Abstract
The arginine dihydrolase (ADH) pathway has an analogous function to the urea cycle in mitochondria-containing cells, by removing nitrogen from amino acids and generating ATP. Subcellular localization of the ADH pathway enzymes in Trichomonas vaginalis revealed that arginine deiminase (ADI) localizes to the hydrogenosome, a mitochondrion-like organelle of anaerobic protists. However the other enzymes of the ADH pathway, ornithine carbamyltransferase and carbamate kinase localize to the cytosol. Three gene sequences of T. vaginalis ADI (ADI 1–3) were identified in the T. vaginalis genome, all having putative mitochondrial targeting sequences. The ADI sequences were cloned and used to probe T. vaginalis using a carboxyterminal di-hemogglutinin epitope tag which demonstrated co-localization with malic enzyme confirming the hydrogenosome localization of this enzyme.
Keywords: Trichomonas vaginalis, hydrogenosome, mitochondrion-like organelle, arginine dihydrolase pathway, arginine deiminase
The arginine dihydrolase (ADH) pathway catalyzes the conversion of arginine to ornithine and ammonia via the enzymes arginine deiminase (ADI), catabolic ornithine carbamyltransferase (OCT), and carbamate kinase. Cumulatively the pathway removes nitrogen from arginine with the generation of ATP, and therefore performs an analogous function to the urea cycle of vertebrates. The ADH pathway is present in some protists such as Trichomonas vaginalis [17] and Giardia intestinalis [15], as well as some Gram positive bacteria e.g. Streptococcus sp., [8], Gram negative bacteria e.g. Pseudomonas sp., [1], and some Mollicutes e.g. Mycoplasma hominis, M. genitalium, [10], where it has been proposed to function as an alternative ATP generating mechanism. The enzymes of the pathway have been characterized in several organisms notably Mycoplasma sp., G. intestinalis, T. vaginalis and Tritrichomonas foetus [12, 17, 20]. Based upon subcellular studies with T. vaginalis all enzymes of the pathway were localized in the cytosol with the exception of arginine deiminase (ADI), which was mainly associated with sedimentable cell components, and its colocalization with the plasma membrane was suggested [18]. The availability of the complete genome sequence for T. vaginalis [6] enables studies for the localization of ADI in T. vaginalis to be confirmed. BLAST search of the T. vaginalis genome database (TrichoDB, http://trichdb.org/trichdb/) using Giardia intestinalis ADI (accession number XP_001705755) as query revealed the presence of three copies of the ADI gene (TvADI-1, TVAG_467820; TvADI-2, TVAG_344520 and TvADI-3, TVAG_183850) coding for proteins with calculated molecular weight 46–47,000. Using ClustalX [14] and BioEdit software the T. vaginalis sequences were aligned with Mycoplasma arginini for which a crystal structure has been determined [2], which revealed the presence of conserved residues involved in the substrate binding and/or the enzyme active site (supplementary Fig. 1). The positional equivalent of the catalytic triad Cys397, His268, Glu213 determined in M. arginine ADI [2] is conserved in the TvADI-2 (Cys408, His283, Glu230) and TvADI-3 (Cys405, His281, Glu228), while the Cys405 is replaced by Ser405 in TvADI-1 (Fig. s1). All three putative TvADI sequences contained mitochondria-like N-terminal targeting pre-sequences with predicted cleavage site for processing peptidase [3] (Fig. s1) and high probability of mitochondrial localization estimated by PSORT II (http://psort.hgc.jp/) (56–65%) and TargetP (mTP values 0.500–0.710).
These predictions suggested the localization of ADI with T. vaginalis hydrogenosomes, an anaerobic form of mitochondria in these parasites. To investigate cellular localization three genes coding for T. vaginalis ADI were amplified by PCR from genomic DNA of T. vaginalis (strain T1) and sub-cloned to TagVag vector, which allows episomal expression of recombinant protein with carboxyterminal di-hemogglutinin epitope tag [4]. The molecular weight of the expressed proteins corresponded well with calculated values (Fig 1A). T. vaginalis cells were transfected as described [4] and probed with a mouse monoclonal anti hem agglutinin antibody [4] (Fig 1B). A hydrogenosome marker protein malic enzyme was used as a positive control and detected using a rabbit polyclonal anti malic enzyme antibody [4]. Images were obtained using immunofluorescence confocal microscopy which revealed that all three gene products localized to organelles organized along the axostyle and costa. A typical result of localization is shown for ADI-3 which colocalizes with the hydrogenosome protein marker, malic enzyme (Fig. 1B), ADI-1 and ADI-2 demonstrated similar results (not shown). To distinguish whether ADI is present inside the organelle or associated with the outer hydrogenosome membrane, hydrogenosomes were isolated from ADI-1 trasfected cells as described [5]. Intact, or disintegrated hydrogenosomes prepared by sonication or 0.5% Triton X-100, were treated with 50 µg/ml proteinase K for 20 min at 0°C. The digestion was inhibited with 1 mM phenylmethylsulfonyl fluoride; the samples were acetone-precipitated and analysed by immuno-bloting. While protease K treatment of intact hydrogenosomes did not affect ADI-1 signal, it was not detected when hydrogenosome membranes were disintegrated (Fig. 1C). This result strongly suggests that ADI-1 is present inside the hydrogenosome.
Figure 1. Expression of ADI-1, ADI-2 and ADI-3 genes in T. vaginalis.
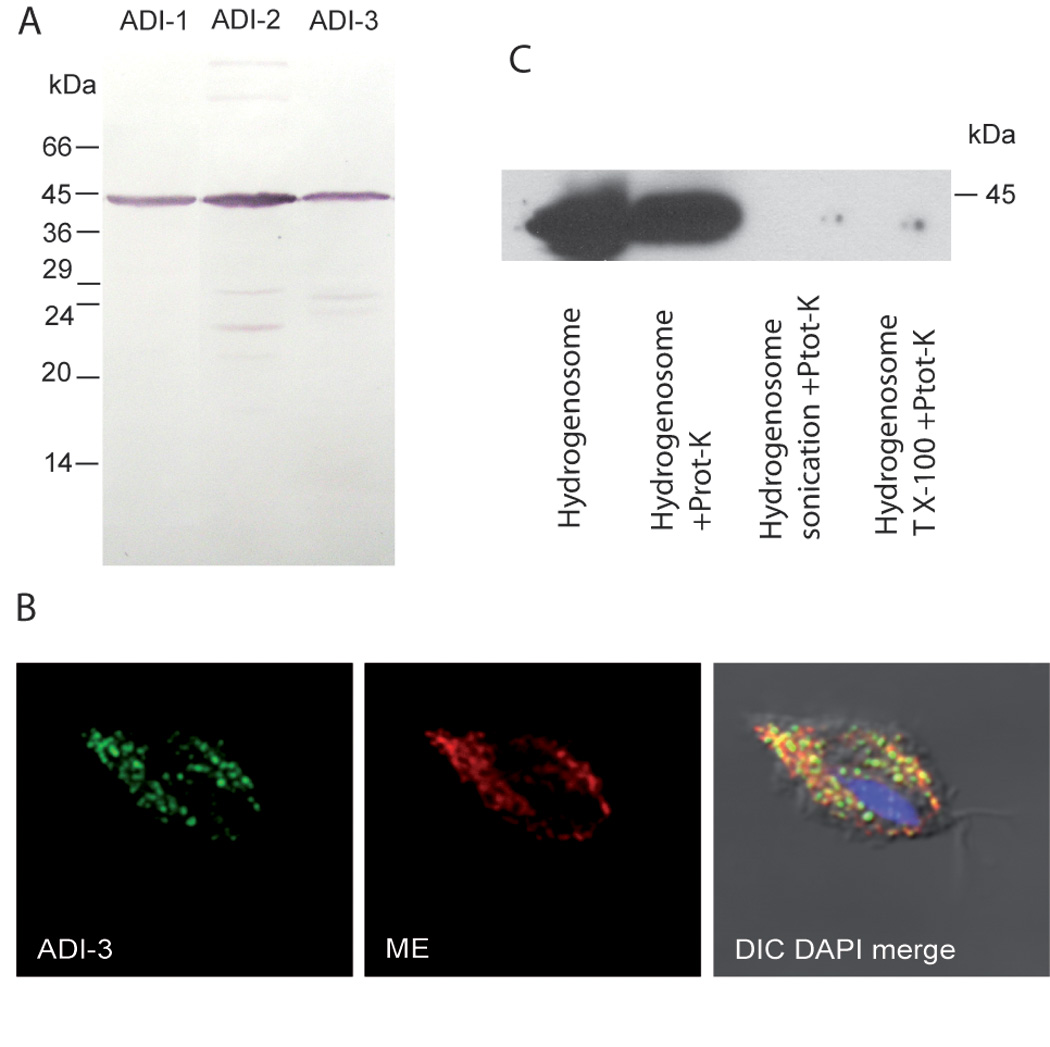
A) Each gene was expressed with C-terminal hemaglutinin tag in T. vaginalis and detected in cellular lysates by immunoblotting with anti-hemaglutinin monoclonal antibody. B) Representative localization of ADI in T. vaginalis hydrogenosomes by confocal fluorescent microscopy. ADI-3, fluorescent microscopy of T. vaginalis expressing ADI-3. Mouse monoclonal anti hemaglutinin (ADI-HA) tag antibody was used to visualize ADI-3 (green); ME, fluorescent microscopy of rabbit polyclonal anti malic enzyme antibody (red); DIC DAPI merge, merged image of ADI-3-HA, ME image stained with DAPI (blue) to show the nucleus. C) ADI localizes to the inside of hydrogenosomes. Treatment of intact hydrogenosomes with proteinase K (Prot-K) did not affect signal for ADI-1, while ADI-1 was not detected by immunobloting when hydrogenosome membranes were disintegrated by sonication or Trition X-100 (T X-100) and subsequently treated with proteinase K.
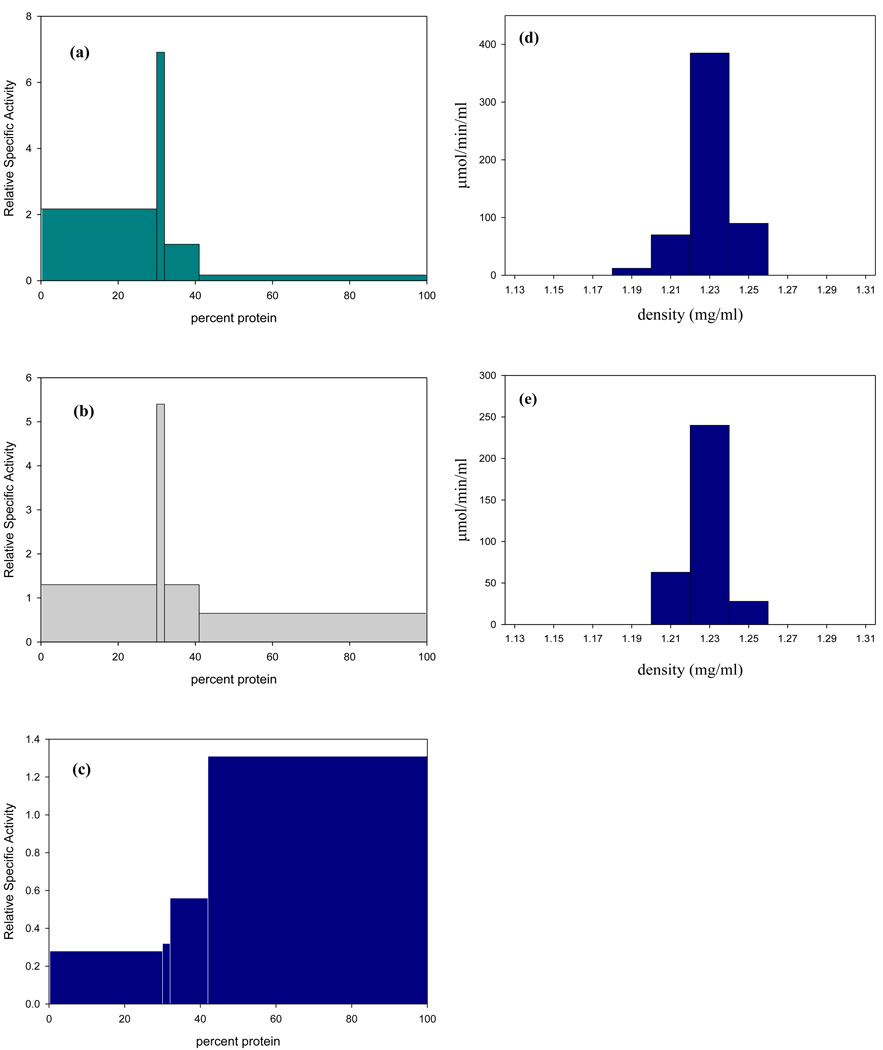
Subcellular localization of ADI activity was performed by differential centrifugation of whole cell extracts using 225 mM sucrose pH 8.0 in 15 mM Tris and 10 mM KCl. ADI activity was determined by measuring the formation of citrulline using 1mM L-arginine in 40 mM HEPPS pH 8.0 [18]. In previous studies a pH of 6.0 was used which is sub-optimal for the T. vaginalis enzyme [18]. Under the assay conditions used in this study ADI was predominantly found in the hydrogenosome enriched fraction (Fig 2a) based upon the distribution of malic enzyme, a marker for hydrogenosome-enriched fractions. The activity of malic enzyme was determined by measuring the absorbance change at 340nm using 6 mM malate, 1 mM NAD and 0.66mM MnCl2 in pH 6.8 Triethanolamine buffer [18] (Fig. 2b). Catabolic OCT was determined by measuring 14CO2 release from L-[14C-carbamyl] citrulline (17.2 mM L-[14C-carbamyl] citrulline, 57.7 mCi/mmol; DuPont, N.E.N. Research Products) in 40 mM MES pH 6.0 [18]. As shown previously, catabolic OCT, which in aerobic eukaryotes localizes to the mitochondrion was found predominantly in the non-sediment able fraction (Fig 2c) [18]. Integrity of the organelles was confirmed by performing all enzyme assays in isotonic buffered solutions (225mM sucrose) to which 0.05% Triton X-100 was added to demonstrate latency of activity. The specific activity of ADI in the hydrogenosome-enriched fraction was 2.0 ± 0.3 µM min−1 (mg of protein)−1 which was statistically unchanged (1.9 ± 0.2 µM min−1 (mg of protein)−1) in the presence of Triton X-100. Further purification of the hydrogenosome-enriched fraction by percoll gradients revealed a single fraction that had >70% of the recovered ADI activity which had an isopycnic density of 1.23 g/ml (Fig 2d) and corresponded to the recovery of malic enzyme activity confirming the identity of the fraction as hydrogenosomes (Fig 2e). Studies aimed at determining whether the ADI is localized to the outside of the membrane or inside were performed by measuring the ADI activity of percoll purified hydrogenosomes before and after a 15 min treatment with 500 µg ml−1 of trypsin in the presence or absence of 50 µg ml−1 saponin. Hydrogenosomes treated with trypsin showed no loss of enzyme activity compared to control incubations without trypsin. However, hydrogenosomes pre-incubated for 15 minutes with saponin, a membrane permeabilizing glycoside, had 30% lowered activity after treatment with trypsin compared to controls with saponin alone or no treatment (data not shown). These results support the immunoblot data confirming the intra-organelle localization of ADI in T. vaginalis hydrogenosomes.
Figure 2. Subcellular localization of ADH enzymes in T. vaginalis isolate (SS-22).
Distribution of enzymes in fractions obtained by differential centrifugation of a cell-free extract of T. vaginalis SS-22 in 225 mM sucrose-10 mM Tris pH 7.4, containing 1 mM calcium chloride and 1 mM magnesium chloride. Relative specific activities (the ratios of specific activities in fractions to those in the cell-free extract) were plotted against cumulative percentage protein recovered in each fraction. The centrifugal force increases from left to right such that the fractions represent the nuclei (400 × g for 10 min), hydrogenosomes (2,200 × g for 10 min), lysosomes (28,000 × g for 30 min), and the non-sedimentable fraction (>28,000 × g for 30 min). Percent recovery for each enzyme after subcellular fractionation was: (a) ADI, 87%; (b) MDH, 93%; (c) cOCT, 98%. Hydrogenosomes obtained by differential centrifugation were further purified using 50% (w/v) percoll gradients containing 225 mM sucrose-10 mM Tris pH 7.4, 1 mM calcium chloride and 1 mM magnesium chloride. One ml of the hydrogenosome-enriched fraction was centrifuged at 46,000 g for 45 min at 4°C. Fractions were assayed for ADI and MDH activity as described; (d) ADI had a single peak of activity at a density of 1.23 g/ml, percent recovery was 84%; (e) The hydrogenosomal marker enzyme MDH had a single peak of activity at the same isopycnic density observed for ADI, percent recovery was 92%.
Phylogenetic analysis of ADI was performed using 66 amino acid sequences of eukaryotic, archeal and bacterial representatives. Sequences were searched using the NCBI and EST TbestDB (http://tbestdb.bcm.umontreal.ca/) databases and aligned using ProbCons [2] and ClustalX [14]. The alignment was manually edited and 288 unambiguously aligned positions were used for phylogenetic reconstruction. Maximum likelihood tree was constructed using PROTGAMMAWAG model implemented in RAxML [12]. The bootstrap support (100 replicates) was calculated using RAxML, using maximum parsimony in PAUP [13] and using distance method (neighbor joining, LogDet distances) in PAUP. This search resulted in 10 eukaryotic sequences from Excavata, Archaeplastida and Amoebozoa groups, in addition to three T. vaginalis ADI sequences (Fig. s2). The analysis revealed three major ADI clusters corresponding to Bacteria, Archea and Eukaryota. Interestingly, the eukaryotic ADI sequences appear as monophyletic group with Archea at sister position, which suggests that ADI-encoding genes were present in common eukaryotic ancestor. Arginine deiminase is present in representatives of all three domains of life, Prokarya, Archaea and Eukarya [19]. Eukaryotic ADI is confined to unicellular organisms, which live under anaerobic or microaerophilic conditions (Trichomonas, Giardia, Mastigamoeba) or sustain prolonged anaerobiosis (Euglena) [9]. In addition ADI is present in green algae (Chlamydomonas, Chlorella) which undergo anaerobic fermentation during the anoxic phase at night when the rate of respiratory oxygen consumption exceeds the rate of photosynthetic oxygen generation [5]. Although the slime mold Dictyostelium is not classically considered an anaerobe it is closely related to the anaerobes Entamoeba and Mastigamoeba [5] and contains both an ADI and an oxygen-independent class II ribonucleotide reductase. These eukaryotes belong to three distinct eukaryotic groups, Excavata, Amebozoa and Plantae, thus independent acquisition of ADI by lateral gene transfer might be expected. However, our phylogenetic analysis suggested monophyletic origin of eukaryotic ADI with Archea as a sister group. Therefore the presence of ADI might be considered as an ancestral eukaryotic feature as previously suggested for G. intestinalis [19].
The reason for ADI localization to the trichomonad hydrogenosome is not clear. In mitochondria-containing eukaryotes arginine is metabolized via the urea cycle in which OCT and carbamate kinase are localized within the organelle. This localization is also found in hydrogenosomes from the anaerobic chytridiomycete, Neocallimastix frontalis [7], which has a urea cycle and lacks an ADH pathway. However the ADH pathway is localized to the cytosol of G. intestinalis, and it has been suggested that ADI may undergo translocation to the nuclei upon encystation [15]. It has been proposed that some prokaryotes may possess a membrane localized ADI, e.g. the cell wall of Streptococcus mitis and Pseudomonas and Mycoplasma hominis [10, 1, 12]. In these organisms OCT and carbamate kinase are located either totally of mainly in the cytoplasmic fractions. In addition bioinformatic analysis of cyanobacterium Synechocystis sp. PCC 6803 revealed the presence of a complete ADH pathway [11], which codes for a 705 amino acid containing protein compared to the typical prokaryotic protein containing 411–418 amino acids [11]. The cyanobacterial ADI contains two transmembrane helixes in their C-terminal region implying that this enzyme is also membrane associated [11]. The reason for the hydrogenosome localization of ADI in T. vaginalis is not known and we could speculate that the ammonium ions released by ADI may buffer the hydrogenosome pH, which based on the hyperfine splitting pattern of 4-nitroimidazole is approximately pH 10 [16]; additionally localization of ADI to an organelle increases the flow through the pathway by sequestering arginine to the hydrogenosome and therefore would increase the yield of citrulline the precursor of ornithine and carbamyl phosphate, which in turn forms ATP via carbamate kinase. We are currently investigating the biochemical and physiological reasons for this localization.
Supplementary Material
Acknowledgements
The authors thank Dr. Thomas E. Gorrell for helpful suggestions. Parts of this study were supported by grants from NIH-NIAID 49785 (NY), and MEYS CR LC07032 and MSM0021620858 (JT).
Abbreviations
- ADH
arginine dihydrolase
- ADI
arginine deiminase
- OCT
ornithine carbamyl transferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bourdineaud J-P, Heierli D, Gamper M, Verhoogt HJC, Driessen AJM, Konings WN, Lazdunski C, Haas D. Characterization of the arcD arginine:ornithine exchanger of Pseudomonas aeroginosa. J Biol Chem. 1993;268:5417–5424. [PubMed] [Google Scholar]
- 2.Das K, Butler GH, Kwiatkowski V, Clark AD, Jr, Yadav P, Arnold E. Crystal structure of arginine deiminase with covalent reaction intermediates: implication for catalytic mechanism. Structure. 2004;12:657–667. doi: 10.1016/j.str.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Dolezal P, Smíd O, Rada P, Zubácová Z, Bursać D, Suták R, Nebesárová J, Lithgow T, Tachezy J. Giardia mitosomes and trichomonad hydrogenosomes share a common mode of protein targeting. Proc Natl Acad Sci USA. 2005;102:10924–10929. doi: 10.1073/pnas.0500349102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drmota T, Proost P, Van Ranst M, Weyda F, Kulda J, Tachezy J. Iron-ascorbate cleavable malic enzyme from hydrogenosomes of Trichomonas vaginalis: purification and characterization. Mol Biochem Parasitol. 1996;83:221–234. doi: 10.1016/s0166-6851(96)02777-6. [DOI] [PubMed] [Google Scholar]
- 5.Sutak R, Doležal P, Fiumera HL, Hrdý I, Dancis A, Delgadillo M, Johnson P, Müller M, Tachezy J. Mitochondrial-type assembly of iron-sulfur centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA. 2004;101:10368–10373. doi: 10.1073/pnas.0401319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlton JM, Hirt RP, Silva JC, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gelius-Dietrich G, Ter Braak M, Henze K. Mitochondrial steps of arginine biosynthesis are conserved in the hydrogenosomes of the Chytridiomycete Neocallimastix frontalis. J Euk Microbiol. 2007;54:42–44. doi: 10.1111/j.1550-7408.2006.00146.x. [DOI] [PubMed] [Google Scholar]
- 8.Hiraoka BY, Harada M, Fukasawa K, Magi M. Intracellular localization of arginine deiminase pathway in Streptococcus mitis. Curr Microbiol. 1987;15:81–84. [Google Scholar]
- 9.Hoffmeister M, van der Klei A, Rotte C, van Grinsven KW, van Hellemond JJ, Henze K, Tielens AG, Martin W. Euglena gracilis rhodoquinone:ubiquinone ratio and mitochondrial proteome differ under aerobic and anaerobic conditions. J Biol Chem. 2004;279:22422–22429. doi: 10.1074/jbc.M400913200. [DOI] [PubMed] [Google Scholar]
- 10.Lin J-SL. Arginine deiminase of M. hominis: cytoplasmic and membrane associated forms. J Gen Micro. 1986;132:1467–1474. doi: 10.1099/00221287-132-6-1467. [DOI] [PubMed] [Google Scholar]
- 11.Schriek S, Ruckert C, Staiger D, Pistorius EK, Michel K-P. Bioinformatic evaluation of L-arginine catabolic pathways in 24 cyanobacteria and transcriptional analysis of genes encoding enzymes of Larginine catabolism in the cyanobacterium Synechocystis sp PCC 6803. BMC Genomics. 2007;8:437–465. doi: 10.1186/1471-2164-8-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 13.Swofford DL. PAUP*: Phylogenetic analysis using parsimony (\and other methods). Version 4. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- 14.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal,X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Touz MC, Ropolo AS, Rivero MR, Vranych CV, Conrad JT, Svard SG, Nash TE. Arginine deiminase has multiple regulatory roles in the biology of Giardia lamblia. J Cell Sci. 2008;121:2930–2938. doi: 10.1242/jcs.026963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarlett N, Rowlands CC, Evans JC, Yarlett NC, Lloyd D. Nitroimidazole and oxygen derived radicals detected by electron spin reseonance in hydrogenosomal and cytosolic fractions from Trichomonas vaginalis. Mol Biochem Parasitol. 1987;24:255–261. doi: 10.1016/0166-6851(87)90157-5. [DOI] [PubMed] [Google Scholar]
- 17.Yarlett N, Martinez MP, Moharrami MA, Tachezy J. The contribution of the arginine dihydrolase pathway to energy metabolism by Trichomonas vaginalis. Mol Biochem Parasitol. 1996;78:117–125. doi: 10.1016/s0166-6851(96)02616-3. [DOI] [PubMed] [Google Scholar]
- 18.Yarlett N, Lindmark DG, Goldberg B, Moharrami MA, Bacchi CJ. Subcellular localization of the enzymes of the arginine dihydrolase pathway in Trichomonas vaginalis and Tritrichomonas foetus. J Euk Microbiol. 1994;41:554–559. doi: 10.1111/j.1550-7408.1994.tb01516.x. [DOI] [PubMed] [Google Scholar]
- 19.Zuniga M, Perez G, Gonzalez-Candelas F. Evolution of arginine deiminase (ADI) pathway genes. Mol Phylogenet Evol. 2002;25:429–444. doi: 10.1016/s1055-7903(02)00277-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.