 2001;235:247–263. doi: 10.1002/0470868694.ch20. [DOI] [PubMed] [Google Scholar]
- 81. Browne SE, Ferrante RJ, Beal MF. Oxidative stress in Huntington’s disease. Brain Pathol. 1999;9(1):147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hersch SM, et al. Creatine in Huntington disease is safe, tolerable, bioavailable in brain and reduces serum 8OH2’dG. Neurology. 2006;66(2):250–252. doi: 10.1212/01.wnl.0000194318.74946.b6. [DOI] [PubMed] [Google Scholar]
- 83. Browne SE, Beal MF. Oxidative damage in Huntington’s disease pathogenesis. Antioxid Redox Signal. 2006;8(11–12):2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 84. Chen CM, et al. Increased oxidative damage and mitochondrial abnormalities in the peripheral blood of Huntington’s disease patients. Biochem Biophys Res Commun. 2007;359(2):335–340. doi: 10.1016/j.bbrc.2007.05.093. [DOI] [PubMed] [Google Scholar]
- 85. Sorolla MA, Reverter-Branchat G, Tamarit J, Ferrer I, Ros J, Cabiscol E. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic Biol Med. 2008;45(5):667–678. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 86. Santamaria A, et al. Comparative analysis of superoxide dismutase activity between acute pharmacological models and a transgenic mouse model of Huntington’s disease. Neurochem Res. 2001;26(4):419–424. doi: 10.1023/A:1010911417383. [DOI] [PubMed] [Google Scholar]
- 87. La Fontaine MA, Geddes JW, Banks A, Butterfield DA. 3-nitropropionic acid induced in vivo protein oxidation in striatal and cortical synaptosomes: insights into Huntington’s disease. Brain Res. 2000;858(2):356–362. doi: 10.1016/S0006-8993(00)01948-X. [DOI] [PubMed] [Google Scholar]
- 88. J Biol Chem. 1995;270(22):13399–13405. doi: 10.1074/jbc.270.22.13399. [DOI] [PubMed] [Google Scholar]
- 89. Martinez A, Portero-Otin M, Pamplona R, Ferrer I. Protein targets of oxidative damage in human neurodegenerative diseases with abnormal protein aggregates. Brain Pathol. 2010;20(2):281–297. doi: 10.1111/j.1750-3639.2009.00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tabrizi SJ, et al. Mitochondrial dysfunction and free radical damage in the Huntington R6/2 transgenic mouse. Ann Neurol. 2000;47(1):80–86. doi: 10.1002/1531-8249(200001)47:1<80::AID-ANA13>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 91. Tretter L, Adam-Vizi V. Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philos Trans R Soc Lond B Biol Sci. 2005;360(1464):2335–2345. doi: 10.1098/rstb.2005.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Nicholls DG. Mitochondrial calcium function and dysfunction in the central nervous system. Biochim Biophys Acta. 2009;1787(11):1416–1424. doi: 10.1016/j.bbabio.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Panov AV, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5(8):731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 94. Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13(14):1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 95. Trushina E, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24(18):8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chang DT, Rintoul GL, Pandipati S, Reynolds IJ. Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol Dis. 2006;22(2):388–400. doi: 10.1016/j.nbd.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 97. Orr AL, et al. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28(11):2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kim J, et al. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum Mol Genet. 2010;19(20):3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sandou F, Humbert S. The biology of Huntington’s disease. In: Duyckaerts C, Litvan I, eds.Handbook of Clinical Neurology . 3rd series. New York, New York, USA: Elsevier Inc.; 2008:619–630. [DOI] [PubMed] [Google Scholar]
- 100. Bae BI, et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron. 2005;47(1):29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 101. Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1(6):361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 102. Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119(1):121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 103. Weydt P, et al. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006;4(5):349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 104. Quintanilla RA, Jin YN, Fuenzalida K, Bronfman M, Johnson GV. Rosiglitazone treatment prevents mitochondrial dysfunction in mutant huntingtin-expressing cells: possible role of peroxisome proliferator-activated receptor-gamma (PPARgamma) in the pathogenesis of Huntington disease. J Biol Chem. 2008;283(37):25628–25637. doi: 10.1074/jbc.M804291200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Taherzadeh-Fard E, Saft C, Andrich J, Wieczorek S, Arning L. PGC-1alpha as modifier of onset age in Huntington disease. Mol Neurodegener. 2009;4:10. doi: 10.1186/1750-1326-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Lee ST, et al. Slowed progression in models of Huntington disease by adipose stem cell transplantation. Ann Neurol. 2009;66(5):671–681. doi: 10.1002/ana.21788. [DOI] [PubMed] [Google Scholar]
- 107. Chaturvedi RK, et al. Impaired PGC-1alpha function in muscle in Huntington’s disease. Hum Mol Genet. 2009;18(16):3048–3065. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lee JM, et al. Unbiased gene expression analysis implicates the huntingtin polyglutamine tract in extra-mitochondrial energy metabolism. PLoS Genet. 2007;3(8):e135. doi: 10.1371/journal.pgen.0030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. O’Gorman E, Beutner G, Wallimann T, Brdiczka D. Differential effects of creatine depletion on the regulation of enzyme activities and on creatine-stimulated mitochondrial respiration in skeletal muscle, heart, and brain. Biochim Biophys Acta. 1996;1276(2):161–170. doi: 10.1016/0005-2728(96)00074-6. [DOI] [PubMed] [Google Scholar]
- 110. Dedeoglu A, et al. Creatine therapy provides neuroprotection after onset of clinical symptoms in Huntington’s disease transgenic mice. J Neurochem. 2003;85(6):1359–1367. doi: 10.1046/j.1471-4159.2003.01706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ferrante RJ, et al. Neuroprotective effects of creatine in a transgenic mouse model of Huntington’s disease. J Neurosci. 2000;20(12):4389–4397. doi: 10.1523/JNEUROSCI.20-12-04389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Andreassen OA, et al. Creatine increase survival and delays motor symptoms in a transgenic animal model of Huntington’s disease. Neurobiol Dis. 2001;8(3):479–491. doi: 10.1006/nbdi.2001.0406. [DOI] [PubMed] [Google Scholar]
- 113. Yang L, et al. Combination therapy with coenzyme Q10 and creatine produces additive neuroprotective effects in models of Parkinson’s and Huntington’s diseases. J Neurochem. 2009;109(5):1427–1439. doi: 10.1111/j.1471-4159.2009.06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Verbessem P, et al. Creatine supplementation in Huntington’s disease: a placebo-controlled pilot trial. Neurology. 2003;61(7):925–930. doi: 10.1212/01.wnl.0000090629.40891.4b. [DOI] [PubMed] [Google Scholar]
- 115. Tabrizi SJ, et al. High-dose creatine therapy for Huntington disease: a 2-year clinical and MRS study. Neurology. 2005;64(9):1655–1656. doi: 10.1212/01.WNL.0000160388.96242.77. [DOI] [PubMed] [Google Scholar]
- 116. Tkac I, Dubinsky JM, Keene CD, Gruetter R, Low WC. Neurochemical changes in Huntington R6/2 mouse striatum detected by in vivo 1H NMR spectroscopy. J Neurochem. 2007;100(5):1397–1406. doi: 10.1111/j.1471-4159.2006.04323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Neurology. 2001;57(3):397–404. doi: 10.1212/wnl.57.3.397. [DOI] [PubMed] [Google Scholar]
- 118. Rebec GV, Barton SJ, Marseilles AM, Collins K. Ascorbate treatment attenuates the Huntington behavioral phenotype in mice. Neuroreport. 2003;14(9):1263–1265. doi: 10.1097/00001756-200307010-00015. [DOI] [PubMed] [Google Scholar]
- 119. Klivenyi P, Ferrante RJ, Gardian G, Browne S, Chabrier PE, Beal MF. Increased survival and neuroprotective effects of BN82451 in a transgenic mouse model of Huntington’s disease. J Neurochem. 2003;86(1):267–272. doi: 10.1046/j.1471-4159.2003.t01-1-01868.x. [DOI] [PubMed] [Google Scholar]
- 120. Mochel F, et al. Dietary anaplerotic therapy improves peripheral tissue energy metabolism in patients with Huntington’s disease. Eur J Hum Genet. 2010;18(9):1057–1060. doi: 10.1038/ejhg.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127(6):1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 122. Parker JA, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37(4):349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 123. Ho DJ, Calingasan NY, Wille E, Dumont M, Beal MF. Resveratrol protects against peripheral deficits in a mouse model of Huntington’s disease. Exp Neurol. 2010;225(1):74–84. doi: 10.1016/j.expneurol.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 124. Kumar P, Padi SS, Naidu PS, Kumar A. Effect of resveratrol on 3-nitropropionic acid-induced biochemical and behavioural changes: possible neuroprotective mechanisms. Behav Pharmacol. 2006;17(5-6):485–492. doi: 10.1097/00008877-200609000-00014. [DOI] [PubMed] [Google Scholar]
- 125. Oliveira JM, et al. Mitochondrial-dependent Ca2+ handling in Huntington’s disease striatal cells: effect of histone deacetylase inhibitors. J Neurosci. 2006;26(43):11174–11186. doi: 10.1523/JNEUROSCI.3004-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]