Abstract
Human glioblastoma multiforme (GBM) is the most common primary brain tumor in adults. The poor prognosis and minimally successful treatments of GBM indicates a need to identify new therapeutic targets. In this study, we examined the role of CXCR3 in glioma progression using the GL261 murine model of malignant glioma. Intracranial GL261 tumors express CXCL9 and CXCL10 in vivo. Glioma-bearing CXCR3-deficient mice had significantly shorter median survival time and reduced numbers of tumor-infiltrated natural killer and natural killer T cells as compared with tumor-bearing wild-type (WT) mice. In contrast, pharmacological antagonism of CXCR3 with NBI-74330 prolonged median survival times of both tumor-bearing WT and CXCR3-deficient mice when compared with vehicle-treated groups. NBI-74330 treatment did not impact tumor infiltration of lymphocytes and microglia. A small percentage of GL261 cells were identified as CXCR3+, which was similar to the expression of CXCR3 in several grade IV human glioma cell lines (A172, T98G, U87, U118 and U138). When cultured as gliomaspheres (GS), the human and murine lines increased CXCR3 expression; CXCR3 expression was also found in a primary human GBM-derived GS. Additionally, CXCR3 isoform A was expressed by all lines, whereas CXCR3-B was detected in T98G-, U118- and U138-GS cells. CXCL9 or CXCL10 induced in vitro glioma cell growth in GL261- and U87-GS as well as inhibited cell loss in U138-GS cells and this effect was antagonized by NBI-74330. The results suggest that CXCR3 antagonism exerts a direct anti-glioma effect and this receptor may be a potential therapeutic target for treating human GBM.
Introduction
Human glioblastoma multiforme (GBM), a grade IV astrocytoma, is the most common and malignant form of human primary brain tumors. Current treatment paradigms for GBM include surgical resection of the tumor mass, followed by adjuvant radiotherapy and chemotherapy, although these approaches are not very successful, having only a modest impact on the survival rate of GBM patients. Due to the relative ineffectiveness of these traditional treatments, studies are focusing on the molecular pathways associated with glioma progression as well as exploring new approaches, such as immunotherapy (1).
Chemokines promote migration of responsive cells and in the context of tumorigenesis, are known to direct intratumoral trafficking of immune cells. Moreover, chemokines also modulate tumor proliferation, metastasis and the angiogenic response associated with tumor growth (2). CXCR3 belongs to the CXC chemokine receptor subfamily and has three endogenous ligands, CXCL9 (MIG), CXCL10 (IP10) and CXCL11 (ITAC). This chemokine system has been reported to be involved in tumor growth, metastasis, angiogenesis and immune cell infiltration into tumors. With the respect to immune cell recruitment, CXCR3 is expressed by activated T cells, natural killer (NK), natural killer T (NKT) cells and, within the central nervous system, microglia (3–5). CXCR3+ lymphocyte recruitment, directed by CXCL10, can promote spontaneous regression of melanoma (6), whereas CXCL11 increases tumor-infiltrating lymphocytes and inhibits tumor growth in both breast cancer and T-cell lymphoma (7,8). Therefore, CXCR3-mediated homing of immune cells represents a potential target for tumor therapy investigation. CXCR3 is also expressed by tumor cells, including melanoma, ovarian and renal carcinoma, breast cancer cells, B-cell leukemia, prostate, colorectal and brain tumor cell lines (9–17). CXCR3 expression has been reported to correlate with poor prognosis of breast cancer patients (18) and with tumor thickness in melanoma (19). CXCR3 activation enhances tumor cell proliferation of myeloma (20) and osteosarcoma (21) and CXCR3 inhibition induces caspase-independent cell death (21). In addition, CXCR3 is involved in matrix metalloproteinase production by colorectal carcinoma (22) and osteosarcoma (21). It also regulates metastatic activity of melanoma (18), breast cancer (9), osteosarcoma (21) and colorectal carcinoma (22).
The recent demonstration that CXCL10 is expressed by murine (23) and human (24) glioma cell lines suggests that this chemokine could play important roles in brain tumor biology. CXCL10 is upregulated in grade III and grade IV human glioma cells as compared with normal astrocytes. Additionally, CXCR3 is also elevated in both grade III and grade IV human glioma cells and its activation can increase DNA synthesis of these cells in vitro. The DNA synthesis enhancing effect of CXCL10 on glioma cells is abolished by CXCL10 neutralizing antibody. While these in vitro results support a role for CXCR3 in malignant glioma, investigations of this receptor in glioma progression in vivo are absent.
In this study, we investigated the role of CXCR3 in glioma progression using the GL261 murine model of malignant glioma (25–27). CXCL9 and CXCL10 expression were determined in GL261 cells and GL261 tumors established in syngeneic C57BL/6 mice. CXCR3-deficient mice and a CXCR3 antagonist, NBI-74330, were utilized to address the role of this receptor in glioma progression. NBI-74330 is a small molecule selective CXCR3 antagonist (28–30) and has been shown to attenuate atherosclerotic plaque formation by blocking the migration of CD4+ T cells and macrophages, as well as enhancing the immune suppression controlled by Foxp3+ T regulatory (Treg) cells (31). We found that CXCR3 deficiency in the host and CXCR3 antagonism with NBI-74330 had different effects on GL261 glioma progression. However, NBI-74330 exerted an antitumor progression effect not dependent on host expression of CXCR3, supporting a role for this receptor directly on glioma cells. The glioma expression of CXCR3 was confirmed through in vitro studies of the murine GL261 cells and then extended by characterization of several human glioma cells. Functional characterization of tumor-expressed revealed a role for CXCR3 in promoting glioma proliferation. Taken together, our results indicate that CXCR3 is involved in glioma progression and is a potential therapeutic target for glioma.
Materials and methods
Cell culture
The A172 and U118 glioma cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1% penicillin–streptomycin, 2 mM l-glutamine. The T98G, U118 and U138 glioma cell lines were maintained in Eagle’s minimum essential medium supplemented with 10% heat-inactivated FBS, 1% penicillin–streptomycin, 1% sodium pyruvate and 2 mM l-glutamine. The GL261 glioma cell line was maintained in RPMI-1640 medium supplemented with 10% heat-inactivated FBS, 1% penicillin–streptomycin, 4 mM l-glutamine. All gliomaspheres (GS) were cultured in DMEM/F12 medium supplemented with 2% B27, 20 ng/ml of epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF), 5 μg/ml of heparin and 1% penicillin–streptomycin. All the cells were grown in a humidified incubator at 37°C with 5% CO2. DMEM, Eagle’s minimum essential medium, RPMI-1640, DMEM/F12 medium, B27, EGF, bFGF, l-glutamine and antibiotics were obtained from Gibco-BRL (Invitrogen, Carlsbad, CA). Sodium pyruvate and heparin were purchased from Sigma–Aldrich (St Louis, MO). FBS was from HyClone (Thermo Scientific, Waltham, MA).
Animals
Wild type (WT) C57BL/6 mice were obtained from either Charles River Laboratories (Wilmington, MA) or Jackson Laboratories (Bar Harbor, ME). CXCR3-deficient mice, backcrossed 16 generations to the C57BL/6 background were generated as described previously (32). All procedures involving mice were carried out in accordance with the guidelines of the University of Florida Institutional Animal Care and Use Committee.
Reverse transcription–polymerase chain reaction
Total RNA was isolated from glioma cells with the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Genomic DNA contamination was removed by RQ1 RNase-free DNase treatment (Promega, Madison, WI). Total RNA was then quantified and stored at −80°C. RNA (1 μg) was retrotranscribed with iScript complementary DNA (cDNA) synthesis kit (Bio-Rad, Hercules, CA). Synthesized cDNA was subjected to polymerase chain reaction analysis. Polymerase chain reaction was performed by heating for 96°C for 2 min, followed by amplification for 35 cycles: 96°C for 30 s, 56°C for 1 min and 72°C for 1 min. The following primers were used: murine CXCL9, 5′-CTCGGATCCGCCATGAAGTCCGCTGTTCTTTTC-3′ (forward) and 5′-TATGAATTCAAATTAACACTTTATGTTTTGTAG-3′ (reverse); murine CXCL10, 5′-CCGGAATTCTCCCCATCAGCACCATGAACCC-3′ (forward) and 5′-CTGCTCGAGGAGTAGCAGCTGATGTGACC-3′ (reverse); murine CXCL11, 5′-GCAGAATTCTGCAGCGGCTGCTGAGATGAACAG-3′ (forward) and 5′-GGACCTTCTAGAAAGTTCTGCAGC-3′ (reverse); murine CXCR3, 5′-GAGGTTAGTGAACGTCAAGTG-3′ (forward) and 5′-GGGGTCCCTGCGGTAGATCTG-3′ (reverse); murine glyceraldehyde 3-phosphate dehydrogenase, 5′-AAATGGTGAAGGTCGGTGTG-3′ (forward) and 5′-TCTCCATGGTGGTGAAGACA-3′ (reverse); human CXCL9, 5′-TGCTGGTTCTGATTGGAGTG-3′ (forward) and 5′-CTGTTGTGAGTGGGATGTGG-3′ (reverse); human CXCL10, 5′-AACCTCCAGTCTCAGCACCA-3′ (forward) and 5′-TTTGAAGCAGGGTCAGAACA-3′ (reverse); human CXCL11, 5′-CCTGGGGTAAAAGCAGTGAA-3′ (forward) and 5′-TGGGGAAAGAAGTGTGTATTTG-3′ (reverse); human CXCR3-A and -B, 5′-ACCCAGCAGCCAGAGCAC-3′ (forward) and 5′-GTTCAGGTAGCGGTCAAAGC-3′ (reverse) and human glyceraldehyde 3-phosphate dehydrogenase, 5′-CGAGATCCCTCCAAAATCAA-3′ (forward) and 5′-TGCTGTAGCCAAATTCGTTG-3′ (reverse). The predicted polymerase chain reaction product sizes are listed in supplementary Table I, available at Carcinogenesis Online.
CXCL10 enzyme-linked immunosorbent assay
To quantitate CXCL10 protein secreted by GL261 cells, 105 cells were plated in a 12-well plate and grown for 24 h. Cells were then washed with phosphate-buffered saline (PBS) twice and incubated with 500 μl serum-free medium. Conditioned medium was collected at 24 and 48 h and the CXCL10 concentration was measured by Mouse CXCL10/IP-10/CRG-2 Quantikine ELISA Kit (R&D Systems, Minneapolis, MN) according to the manufacturer’s protocol. CXCL10 concentration was normalized to the number of cells in the well and expressed as nanograms per milliliter per 106 cells.
Intracranial injection of GL261 glioma cells and NBI-74330 treatment
For implantation, GL261 glioma cells (1.6 × 105) in a total volume not exceeding 3 μl were injected 3 mm deep into the right cerebral hemisphere (1 mm posterior and 2 mm lateral from Bregma) of WT and CXCR3-deficient mice. NBI-74330 was synthesized according to Medina et al. (Patent WO02083143, USA, 24 October 2002) and the dosing protocol was performed as described previously (31). Briefly, animals received 100 mg/kg/day of NBI-74330 in 1% sodium docusate in 0.5% 400Cp methylcellulose, injected subcutaneously, beginning from day 3 after surgery, for 12 days. A control group of mice were treated with vehicle only. Glioma-bearing mice were euthanized using sodium pentobarbital (32 mg/kg) and subsequently perfused with 0.9% saline followed by buffered 4% paraformaldehyde. Brains were surgically removed and post-fixed with 4% paraformaldehyde. After fixation, tissues were incubated in 30% sucrose solution at 4°C overnight followed by liquid nitrogen freezing. Frozen brains were then sectioned and subjected to either in situ hybridization analysis or immunohistochemistry. For Kaplan–Meier survival rate analysis, percentages of surviving mice in the specific groups of animals were recorded daily after GL261 glioma implantation. The endpoint was defined by a lack of physical activity and a body weight reduction of >15%. The data were subjected to log-rank test in order to determine if significant differences existed in survival between the experimental groups.
In situ hybridization analysis
In situ hybridization was performed as described previously (33). Plasmid constructs were generated that contained either a 530 bp cDNA fragment of murine CXCL9 or 470 bp cDNA fragment of murine CXCL10. The primers for synthesis of CXCL9 and CXCL10 cDNA were the same as used in reverse transcription–polymerase chain reaction (RT–PCR).
Immunohistochemistry
For immunohistochemistry, brain sections were permeabilized with 0.5% Triton X-100 in PBS for 15 min at room temperature followed by blocking with 10% goat serum in PBS for 30 min. In some cases (anti-Foxp3 immunohistochemistry), sections underwent an antigen retrieval treatment. In brief, slides were first permeabilized with 0.5% Triton X-100 followed by heating slides (immersed in a boiling water bath for 25 min) in a buffer containing 10 mM sodium citrate, 0.05% Tween 20, pH 6.0. Slides were then cooled to room temperature for 20 min, washed with PBS three times and finally subjected to standard immunohistochemistry procedures. The sections were incubated in primary antibodies at 4°C overnight. The following antibodies were used: rat anti-CD11b (dilution 1:500, Serotec, MorphoSys US, Raleigh, NC), rat anti-CD4 (dilution 1:50; BD Biosciences, San Jose, CA), rat anti-CD8 (dilution 1:50, Serotec, MorphoSys US), rat anti-Foxp3 (dilution 1:50; eBioscience, San Diego, CA) and rat anti-Ly49G2 (dilution 1:50; BD Biosciences). The following day, sections were washed three times with PBS and incubated subsequently in goat anti-rat Alexa 594 (dilution 1:1000; Invitrogen). The sections were then washed three times with PBS and finally counterstained with 4′,6-diamidino-2-phenylindole. For quantification of CD4+, CD8+, CD11b+, Foxp3+ and Ly49G2+ cells, the number of cells per high-powered field in several sections from multiple animals were determined and the mean and standard error of the means calculated. The data were subjected to statistical analysis.
FACS analysis
Adherent (AD) glioma cells and GS were harvested with 0.01% ethylenediaminetetraacetic acid in PBS, pH 7.4, washed with ice cold 0.5% bovine serum albumin in PBS and subsequently blocked with 5 μg/ml of mouse and rat IgG mixture for 15 min at room temperature. Cells were then incubated with specific antibody for 30 min on ice. Mouse anti-human CXCR3-PE (1:12; BD Biosciences), rat anti-mouse CXCR3-APC (dilution 1:12; R&D Systems), mouse anti-human Nestin-APC (dilution 1:40; R&D Systems), mouse anti-Nestin-Alexa 647 (1:12; BD Biosciences), mouse anti-human/mouse SOX2-APC (dilution 1:12; R&D Systems) were used. Samples were then washed and analyzed with BD LSR II system (BD Biosciences). Dead cells were excluded by 7-AAD (eBioscience) or 4′,6-diamidino-2-phenylindole (Sigma–Aldrich) staining. All data were analyzed by FlowJo software version 7.6 (Tree Star, Ashland, OR).
In vitro growth analysis
GS cells were plated in 12-well plates at different density and treated with either CXCL9 (1 or 10 nM), CXCL10 (1 or 10 nM) or the combination of EGF and bFGF (each at 20 ng/ml). Cells cultured in medium without growth factor supplements served as the control. Cell numbers were determined at days 3, 6 and 9. To investigate the effect of CXCR3 inhibition, cells cultured as described above with 1 μM of NBI-74330 were compared with samples without NBI-74330. Cell numbers were determined on day 6 or 9. All experiments were performed in triplicate and are representative of three independent experiments. Recombinant mouse and human CXCL9 and CXCL10 were purchased from R&D Systems.
Statistical analysis
All statistical analyses were calculated using either Microsoft Excel or GraphPad Prism 5 software (GraphPad Software, La Jolla, CA). All data are presented as mean ± standard error of the mean. P-values were calculated using Student’s t-test with two-tailed distribution. Survival data were subjected to log-rank test to determine statistically significant differences between groups. A P value <0.05 was considered significant and is indicated by symbols depicted in the figures and figure legends.
Results
Murine glioma GL261 cells express CXCL10 in vitro and GL261 tumors express CXCL9 and CXCL10 in vivo
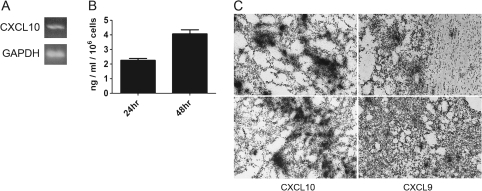
Murine GL261 glioma cells are known to express CXCL10 as previously established using microarray analysis (23). Thus, we utilized the GL261 murine model of malignant glioma to address the role of CXCR3 in glioma progression in vitro and in vivo. By using RT–PCR, we determined that CXCL10 messenger RNA (mRNA) was expressed by cultured GL261 cells (Figure 1A); CXCL9 mRNA was undetectable. Since the in vivo expression of the CXCR3 system has not been reported, we implanted GL261 cells into WT C57BL/6 mice and the expression of CXCL9 and CXCL10 mRNAs were detected by in situ hybridization analysis (Figure 1C). These data indicate that CXCL9 and 10 are expressed in GL261 glioma and might be involved in GL261 glioma progression.
Fig. 1.
CXCL9 and CXCL10 are expressed in GL261 glioma cells and/or tumors. (A) RT–PCR identified CXCL10 mRNA in GL261 cells in vitro. GAPDH was used as a control. (B) CXCL10 ELISA showing CXCL10 protein secretion by GL261 cells in vitro at 24 and 48 h (C) CXCL9 and CXCL10 are expressed in intracranial GL261 tumors in vivo as determined by in situ hybridization analysis. Two representative sections, depicting expression of each chemokine, are shown. The colour version of this figure can be found at www.carcin.oxfordjournals.org.
Reduced numbers of tumor-infiltrated Ly49G2+ NK and NKT cells in GL261 gliomas from CXCR3-deficient mice is associated with decreased animal survival
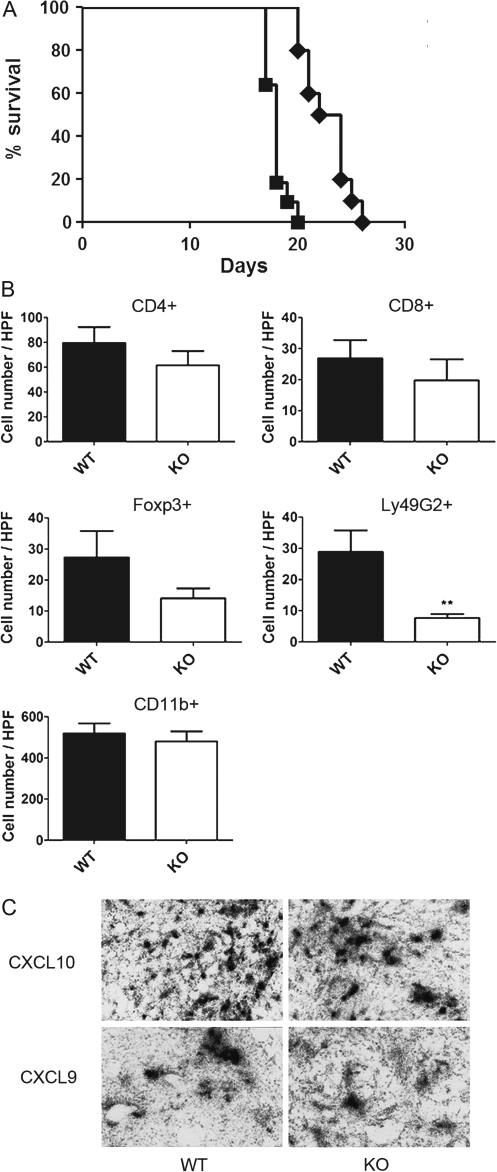
To address the functional significance of host-expressed CXCR3 in glioma progression, we evaluated the effect of CXCR3 deficiency on GL261 tumor growth and tumor-bearing animal survival using CXCR3-deficient mice. Kaplan–Meier survival analysis was carried out by comparing animal survival rates of WT and CXCR3-deficient mice implanted with GL261 cells. The results demonstrated that tumor-bearing CXCR3-deficient mice succumbed to tumor growth more rapidly than WT mice (Figure 2A, P < 0.0001). The median survival time of tumor-bearing CXCR3-deficient mice was significantly decreased to 18 days as compared with WT mice (23 days).
Fig. 2.
GL261 tumor-bearing CXCR3-deficient mice have decreased survival rates and tumor-infiltrated NK and NKT cells. (A) Kaplan–Meier survival analysis indicated that CXCR3-deficient mice (n = 10) have shorter life span than WT mice (n = 10, P < 0.0001). Filled squares: CXCR3-deficient mice; filled diamonds: WT mice. (B) Numbers of tumor-infiltrated CD4+, CD8+, Foxp3+, Ly49G2+ and CD11b+ cells were evaluated by immunohistochemistry. Gliomas from CXCR3-deficient mice had a significant reduction of Ly49G2+ (NK and NKT) cells in the tumor as compared with WT mice (**P < 0.01). WT: wild-type; KO: CXCR3-deficient mice. (C) Intratumoral expression of CXCL9 and CXCL10 mRNA was not altered by host CXCR3 deficiency. Shown are representative sections from WT and CXCR3-deficient glioma-bearing mice subjected to in situ hybridization analysis. The colour version of this figure can be found at www.carcin.oxfordjournals.org.
To access the possibility that the augmented GL261 tumor growth in the CXCR3-deficient mice resulted from malfunction of CXCR3-mediated homing of lymphocytes and/or microglia into the gliomas, we investigated numbers of tumor-infiltrated lymphocytes (CD4+, CD8+, Ly49G2+ and Foxp3+ cells) as well as tumor-infiltrated CD11b+ microglia. The numbers of CD4+, CD8+ cells and microglia inside the tumor were not significantly different between WT and CXCR3-deficient groups (Figure 2B). However, the number of Ly49G2+ cells inside the tumors from CXCR3-deficient mice was significantly reduced when compared with cell numbers in tumor sections from the WT group (CXCR3-deficient: 7.7 ± 1.3 cells per high-powered field, n = 9; WT: 28.9 ± 6.9 cells per high-powered field, n = 7, P = 0.0041) (Figure 2B). While there was a tendency for decreased numbers of Foxp3+ cells in tumors from CXCR3-deficient mice, the difference did not reach statistical significance (Figure 2B). In situ hybridization analysis determined that CXCL9 and CXCL10 mRNAs were detected in tumors from CXCR3-deficient mice, indicating that CXCR3 deficiency did not affect CXCL9 and CXCL10 expression (Figure 2C).
CXCR3 antagonism suppressed GL261 tumor growth and increased animal survival independent of host CXCR3 expression
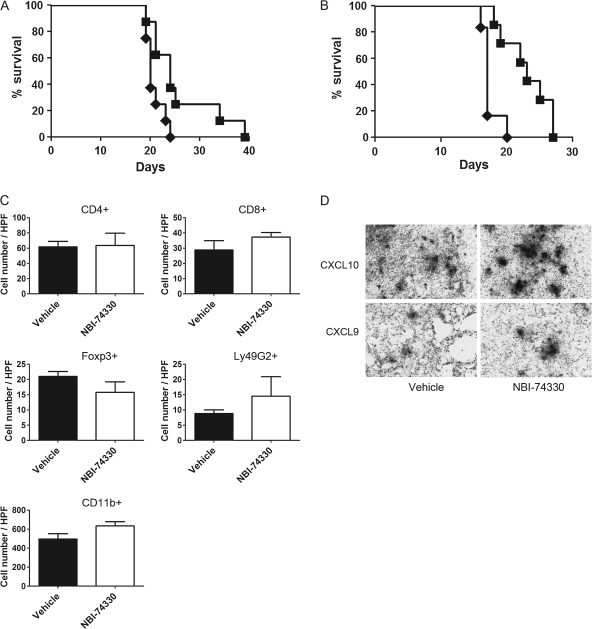
To determine if pharmacological antagonism of CXCR3 mimicked CXCR3 deficiency in promoting GL261 glioma growth, we evaluated the effect of a CXCR3 antagonist, NBI-74330, on GL261 glioma progression. WT and CXCR3-deficient mice implanted with GL261 cells were treated with either NBI-74330 or vehicle as a control. Kaplan–Meier survival analysis showed that this CXCR3 antagonist prolonged survival of glioma-bearing WT animals (Figure 3A, P = 0.0212) and increased median survival days from 20 days (vehicle) to 24 days (NBI-74330). Moreover, in CXCR3-deficient animals, CXCR3 antagonism overcame the effect of CXCR3-deficiency and increased the rate of animal survival (Figure 3B, P = 0.0028). The median survival time for CXCR3 antagonist-treated group was increased to 23 days from a median survival time for vehicle-treated group of 17 days. Tumor-infiltrated cells in tumors from vehicle- and NBI-74330-treated WT mice were also evaluated, and no significant differences in the numbers of tumor-infiltrated CD4+, CD8+, Foxp3+, Ly49G2+ cells or CD11b+-microglia were found (Figure 3C). In addition, expression of CXCL9 and CXCL10 was unaltered by NBI-74330 treatment (Figure 3D).
Fig. 3.
NBI-74330 suppresses tumor growth in both WT and CXCR3-deficient mice. (A) Kaplan–Meier survival analysis of glioma-bearing WT mice shows that NBI-74330 prolonged animal survival (n = 8), as compared with vehicle-treated mice (n = 8) (P = 0.0212). Filled squares: NBI-74330 treated; filled diamonds: vehicle treated. (B) Kaplan–Meier survival analysis shows that glioma-bearing CXCR3-deficient mice treated with NBI-74330 (n = 7) had a higher survival rate than vehicle-treated glioma-bearing CXCR3-deficient mice (n = 6, P = 0.0028). Filled squares: NBI-74330 treated; filled diamonds: vehicle treated. (C) Similar numbers of tumor-infiltrated lymphocytes and microglia in GL261 gliomas from NBI-74330- and vehicle-treated WT mice. Numbers of tumor-infiltrated CD4+, CD8+, Foxp3+, Ly49G2+ and CD11b+ cells were not affected by NBI-74330 when compared with vehicle treatment. (D) In vivo expression of CXCL9 and CXCL10 was not altered by NBI-74330 treatment. Shown are representative sections from vehicle- and NBI-74330-treated glioma-bearing mice subjected to in situ hybridization analysis. The colour version of this figure can be found at www.carcin.oxfordjournals.org.
CXCR3 and its ligands are expressed by murine and human glioma cells
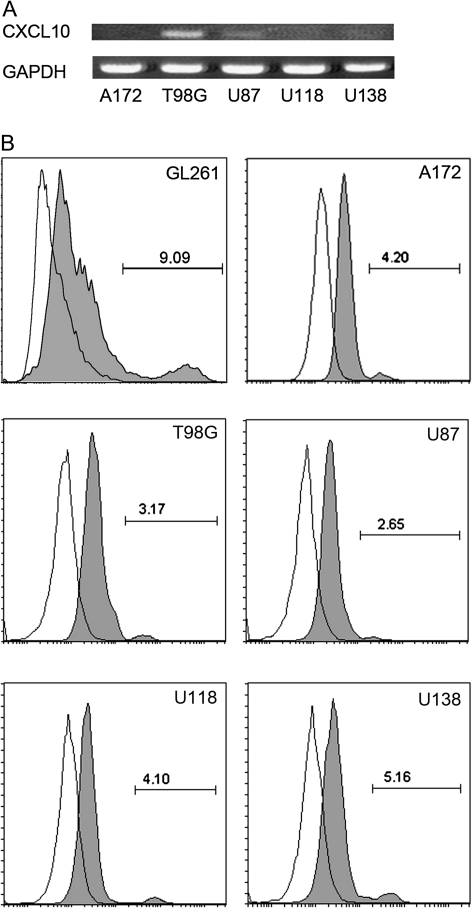
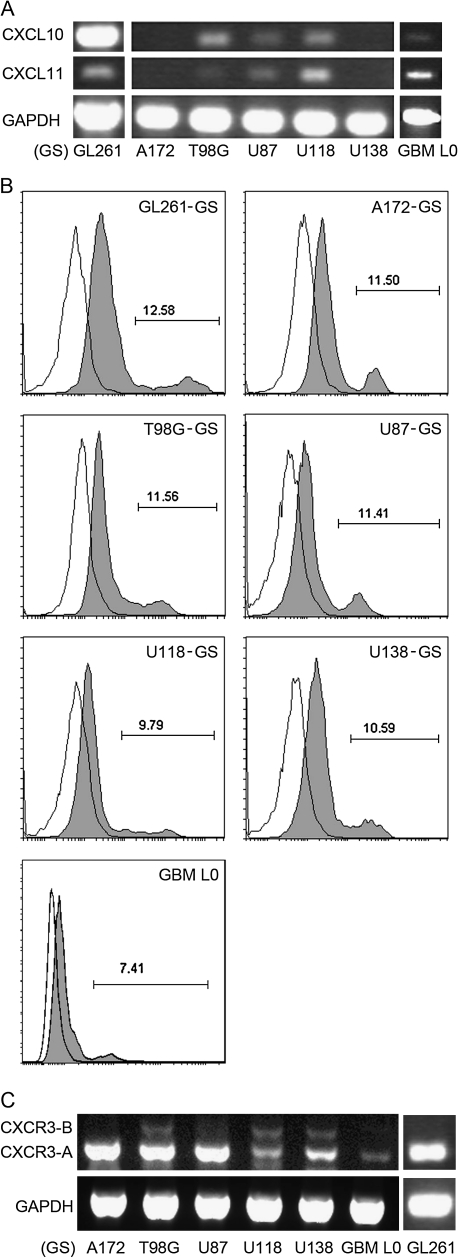
The effect of CXCR3 antagonism on tumor-bearing animal survival, which was independent of host CXCR3 expression, lead to the hypothesis that NBI-74330 inhibits glioma growth by exerting its effect on GL261 cells. Thus, we elucidated the expression of CXCR3 in GL261 cells by fluorescence-activated cell sorting analysis. A small fraction (8.4 ± 0.5%) of GL261 cells were determined to express CXCR3 (Figure 4B). To extend the results in the murine model to human GBM, we examined the expression of CXCR3 and its ligands, in five commonly studied grade IV human glioma cell lines, namely the A172, T98G, U87, U118 and U138 cell lines. With RT–PCR, we determined that two human glioma cell lines, T98G and U87, expressed CXCL10 mRNA (Figure 4A). T98G showed the highest level of CXCL10 mRNA expression and U87 had a lower level of CXCL10. CXCL10 was undetectable in the other three human cell lines and all lines lacked both CXCL9 and CXCL11 mRNAs. In addition, CXCR3 was expressed on all of the human glioma cells, as assessed by fluorescence-activated cell sorting analysis (Figure 4B). The percentage of CXCR3+ cells in the various lines ranged from approximately 3–8%.
Fig. 4.
CXCR3 and CXCL10 expression in murine and human glioma cell lines cultured in serum-containing media. (A) RT–PCR identified CXCL10 mRNA in T98G and U87 cells in vitro. GAPDH was used as a control. (B) Representative histograms from fluorescence-activated cell sorting analysis showing CXCR3 expression by GL261, A172, T98G, U87, U118 and U138 cells in vitro. Gray filled area, anti-CXCR3-APC (mouse) or anti-CXCR3-PE (human) staining; blank area: isotype controls. GL261 has the highest CXCR3 expression level among all the glioma cell lines.
It has been reported that human glioma cell lines, cultured as AD cells in the presence of fetal calf serum, are phenotypically different from their matched primary human tumor-derived tumor stem cells (34). In contrast, GS, derived from culturing cells under more defined conditions that include bFGF and EGF, have higher resemblance to primary human GBM (35) and exhibit a stem cell phenotype characterized by nestin and SOX2 expression (supplementary Figure 1 is available at Carcinogenesis Online). Thus, we evaluated the expression of CXCR3, its ligands (CXCL9/10/11) in a patient GBM tissue-derived GS, GBM L0, and compared it with GL261 and other human glioma cell lines cultured as GS. CXCL10 was expressed by GL261-GS and three of the human GS lines, including T98G-, U87- and U118-GS (Figure 5A). In addition, CXCL11, while undetectable in cells grown in media containing serum, was expressed by GL261-, T98G-, U87- and U118-GS (Figure 5A). CXCR3 expression in the various GS was determined by fluorescence-activated cell sorting analysis. All GS cell lines as well as the GBM L0 expressed CXCR3, albeit at various levels (Figure 5B). When compared with their matched serum-supplemented cell lines, the percentage of CXCR3-expressing cells significantly increased in GL261, A172, T98G, U87, U118 and U138 GS (supplementary Table II is available at Carcinogenesis Online). Two CXCR3 isoforms (CXCR3-A and -B) have been identified in human (36,37), whereas only one form of CXCR3, most similar to the human A isoform, exists in the mouse genome. The presence of CXCR3 isoforms in human GS was characterized by RT–PCR. CXCR3-A was expressed by all cell lines examined with A172-, T98G- and U87-GS expressing the highest levels of CXCR3-A (Figure 5C). CXCR3-B was detected in T98G-, U118- and U138-GS cells (Figure 5C).
Fig. 5.
CXCL10, CXCL11 and CXCR3 expression in GS cells. (A) RT–PCR identified in vitro expression of CXCL10 mRNA in GL261-, T98G-, U87- and U118-GS cells and a patient GBM tissue-derived primary GS (GBM L0). In addition, CXCL11 is expressed by GL261-, T98G-, U87-, U118-GS and GBM L0 cells. GAPDH was used as a control. (B) Representative histograms from fluorescence-activated cell sorting analysis showing CXCR3 expression by GL261-, A172-, T98G-, U87-, U118-, U138-GS and GBM L0 cells. Gray filled area: anti-CXCR3-APC (mouse) or anti-CXCR3-PE (human) staining. Blank area: isotype controls. (C) RT–PCR analysis identified in vitro expression of CXCR3 mRNA isoforms in murine and human GS cells. CXCR3-A was detected in all cells with A172-, T98G- and U87-GS cells expressing the highest levels. CXCR3-B was detected in T98G-, U118- and U138-GS cells. One form of CXCR3 mRNA was detected in GL261-GS cells; only the equivalent of CXCR3-A exists in mouse.
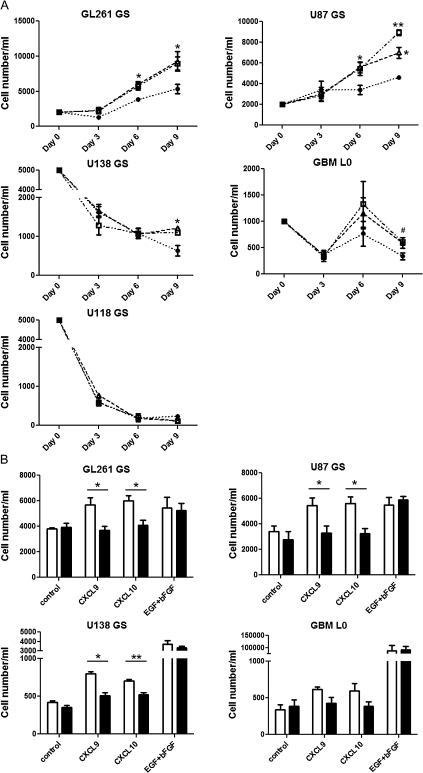
CXCL9 and CXCL10 stimulated growth of murine and human GS
Because expression of CXCR3 was enhanced in GS as compared with serum-supplemented cell lines and differential expression of CXCR3 isoforms were found in these cells, we compared the effects of CXCR3 activation on the cell growth of GL261-, U87-, U118-, U138-GS and GBM L0 cells. Both CXCL9 and CXCL10 stimulated growth of GL261- and U87-GS by day 6 of incubation as compared with control (Figure 6A); an impact on proliferation was not evident at the earliest time point measured (3 days). The growth effect of these chemokines was sustained through day 9 (Figure 6A). Cell numbers of U118- and U138-GS continuously decreased through day 9, and both CXCL9 and CXCL10 attenuated the cell loss of U138-GS by day 9, whereas no significant effect was observed in U118-GS cells (Figure 6A). In GBM L0, the CXCL9-treated group had significantly higher cell numbers by day 9 when compared with the control group (Figure 6A). The CXCL10-treated group, showed a similar tendency as the CXCL9-treated group, but the difference from control did not reach statistical significance (P = 0.11). AD T98G cells were able to form spheres when initially seeded into the serum-free defined growth factor conditions but did not survive subsequently in the presence of CXCL9, CXCL10 or EGF/bFGF. A172 cells also formed spheres but did not proliferate in the presence of the CXCR3 ligands or EGF/bFGF. Co-incubation of CXCL9 or CXCL10 with NBI-74330 blocked the response to either CXCL9 or CXCL10 stimulation in GL261- and U87-GS cells by day 6 and U138-GS cells by day 9 (Figure 6B). A trend for NBI-74330 to attenuate the CXCL9 or CXCL10 responses in GBM L0 cells was observed (CXCL9: P = 0.14; CXCL10: P = 0.16). NBI-74330 did not affect cell growth in either the control or growth factor(s)-supplemented groups, suggesting that NBI-74330 selectively inhibited CXCR3 activation.
Fig. 6.
NBI-74330 inhibits CXCL9 and CXCL10 stimulation of GS growth or survival in vitro. (A) GL261- and U87-GS cells (2000 cells/ml) were incubated with 1 nM CXCL9 (open squares) or 1 nM CXCL10 (open triangles); U118-, U138-GS cells (5000 cells/ml) and GBM L0 (1000 cells/ml) were incubated with 10 nM CXCL9 (open squares) or 10 nM CXCL10 (open triangles). The control group was cultured in medium without chemokines or growth factors (filled circles). All conditions contained 0.1% dimethyl sulfoxide (NBI-74330 vehicle). Representative results of three individual experiments performed in triplicate are shown. CXCL9 and CXCL10 significantly enhanced GL261- and U87-GS growth at day 6 and 9 (*P < 0.05, **P < 0.01) and prevented U138-GS cell loss at day 9. CXCL9 stimulation significantly increased cell numbers of GBM L0 at day 9 (#P < 0.05); CXCL10-stimulated group was not statistically significant as compared with control. (B) The effect of 1 μM NBI-74330 (black filled bars) and 0.1% dimethyl sulfoxide (open bars) on chemokine- and growth factor-stimulated GS growth. Representative results of three individual experiments performed in triplicate are shown. NBI-74330 attenuated response of GL261-, U87- and U138-GS to CXCL9 and CXCL10 but did not affect either control or growth factor-supplemented groups.
Discussion
The role of CXCR3 in a variety of cancers has gained considerable attention. However, the importance of CXCR3 and its ligands in human GBM and mouse models of the disease is still unclear. Previous studies have shown that CXCR3 and CXCL10 are expressed by several human glioma cells lines (24), whereas CXCL10 mRNA is also detected in the murine GL261 glioma cell line (23). In human glioma cells, CXCL10 stimulates DNA synthesis and cell proliferation in vitro (24). These data suggest an involvement of CXCR3 in glioma formation and progression, although an in vivo relationship of this chemokine system and glioma progression has not yet been established. The results reported here indicate that human and murine glioma cell lines and tumors express components of the CXCR3 chemokine system. More important, the increased presence of CXCR3+ cells in cultures enriched in glioma-initiating cells and suppression of the in vitro and in vivo growth of glioma by pharmacological antagonism of CXCR3 supports future consideration of this receptor as a target for GBM therapy.
To investigate the in vivo function of CXCR3 in glioma progression, we initially established that GL261 cells expressed CXCL10 in vitro and CXCL9 and CXCL10 in vivo. The intra-glioma expression of these chemokines prompted us to study the role of CXCR3 in glioma progression using two approaches, specifically CXCR3-deficeint mice and pharmacological antagonism. Glioma-bearing CXCR3-deficient mice had lower survival rate and significantly fewer numbers of NK and NKT cells inside the tumor when compared with WT mice. The absence of tumor-infiltrating NK and NKT cells is the likely reason for the enhanced tumor growth and shorter median survival time in the tumor-bearing CXCR3-deficient animals. The reduction of NK and NKT cells in CXCR3-deficient mice has also been documented in ocular herpes simplex virus type 1 infection (38) as well as pulmonary fibrosis (39). However, it has been reported that CXCR3-deficiency results in an impaired homeostasis of NK and NKT cells, with unchallenged CXCR3-deficient mice having significantly reduced numbers of NK and NKT cells in lung, liver and peripheral blood (39). Thus, the reduction of NK and NKT cells in GL261 gliomas we observed is likely a result from a defect in NK and NKT cell homeostasis and not from a specific alteration of CXCR3-mediated cell migration into the tumor. Altered migration of Foxp3+ Treg cells in CXCR3-deficient mice has also been reported. In an experimental autoimmune encephalomyelitis model of multiple sclerosis, Treg cells are decreased in number and dispersed in lesions from CXCR3-deficient mice (40). These data, coupled with results showing an involvement of CD4+CD25+ Treg cells in suppression of anti-glioma immune responses in the GL261 model(40,41), suggested a Treg phenotype in the glioma-bearing CXCR3-deficient animals might have been observed. While we found a tendency of Foxp3+ Treg cell reduction in tumors from CXCR3-deficient mice, the difference was not statistically significant.
To circumvent the NK/NKT defect in the CXCR3-deficient mice, a pharmacological approach, using NBI-74330, was undertaken. The selectivity of NBI-74330 for CXCR3 has been determined in previous studies. NBI-74330 inhibits CXCR3-agonist binding and CXCR3-mediated phospholipase C activation but has little or no effect on other chemokine and histamine receptors (28–30). Our results are consistent with the pharmacological properties of this antagonist since NBI-74330 attenuated the responses of the GS cells to both CXCL9 and CXCL10 but had no effect on either control or EGF/bFGF-supplemented groups. In contrast to the outcomes from CXCR3-deficient mice, NBI-74330 enhanced the survival rate of tumor-bearing WT mice with no impact on microglia and lymphocyte(s) infiltration. Tumor-bearing CXCR3-deficient mice also displayed prolonged survival with NBI-74330 treatment, a result that suggested a CXCR3 inhibitory effect directly on the tumor cells. Indeed, we found that GL261 cells express CXCR3 as do several human glioma cells. The lack of effect of CXCR3 antagonism on the numbers of tumor-infiltrating microglia and lymphocytes, cells known to express CXCR3, suggests that this receptor system is not the primary means by which these immune cells traffic into glioma. Given that multiple chemokine systems have been shown to mediate microglia and lymphocytes infiltration into the brain (42–45), the influx of these cells into the glioma is probably mediated by other pro-migratory systems.
The applicability of the murine GL261 model to human GBM was further validated with analysis of five different human GBM cell lines, A172, T98G, U87, U118 and U138. All of the human lines express CXCR3 protein but showed varying levels of expression of CXCL10. CXCL10 was only detectable in T98G and U87. The variation of CXCL10 expression in human glioma lines had been documented that the lack of constitutive nuclear factor-kappaB activity in A172 results in undetectable CXCL10 expression, even with interferon gamma stimulation (46). On the other hand, T98G has been shown to have constitutive nuclear factor-kappaB activity and interferon gamma treatment enhances CXCL10 expression in these cells (46). In addition, we analyzed the expression patterns of CXCR3 and its ligands in GL261 and human GBM lines cultured in a serum-free stem cell-enriched condition. It has been suggested that phenotypes of these cells better recapitulate the cells in the glioma environment than cell lines cultured in the presence of serum (34,35). Interestingly, we found that GL261 GS and all of the human GS had greater CXCR3+ population when compared with their matched serum-supplemented cultures; a patient GBM tissue-derived primary GS, GBM L0, also contained a CXCR3 population. The positive correlation of CXCR3 expression and glioma malignancy has been suggested in human gliomas (24), and GL261 GS were confirmed to be more malignant than their matched AD cell lines with serum supplement (23). Moreover, the expression of CXCR3 ligands was also enhanced in some of the GS cell lines. For example, CXCL10 expression was detected in U118-GS but not in U118-AD cells. Furthermore, CXCL11 expression was detected in GL261, T98G, U87 and U118 GS as well as the patient-derived GBM L0 cells, whereas this chemokine was undetectable in all of the AD cells. Thus, the CXCR3 system may contribute to glioma malignancy.
Insights into the role of CXCR3 in glioma progression came from in vitro studies where we determined that both CXCL9 and CXCL10 were able to enhance GL261 and U87 GS cell growth. This result is consistent with the DNA synthesis-promoting effect found in other AD human glioma cell lines (24). In addition, CXCL9 and CXCL10 prevented the loss of cells in U138 GS by day 9. The growth and/or survival effects of CXCR3 activation on human GS are correlated with their expression pattern of CXCR3 mRNA isoforms. Two human CXCR3 isoforms, CXCR3-A and -B, have been identified (36,37). Stimulation of CXCR3-A activates extracellular signal-regulated kinase and AKT pathway, enhancing cell proliferation, whereas CXCR3-B activation exerts proliferation inhibitory and angiostatic effects through activation of p38 mitogen-activated protein kinase (36,47,48). In human myeloma cells, CXCL10 stimulation results in an anti-apoptotic effect on cell lines that have high CXCR3-A expression but not on cell lines with a predominance of CXCR3-B (20). Our data, consistent with previous studies, indicates that CXCR3 enhances cell growth in cell lines that express only CXCR3-A (U87-GS). In lines expressing both isoforms, the functional response to CXCR3 agonists is more complex. The ratio of CXCR3-A and -B isoforms has been postulated to determine the outcome. For example, CXCR3 activation inhibits the loss of cells in U138-GS cells (expresses higher level of CXCR3-A than CXCR3-B) but has little or no effect on U118-GS cells (comparable expression of CXCR3-A and -B). CXCL9 and CXCL10 stimulation also showed a tendency to increase numbers of GBM L0 cells although the difference from control was not statistically significant. While GBM L0 expresses CXCR3-A and not CXCR3-B, mRNA levels of CXCR3-A were relatively lower as compared with the other human GS, which may explain the difficulty of observing an effect of CXCL9 and CXCL10 on the GBM L0 GS.
In summary, components of the CXCR3 system are expressed by glioma cells in vitro and in vivo. The increased expression of CXCR3 in the more malignant population of GS cells, and the CXCR3 antagonist-sensitive effects on the in vitro and in vivo growth of glioma, suggest that this chemokine system could be a unique target for human GBM therapy.
Supplementary material
Supplementary Tables I and II, supplementary Figure 1 and the color version all in situ hybridization data panels (Figures 1C, 2C, 3D) can be found at http://carcin.oxfordjournals.org/
Funding
This work was supported by a grant to J.K.H. from the National Institutes of Health (AI058256) as well as additional funding from the HHMI-UF Science for Life program.
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- bFGF
basic fibroblast growth factor
- AD
adherent
- cDNA
complementary DNA
- DMEM
Dulbecco’s modified Eagle’s medium
- EGF
epidermal growth factor
- FBS
fetal bovine serum
- GBM
glioblastoma multiforme
- GS
gliomasphere
- mRNA
messenger RNA
- NK
natural killer
- NKT
natural killer T
- PBS
phosphate-buffered saline
- RT
reverse transcription
- Treg
Tregulatory
- WT
wild-type
References
- 1.Kawakami Y, et al. Dendritic cell based personalized immunotherapy based on cancer antigen research. Front. Biosci. 2008;13:1952–1958. doi: 10.2741/2814. [DOI] [PubMed] [Google Scholar]
- 2.Vandercappellen J, et al. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008;267:226–244. doi: 10.1016/j.canlet.2008.04.050. [DOI] [PubMed] [Google Scholar]
- 3.Curbishley SM, et al. CXCR 3 activation promotes lymphocyte transendothelial migration across human hepatic endothelium under fluid flow. Am. J. Pathol. 2005;167:887–899. doi: 10.1016/S0002-9440(10)62060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wendel M, et al. Natural killer cell accumulation in tumors is dependent on IFN-gamma and CXCR3 ligands. Cancer Res. 2008;68:8437–8445. doi: 10.1158/0008-5472.CAN-08-1440. [DOI] [PubMed] [Google Scholar]
- 5.Rappert A, et al. CXCR3-dependent microglial recruitment is essential for dendrite loss after brain lesion. J. Neurosci. 2004;24:8500–8509. doi: 10.1523/JNEUROSCI.2451-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wenzel J, et al. Type I interferon-associated recruitment of cytotoxic lymphocytes: a common mechanism in regressive melanocytic lesions. Am. J. Clin. Pathol. 2005;124:37–48. doi: 10.1309/4EJ9KL7CGDENVVLE. [DOI] [PubMed] [Google Scholar]
- 7.Chu Y, et al. In situ expression of IFN-gamma-inducible T cell alpha chemoattractant in breast cancer mounts an enhanced specific anti-tumor immunity, which leads to tumor regression. Cancer Immunol. Immunother. 2007;56:1539–1549. doi: 10.1007/s00262-007-0296-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hensbergen PJ, et al. The CXCR3 targeting chemokine CXCL11 has potent antitumor activity in vivo involving attraction of CD8+ T lymphocytes but not inhibition of angiogenesis. J. Immunother. 2005;28:343–351. doi: 10.1097/01.cji.0000165355.26795.27. [DOI] [PubMed] [Google Scholar]
- 9.Walser TC, et al. Antagonism of CXCR3 inhibits lung metastasis in a murine model of metastatic breast cancer. Cancer Res. 2006;66:7701–7707. doi: 10.1158/0008-5472.CAN-06-0709. [DOI] [PubMed] [Google Scholar]
- 10.Datta D, et al. Ras-induced modulation of CXCL10 and its receptor splice variant CXCR3-B in MDA-MB-435 and MCF-7 cells: relevance for the development of human breast cancer. Cancer Res. 2006;66:9509–9518. doi: 10.1158/0008-5472.CAN-05-4345. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg-Bittman L, et al. The expression of the chemokine receptor CXCR3 and its ligand, CXCL10, in human breast adenocarcinoma cell lines. Immunol. Lett. 2004;92:171–178. doi: 10.1016/j.imlet.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 12.Kawada K, et al. Pivotal role of CXCR3 in melanoma cell metastasis to lymph nodes. Cancer Res. 2004;64:4010–4017. doi: 10.1158/0008-5472.CAN-03-1757. [DOI] [PubMed] [Google Scholar]
- 13.Kawada K, et al. Chemokine receptor CXCR3 promotes colon cancer metastasis to lymph nodes. Oncogene. 2007;26:4679–4688. doi: 10.1038/sj.onc.1210267. [DOI] [PubMed] [Google Scholar]
- 14.Suyama T, et al. Up-regulation of the interferon gamma (IFN-gamma)-inducible chemokines IFN-inducible T-cell alpha chemoattractant and monokine induced by IFN-gamma and of their receptor CXC receptor 3 in human renal cell carcinoma. Cancer. 2005;103:258–267. doi: 10.1002/cncr.20747. [DOI] [PubMed] [Google Scholar]
- 15.Furuya M, et al. Up-regulation of CXC chemokines and their receptors: implications for proinflammatory microenvironments of ovarian carcinomas and endometriosis. Hum. Pathol. 2007;38:1676–1687. doi: 10.1016/j.humpath.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Jones D, et al. The chemokine receptor CXCR3 is expressed in a subset of B-cell lymphomas and is a marker of B-cell chronic lymphocytic leukemia. Blood. 2000;95:627–632. [PubMed] [Google Scholar]
- 17.Engl T, et al. Prostate tumor CXC-chemokine profile correlates with cell adhesion to endothelium and extracellular matrix. Life Sci. 2006;78:1784–1793. doi: 10.1016/j.lfs.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 18.Ma X, et al. CXCR3 expression is associated with poor survival in breast cancer and promotes metastasis in a murine model. Mol. Cancer Ther. 2009;8:490–498. doi: 10.1158/1535-7163.MCT-08-0485. [DOI] [PubMed] [Google Scholar]
- 19.Monteagudo C, et al. CXCR3 chemokine receptor immunoreactivity in primary cutaneous malignant melanoma: correlation with clinicopathological prognostic factors. J. Clin. Pathol. 2007;60:596–599. doi: 10.1136/jcp.2005.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giuliani N, et al. CXCR3 and its binding chemokines in myeloma cells: expression of isoforms and potential relationships with myeloma cell proliferation and survival. Haematologica. 2006;91:1489–1497. [PubMed] [Google Scholar]
- 21.Pradelli E, et al. Antagonism of chemokine receptor CXCR3 inhibits osteosarcoma metastasis to lungs. Int. J. Cancer. 2009;125:2586–2594. doi: 10.1002/ijc.24665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zipin-Roitman A, et al. CXCL10 promotes invasion-related properties in human colorectal carcinoma cells. Cancer Res. 2007;67:3396–3405. doi: 10.1158/0008-5472.CAN-06-3087. [DOI] [PubMed] [Google Scholar]
- 23.Pellegatta S, et al. Neurospheres enriched in cancer stem-like cells are highly effective in eliciting a dendritic cell-mediated immune response against malignant gliomas. Cancer Res. 2006;66:10247–10252. doi: 10.1158/0008-5472.CAN-06-2048. [DOI] [PubMed] [Google Scholar]
- 24.Maru SV, et al. Chemokine production and chemokine receptor expression by human glioma cells: role of CXCL10 in tumour cell proliferation. J. Neuroimmunol. 2008;199:35–45. doi: 10.1016/j.jneuroim.2008.04.029. [DOI] [PubMed] [Google Scholar]
- 25.Ausman JI, et al. Studies on the chemotherapy of experimental brain tumors: development of an experimental model. Cancer Res. 1970;30:2394–2400. [PubMed] [Google Scholar]
- 26.Szatmari T, et al. Detailed characterization of the mouse glioma 261 tumor model for experimental glioblastoma therapy. Cancer Sci. 2006;97:546–553. doi: 10.1111/j.1349-7006.2006.00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C, et al. CX3CL1 and CX3CR1 in the GL261 murine model of glioma: CX3CR1 deficiency does not impact tumor growth or infiltration of microglia and lymphocytes. J. Neuroimmunol. 2008;198:98–105. doi: 10.1016/j.jneuroim.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heise CE, et al. Pharmacological characterization of CXC chemokine receptor 3 ligands and a small molecule antagonist. J. Pharmacol. Exp. Ther. 2005;313:1263–1271. doi: 10.1124/jpet.105.083683. [DOI] [PubMed] [Google Scholar]
- 29.Jopling LA, et al. Analysis of the pharmacokinetic/pharmacodynamic relationship of a small molecule CXCR3 antagonist, NBI-74330, using a murine CXCR3 internalization assay. Br. J. Pharmacol. 2007;152:1260–1271. doi: 10.1038/sj.bjp.0707519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verzijl D, et al. Noncompetitive antagonism and inverse agonism as mechanism of action of nonpeptidergic antagonists at primate and rodent CXCR3 chemokine receptors. J. Pharmacol. Exp. Ther. 2008;325:544–555. doi: 10.1124/jpet.107.134783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Wanrooij EJ, et al. CXCR3 antagonist NBI-74330 attenuates atherosclerotic plaque formation in LDL receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2008;28:251–257. doi: 10.1161/ATVBAHA.107.147827. [DOI] [PubMed] [Google Scholar]
- 32.Hancock WW, et al. Requirement of the chemokine receptor CXCR3 for acute allograft rejection. J. Exp. Med. 2000;192:1515–1520. doi: 10.1084/jem.192.10.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrison JK, et al. In situ hybridization analysis of chemokines and chemokine receptors in the central nervous system. Methods. 2003;29:312–318. doi: 10.1016/s1046-2023(02)00354-7. [DOI] [PubMed] [Google Scholar]
- 34.De Witt Hamer PC, et al. The genomic profile of human malignant glioma is altered early in primary cell culture and preserved in spheroids. Oncogene. 2008;27:2091–2096. doi: 10.1038/sj.onc.1210850. [DOI] [PubMed] [Google Scholar]
- 35.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 36.Lasagni L, et al. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J. Exp. Med. 2003;197:537–549. doi: 10.1084/jem.20021897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehlert JE, et al. Identification and partial characterization of a variant of human CXCR3 generated by posttranscriptional exon skipping. J. Immunol. 2004;173:6234–6240. doi: 10.4049/jimmunol.173.10.6234. [DOI] [PubMed] [Google Scholar]
- 38.Carr DJ, et al. An increase in herpes simplex virus type 1 in the anterior segment of the eye is linked to a deficiency in NK cell infiltration in mice deficient in CXCR3. J. Interferon Cytokine Res. 2008;28:245–251. doi: 10.1089/jir.2007.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang D, et al. Regulation of pulmonary fibrosis by chemokine receptor CXCR3. J. Clin. Invest. 2004;114:291–299. doi: 10.1172/JCI16861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller M, et al. CXCR3 signaling reduces the severity of experimental autoimmune encephalomyelitis by controlling the parenchymal distribution of effector and regulatory T cells in the central nervous system. J. Immunol. 2007;179:2774–2786. doi: 10.4049/jimmunol.179.5.2774. [DOI] [PubMed] [Google Scholar]
- 41.Grauer OM, et al. CD4+FoxP3+ regulatory T cells gradually accumulate in gliomas during tumor growth and efficiently suppress antiglioma immune responses in vivo. Int. J. Cancer. 2007;121:95–105. doi: 10.1002/ijc.22607. [DOI] [PubMed] [Google Scholar]
- 42.Kim JV, et al. Two-photon laser scanning microscopy imaging of intact spinal cord and cerebral cortex reveals requirement for CXCR6 and neuroinflammation in immune cell infiltration of cortical injury sites. J. Immunol. Methods. 2010;352:89–100. doi: 10.1016/j.jim.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cross AK, et al. Chemokines induce migration and changes in actin polymerization in adult rat brain microglia and a human fetal microglial cell line in vitro. J. Neurosci. Res. 1999;55:17–23. doi: 10.1002/(SICI)1097-4547(19990101)55:1<17::AID-JNR3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 44.Kohler RE, et al. Antagonism of the chemokine receptors CXCR3 and CXCR4 reduces the pathology of experimental autoimmune encephalomyelitis. Brain Pathol. 2008;18:504–516. doi: 10.1111/j.1750-3639.2008.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balashov KE, et al. CCR5(+) and CXCR3(+) T cells are increased in multiple sclerosis and their ligands MIP-1alpha and IP-10 are expressed in demyelinating brain lesions. Proc. Natl Acad. Sci. USA. 1999;96:6873–6878. doi: 10.1073/pnas.96.12.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hiroi M, et al. Constitutive nuclear factor kappaB activity is required to elicit interferon-gamma-induced expression of chemokine CXC ligand 9 (CXCL9) and CXCL10 in human tumour cell lines. Biochem. J. 2003;376:393–402. doi: 10.1042/BJ20030842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrai I, et al. Activation of p38(MAPK) mediates the angiostatic effect of the chemokine receptor CXCR3-B. Int. J. Biochem. Cell Biol. 2008;40:1764–1774. doi: 10.1016/j.biocel.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Romagnani P, et al. Cell cycle-dependent expression of CXC chemokine receptor 3 by endothelial cells mediates angiostatic activity. J. Clin. Invest. 2001;107:53–63. doi: 10.1172/JCI9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.