Abstract
The DNA amplification process can be a source of bias and artifacts, especially when amplifying genomic areas with extreme AT or GC content. The human malaria parasite Plasmodium falciparum has an AT-rich genome, and some of its highly AT-rich regions have been shown to be refractory to polymerase chain reaction (PCR) amplification. Biased amplification may lead to erroneous conclusions for studies investigating genome-wide gene expression, nucleosome position, and copy number variation. Here we compare genome-wide nucleosome coverage in libraries amplified at three different extension temperatures and show that reduction in PCR extension temperature from 70ºC to 60ºC can greatly increase the fraction of coverage at AT-rich regions of the P. falciparum genome. Our method will improve the efficiency and coverage in sequencing an AT-rich genome.
Keywords: new generation sequencing, malaria, genome, amplification bias, nucleosome
Polymerase chain reaction (PCR) is widely employed to amplify DNA fragments before they are hybridized to a microarray chip or are processed for parallel sequencing. Indeed, the majority of current high-throughput parallel sequencing methods involves a step of PCR amplification [1] that can introduce bias in sequence coverage in DNA regions with different GC contents [2]. With commonly used PCR conditions, repetitive AT-rich regions may not be amplified properly or not amplified at all, leading to an artificial lack of coverage in AT-rich regions, whereas more GC-rich regions may be excessively amplified [3]. Biased amplification can result in erroneous conclusions for studies investigating gene expression level, nucleosome position, and copy number variation [4]. Lack of sequence amplification will also produce sequence gaps that can prevent assembly of genome sequences. To overcome the problem, procedures without PCR amplification have been developed [5–7]; however, it may be necessary to amplify the DNA or RNA samples before large-scale sequencing or array hybridization can be performed, because the quantity of genetic material is often limited.
Many organisms—such as the human malaria parasite Plasmodium falciparum and free-living protozoan Paramecium tetraurelia—have AT-rich genomes [8,9]. For P. falciparum, highly AT-rich regions (> 90% AT) are usually present in non-coding regions and highly repetitive. They have a very low melting temperature and are difficult to amplify using standard PCR conditions. Use of a 60°C extension temperature has been shown to be necessary in order to amplify regions with AT content 90% or higher because the DNA segments are already denatured at a 72°C extension temperature [10].
To improve sequencing coverage over AT-rich regions of the P. falciparum genome in efforts to study genome-wide nucleosome positioning, we investigated the effects of the PCR extension temperature on sequence coverage obtained from Illumina parallel sequencing. We used nucleosomal DNA obtained from the P. falciparum schizont stage to construct three libraries using extension temperatures of 60°C, 65°C, and 70°C, respectively. P. falciparum strain 3D7 was cultured in vitro as described in Trager and Jensen [11]. The schizont stage of the parasite was purified using Percoll-sorbitol gradient (60–40%) and cultured for 6 h before treatment with 5% sorbitol at 37ºC for 15 min. Synchronized parasites were harvested at 44 h, treated with 0.06% saponin, and washed twice with ice-cold PBS.
Saponin-treated parasites were lyzed using a ChIP-IT Express kit according to manufacturer's instruction (Active Motif). Briefly, a pellet was collected after centrifugation at 14,000 rpm for 40 min and was re-suspended in digestion buffer in the presence of protease inhibitors cocktail and PMSF (1 mM final). To facilitate re-suspension of the nuclei in digestion buffer, a brief sonication (3 cycles of 5 sec at medium power) was performed at 4ºC in a Bioruptor (Diagenode®). The re-suspended nuclei were incubated on ice for 15 min, with flicking the tube occasionally, and then warmed at 37ºC for 5 min. After adding 5 U of micrococcal nuclease (MNase, Active Motif), the sample was incubated at 37ºC for 25 min. MNase digestion was stopped by addition of 5 mM EDTA. Nuclear debris was removed by centrifugation at 14,000 rpm for 20 min, and the chromatin present at the supernatant was treated with RNaseA at 37ºC for 1 h to remove any contaminant RNA. Proteins were removed from digested chromatin with treatment of proteinase K at 42ºC for 2 h. DNA was phenol/chloroform extracted, ethanol precipitated, and separated in a 3% agarose gel. The DNA band corresponding to mononucleosome was purified using the QIAquick gel extraction kit (Qiagen).
Mononucleosomal DNA fragments were blunt-ended after Taq DNA polymerase (New England BioLabs) treatment and purified using QIAquick PCR purification kit (QIAGEN). Blunt-ended DNA fragments were ligated to paired-end adapters (Illumina) and further purified using QIAquick PCR purification kit. The ligated DNA was PCR amplified using Finnzymes high-fidelity DNA polymerase master mix (New England BioLabs) and the PCR primers PE 1.0 and 2.0 (Illumina). DNA fragments were amplified with PCR cycles of 98°C for 10 sec, 65°C for 30 sec, and extension at either 70°C, 65°C, or 60°C for 30 sec for 19 cycles. PCR products were purified as described above and sequenced using the Illumina IIG genome analyzer and methods described previously [12].
Prior to mapping of DNA sequence reads to the 3D7 reference genome, each of the three datasets containing 36-bp reads obtained from the Illumina Sequencing Pipeline was examined for quality scores (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/) to ensure good and comparable quality between datasets. The Bowtie short-read alignment tool [13] was used to align the 36-bp reads to the reference genome P. falciparum 3D7 (version 2.1.4, GeneDB April 2010) with parameters of 0 mismatches along the entire read allowed and only one possible match in the genome. The output was converted to bam files using Samtools [14] and uploaded into the IGV browser (http://www.broadinstitute.org/igv) for visual inspection of the coverage. A plot of AT percentage generated from calculations of AT content in 10-bp sliding windows using emboss isochore [15] was added to the IGV browser as a wig file.
The AT percentage for each read in the datasets and for each read found to align to the reference genome was determined. Custom scripts were used to group and count the AT percentage of the reads with 1% increments from 60% to 95%. The fraction of coverage along the genome was calculated from the bases overlapped by reads in each of the 100-bp fragments that was previously clustered in groups of 1% increments between 60% and 95% AT using BEDTools [16], which also allowed us to generate histograms of coverage in each 100-bp fragment, to calculate fold coverage, and to count reads overlapping introns, exons, and intergenic regions. To obtain the ratio of fraction of coverage, we divided the values obtained from fraction of coverage in the 60°C dataset at each of the 100-bp AT percent groups by the value of the fraction of coverage obtained in the same group of 100-bp fragments in the 70°C dataset.
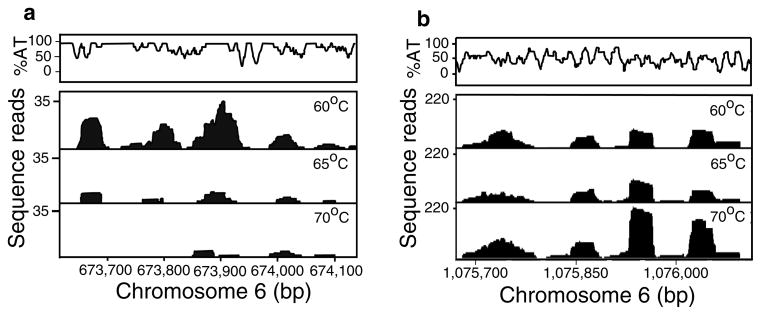
We obtained approximately 15 million 36-bp reads from each library, from which nearly 12 million reads were mapped to the 3D7v2.1.4 reference genome (GeneDB) with cutoffs of 0 mismatches and single hit in the genome (Supplementary Table 1). The total numbers of both raw and mapped sequence reads obtained from the three libraries were similar, with an average of 4- to 10-fold higher genome coverage than those reported in a recent study [17]. Mapped reads were visualized using the IGV genome browser (http://www.broadinstitute.org/igv/), and differential coverage was observed at the three libraries. We detected consistently better sequence coverage within intergenic areas amplified at the 60°C library compared with those obtained from the 65°C and 70°C libraries (Fig. 1a). On the contrary, some areas of the genome with lower AT content often had increased fold coverage at the 70°C library (Fig. 1b), which may represent preferential amplification of genomic regions of high CG content at a 70°C extension temperature, as more amplification resources are directed to fewer application sites at 70°C.
Fig. 1.
Coverage of sequence reads at AT-rich and GC-rich regions amplified under different extension temperatures. Images of coverage plot from IGV genome browser (http://www.broadinstitute.org/igv/) displaying (a) a 530-bp AT-rich intergenic region on chromosome 6 (673,610– 674,140 bp) with good coverage when amplified at 60°C, with reduced coverage at 65°C, and with almost no coverage at 70°C, and (b) a 400-bp coding region on chromosome 6 (1,075,670–1,076,110 bp) with increased fold of coverage at two regions when amplified at 70°C. The AT contents of both the regions are indicated at the top of the figures.
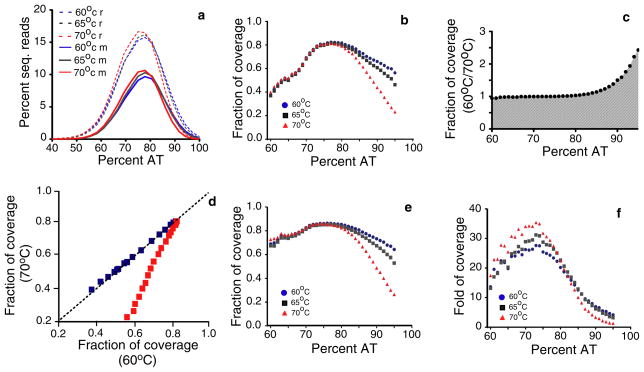
All three libraries had a similar distribution of sequence reads based on their AT content, peaking at ~77% AT (Fig. 2a). To estimate the fraction and depth of sequence coverage over DNA regions with different AT content, we divided the parasite genome into 100-bp non-overlapping fragments and grouped them into clusters based on the mean values of their AT content (Supplementary Table 2). A total of 211,812 genomic fragments were generated, of which ~50% had AT contents of 78% to 87%. Alignment of the sequence reads from the three libraries to the 100-bp fragments showed that decrease in extension temperature from 70°C to 60°C significantly increased the fraction of coverage at AT-rich regions, particularly when AT content was 90% or higher (Fig. 2b). The ratios of fraction of coverage (60°C over 70°C) remained around 1, but began to increase at 80% AT, showing a maximum ratio of ~2.8 when AT > 95% (Fig. 2c and d). These results showed a high correlation of sequence coverage among all three libraries for genomic areas with AT content lower than 80%, but for regions with AT content higher than 80%, better sequence coverage was obtained when amplified at 60°C. There was only a slight decrease in the mean fraction of coverage with the increase of AT content from 70% to 95% when amplified at 60°C (Fig. 2b), suggesting that DNA with a wide range of AT content can be amplified reliably using an extension temperature of 60°C.
Fig. 2.
Coverage of sequence reads over DNA fragments with different AT contents obtained under different extension temperatures. (a) distribution of sequenced reads with different AT contents. Temperatures labeled with ‘r’ are plots from raw sequence reads; those with ‘m’ are plots from mapped reads. (b) mean fraction of coverage of 100-bp chromosomal segments with different AT contents; (c) ratios of fraction of coverage (60°C/70°C) at various AT contents; (d) plots of ratios of fraction of coverage (60°C/70°C) within 100-bp fragments having AT content < 80% (blue) and fragments with AT > 80% (red); (e) the same plots as those in b after excluding the fragments without any sequence read coverage; (f) depth of coverage of 100-bp genome fragments with different AT contents.
We also excluded the 100-bp DNA fragments that had no sequence coverage and plotted the fraction of sequence-read coverage against AT content. Removal of the 100-bp sequences without read coverage increased the fraction of coverage at AT content below 70% dramatically (Fig. 2e), suggesting that the majority of 100-bp fragments not covered by sequence reads are relatively GC rich. Because there are large numbers of repetitive sequences and GC-rich gene families in the P. falciparum genome such as the var genes [18] and we used strict cutoff criteria (one single hit in the genome with no mismatches) to remove sequence reads that may align to more than one position, many GC-rich reads could be removed because they might align with more than one position. Fragments without read coverage could be due to the removal of the GC-rich reads from the gene families, which could explain the relatively fewer reads and lower coverage at regions with 70% < AT (Fig. 2b).
We next investigate the effect of extension temperature on the depth of coverage or the numbers of times each base pair is covered by the reads. The fold of coverage was slightly higher when amplified at 70°C for fragments with an average 80% AT or lower (Fig. 2f). The higher fold of coverage seen at low AT content regions can be explained by preferential amplification of some relatively GC-rich segments in the genome (Fig. 1b); however, the depth of coverage amplified at 70°C decreased when the fragment AT content averaged 84% or higher. It is clear that for high AT regions, both fraction and depth of coverage can be greatly improved by amplifying the DNA at a 60°C extension temperature.
As the introns and intergenic sequences of this parasite have higher AT content than the exons, the highest numbers of reads covering introns and intergenic regions were also obtained when amplified using the 60°C extension temperature (Supplementary Table 1). Indeed, many AT introns/intergenic regions were completely refractory for amplification using a 70°C extension temperature (Fig. 1a). Although we cannot conclude that the sequence coverage from 60°C represents the true state of nucleosome coverage in P. falciparum, our data demonstrate that nucleosomes are present in highly AT-rich regions in the P. falciparum genome. Improved genome coverage for highly AT-rich genomes can be obtained if DNA samples are amplified at a lower extension temperature. Our method provides an alternative to the amplification free procedures [5–7], particularly when small amount of DNA or RNA is available.
Research highlights.
Sequence coverage from libraries amplified at extension temperatures of 70°C, 65°C, and 60°C were compared.
Significantly increased sequence coverage at AT-rich regions when amplified with an extension temperature of 60°C, compared with those amplified at 70°C.
Only a slight decrease in the mean fraction of coverage with the increase of AT content from 70% to 95% when amplified at 60°C, suggesting that DNA with a wide range of AT content can be amplified reliably using an extension temperature of 60°C.
Supplementary Material
Acknowledgments
We thank Artem Barski, Qingsong Tang, and Gang Wei for assistance with the Illumina sequencing. This work was supported by the Divisions of Intramural Research at the National Institute of Allergy and Infectious Diseases and National Heart, Lung and Blood Institute. We thank NIAID intramural editor Brenda Rae Marshall for assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shendure JA, Porreca GJ, Church GM. Overview of DNA sequencing strategies. Curr Protoc Mol Biol. 2008;Chapter 7(Unit 7):1. doi: 10.1002/0471142727.mb0701s81. [DOI] [PubMed] [Google Scholar]
- 2.Pinard R, de Winter A, Sarkis GJ, et al. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics. 2006;7:216. doi: 10.1186/1471-2164-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pugh TJ, Delaney AD, Farnoud N, et al. Impact of whole genome amplification on analysis of copy number variants. Nucleic Acids Res. 2008;36:e80. doi: 10.1093/nar/gkn378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mamanova L, Andrews RM, James KD, et al. Frt-seq: Amplification-free, strand-specific transcriptome sequencing. Nat Methods. 2010;7:130–2. doi: 10.1038/nmeth.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris TD, Buzby PR, Babcock H, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–9. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 7.Kozarewa I, Ning Z, Quail MA, et al. Amplification-free illumina sequencing-library preparation facilitates improved mapping and assembly of (g+c)-biased genomes. Nat Methods. 2009;6:291–5. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner MJ, Hall N, Fung E, et al. Genome sequence of the human malaria parasite plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duret L, Cohen J, Jubin C, et al. Analysis of sequence variability in the macronuclear DNA of paramecium tetraurelia: A somatic view of the germline. Genome Res. 2008;18:585–96. doi: 10.1101/gr.074534.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su X-z, Wu Y, Sifri CD, Wellems TE. Reduced extension temperatures required for pcr amplification of extremely a+t-rich DNA. Nucleic Acids Res. 1996;24:1574–5. doi: 10.1093/nar/24.8.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–5. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 12.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice P, Longden I, Bleasby A. Emboss: The european molecular biology open software suite. Trends Genet. 2000;16:276–7. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 16.Quinlan AR, Hall IM. Bedtools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponts N, Harris EY, Prudhomme J, et al. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 2010;20:228–38. doi: 10.1101/gr.101063.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su X-z, Heatwole VM, Wertheimer SP, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of plasmodium falciparum-infected erythrocytes [see comments] Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.