Abstract
Isothiocyanates (ITCs), including benzyl isothiocyanate (BITC), phenethyl isothiocyanate (PEITC) and sulforaphane, compounds found in cruciferous vegetable, are highly effective in inducing cell cycle arrest and apoptosis in a variety of cancer cells and animal models. Although some studies indicate that ITC-induced reactive oxygen species (ROS) generation may underlie apoptosis induction, our recent studies show that covalent binding to target proteins may be an important event triggering apoptosis. In this study, we report that BITC and PEITC significantly inhibit proteasome activity in a variety of cell types. Further studies show that ITCs inhibit both the 26S and 20S proteasomes, presumably through direct binding, and that this inhibition is unrelated to either ROS generation or ITC-induced protein aggregation. The potency of ITC-induced proteasome inhibition correlates with the rapid accumulation of p53 (tumor suppressor) and IκB nuclear factor-kappaB (nuclear factor-kappaB inhibitor). Finally, our results demonstrate that BITC and PEITC, the two strongest proteasome inhibitors, significantly suppress growth of multiple myeloma (MM) cells through induction of cell cycle arrest at G2/M phase and apoptosis. This study suggests that proteasome, like tubulin, is a potential molecular target of ITCs, thus providing a novel mechanism by which ITCs strongly inhibit growth of MM cells and new leads in identifying compounds with therapeutic and preventative efficacies for MM. It also supports the future studies of ITCs as therapeutic and preventive agents for MM.
Introduction
Multiple myeloma (MM), a neoplastic proliferation of plasma cells, is currently the second most prevalent blood cancer (10% of all blood cancers), after non-Hodgkin’s lymphoma, in the USA. The incidence of myeloma is 9.5 cases per 100 000 African–Americans and 4.1 cases per 100 000 Caucasian–Americans (1). Among African–Americans, myeloma is one of the top 10 leading causes of cancer death. Almost all patients with MM who survive initial treatment will eventually relapse and require further therapy (2). Since the Food and Drug Administration’s approval in 2003, Bortezomib, a proteasome inhibitor, has become a standard treatment of patients with relapsed and resistant MM (2,3). However, its clinical application is still plagued by relapse, drug resistance and adverse side effects (4). Compounds with better therapeutic efficacy and lesser side effects are in critical need. Also, the area of chemoprevention against MM has remained largely unexplored. Yet, given that the average age of onset of MM is >60 years, there is potentially a large timeframe for preventive intervention.
The promising applications of Bortezomib in MM and other malignancies have validated the proteasome as an important target in the treatment of cancer (2,3,5). The ubiquitin–proteasome system (UPS) is a non-lysosomal protein degradation mechanism that plays a primary role in the control of protein turnover in mammalian cells, as well as in the removal of abnormal proteins (6). This tightly regulated proteasome complex plays a pivotal role in the cell cycle, cellular signal transduction, transcriptional regulation, stress responses, cell differentiation and metabolic adaptation. Proteasome inhibition results in the dysregulation of cell cycle progression and, ultimately, apoptosis (3,5,6). Additionally, accumulation of ubiquitinated proteins due to proteasome inhibition also leads to G2/M phase arrest (7,8).
Electrophilic benzyl isothiocyanate (BITC) and phenethyl isothiocyanate (PEITC) are two of the most-studied dietary isothiocyanates (ITCs). They are highly effective in protecting against a variety of chemical carcinogen-induced cancers in animal models (9). Epidemiological studies have also shown that dietary intake of ITCs is associated with reduced risk of human cancers (9). Evidence obtained from both in vitro and in vivo studies supports that they exert their antiproliferative effects through inducing cell cycle arrest and apoptosis, functions vital for their antitumor activities (9). Although the upstream biochemical events underlying ITC-induced apoptosis remain unclear, studies indicate that ITC-induced reactive oxygen species (ROS) generation may underlie apoptosis induction (10–12). However, our previous results show that covalent binding to target proteins may be an important event triggering apoptosis (13). For example, differential affinities of binding to cysteine residues in tubulin by BITC, PEITC and sulforaphane (SFN) correlate well with tubulin conformation and functional changes, microtubule disruption, tubulin precipitation and degradation and eventually cell cycle arrest and apoptosis induction (14,15). More importantly, our results show that the tubulin-related effects were ROS independent, raising questions on the relationship and significance of ROS generation and protein binding in apoptosis induction (14–16).
In this study, we report that ITCs effectively inhibit the activity of the UPS presumably through direct binding. The degradation of some UPS-dependent substrates that are crucial to signal transduction, such as p53 and IκBα, was inhibited. Finally, our data show that BITC and PEITC induce significant cell cycle arrest at G2/M phase, apoptosis and growth inhibition in MM cells at physiologically relevant concentrations.
Materials and methods
Cells and chemicals
The human MM cell lines U266 and RPMI-8226 (American Type Culture Collection Manassas, VA) were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) at 37°C in 5% CO2. HeLa and A549 cells (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Invitrogen) at 37°C in 5% CO2. Bortezomib and MG132 were purchased from LC Labs (Woburn, MA) and Biomol (Plymouth Meeting, PA), respectively. SFN was provided by Dr Stephen Hecht (University of Minnesota). PEITC, BITC, N-methyl phenethylamine (NMPEA), dimethyl sulfoxide, cycloheximide and all other reagents were the highest grade available from Sigma–Aldrich (St Louis, MO), unless otherwise noted.
Assay for proteasome activity
The assay was based on a previously published method (17). After treatment, cells were harvested and lysed with proteasome lysis buffer (50 mM Tris–HCl, pH 7.8, 20 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP, 10% glycerol and 0.04% NP-40) by repeated pipetting, followed by incubation on ice for 20 min. Cell lysate with 30 μg proteins was incubated with 20 μM fluorogenic peptide substrates in 100 μl of volume for 60 min at 37°C in the dark. Cold phosphate-buffered saline was used to dilute samples to 200 μl before fluorescence was measured by a Synergy 96-well plate reader (Bio-Tek, Winooski, VT) with an excitation filter of 365 ± 20 nm and an emission filter of 485 ± 25 nm.
Purified 20S proteasome extracted from rabbit erythrocytes (Boston Biochem;, Cambridge, MA) was further purified by size exclusion column (Sephadex G-25; GE Healthcare, Piscataway, NJ) to remove dithiothreitol before incubation with up to 40 μM PEITC, SFN and NMPEA individually for 2 h at room temperature in the dark.
Cell lysate preparation and western blotting
The assay was based on a previously published method (14). Treated cells were lysed in a buffer containing 20 mM Tris–HCl, 0.5% NP-40, on ice for 20 min before centrifugation at 15 000g for 10 min. The supernatants were collected as the soluble fraction, whereas the pellets were redissolved in sodium dodecyl sulfate buffer (65 mm Tris, pH 7.0, 2% sodium dodecyl sulfate, 50 mm dithiothreitol, 10% glycerol and 0.001% bromophenol blue) and used as the insoluble fraction. Twenty micrograms of proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore). Immunoblot analysis was performed with specific antibodies against p38–MAPK and phosphorylated p38 (Cell Signaling, Danvers, MA) and enhanced chemiluminescence-based detection (GE Healthcare).
Native gel electrophoresis and substrate overlay assay
The assay was based on a previously published method (18). Samples with 50 ng proteins were analyzed on mini gels using a Mini-Protean gel apparatus (Bio-Rad, Hercules, CA). Non-denaturing gels consisted of 2.5% stacking and 4.5% resolving gels cast in 90 mM Tris, pH 8.3, 1.6 mM borate and 0.08 mM ethylenediaminetetraacetic acid. Samples were electrophoresed for 280 Vh in a cold room. After electrophoresis, protease activity was detected in non-denaturing gels by overlaying the gels with 20 mM Tris–HCl, pH 7.8, 5 mM MgC12, 10 mM KC1, 1 mM dithiothreitol, 2 mM ATP and 200 pM fluorogenic peptide and incubating the gels at 37°C for 30–60 min. The fluorescent gels were transilluminated by a ultraviolet light and photographed. The density of the fluorescent bands was determined by ImageJ software.
Measurement of reactive oxidative species production
The assay was based on a previously published method (13). After treatment with a variety of agents, cells were further stained with 10 μmol/l 5-(and-6)-carboxy-2′,7′-difluorodihydro-fluorescein diacetate (Invitrogen) at 37°C for 30 min before being washed with phosphate-buffered saline for three times. The intracellular ROS generation was measured using FACScalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ) using appropriate filters.
Determination of cellular glutathione
The Ellman assay was based on a previously published method (19). Briefly, U266 cells treated with dimethyl sulfoxide, ITCs and NMPEA were lysed in 5% trichloroacetic acid to precipitate proteins. The supernatant fraction (100 μl) was mixed with 100 μl of 10 mM 5,5′-dithiobis(2-nitrobenzoic acid) and 800 μl buffer containing 100 mM potassium phosphate, pH 7.4. The absorbance at 412 nm was read within 30 s at room temperature with a Shimadzu UV-1700 spectrophotometer. Cells treated with solvent dimethyl sulfoxide were used as a control.
Cell cycle analysis
The assay was based on a previously published method (14). After treatment, cells were fixed with ice-cold 80% ethanol, treated with 500 μg/ml RNase A (Sigma-Aldrich) and subsequently stained with 25 μg/ml propidium iodide (Sigma-Aldrich). Cell cycle distributions were measured by flow cytometry on a FACSCalibur (BD Biosciences).
Cell proliferation assay
The assay was based on a previously published method (14). CellTiter 96 Aqueous One Solution cell proliferation assay kit (Promega, Madison, WI) was used. Briefly, cells were plated in 96-well plates at a density of 104 cells per well (200 μl). Twenty-four hours after plating, cells were treated with different doses of each ITC for 24 h. Then, 20 μl of the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium solution was added. After 2 h, the absorbance was measured at 490 nm with a Synergy HT fluorescent microplate reader (BIO-TEK Instruments).
Data analysis
At least three independent experiments were conducted for all analyses. Values are expressed as means ± standard deviations. A Student’s t-test was used to estimate statistical significance.
Results
BITC and PEITC are effective proteasome inhibitors in a variety of cancer cells
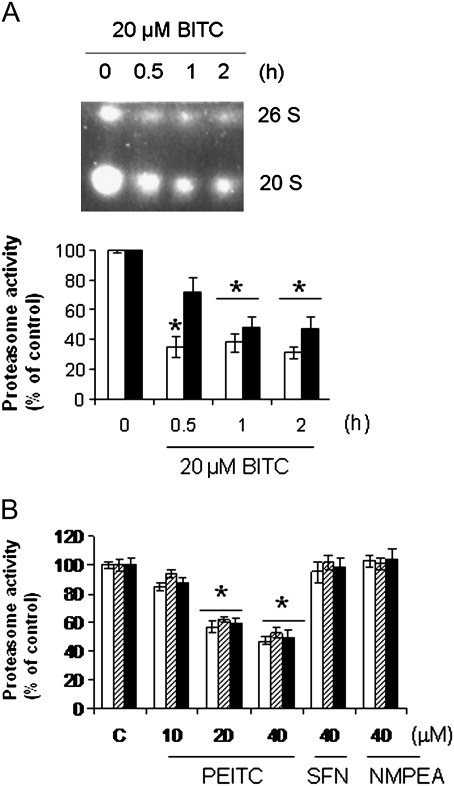
To study whether ITCs (structures are shown in Figure 1A) inhibit UPS, we treated U266 cells with a series of concentrations of BITC, PEITC and SFN for 4 h. Proteasome activities, including chymotrypsin-like, trypsin-like and caspase-like, were measured using fluorescent-labeled peptides. The results (Figure 1B) show that both BITC and PEITC significantly inhibited all three proteasome activities in a concentration-dependent fashion. SFN, however, was unable to inhibit proteasome activities at concentrations up to 30 μM. Instead, it promoted both chymotrypsin-like and caspase-like activities at lower concentrations, consistent with a previous study that SFN enhances proteasome activity in murine neuroblastoma cells (17). MG132, a known proteasome inhibitor, was more effective in inhibiting chymotrypsin-like and caspase-like, but not trypsin-like, activities compared with BITC and PEITC. The inhibition was rapid—observed as early as 30 min after PEITC treatment (Figure 1C). It was time dependent within the first 4 h and the effects lasted up to 24 h. NMPEA, a structural analog of PEITC without ITC functionality, did not inhibit proteasome activity under the same conditions, suggesting that the ITC functional group is essential for inhibiting proteasome activity. The potency order of BITC > PEITC > SFN suggested that the side chain moiety dictates the inhibitory potency.
Fig. 1.
ITCs significantly inhibit proteasome activity in a variety of cells. (A) Structures of BITC, PEITC, SFN and NMPEA, a structural analog of PEITC without ITC functionality. (B) U266 cells were treated with a series of concentrations of BITC, PEITC and SFN for 4 h. Chymotrypsin-like, caspase-like and trypsin-like proteasome activities were measured using Suc-LLVY-AMC, Z-LLE-AMC and Boc-LRR-AMC substrates, respectively. White bars: chymotrypsin-like activity; striped bars: trypsin-like activity; black bars: caspase-like activity. (C) Inhibiting proteasome activity by PEITC is time-dependent and long lasting. U266 cells were treated with 10 μM PEITC for up to 24 h. NMPEA was used as a negative control. (D) A variety of cancer cells, including HeLa, A549, HT-29, PC-3 and MCF-7, were treated with 10 μM BITC for 4 h. Chymotrypsin-like activity was measured in cell lysates before (white bars) and after (black bars) the treatment (upper panel). The ubiquitinated proteins in the same cell lysates were immunoblotted (lower panel). *P < 0.05; **P < 0.01.
To study whether ITC-induced proteasome inhibition is cell type specific, we treated a variety of cell types, including HeLa (cervical cancer), A549 (non-small cell lung cancer), HT-29 (colon cancer), PC-3 (prostate cancer) and MCF-7 (breast cancer), with 10 μM BITC for 4 h. Results (Figure 1D) show that the chymotrypsin-like activity in all five cell lines was significantly (∼50%) inhibited, indicating that ITC-induced proteasome inhibition is not cell type specific.
BITC inhibits both 20S and 26S proteasomes
Next, we studied the mechanisms of ITC-induced proteasome inhibition. The 26S and 20S proteasome complexes are responsible for ATP-dependent and -independent protein degradation, respectively (5,6). The 26S complex is composed of one or two 19S regulatory particles and a 20S catalytic particle. To study which complexes were affected by ITC treatment, we treated HeLa cells with 20 μM BITC and purified both 26S and 20S from cell lysates using native gel electrophoresis (18). The results of substrate overlay assay (Figure 2A) show that chymotrypsin-like activity in both the 26S and 20S complexes was significantly inhibited by BITC treatment. The inhibition occurred as rapid as 30 min. The inhibition of the 26S complex, indicated by the densitometry, was more substantial than that of 20S, suggesting that the functions of both the 19S regulatory particle and 20S catalytic particle were affected by ITC treatment.
Fig. 2.
ITCs inhibit both 26S and 20S proteasome complexes through direct binding. (A) Chymotrypsin-like activity in both 26S and 20S proteasome complexes was inhibited by BITC. HeLa cells were treated with 20 μM BITC for up to 2 h. Cell lysate was extracted and separated using native gel electrophoresis. The proteasome activity was determined by the substrate overlay assay (see Materials and methods). The panel below is the densitometry analysis of the results. White bars: 26S; black bars: 20S. (B) Purified proteasome was inhibited by PEITC, but not SFN and NMPEA. Purified 20S proteasome from rabbit erythrocytes was incubated with PEITC, SFN and NMPEA for 4 h at room temperature in the dark before the activity was determined using the fluorogenic substrates. *P < 0.01.
ITCs inhibit proteasome activity through direct binding
To further study the interaction between ITCs and proteasome activity, we incubated PEITC, SFN and NMPEA individually with purified 20S proteasome at room temperature for 4 h. The results (Figure 2B) show that similar to the cultured cell data, PEITC significantly inhibited all three activities of purified proteasome, suggesting that ITC-induced proteasome inhibition is through direct binding. Furthermore, both SFN and NMPEA had little effect on proteasome activity, suggesting that both the ITC moiety and side-chain structure play important roles in ITC activity.
ITC-induced proteasome inhibition is unrelated to protein aggregation
Previously, we found that BITC- and PEITC -induced tubulin-containing aggresome-like protein aggregates that are insoluble in buffers that are often used to extract whole cell lysate (15,16). These buffers usually contain 1% non-ionic detergents, such as NP-40, Triton X-100 or Tween 20. We also showed that 10 μM of colchicine, vinblastine or taxol, three tubulin binding agents, can almost completely block the binding of ITCs to tubulin and the consequential formation of the protein aggregates (16). Among these three agents, colchicine and vinblastine effectively disrupt microtubules, whereas taxol promotes microtubule formation. To study if ITC-induced tubulin-containing protein aggregates contribute to proteasome inhibition, we treated HeLa cells with 1 and 10 μM colchicine, vinblastine or taxol for 1 h followed by treatment with 10 μM BITC for 4 h. The results (Figure 3A–C) show that 10 μM colchicine and vinblastine, but not taxol, alone caused moderate proteasome inhibition (<15%). However, pretreatments with these agents had little effects on proteasome inhibition by BITC, suggesting that BITC-induced proteasome inhibition may be unrelated to ITC-induced protein aggregation, supporting again that ITC direct binding may be the underlying mechanism of proteasome inhibition.
Fig. 3.
ITC-induced proteasome inhibition is unrelated to protein aggregation and ROS generation. Pretreatments with colchicine (A), vinblastine (B) and paclitaxel (C) did not have substantial effects on proteasome inhibition by BITC. HeLa cells were pretreated with 1 or 10 μM colchicine, vinblastine or paclitaxel for 1 h followed by treatment with 10 μM BITC for another 4 h. White bars: chymotrypsin-like activity; striped bars: trypsin-like activity; black bars: caspase-like activity. (D) Proteasome was inhibited by 1 mM H2O2 or 10 μM PEITC. U266 cells were treated with various concentrations of H2O2 or 10 μM PEITC for 4 h. (E) Polyethylene glycol-linked catalase did not affect ITC-induced proteasome inhibition. U266 cells were pretreated with 500 U (low cat., lower amount of catalase) or 1000 U (high cat., higher amount of catalase) polyethylene glycol catalase for 1 h followed by 10 μM BITC for 4 h. (F) 3-Amino-1,2,4-triazole (ATZ), a catalase inhibitor, did not aggravate BITC-induced proteasome inhibition. U266 cells were treated with 10 μM ATZ alone or in combination with 10 μM BITC for 4 h. White bars: chymotrypsin-like activity; striped bars: trypsin-like activity; black bars: caspase-like activity. (G) Three ITCs depleted GSH at similar rates. U266 cells were treated with 10 μM (white bars) and 20 μM (black bars) of BITC, PEITC, SFN and NMPEA for 1 h. Cellular GSH concentration was measured and the relative amount was calculated against the dimethyl sulfoxide-treated control. *P < 0.01; **P < 0.001.
ITC-induced proteasome inhibition is unrelated to ROS generation
ITCs are known to induce ROS generation at least partially due to conjugation and depletion of intracellular glutathione (GSH) (19,20). Therefore, an important aspect of this mechanistic study is to understand the role of ROS in ITC-induced proteasome inhibition. To investigate this, we treated U266 cells with either 10 μM PEITC or up to 1 mM H2O2 for 4 h. Results (Figure 3D) show that up to 250 μM, H2O2 did not significantly induce proteasome inhibition. H2O2 at 1 mM was needed to achieve comparable levels of proteasome inhibition to that of 10 μM PEITC. A separate experiment indicated that the ROS level in U266 cells treated with 10 μM PEITC was much less than that of treatment with 100 μM H2O2 at all three time points of 1, 4 and 24 h (data not shown). Next, we treated U266 cells with 10 μM BITC in the presence and absence of cell membrane-permeable polyethylene glycol-linked catalase, which is effective in quenching intracellular ROS (21). The results (Figure 3E) show that polyethylene glycol catalase, at both low and high doses, did not affect BITC-induced proteasome inhibition, confirming that ITC-induced ROS do not play a significant role in proteasome inhibition. Additionally, we treated U266 cells with up to 1 mM 3-amino-1,2,4-triazole (22), a specific catalase inhibitor, followed by treatment with 10 μM BITC. Results (Figure 3F) show that 3-amino-1,2,4-triazole alone failed to induce substantial proteasome inhibition and 3-amino-1,2,4-triazole did not aggravate proteasome inhibition by BITC. Lastly, we measured cellular GSH concentration levels after cells were treated with ITCs and NMPEA for 1 h. The results (Figure 3G) show that at both concentrations of 10 and 20 μM, SFN depleted GSH at a slightly higher rate than PEITC and BITC. The results agree with a previous study in which BITC and SFN at several concentration levels depleted GSH at similar rates (19). In an earlier study (13), we demonstrated that SFN conjugates faster to GSH than PEITC. The inconsistency between the potency of ITC depleting GSH and that of proteasome inhibition suggests that proteasome inhibition by ITCs is not linked to GSH depletion. Taken together, these data suggest that ITC-induced proteasome inhibition is unrelated to ITC-induced ROS generation.
Degradation of p53 and IκBα is inhibited in ITC-treated cells
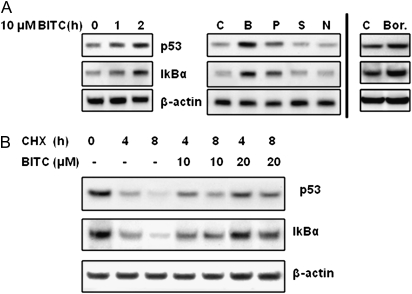
It has been shown that proteasome inhibitors induce accumulation of wild-type p53, the most-studied tumor suppressor protein, and IκBα, a natural inhibitor of nuclear factor-kappaB (NF-κB), because they are known UPS substrates (2,5). To investigate the functional consequences of ITC-induced proteasome inhibition, we treated A549 cells with 10 μM BITC for up to 2 h and determined the expression of p53 and IκBα. A549 was chosen because of its wild-type p53 status, whereas both U266 and RPMI-8226 harbor mutant p53. The results (Figure 4A left) show that, as expected, both p53 and IκBα proteins accumulated in a time-dependent manner after BITC treatment. Also, ITCs displayed a potency order of inducing p53 accumulation of BITC>PEITC>SFN (Figure 4A) that is consistent with the order of proteasome inhibition potency, suggesting that the accumulation of p53 and IκBα is related to ITC-induced proteasome inhibition. Additionally, the accumulation of p53 and IκBα was also observed in MCF-7 cells (data not shown), another cell line containing wild-type p53.
Fig. 4.
ITCs induce accumulation of proteasome substrates p53 and IκBα. (A) ITCs induce accumulation of proteasome substrates p53 and IκBα. Left: A549 cells were treated with 10 μM BITC for up to 2 h. Middle: A549 cells were treated with dimethyl sulfoxide (denoted as C), 10 μM BITC (denoted as B), PEITC (denoted as P), SFN (denoted as S) and NMPEA (denoted as N) for 2 h. Both p53 and IκBα was analyzed by immunoblots. Right: Bortezomib (40 nM for 2 h) was used as a positive control. (B) Degradation of p53 and IκBα was inhibited by BITC treatment. A549 cells were treated with 20 μg/ml cycloheximide (a protein synthesis inhibitor) and up to 20 μM BITC for 4 and 8 h.
To study the mechanism of the accumulation of p53 and IκBα by BITC, we treated A549 cells with 10 and 20 μM BITC in the absence and presence of 20 μg/ml cycloheximide (cycloheximide, a protein synthesis inhibitor) for up to 8 h. The results (Figure 4B) show a time- dependent decrease in p53 and IκBα expression in cells treated with cycloheximide alone due to its rapid degradation; however, the degradation was inhibited by BITC. The inhibition by 20 μM BITC is more substantial than that of 10 μM. This is consistent with the concentration dependence of proteasome inhibition by BITC, suggesting that proteasome inhibition is responsible for accumulation of p53 and IκBα.
BITC and PEITC induce significant G2/M arrest and apoptosis in MM cells
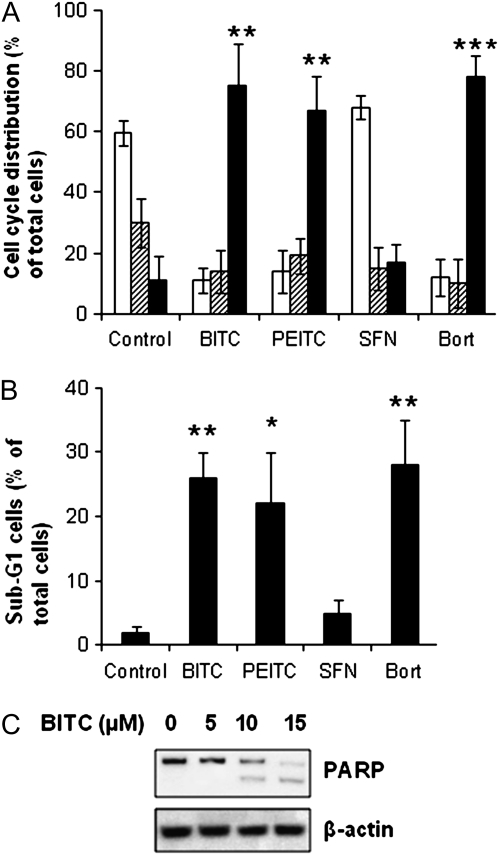
The efficacy of Bortezomib in MM suggests that it is a cancer-type sensitive to proteasome inhibitors (2–4). To study whether ITCs are effective in inducing cell cycle arrest and apoptosis in MM cells, we treated U266 cells with 10 μM BITC, PEITC and SFN for 24 h. The results (Figure 5A) show that both BITC and PEITC induced significant amounts of G2/M arrest and reduced G1 and S phase populations. In contrast, G1 reduction and G2/M arrest were not substantial in cells treated with SFN. Cell cycle arrest at G2/M phase was also observed in cells treated with Bortezomib at 40 nM.
Fig. 5.
ITCs induce cell cycle arrest and apoptosis in MM cells. (A) U266 cells were treated with 10 μM ITCs or 40 nM Bortezomib for 24 h followed by cell cycle analysis. White bars: G1 phase; striped bars: S phase; black bars: G2/M phase. (B) ITCs induce substantial amount of sub-G1 (apoptotic) cells. (C) BITC induced poly (ADP-ribose) polymerase cleavage. U266 cells were treated with up to 15 μM BITC for 24 h before detection of both full length and cleaved poly (ADP-ribose) polymerase by immunoblots. *P < 0.05; **P < 0.01; ***P < 0.001.
The percentage of sub-G1 cells, a marker for apoptosis induction, was also drastically increased in BITC- and PEITC-treated cells compared with SFN-treated cells (Figure 5B). The induction of apoptosis was further confirmed by poly (ADP-ribose) polymerase cleavage, which was proportional to BITC concentration (Figure 5C). Taken together, these data suggest that both BITC and PEITC, but not SFN, are strong inducers of cell cycle arrest and apoptosis in MM cells. The potency of their apoptosis induction correlated well with their ability to inhibit proteasome activity.
BITC inhibits proliferation of MM cells
To study whether BITC inhibits growth of MM cells, U266 cells were treated with BITC (0–40 μM) for 24 and 48 h. The results (Figure 6A) show that BITC was effective in inhibiting cell proliferation at both time points. The inhibition was concentration dependent. The half maximal inhibitory concentration values for BITC at 24 and 48 h were 8.3 ± 1.7 and 4.7 ± 1.3 μM, respectively. Figure 6B shows that the potency order of inhibiting cell growth among three ITCs was BITC>PEITC>SFN, in agreement with their potency of inhibiting proteasome activity. It should be noted that growth of another MM cell line RPMI-8226 was also inhibited by BITC and PEITC, with similar half maximal inhibitory concentration values at both time points and with a similar the potency order among ITCs (data not shown).
Fig. 6.
ITCs inhibit proliferation of MM cells. (A) U266 cells were treated with a series of concentrations of BITC or Bortezomib for 24 h (white bars) and 48 h (black bars). The viable cells were assayed by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium assay as triplicates. (B) The potency order of inhibiting cell growth was BITC > PEITC > SFN. U266 cells were treated with dimethyl sulfoxide (white bars), 5 μM (striped bars), 10 μM (gray bars), 15 μM (black bars) BITC, PEITC, SFN and NMPEA for 48 h. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
The approval of Bortezomib for the clinical treatment of MM has led to a surge in the research and development of proteasome inhibitors (2–6). The current pool of proteasome inhibitors includes numerous compounds with diverse structures. What is common in these compounds is that they are all short peptides capped with an electrophilic C-terminus (5,23,24). The C-terminal modifications include boronic acid (such as Bortezomib), aldehyde (MG132), epoxyketone (epoxomycin), vinyl sulfone (z-Leu-Leu-Leu-vinyl sulfone) and others (5,23,24). The findings in this study may suggest ITCs as new additions to the family of electrophilic proteasome inhibitors. Compared with peptidal proteasome inhibitors, dietary ITCs have advantages of improved uptake rates and levels due to their smaller size.
In our earlier proteomic study using radiolabeled ITCs and 2-dimensional gel electrophoresis (14), some proteasome subunits have been identified in gel spots containing radioactivity, suggesting that proteasome is a potential target of ITCs. These subunits include proteasome subunit alpha type 3 (PSA3), proteasome subunit alpha type 4 (PSA4), proteasome subunit beta type 9 (PSB9), 26S protease regulatory subunit 6A (PRS6A) and 26S protease regulatory subunit 8 (PRS8). In this study using functional assays, we showed that ITCs inhibit proteasome activity in cells and purified proteasome. Proteasome inhibition by ITCs may contribute to ITC-induced cell cycle arrest and apoptosis.
Our study indicates that ITCs inhibit proteasome activity presumably through direct binding. Previously, we identified an in vitro adduct of BITC with Cys303 in β-tubulin and an in vivo adduct of BITC with Cys347 in α-tubulin. The ITC-binding affinities correlate well with not only cell cycle arrest and apoptosis induction in A549 cells (13) but also tubulin precipitation and degradation in a variety of cells (14–16). Recently, BITC has been shown to form a covalent adduct with the N-terminal proline of macrophage migration inhibitory factor and inhibit the migration inhibitory factor-mediated inflammatory response (25–27). These results, together with previous studies on cytochrome P450 (28) and Keap-Nrf2 (29), support the notion that the binding of ITCs to nucleophilic residues in target proteins constitutes an important mechanism for their downstream molecular/cellular effects. A similar mechanism by which peptidal electrophiles inhibit proteasome activity was reported: the electrophilic cap binds with the γ-hydroxyl of the N-terminal threonine within the active site of the catalytic subunits to form a covalent and (in most cases) irreversible bond (5,23). Additionally, X-ray structure reveals that the cysteine 118 of β3 subunit, which protrudes into the β2 active site, is also susceptible to electrophilic attack (24). In this study, we found that ITCs are able to inhibit proteasome activity, including both 26S and 20S complexes. More importantly, ITCs can directly inhibit the activity of the purified proteasome, suggesting that the proteasome, like tubulin, may serve as a molecular target of ITCs. The notion of direct binding is also supported by the observation that BITC-induced proteasome inhibition is unrelated to ITC-induced protein aggregation and ROS generation. Further investigations are needed to identify the binding site(s).
Another major finding is that not all ITCs are equal. Results showed that the ITC functional group is required for proteasome inhibition and the side chain also affects the scale of inhibition. Interestingly, the potency order of inhibiting proteasome is consistent with that of inducing cell cycle arrest at G2/M phase, apoptosis and ultimately cell growth inhibition. The structure–activity relationship among three ITCs, which has been also observed in our previous studies (13–16), confirming that binding to target proteins for hydrophobic ITCs such as BITC and PEITC is an important biological event and suggesting that the activity of ITCs can be optimized through structure design of the agents. Contrary to BITC and PEITC, SFN has been demonstrated to upregulate proteasome activity and expressions of some proteasome subunits, such as PSMB5 and PSMB6, through activation of Keap1–Nrf2 pathway (17,30). Since previous studies have shown that SFN induces direct and indirect antioxidant response in cells through mild oxidative stress (29,31), the findings in the current study may suggest that stress on proteasome activity is a triggering event leading to proteasome activity enhancement by SFN. However, whether SFN inhibits proteasome activity, especially at early time points, requires further studies.
Proteasome inhibitors modulate the activity of transcription factors such as p53 (32). Wild-type p53 expression is low in most cells under normal conditions and it has a short half-life. Its level is tightly regulated by Mdm2 (or Hdm2), the product of an inducible gene by p53. Mdm2, a RING finger-containing ubiquitin E3 ligase, binds to the N-terminal domain of p53 and labels it with ubiquitin before its degradation by the 26S proteasome. Rapid accumulation of p53, resulting from proteasome inhibition, triggers a variety of cellular responses including cell cycle arrest, apoptosis induction, DNA repair and differentiation (32,33). PEITC has been shown to induce p53 accumulation in a variety of cells and apoptosis induction by PEITC occurs through a p53-dependent pathway (34,35). Our results in this study show that p53 accumulation correlates well with the potency order of proteasome inhibition by ITCs, suggesting a possible link between these two events. However, it is unlikely that p53 plays a role in ITC-induced apoptosis in U266 cells because of its mutant p53 status. We believe that inhibition of NF-κB through accumulation of IκB may contribute to apoptosis induction in U266 cells (36–38). NF-κB regulates various immune and inflammatory responses and plays an important role in tumorigenesis by stimulating cell survival pathways, blocking apoptosis, inducing angiogenesis and increasing metastatic potential. IκB, the natural inhibitor of NF-κB, is a UPS substrate, so its level depends on proteasome activity. Inhibition of IκB degradation by proteasome inhibitors inhibits NF-κB activity through sequestering NF-κB in the cytoplasm. Additionally, proteasome inhibition blocks NF-κB activation through inhibiting proteasome-mediated cleavage of NF-κB family members p100 and p105 to the activated forms p50 and p52 (39). Both BITC and PEITC have been shown to inhibit NF-κB and inflammatory responses (40,41). In this study, we showed that treatment with BITC or PEITC resulted in delayed degradation of IκB. However, whether ITC-induced proteasome inhibition is responsible for NF-κB inhibition and apoptosis induction needs further investigation.
BITC and PEITC have been shown to induce apoptosis in leukemia and lymphoma cells (42–46). Currently, PEITC is being used in a clinical trial for the treatment of leukemia (47). These results are consistent with our finding that BITC and PEITC are potent inhibitors of MM cell growth. In fact, cells derived from hematologic cancers, such as MM and lymphoma, are among the most sensitive to proteasome inhibition (2,4,42). Previously, our laboratory studied the pharmacokinetics of orally administered PEITC in lung cancer prevention using F344 rats (48). The results indicate that PEITC can reach a peak concentration of 18.8 μM in blood after 2.9 h and PEITC concentration stays >5 μM for >12 h, suggesting that the effective concentrations for proteasome inhibition by BITC and PEITC are physiologically attainable. Additionally, it has been shown that dietary ITCs, including BITC and PEITC, have markedly low toxicity in a variety of animal studies (9). The findings in this study support the future investigation of ITCs as promising agents for prevention and treatment of MM.
Funding
National Institutes of Health (CA-100853) to F-L Chung.
Acknowledgments
The authors thank Martin Rechsteiner and Greg Pratt at University of Utah for technical assistance and fruitful discussion.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BITC
benzyl isothiocyanate
- GSH
glutathione
- ITC
isothiocyanate
- MM
multiple myeloma
- NF-κB
nuclear factor-kappaB
- NMPEA
N-methyl phenethylamine
- PEITC
phenethyl isothiocyanate
- ROS
reactive oxygen species
- SFN
sulforaphane
- UPS
ubiquitin–proteasome system
References
- 1.Horner MJ, et al., editors. SEER Cancer Statistics Review, NIH, Bethesda, MD. 1975–2006. [Google Scholar]
- 2.Chauhan D, et al. Targeting proteasomes as therapy in multiple myeloma. Adv. Exp. Med. Biol. 2008;615:251–260. doi: 10.1007/978-1-4020-6554-5_12. [DOI] [PubMed] [Google Scholar]
- 3.Orlowski RZ, et al. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin. Cancer Res. 2008;14:1649–1657. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- 4.Oakervee HE, et al. PAD combination therapy (PS-341/bortezomib, doxorubicin and dexamethasone) for previously untreated patients with multiple myeloma. Br. J. Haematol. 2005;129:755–762. doi: 10.1111/j.1365-2141.2005.05519.x. [DOI] [PubMed] [Google Scholar]
- 5.Kisselev AF, et al. Proteasome inhibitors: from research tools to drug candidates. Chem. Biol. 2001;8:739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem. Soc. Trans. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- 7.Gottesman S, et al. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 8.Bence NF, et al. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 9.WHO. Cruciferous Vegetables, Isothiocyanates and Indoles. IARC Handbook On Cancer Prevention. Vol. 9. Lyon, France: IARC; 2004. [Google Scholar]
- 10.Singh SV, et al. Sulforaphane-induced cell death in human prostate cancer cells is initiated by reactive oxygen species. J. Biol. Chem. 2005;280:19911–19924. doi: 10.1074/jbc.M412443200. [DOI] [PubMed] [Google Scholar]
- 11.Trachootham D, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Xiao D, et al. Benzyl isothiocyanate targets mitochondrial respiratory chain to trigger reactive oxygen species-dependent apoptosis in human breast cancer cells. J. Biol. Chem. 2008;283:30151–30163. doi: 10.1074/jbc.M802529200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mi L, et al. The role of protein binding in induction of apoptosis by phenethyl isothiocyanate and sulforaphane in human non-small lung cancer cells. Cancer Res. 2007;67:6409–6416. doi: 10.1158/0008-5472.CAN-07-0340. [DOI] [PubMed] [Google Scholar]
- 14.Mi L, et al. Covalent binding to tubulin by isothiocyanates. A mechanism of cell growth arrest and apoptosis. J. Biol. Chem. 2008;283:22136–22146. doi: 10.1074/jbc.M802330200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi L, et al. Cancer preventive isothiocyanates induce selective degradation of cellular {alpha}- and {beta}-tubulins by proteasomes. J. Biol. Chem. 2009;284:17039–17051. doi: 10.1074/jbc.M901789200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mi L, et al. Aggresome-like structure induced by isothiocyanates is novel proteasome-dependent degradation machinery. Biochem. Biophys. Res. Commun. 2009;388:456–462. doi: 10.1016/j.bbrc.2009.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwak MK, et al. Role of increased expression of the proteasome in the protective effects of sulforaphane against hydrogen peroxide-mediated cytotoxicity in murine neuroblastoma cells. Free Radic. Biol. Med. 2007;43:809–817. doi: 10.1016/j.freeradbiomed.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman L, et al. Multiple forms of the 20 S multicatalytic and the 26 S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J. Biol. Chem. 1992;267:22362–22368. [PubMed] [Google Scholar]
- 19.Zhang Y. Role of glutathione in the accumulation of anticarcinogenic isothiocyanates and their glutathione conjugates by murine hepatoma cells. Carcinogenesis. 2000;21:1175–1182. [PubMed] [Google Scholar]
- 20.Zhang Y, et al. Mechanism of differential potencies of isothiocyanates as inducers of anticarcinogenic Phase 2 enzyme. Cancer Res. 1998;58:4632–4639. [PubMed] [Google Scholar]
- 21.Liang HL, et al. Partial attenuation of cytotoxicity and apoptosis by SOD1 in ischemic renal epithelial cells. Apoptosis. 2009;14:1176–1189. doi: 10.1007/s10495-009-0393-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardoso LM, et al. Cardiovascular responses to hydrogen peroxide into the nucleus tractus solitarius. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009;297:R462–R469. doi: 10.1152/ajpregu.90796.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borissenko L, et al. 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem. Rev. 2007;107:687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- 24.Loidl G, et al. Bifunctional inhibitors of the trypsin-like activity of eukaryotic proteasomes. Chem. Biol. 1999;6:197–204. doi: 10.1016/S1074-5521(99)80036-2. [DOI] [PubMed] [Google Scholar]
- 25.Cross JV, et al. Nutrient isothiocyanates covalently modify and inhibit the inflammatory cytokine macrophage migration inhibitory factor (MIF) Biochem. J. 2009;423:315–321. doi: 10.1042/BJ20091170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouertatani-Sakouhi H, et al. A new class of isothiocyanate-based irreversible inhibitors of macrophage migration inhibitory factor. Biochemistry. 2009;48:9858–9870. doi: 10.1021/bi900957e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown KK, et al. Direct modification of the proinflammatory cytokine macrophage migration inhibitory factor by dietary isothiocyanates. J. Biol. Chem. 2009;284:32425–32433. doi: 10.1074/jbc.M109.047092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goosen TC, et al. Inactivation of cytochrome P450 2B1 by benzyl isothiocyanate, a chemopreventative agent from cruciferous vegetables. Chem. Res. Toxicol. 2000;13:1349–1359. doi: 10.1021/tx000133y. [DOI] [PubMed] [Google Scholar]
- 29.Dinkova-Kostova AT, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl Acad. Sci. USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwak MK, et al. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1–Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 31.Dinkova-Kostova AT, et al. Direct and indirect antioxidant properties of inducers of cytoprotective proteins. Mol. Nutr. Food Res. 2008;52:S128–S138. doi: 10.1002/mnfr.200700195. [DOI] [PubMed] [Google Scholar]
- 32.Maki CG, et al. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 33.Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat. Rev. Cancer. 2003;3:102–109. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- 34.Huang C, et al. Essential role of p53 in phenethyl isothiocyanate-induced apoptosis. Cancer Res. 1998;58:4102–4106. [PubMed] [Google Scholar]
- 35.Kuang YF, et al. Induction of apoptosis in a non-small cell human lung cancer cell line by isothiocyanates is associated with P53 and P21. Food Chem. Toxicol. 2004;42:1711–1718. doi: 10.1016/j.fct.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Karin M, et al. The IKK NF-kappa B system: a treasure trove for drug development. Nat. Rev. Drug Discov. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 37.Haefner B. NF-kappa B: arresting a major culprit in cancer. Drug Discov. Today. 2002;7:653–663. doi: 10.1016/s1359-6446(02)02309-7. [DOI] [PubMed] [Google Scholar]
- 38.Van Waes C. Nuclear factor-kappaB in development, prevention, and therapy of cancer. Clin. Cancer Res. 2007;13:1076–1082. doi: 10.1158/1078-0432.CCR-06-2221. [DOI] [PubMed] [Google Scholar]
- 39.Chen ZJ. Ubiquitin signaling in the NF-kB pathway. Nat. Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong WS, et al. Modulatory properties of various natural chemopreventive agents on the activation of NF-kappaB signaling pathway. Pharm. Res. 2004;21:661–670. doi: 10.1023/b:pham.0000022413.43212.cf. [DOI] [PubMed] [Google Scholar]
- 41.Srivastava SK, et al. Cell cycle arrest, apoptosis induction and inhibition of nuclear factor kappa B activation in anti-proliferative activity of benzyl isothiocyanate against human pancreatic cancer cells. Carcinogenesis. 2004;25:1701–1709. doi: 10.1093/carcin/bgh179. [DOI] [PubMed] [Google Scholar]
- 42.Drexler HC. Activation of the cell death program by inhibition of proteasome function. Proc. Natl Acad. Sci. USA. 1997;94:855–860. doi: 10.1073/pnas.94.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu K, et al. Studies on the mechanism of the inhibition of human leukaemia cell growth by dietary isothiocyanates and their cysteine adducts in vitro. Biochem. Pharmacol. 2000;60:221–231. doi: 10.1016/s0006-2952(00)00319-1. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Selected isothiocyanates rapidly induce growth inhibition of cancer cells. Mol. Cancer Ther. 2003;2:1045–1052. [PubMed] [Google Scholar]
- 45.Johnson CR, et al. Intrinsic cytotoxicity and chemomodulatory actions of novel phenethylisothiocyanate sphingoid base derivatives in HL-60 human promyelocytic leukemia cells. J. Pharmacol. Exp. Ther. 2004;309:452–461. doi: 10.1124/jpet.103.060665. [DOI] [PubMed] [Google Scholar]
- 46.Thomson SJ, et al. Phenethyl isothiocyanate triggers apoptosis in Jurkat cells made resistant by the overexpression of Bcl-2. Cancer Res. 2006;66:6772–6777. doi: 10.1158/0008-5472.CAN-05-3809. [DOI] [PubMed] [Google Scholar]
- 47. Tsimberidou, A.M. (2009) Study of Phenethyl Isothiocyanate in Lymphoproliferative Disorders. http://clinicaltrials.gov/ct2/show/NCT00968461?term=phenethyl+isothiocyanate&rank=1. [Google Scholar]
- 48.Conaway CC, et al. Disposition and pharmacokinetics of phenethyl isothiocyanate and 6-phenylhexyl isothiocyanate in F344 rats. Drug Metab. Dispos. 1999;27:13–20. [PubMed] [Google Scholar]