Abstract
Malaria is caused by protozoan parasites of the genus Plasmodium and involves infection of multiple hosts and cell types during the course of an infection. To complete its complex life cycle the parasite requires strict control of gene regulation for survival and successful propagation. Thus far, the Apicomplexan AP2 (ApiAP2) family of DNA-binding proteins is the sole family of proteins to have surfaced as candidate transcription factors in all apicomplexan species. Work from several laboratories is beginning to shed light on how the ApiAP2 proteins from Plasmodium spp. contribute to the regulation of gene expression at various stages of parasite development. Here we highlight recent progress toward understanding the role of Plasmodium ApiAP2 proteins in DNA related regulatory processes including transcriptional regulation and gene silencing.
Keywords: Gene regulation, Transcription, ApiAP2, Malaria, Plasmodium, Apicomplexan
Plasmodium gene regulation: What do we know?
The various developmental stage transitions in the malaria parasite lifecycle and the observation of highly coordinated gene expression [1, 2] both imply a need for precise control of mRNA transcript levels, which is mediated through various forms of regulation including transcriptional, post-transcriptional, and translational repression mechanisms. Currently, the best-characterized contributor to transcriptional regulation in Plasmodium is the effect of chromatin modifications. Considerable work has been carried out to identify and characterize the chromatin remodeling machinery of Plasmodium, and many components homologous to those in model eukaryotic organisms have been found (reviewed in [3]). Recently, several groups have demonstrated that specific histone modifications are associated with gene silencing or activation during intraerythrocytic development, suggesting that these modifications are involved in transcriptional control [4, 5]. Of particular note, the trimethylation gene repression mark at histone 3 lysine 9 (H3K9me3) has been shown to correlate with the repression of clonally variant genes including the subtelomeric var genes [4, 5]. Two studies have further demonstrated that the Plasmodium falciparum heterochromatin protein 1 (PfHP1, PFL1005c) is intimately associated with the H3K9me3 mark [6, 7]. In addition to histone modifications, other factors such as var promoters, subnuclear localization, and non-coding RNA, are also likely to play a role in maintaining epigenetic memory [3]. Meanwhile, transcriptional regulation of other subtelomeric gene families such as rifin, stevor, or Pfmc-2tm, are not as well characterized; however, the expression of members of these gene families is also clonally variant and highly regulated throughout intraerythrocytic development (reviewed in [8]). Although var genes are transcribed during the ring stage, rifins during early trophozoite, and stevors and Pfmc-2tm during the mature trophozoite stage, the precise mechanisms and factors controlling their temporal activation are still under intense investigation [8].
Beyond epigenetic regulation of subtelomeric gene expression, post-transcriptional gene regulation plays a role in the regulation of Plasmodium development. During the sexual stages of the lifecycle, translational repression of specific mRNA transcripts plays an important role in the gametocyte fertilization process [9]. In female gametocytes a translational repression complex has been identified that includes the DEAD-box RNA helicase PfDOZI (PFC0915w), which serves to repress translation of specific proteins until a precise time during sexual development [10]. It remains to be seen whether similar mechanisms play a role during other developmental stage transitions such as merozoite invasion of erythrocytes and sporozoite invasion of hepatocytes [11–13]. Similarly, stage-specific stabilization (or degradation) of mRNA plays a role in gene regulation, since mRNA decay rates vary dramatically during the blood stages with an overall increase in transcript half-lives as the parasite progresses from rings to schizonts, a phenomenon not seen in any other Apicomplexa [14]. Lastly, gene-specific nuclear run-on assays have identified discrepancies between transcriptional activity and mRNA abundance for a number of transcripts [15]. Taken together these results have led to the idea that post-transcriptional regulation may be a dominant mechanism controlling gene expression in Plasmodium; however, current work on a novel apicomplexan family of DNA-binding proteins is beginning to challenge this hypothesis.
Filling the void: ApiAP2 family of DNA-binding proteins
Until recently, little progress had been made toward understanding the details of transcription factor-based gene regulation in the complex lifecycle of malaria parasites. With the completion of the P. falciparum genome sequence, it became clear that there was a significant gap in our understanding of transcriptional regulation as no annotations for specific transcription factors were reported [16]. Despite this, the core transcriptional machinery for RNA polymerase II transcription was identified and a full complement of factors involved in chromatin remodeling has been reported [17–20]. Searches for specific transcription factors in the Plasmodium spp. have involved extensive bioinformatic analyses focusing mainly on sequence similarity to known eukaryotic transcription factors [18–21]. The majority of these searches have suggested a paucity of transcription factors, until Balaji et al. described a group of conserved proteins containing putative AP2 DNA-binding domains, now known as the Apicomplexan AP2 (ApiAP2) protein family (Figure 1) [22]. This study provided the first indication that apicomplexan parasites encode a family of regulatory proteins unlike those of their hosts, sparking renewed interest in transcriptional regulation.
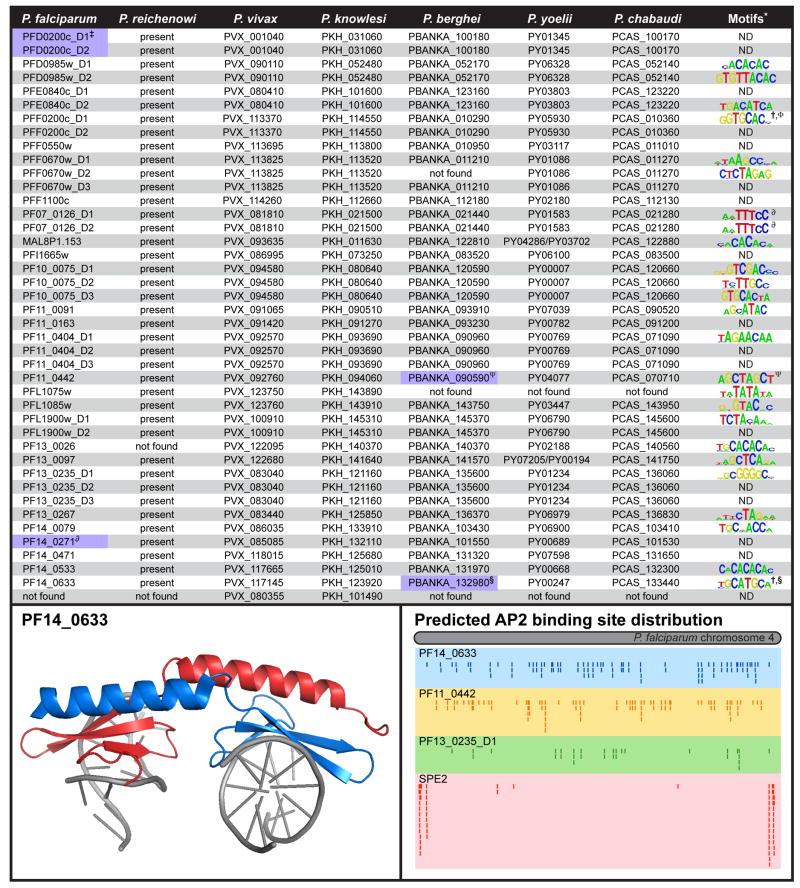
Figure 1. Conservation, motifs, structure and binding sites of Plasmodium AP2 domains.
The top panel lists the gene IDs and DNA motifs for each P. falciparum AP2 domain. The best match for each domain in the other Plasmodium species was determined by BLAST results for each individual domain. The presence of an orthologue in P. reichenowi is based on contig evidence from partial genome shotgun sequencing by the Wellcome Trust Sanger Institute. Purple shading indicates a viable knock-out exists based on data from ‡ [40], ∂ [41], ψ [39], and § [38]. Motifs taken from * [42], † [37], Φ[28], ψ [39], § [38]; ND indicates no motif has been found to date. ∂ The PF07_0126 motif is only observed when both domains are present in tandem. The bottom left panel shows a model of the crystal structure of the PF14_0633 AP2 domain [27]. Individual monomers are shown in blue and red, which demonstrates that the helix is swapped between the two domains. The bottom right panel shows the distribution of four P. falciparum AP2 motifs on chromosome 4. Binding sites for PF14_0633, PF13_0235_D1 and PF11_0442 are based on predictions from [42], and the SPE2 chromatin immunoprecipitation data (ChIP-chip) was taken from [37]. PF14_0633 and PF11_442 have been demonstrated to function as transcription factors [38, 39] and their motifs are distributed across the entire chromosome. Conversely the SPE2 binding sites are restricted to chromosome ends and serve as DNA tethering sites [37].
The ApiAP2 family is homologous to the plant Apetela2/Ethylene Response Factor (AP2/ERF) DNA-binding proteins, which comprise the second largest class of transcription factors in Arapidopsis thaliana [23]. In plants, these AP2/ERF proteins function as either activators or repressors of transcription [23] and contain one or two 60 amino acid AP2 DNA-binding domains that bind DNA using a triple stranded β-sheet stabilized by a C-terminal α-helix [24]. AP2 domain architecture in plants is intimately linked to protein function; AP2/ERF proteins with one AP2 domain regulate genes involved in pathogenesis and environmental response pathways [25], while proteins with two tandem AP2 domains, separated by a short, conserved linker sequence of 25 amino acids, are involved in regulating plant development [26]. Similar to plants, the AP2 domains in ApiAP2 proteins are also approximately 60 amino acids in length and are found in both single and tandem domain arrangements. However, unlike the limited number of domains found in plant AP2/ERF proteins, some members of the ApiAP2 family are predicted to contain more two AP2 domains in a given protein [22]. It is of great interest to determine if such unique AP2 domain architectures are related to the functional role(s) of the ApiAP2 proteins. Full-length ApiAP2 proteins vary in size from a few hundred to several thousand amino acids. This large variability in size raises an intriguing question as to what other functional domains are present in these proteins. Recently the crystal structure for the P. falciparum AP2 domain from PF14_0633 bound to DNA was determined (Figure 1, bottom left), revealing that the protein fold (triple stranded β-sheet stabilized by an α-helix) has been maintained between plants and the Apicomplexa. The structure identified four important residues within the β-strand region that directly contact the DNA [27]. These four amino acids are highly conserved among all apicomplexan orthologues of PF14_0633 suggesting that the DNA sequence specificity is well-conserved [27, 28].
The P. falciparum ApiAP2 gene family has 27 members, which are largely conserved across Plasmodium species, with nearly identical AP2 DNA-binding domains in orthologues from different species (Figure 1) [22, 29]. Additionally, ApiAP2 proteins are also found in all other Apicomplexa including Theileria, Cryptosporidium [22] and Toxoplasma which has a lineage specific expansion of this family (up to 68 predicted family members) [30, 31]. Although the majority of ApiAP2 proteins are conserved among Plasmodium spp., two family members appear to be species specific. The first is PFL1075w which is found only in the primate malarias P. falciparum, P. vivax (PVX_123750), P. knowlsei (PKH_143890), and P. reichenowi (reich237a11.qlk) (Figure 1). The conservation of PFL1075w in these four species implies a role in the regulation of genes that are involved in parasite biology specific to the primate host. The second species-specific ApiAP2 family member is the recently annotated P. vivax gene PVX_080355, which has a single orthologue in P. knowlesi (PKH_101490) and is not found in any other Plasmodium species (Figure 1). The evolutionary conservation of this ApiAP2 in two closely related human malaria parasites suggests that it may play a role in processes unique to these two parasites such as hypnozoite formation. Selective conservation of these species-specific ApiAP2 proteins presents an exciting opportunity to expand our understanding of parasite biology within different hosts through the elucidation of their individual functions.
Plasmodium ApiAP2 proteins: Developmental regulators?
ApiAP2 expression throughout Plasmodium development
Global transcriptional analysis of a number of Plasmodium species and strains has revealed a temporal pattern of gene expression throughout the asexual intraerythrocytic development cycle (IDC) and members of the ApiAP2 family are no exception [1, 2, 22, 32–34]. Microarray data has demonstrated that 21 of the 27 P. falciparum ApiAP2 genes are transcribed during the IDC [1, 2], and recently, a more sensitive RNA-sequencing approach has provided evidence for the expression of several additional ApiAP2 genes [35]. The timing of ApiAP2 gene expression can be clustered into four major classes corresponding to the ring, early trophozoite, early schizont, and schizont stages of intraerythrocytic development [22]. Similarly, several Toxoplasma gondii ApiAP2 genes are cell cycle regulated and expressed during restricted timeframes, suggesting a role in regulating progression through the tachyzoite lytic cycle [36]. In Plasmodium, blood stage gene expression suggests that the ApiAP2 proteins are likely to be major transcriptional regulators functioning during this stage of development. Furthermore, very few ApiAP2 genes have been successfully knocked out, emphasizing their likely essentiality to blood-stage development ([36, 37] M. Llinás, unpublished). This being said, four of the 27 Plasmodium ApiAP2 proteins have been genetically disrupted with varying degrees of developmental effects on the parasite (Figure 1) [38–41]. Targeted knockout studies have successfully demonstrated that PF14_0271 is not essential for P. falciparum blood-stage development [41]. Similarly, a genome-wide transposon mutagenesis survey utilizing the piggyBac system disrupted pfd0200c without any discernable phenotypic changes in intraerythrocytic development [40]. Interestingly, the individually expressed AP2 domains from these two proteins do not appear to bind DNA as determined by protein binding microarrays (Figure 1) [42]. Lastly, despite data showing that pf14_0633 and pf11_0442 mRNA is transcribed during the P. falciparum blood stages, their P. berghei orthologues (pbanka_132980 and pbanka_090590) are dispensable for intraerythrocytic development in the rodent model, but play key roles during other developmental stages (see below) [38, 39]. A global analysis of the essentiality of all ApiAP2 proteins will further our understanding of the in vivo role of each protein during specific stages of Plasmodium development.
While many of the Plasmodium ApiAP2 genes are expressed during the asexual blood stage of development, transcriptional and proteomic evidence suggests expression in other developmental stages as well. Therefore, these factors likely play a role throughout the complete life cycle of the parasite. Analysis of the progression from the asexual IDC to sexual development reveals transcript evidence for the ApiAP2 genes pff1100c, pf11_0091, pfd0985w, pf11_0442, and pff0200c during the early stages of gametocytogenesis [43, 44]. Furthermore, transcripts for five ApiAP2 genes are found in the gametocyte associated DOZI repression complex: pf13_0026, pf11_0091, pff0200c, pfd0200c and pf11_0442 [9]. Recently, the P. berghei orthologue of PF11_0442 (PBANKA_090590) has been demonstrated to be important in ookinete development (summarized below) [38]. During oocyst development, py00689 (pf14_0271) transcripts are upregulated in P. yoelii midgut sporozoites [45], while py01234 (pf13_0235) [45] and pbanka_132980 (pf14_0633) [39] transcripts are upregulated in salivary gland sporozoites. Additionally, transcriptional analysis from P. yoelii has identified orthologues of pfi1665w, pff0550w, pff0200c, pff1100c, and pf13_0235 (see Figure 1) that are expressed during the liver stage of Plasmodium development [46]. Taken together, there is evidence for expression of multiple ApiAP2 proteins throughout the lifecycle, again implicating this family as major regulators of gene expression at all stages of Plasmodium development.
Perturbation of ApiAP2 gene expression
Recently, various groups have examined the global transcriptional responses of Plasmodium to various conditions and several genetic backgrounds [6, 32, 47, 48]. Chemical perturbation data from P. falciparum has been shown to result in the alteration of the expression of several ApiAP2 genes. For example, treatment of blood-stage parasites with apicidin, a class I and II histone deacetylase inhibitor, resulted in the up-regulation of a number of ApiAP2 genes in stages during which they would normally be down-regulated [48]. This raises an interesting, and indeed likely, possibility that there may be overlap between chromatin remodeling and transcription factor-based regulation of gene expression in Plasmodium. Further evidence for such overlap comes from the high prevalence of the H3K9me3 mark at the pfl1085w ApiAP2 gene locus, which likely leads to silencing of this gene [4, 5]. Moreover, the inhibition of P. falciparum histone deacetylases by apicidin seems to interfere with the methylation status of H3K9, resulting in the overexpression of pfl1085w [48]. In an organism where precise timing of gene expression appears to be vitally important [1, 2, 33], it is not surprising that multiple mechanisms may function together to control transcriptional regulation.
Expression of invasion related genes in ookinetes is controlled by PBANKA_090590 (AP2-O)
Several recent reports are beginning to reveal the functional roles for ApiAP2 proteins and their relevance in controlling Plasmodium development. Seminal work from Yuda et al. characterized the ApiAP2 protein PBANKA_090590 from P. berghei (AP2-O; orthologue of PF11_0442), which is highly expressed in ookinetes [38]. Interestingly, the mRNA transcript encoding pbanka_090590, which is abundant in the gametocyte stage, is subject to translational repression via the DOZI complex [9]. Upon ookinete formation this translational repression is relieved and AP2-O protein is made, activating transcription of ookinete genes. The characterization of AP2-O has provided the first direct link between an ApiAP2 protein and transcriptional regulation. This factor was shown to regulate the expression of a large set of ookinete stage-specific genes by binding directly to a cis-acting control element TAGCTA in their 5′ upstream regions (Figure 1) and a knockout of pbanka_090590 was unable to develop oocysts [38]. The TAGCTA target sequence is also conserved in the upstream sequences of P. falciparum and P. vivax ookinete genes [38], and the AP2 domain from the P. falciparum orthologue of AP2-O, PF11_0442, has been shown to bind the same sequence [42], suggesting that AP2-O may be a major regulator of ookinete genes in all Plasmodium spp. Surprisingly, AP2-O is also expressed during the intraerythrocytic developmental cycle of P. falciparum [1, 2, 35], implying that it may have additional roles outside of ookinete development in the human parasite.
Sporozoite development is regulated by PBANKA_132980 (AP2-Sp)
A second study by Yuda et al. demonstrated that the P. berghei ApiAP2 protein PBANKA_132980 (AP2-Sp; PF14_0633) is essential for sporozoite development in the mosquito vector [39]. AP2-Sp is expressed in the late oocyst and sporozoites localizes to the nucleus [39]. Deletion of pbanka_132980 blocks sporozoite formation during oocyst development. The protein contains one AP2 domain that interacts with the cis-element GCATGCA (Figure 1), is found in the promoter regions of sporozoite specific genes [27, 28], and induces their transcription during sporozoite formation [39]. The GCATGCA motif was previously shown to be enriched in the upstream regions of P. falciparum sporozoite genes and significant overlap was seen between the experimentally identified and computationally predicted targeted genes [39, 49]. Together these data confirm AP2-Sp as a sporozoite specific transcriptional regulator. However, similar to AP2-O, there is transcript evidence for AP2-Sp during the P. falciparum blood stages [1, 2]. Although AP2-Sp protein was not observed in P. berghei blood stages, based on an absence of fluorescence from a GFP-tagged version of the protein [39], proteomic data supports expression of AP2-Sp during the P. falciparum trophozoite stage of intraerythrocytic development [50]. Again, what role, if any, this protein plays during the blood stages of development in P. falciparum remains to be determined.
PFF0200c (PfSIP2) is involved in var gene silencing
A third in vivo ApiAP2 study by Flueck et al. [37] has shown that, in P. falciparum, the tandem AP2 domains from PFF0200c (PfSIP2) interact with the SPE2 motif ((T/G)GTGC(A/G)(N)4(T/G)GTGC(A/G)) (half-site represented in Figure 1) located approximately 2.0 kilobases upstream of the subtelomeric UpsB var genes [51]. The authors also found that full length PFF0200c was processed to an active N-terminal fragment that co-localizes with chromosome ends via interactions with SPE2 motifs (Figure 1, bottom right) [37]. Overexpression of this N-terminal fragment had no effect on global transcriptional profiles compared to wild-type gene expression, suggesting that this ApiAP2 protein may not function as a transcription factor but rather acts as a DNA tethering protein playing a role in the formation and maintenance of heterochromatin [37]. It is interesting that PFF0200c appears to have a very specific function in P. falciparum related to var gene regulation, since the tandem AP2 domains from this protein are highly conserved in all other Plasmodium species where no var genes are present (Figure 1). This implies that homologues of PfSIP2 likely associate with the same GTGCAC DNA motifs in these other species, but serve other functional roles.
In vitro characterization of ApiAP2 DNA-binding domains
Currently only three ApiAP2 proteins have been characterized in depth (described above), but biochemical experiments directed at characterizing the functional role of all AP2 domains are providing insight into the role of this protein family as potential transcriptional regulators. DNA-binding activity has been characterized in vitro for all P. falciparum AP2 domains using protein binding microarrays (PBMs) and electrophoretic mobility shift assays (EMSA) [28, 42]. These studies demonstrate that most ApiAP2 proteins (20 of 27) bind specific DNA sequence elements (Figure 1). Among the identified DNA motifs there is little overlap in binding specificity from different AP2 domains even when more than one domain is present in a single protein. Genome-wide mapping of the DNA sequence motifs shows broad distribution in upstream regions (Figure 1, bottom right). Indeed many upstream sequence elements have more than one ApiAP2 binding site suggesting combinatorial gene regulation [42, 52]. Such multifactorial regulation is common in other eukaryotes and may suffice to provide the diversity required to control a large number of genes using a small number of factors.
The identification of 24 DNA sequence motifs bound by the ApiAP2 factors allows for the prediction of putative target genes based on motif occurrences in upstream regions (Figure 1, bottom right). To this end, Campbell et al. [42] analyzed motif occurrences in the 2.0 kb upstream regions of all P. falciparum genes, providing putative target gene lists. Additionally, a number of in silico studies have identified motifs enriched upstream of co-expressed or functionally related genes in various Plasmodium species [49, 52–58]. These targets further expand the catalogue of functional DNA motifs in Plasmodium and provide a good starting point for characterizing the role of Plasmodium ApiAP2 proteins in transcriptional regulation. The expression profiles of putative co-expressed targets are both positively and negatively correlated to the expression profile of the corresponding ApiAP2 gene [42] indicating that these factors may function either as activators or repressors of transcription. However, a complete understanding of the cellular roles of ApiAP2 proteins will also require characterizing stage-specific DNA interactions in vivo as well as their possible interaction with other proteins.
ApiAP2 protein interactions
Although functional domains outside of the AP2 DNA-binding region have not been identified for the majority of ApiAP2 proteins, there is some evidence for interactions with other proteins. Yeast two-hybrid assays in P. falciparum have identified potential homo and heterotypic interactions between different ApiAP2 family members as well as interactions with other potential transcription associated proteins [59]. Using two-hybrid data, Bougdour et al. have created a preliminary protein interaction network centered on the ApiAP2 factors thereby highlighting both direct and indirect links to a number of DNA-binding proteins [60]. The resulting in silico predictions revealed Plasmodium ApiAP2 protein interaction with chromatin or transcription related proteins, including the PfGCN5 histone acetyltransferase (PF08_0034), a high mobility group (HMG) protein (MAL8P1.72), a fork head domain protein (PF13_0042), and a plant homeodomain (PHD)-containing protein (PF14_0315) [59, 60]. In Toxoplasma gondii, the association of an ApiAP2 protein with GCN5 has been successfully demonstrated [61]. Precedence for such interactions exists in plants where CBF1, an Arapidopsis AP2 factor, has been demonstrated to interact with GCN5 [62]. In P. falciparum, Flueck et al. have demonstrated that the ApiAP2 protein PfSIP2 colocalizes with heterochromatin protein 1 (PfHP1) at perinuclear chromosome end clusters and upstream of upsB var genes [37]. Such interactions of ApiAP2 proteins with the epigenetic machinery is likely to be conserved across all Plasmodium spp..
In the absence of any structural information beyond the DNA-binding domain, it has been suggested that dimerization may be mediated by the Plasmodium AP2 domain itself. The crystal structure of PF14_0633 AP2 domain reveals the formation of homodimers where domain swapping was observed with the α-helix from one domain monomer associating with the β-sheet of a second monomer (Figure 1, bottom left) [27]. Lindner et al. have proposed that the binding of one AP2 monomer to DNA induces a conformational change that recruits a second AP2 domain, with the dimer forming a more stable interaction with the DNA [27]. This model suggests that multiple DNA-binding sites upstream of target genes would enhance the recruitment of such dimeric ApiAP2 protein complexes. Furthermore, the ability of ApiAP2 proteins to form homo and heterodimers increases the potential number of target genes that could be differentially regulated by a small number of factors. In support of this idea, genome-wide bioinformatic predictions of AP2 domain recognition elements define multiple potential binding sites upstream of virtually all open reading frames [42]. Of course additional DNA binding proteins, some of which have already been identified such as Myb1 (PF13_0088) [63, 64], Myb2 (PF10_0327) [21] and the high mobility group (HMGB) proteins (MAL8P1.72 and PFL0145c) [65, 66], will contribute to the overall picture of transcriptional regulation. In summary, ApiAP2 proteins may function in protein complexes to regulate transcription, and the identification of such complexes will be a key step in unraveling transcription factor-based control of gene expression in Plasmodium.
Remaining questions and future directions
The identification and initial characterization of the ApiAP2 family of transcriptional regulators is a major step toward understanding gene regulation in Plasmodium spp. It has been established that the majority of Plasmodium ApiAP2 family members interact with sequence specific elements and have the potential to function as trans-acting factors [42]. Experiments on individual ApiAP2 factors, in both the mosquito and blood stages of development, have begun to answer some fundamental questions regarding the in vivo function of this protein family [37–39]. However, the exact role that each ApiAP2 protein is playing in the biology of the parasite largely remains to be determined, emphasizing the necessity for more in vivo experimentation.
Although the 60 amino acid AP2 domains are highly conserved among the Apicomplexa, the sequence similarity does not extend to the rest of the protein and homology outside of the AP2 DNA-binding domain is low [22]. With the wide range of sizes predicted for ApiAP2 proteins, it can be anticipated that there are additional domains that activate transcription or promote interaction with other proteins required for regulation. Furthermore, there is evidence to suggest that some ApiAP2 proteins may be processed during parasite development implying that multiple forms may exist for individual proteins [37]. PFF0200c (PfSIP2) has been shown to be proteolytically processed from the full length 230 kDa form to a 50–60 kDa N-terminal segment containing the two AP2 DNA-binding domains [37]. Regarding other active regions outside of the AP2 domains, a portion of PFF0200c (amino acids 177–313) has been demonstrated to act as a transactivation domain in a yeast system and has subsequently been exploited in a series of tetracycline transactivator-based inducible conditional knockout vectors for P. berghei ([67, 68], P. Pino, E. Bush, O. Billker, M. Llinás, D. Soldati, unpublished data).
Beyond these experiments, it is unknown what other domains may exist and how they participate in coordinating repression or activation of target genes. Furthermore, the identification of specific interacting partners of ApiAP2 proteins will be a major advance in understanding in vivo functions for many of these factors. As shown by the work summarized herein, specific ApiAP2 proteins can directly regulate subsets of genes involved in Plasmodium developmental transitions or promote the formation of heterochromatin in subtelomeric regions. Although the majority of AP2 domains bind DNA in vitro, it is currently unknown how the DNA-tethered ApiAP2 proteins mediate interactions with proteins of the general transcription complexes or chromatin remodeling machinery. Demonstration of protein-protein interactions, such as with RNA polymerase II or GCN5, a heterochromatin remodeler, would further support the role of ApiAP2 proteins in transcriptional regulation.
In addition to the specific functional questions for individual ApiAP2 factors lie broad questions regarding the family as a whole. Foremost is the issue of whether the ApiAP2 family will perform parallel functions in all Apicomplexa, or whether there is species-specific variability. Given the virtually complete sequence identity of individual AP2 DNA-binding domains across the Apicomplexa, it is likely that the same DNA sequence elements will be bound in different organisms, however, target genes under the control of these DNA motifs may vary greatly from species to species [28]. Preliminary evidence shows that the AP2-O motif (TAGCTA) is conserved upstream of common target genes in P. berghei, P. vivax, and P. falciparum [38], but other motifs share little overlap in target gene predictions between different Plasmodium species [42]. Dissecting such differences in target regulons will ultimately illuminate the species-specific regulatory functions served by this protein family.
In conclusion, the ApiA2 proteins are excellent candidate regulators of the precisely timed and coordinated gene expression governing multi-stage development and inter-host transitions required for the successful propagation of Plasmodium parasites and the other Apicomplexa. We anticipate that a better understanding of these factors will facilitate the exploration of new therapeutic interventions in the foreseeable future. Such strategies might be aimed at disrupting the interaction of ApiAP2 proteins and their target DNAs or interfering with protein-protein interactions crucial for their downstream function, with the ultimate goal of arresting parasite development. The in-depth characterization of ApiAP2 proteins will fill a void in apicomplexan biology that is crucial to fully understanding parasite development and pathogenesis and may reveal mechanisms underlying host selection and disease severity.
Research Highlights.
The ApiAP2 protein family are candidate transcription regulators in Plasmodium spp.
ApiAP2 proteins play a role in gene regulation at various stages of Plasmodium development
Most of the P. falciparum ApiAP2 have been shown to bind sequence specific DNA elements
Acknowledgments
We would like to thank Erandi K. De Silva for the critical reading of this manuscript and Rohan Bansal for graphical display of ApiAP2 predicted binding sites. This work was funded by the NIH (R01 AI076276) and the Arnold and Mabel Beckman Foundation with support from the Center for Quantitative Biology (P50 GM071508) (ML). TLC was funded by an NSERC Postdoctoral Fellowship.
Abbreviations
- ApiAP2
Apicomplexan AP2
- ERF
ethylene response factor
- AP2
Apetela2
- IDC
intraerythrocytic development cycle
- SPE2
subtelomeric var promoter element 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–8. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 3.Cui L, Miao J. Chromatin-mediated epigenetic regulation in the malaria parasite Plasmodium falciparum. Eukaryotic Cell. 2010;9:1138–49. doi: 10.1128/EC.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009;5:179–90. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Salcedo-Amaya AM, van Driel MA, Alako BT, Trelle MB, van den Elzen AM, Cohen AM, et al. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc Natl Acad Sci U S A. 2009;106:9655–60. doi: 10.1073/pnas.0902515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, Alako BT, et al. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 2009;5:e1000569. doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Toledo K, Rojas-Meza AP, Mancio-Silva L, Hernandez-Cuevas NA, Delgadillo DM, Vargas M, et al. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 2009;37:2596–606. doi: 10.1093/nar/gkp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–70. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 9.Mair GR, Braks JA, Garver LS, Wiegant JC, Hall N, Dirks RW, et al. Regulation of sexual development of Plasmodium by translational repression. Science. 2006;313:667–9. doi: 10.1126/science.1125129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mair GR, Lasonder E, Garver LS, Franke-Fayard BM, Carret CK, Wiegant JC, et al. Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 2010;6:e1000767. doi: 10.1371/journal.ppat.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang M, Fennell C, Ranford-Cartwright L, Sakthivel R, Gueirard P, Meister S, et al. The Plasmodium eukaryotic initiation factor-2a kinase IK2 controls the latency of sporozoites in the mosquito salivary glands. The Journal of Experimental Medicine. 2010;207:1465–74. doi: 10.1084/jem.20091975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aly ASI, Mikolajczak SA, Rivera HS, Camargo N, Jacobs-Lorena V, Labaied M, et al. Targeted deletion of SAP1 abolishes the expression of infectivity factors necessary for successful malaria parasite liver infection. Molecular Microbiology. 2008;69:152–63. doi: 10.1111/j.1365-2958.2008.06271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silvie O, Goetz K, Matuschewski K. A sporozoite asparagine-rich protein controls initiation of Plasmodium liver stage development. PLoS Pathog. 2008;4:e1000086. doi: 10.1371/journal.ppat.1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shock JL, Fischer KF, DeRisi JL. Whole-genome analysis of mRNA decay in Plasmodium falciparum reveals a global lengthening of mRNA half-life during the intra-erythrocytic development cycle. Genome Biol. 2007;8:R134. doi: 10.1186/gb-2007-8-7-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sims JS, Militello KT, Sims PA, Patel VP, Kasper JM, Wirth DF. Patterns of gene-specific and total transcriptional activity during the Plasmodium falciparum intraerythrocytic developmental cycle. Eukaryot Cell. 2009;8:327–38. doi: 10.1128/EC.00340-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callebaut I, Prat K, Meurice E, Mornon JP, Tomavo S. Prediction of the general transcription factors associated with RNA polymerase II in Plasmodium falciparum: conserved features and differences relative to other eukaryotes. BMC Genomics. 2005;6:100. doi: 10.1186/1471-2164-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coulson RM, Hall N, Ouzounis CA. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res. 2004;14:1548–54. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Templeton TJ, Iyer LM, Anantharaman V, Enomoto S, Abrahante JE, Subramanian GM, et al. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 2004;14:1686–95. doi: 10.1101/gr.2615304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: genomic gleanings. Cell. 2003;115:771–85. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- 21.Bischoff E, Vaquero C. In silico and biological survey of transcription-associated proteins implicated in the transcriptional machinery during the erythrocytic development of Plasmodium falciparum. BMC Genomics. 2010;11:34. doi: 10.1186/1471-2164-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balaji S, Babu MM, Iyer LM, Aravind L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379:633–46. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- 24.Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998;17:5484–96. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–82. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–25. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindner SE, De Silva EK, Keck JL, Llinas M. Structural determinants of DNA binding by a P. falciparum ApiAP2 transcriptional regulator. J Mol Biol. 2010;395:558–67. doi: 10.1016/j.jmb.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Silva EK, Gehrke AR, Olszewski K, Leon I, Chahal JS, Bulyk ML, et al. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc Natl Acad Sci U S A. 2008;105:8393–8. doi: 10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int J Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 31.Altschul SF, Wootton JC, Zaslavsky E, Yu Y-K. The Construction and Use of Log-Odds Substitution Scores for Multiple Sequence Alignment. PLoS Comput Biol. 2010;6:e1000852. doi: 10.1371/journal.pcbi.1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackinnon MJ, Li J, Mok S, Kortok MM, Marsh K, Preiser PR, et al. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. PLoS Pathog. 2009;5:e1000644. doi: 10.1371/journal.ppat.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llinas M, Bozdech Z, Wong ED, Adai AT, DeRisi JL. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 2006;34:1166–73. doi: 10.1093/nar/gkj517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bozdech Z, Mok S, Hu G, Imwong M, Jaidee A, Russell B, et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci U S A. 2008;105:16290–5. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otto TD, Wilinski D, Assefa S, Keane TM, Sarry LR, Böhme U, et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Molecular Microbiology. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Behnke MS, Wootton JC, Lehmann MM, Radke JB, Lucas O, Nawas J, et al. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS One. 5:e12354. doi: 10.1371/journal.pone.0012354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flueck C, Bartfai R, Neiderwieser I, Witmer K, Alako BTF, Moes S, et al. A major role for the Plasmodium falciparum ApiAP2 protein PFSIP2 in chromosome end biology. PLoS Pathog. 2010;6:e1000784. doi: 10.1371/journal.ppat.1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuda M, Iwanaga S, Shigenobu S, Mair GR, Janse CJ, Waters AP, et al. Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol Microbiol. 2009;71:1402–14. doi: 10.1111/j.1365-2958.2009.06609.x. [DOI] [PubMed] [Google Scholar]
- 39.Yuda M, Iwanaga S, Shigenobu S, Kato T, Kaneko I. Transcription factor AP2-Sp and its target genes in malarial sporozoites. Mol Microbiol. 2010;75:854–63. doi: 10.1111/j.1365-2958.2009.07005.x. [DOI] [PubMed] [Google Scholar]
- 40.Balu B, Chauhan C, Maher S, Shoue D, Kissinger J, Fraser M, et al. piggyBac is an effective tool for functional analysis of the Plasmodium falciparum genome. BMC Microbiology. 2009;9:83. doi: 10.1186/1471-2180-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maier AG, Rug M, O’Neill MT, Brown M, Chakravorty S, Szestak T, et al. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell. 2008;134:48–61. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell TL, De Silva EK, Olszewski KL, Elemento O, Llinás M. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathogens. 2010:6. doi: 10.1371/journal.ppat.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silvestrini F, Bozdech Z, Lanfrancotti A, Di Giulio E, Bultrini E, Picci L, et al. Genome-wide identification of genes upregulated at the onset of gametocytogenesis in Plasmodium falciparum. Mol Biochem Parasitol. 2005;143:100–10. doi: 10.1016/j.molbiopara.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 44.Young JA, Fivelman QL, Blair PL, de la Vega P, Le Roch KG, Zhou Y, et al. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol Biochem Parasitol. 2005;143:67–79. doi: 10.1016/j.molbiopara.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Mikolajczak SA, Silva-Rivera H, Peng X, Tarun AS, Camargo N, Jacobs-Lorena V, et al. Distinct malaria parasite sporozoites reveal transcriptional changes that cause differential tissue infection competence in the mosquito vector and mammalian host. Mol Cell Biol. 2008;28:6196–207. doi: 10.1128/MCB.00553-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva-Rivera H, Camargo N, et al. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc Natl Acad Sci U S A. 2008;105:305–10. doi: 10.1073/pnas.0710780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu G, Cabrera A, Kono M, Mok S, Chaal BK, Haase S, et al. Transcriptional profiling of growth perturbations of the human malaria parasite Plasmodium falciparum. Nat Biotechnol. 2010;28:91–8. doi: 10.1038/nbt.1597. [DOI] [PubMed] [Google Scholar]
- 48.Chaal BK, Gupta AP, Wastuwidyaningtyas BD, Luah YH, Bozdech Z. Histone deacetylases play a major role in the transcriptional regulation of the Plasmodium falciparum life cycle. PLoS Pathog. 2010;6:e1000737. doi: 10.1371/journal.ppat.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young JA, Johnson JR, Benner C, Yan SF, Chen K, Le Roch KG, et al. In silico discovery of transcription regulatory elements in Plasmodium falciparum. BMC Genomics. 2008;9:70. doi: 10.1186/1471-2164-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, Berriman M, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–6. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 51.Voss TS, Kaestli M, Vogel D, Bopp S, Beck HP. Identification of nuclear proteins that interact differentially with Plasmodium falciparum var gene promoters. Mol Microbiol. 2003;48:1593–607. doi: 10.1046/j.1365-2958.2003.03528.x. [DOI] [PubMed] [Google Scholar]
- 52.van Noort V, Huynen MA. Combinatorial gene regulation in Plasmodium falciparum. Trends Genet. 2006;22:73–8. doi: 10.1016/j.tig.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28:337–50. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J, Sieglaff DH, Gervin J, Xie XS. Discovering regulatory motifs in the Plasmodium genome using comparative genomics. Bioinformatics. 2008;24:1843–9. doi: 10.1093/bioinformatics/btn348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jurgelenaite R, Dijkstra TM, Kocken CH, Heskes T. Gene regulation in the intraerythrocytic cycle of Plasmodium falciparum. Bioinformatics. 2009;25:1484–91. doi: 10.1093/bioinformatics/btp179. [DOI] [PubMed] [Google Scholar]
- 56.Iengar P, Joshi NV. Identification of putative regulatory motifs in the upstream regions of co-expressed functional groups of genes in Plasmodium falciparum. BMC Genomics. 2009;10:18. doi: 10.1186/1471-2164-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Essien K, Stoeckert CJ., Jr Conservation and divergence of known apicomplexan transcriptional regulons. BMC Genomics. 2010;11:147. doi: 10.1186/1471-2164-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Westenberger SJ, McClean CM, Chattopadhyay R, Dharia NV, Carlton JM, Barnwell JW, et al. A systems-based analysis of Plasmodium vivax lifecycle transcription from human to mosquito. PLoS Negl Trop Dis. 2010;4:e653. doi: 10.1371/journal.pntd.0000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, Hesselberth JR, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103–7. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 60.Bougdour A, Braun L, Cannella D, Hakimi MA. Chromatin modifications: implications in the regulation of gene expression in Toxoplasma gondii. Cell Microbiol. 2010;12:413–23. doi: 10.1111/j.1462-5822.2010.01446.x. [DOI] [PubMed] [Google Scholar]
- 61.Dixon SE, Stilger KL, Elias EV, Naguleswaran A, Sullivan WJ., Jr A decade of epigenetic research in Toxoplasma gondii. Mol Biochem Parasitol. 2010;173:1–9. doi: 10.1016/j.molbiopara.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci U S A. 1997;94:1035–40. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boschet C, Gissot M, Briquet S, Hamid Z, Claudel-Renard C, Vaquero C. Characterization of PfMyb1 transcription factor during erythrocytic development of 3D7 and F12 Plasmodium falciparum clones. Mol Biochem Parasitol. 2004;138:159–63. doi: 10.1016/j.molbiopara.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 64.Gissot M, Briquet S, Refour P, Boschet C, Vaquero C. PfMyb1, a Plasmodium falciparum transcription factor, is required for intra-erythrocytic growth and controls key genes for cell cycle regulation. J Mol Biol. 2005;346:29–42. doi: 10.1016/j.jmb.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 65.Briquet S, Boschet C, Gissot M, Tissandie E, Sevilla E, Franetich JF, et al. High-mobility-group box nuclear factors of Plasmodium falciparum. Eukaryot Cell. 2006;5:672–82. doi: 10.1128/EC.5.4.672-682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gissot M, Ting LM, Daly TM, Bergman LW, Sinnis P, Kim K. High mobility group protein HMGB2 is a critical regulator of Plasmodium oocyst development. J Biol Chem. 2008;283:17030–8. doi: 10.1074/jbc.M801637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krizek BA, Sulli C. Mapping sequences required for nuclear localization and the transcriptional activation function of the Arabidopsis protein AINTEGUMENTA. Planta. 2006;224:612–21. doi: 10.1007/s00425-006-0253-9. [DOI] [PubMed] [Google Scholar]
- 68.Meissner M, Krejany E, Gilson PR, de Koning-Ward TF, Soldati D, Crabb BS. Tetracycline analogue-regulated transgene expression in Plasmodium falciparum blood stages using Toxoplasma gondii transactivators. Proc Natl Acad Sci U S A. 2005;102:2980–5. doi: 10.1073/pnas.0500112102. [DOI] [PMC free article] [PubMed] [Google Scholar]