Abstract
Mantle cell lymphoma (MCL) is a specific type of aggressive B-cell non-Hodgkin lymphoma. We recently found that IL-22RA1, one of the two subunits of the interleukin 22 (IL-22) receptor, is expressed in MCL cell lines but not benign lymphocytes. In view of normal functions of IL-22 signaling, we hypothesized that the aberrant expression of IL-22RA1 may contribute to the deregulation of various cell signaling pathways, thereby promoting cell growth in MCL. In this study, we first demonstrated the expression of IL-22RA1 in all three MCL cell lines and eight frozen tumors examined using reverse transcription-polymerase chain reaction and Western blot analysis. In support of the concept that IL-22 signaling is biologically important in MCL, we found that MCL cells treated with recombinant IL-22 had a significant increase in cell growth that was associated with STAT3 activation. To investigate the mechanism underlying the aberrant expression of IL-22RA1, we analyzed the gene promoter of IL-22RA1, and we found multiple binding sites for NF-κB, a transcriptional factor strongly implicated in the pathogenesis of MCL. Pharmacologic inhibition of NF-κB resulted in a substantial reduction in the level of IL-22RA1 protein expression in MCL cells. To conclude, IL-22RA is aberrantly expressed in MCL, and we have provided evidence that IL-22 signaling contributes to the pathogenesis of MCL.
Introduction
Mantle cell lymphoma (MCL) is a distinct clinicopathologic entity recognized by the World Health Organization Classification Scheme [1]. Development of resistance to conventional chemotherapeutic agents is frequent, and the median survival is approximately 3 years [2–5]. The genetic hallmark of this disease is the chromosomal abnormality, t(11;14)(q13;q32), which results in aberrant cyclin D1 expression in these neoplastic B cells [2]. Experimental results derived from studies of the cyclin D1 transgenic mouse models support the concept that enforced cyclin D1 expression in B cells is not sufficient for lymphomagenesis [6]. Consistent with this view, an in vitro study recently showed that the knockdown of cyclin D1 using small hairpin RNA has minimal effects on the survival of MCL cells [7]. Accumulating evidence has suggested that MCL tumors often carry a relatively large number of biochemical abnormalities, including multiple defects in the regulation of the apoptotic pathway and cell cycle progression [8–17]. These findings have highlighted the biological complexity of MCL.
Interleukin 22 (IL-22) belongs to the family of IL-10-related proteins, which includes IL-19, IL-20, IL-24/MDA-7, IL-26/AK155, IL-28, and IL-29 [18–20]. IL-22 is normally produced by T lymphocytes and mucosal epithelial cells in various anatomic sites [21–27]. It has been shown that IL-22 triggers intracellular signals by binding to a heterodimeric receptor complex that is composed of IL-22RA1 and IL-10R2 [28–31]. Although IL-10R2 is ubiquitously expressed, IL-22RA1 is expressed in a relatively restricted pattern, being found at relatively high levels in the pancreas, small intestine, colon, kidney, and liver [32–35]. Importantly, IL-22RA1 is not detectable in immune cells including monocytes, resting or activated B/T cells, natural killer cells, macrophages, and dendritic cells [36,37]. IL-22 is known to activate a number of signaling pathways including that of STAT3 and mitogen-activated protein kinase [29,38–41]. On the basis of the current understanding of the biology of IL-22, it is believed that IL-22 produced by T cells plays an important role in enhancing innate immunity and tissue repair [26].
We have previously reported that the IL-22 signaling pathway carry biological significance in the pathogenesis of ALK-positive anaplastic large cell lymphoma, a lymphoma of mature T-cell immunophenotype [42]. We hypothesized that the IL-22 signaling may also play a role in the pathogenesis of MCL by contributing to the constitutive activation of STAT3 in MCL [17]. In this study, we first demonstrated that the aberrant expression of IL-22RA1 is a consistent phenomenon found in MCL cell lines and tumors. We then provided evidence that the IL-22 signaling is biologically important in MCL.
Materials and Methods
Cell Culture and Chemicals
The characteristics of the three MCL cell lines, Jeko-1, Mino, and SP53, have been previously described [43]. Briefly, all of these three cell lines have the mature B-cell immunophenotype, carry the t(11;14)(q13;q32) cytogenetic abnormality, and overexpress cyclin D1. All three cell lines are negative for the Epstein-Barr virus nuclear antigen. MCL cells were treated with 20 ng/ml of human recombinant IL-22 protein (rIL-22; R&D Systems, Minneapolis, MN) for 0 and 30 minutes and harvested for Western blot analysis. To obtain highly purified peripheral blood B cells from healthy donors, we first collected peripheral blood mononuclear cells by centrifugation over Ficoll-Hypaque. CD19-positive B cells were isolated by positive selection using specific monoclonal antibody-coated magnetic beads and a preparative magnetic cell sorter (Miltenyi, Bergisch Gladbach, Germany) in accordance with the manufacturer's recommended protocol. The purity of the isolated B-cell population was analyzed by flow cytometry (FACScan; Becton Dickinson, San Jose, CA) and confirmed to be greater than 98%. NF-κB activation inhibitor 6-amino-4-(4-phenoxyphenylethylamino quinazoline (catalog no. EI-352) was purchased from Enzo Life Sciences International (Farmingdale, NY) and E-2-fluoro-4′-methoxystilbene (catalog no. 481412) was purchased from EMD (Gibbstown, NJ).
Western Blot Analysis and Antibodies
Western blot analysis was performed using standard techniques. Briefly, cells were lysed in a buffer containing 20 mM Tris-HCl, pH 7.5, 5 mM EDTA, 40 µg/ml leupeptin, 1 µM pepstatin, and 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride and centrifuged at 15,000g for 15 minutes at 4°C. The supernatant was removed, and 50 to 100 µg of protein was run on an SDS-polyacrylamide gel. After the proteins were transferred to nitrocellulose membranes, the membranes were blocked with 5% milk in TBS buffer (20 mM Tris-HCl, pH 7.6, 150 mM NaCl) and then incubated with primary antibodies overnight followed by 1 hour of incubation with horseradish peroxidase-conjugated secondary antibody ( Jackson Immunoresearch Laboratories, Inc, West Grove, PA). Membranes were washed in PBS with 0.05% Tween-20 for 30 minutes between steps. Proteins were detected using the enhanced chemiluminescence detection kit (Amersham Life Sciences, Arlington Heights, IL). Antibodies used were anti-STAT3 (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA), antipSTAT3 (1:500; Santa Cruz Biotechnology), anti-IL-22RA1 (1:1000; Sigma-Aldrich, Oakville, Ontario, Canada), and anti-β-actin (1:3000; Sigma-Aldrich).
Immunofluorescence Staining and Confocal Microscopy
Immunofluorescence was performed using standard techniques. Briefly, cells grown on coverslip in a six-well plate were fixed with 4% paraformaldehyde in PBS. Cells were rinsed three times with 1x PBS, incubated with 30 µl of anti-IL-22RA1 (1:50; Sigma-Aldrich) antibody overnight followed by rinsing three times with 1x PBS. After incubating with 200 µl of Alexa Fluor 488 secondary antibody (1:250; Invitrogen, Burlington, Ontario, Canada) for 1 hour at room temperature, cells were rinsed with PBS, and the procedure was completed with a mounting medium (Dako, Mississauga, Ontario, Canada) added to the slides. Cells were visualized with a Zeiss LSM 510 (Carl Zeiss, Toronto, Canada) confocal microscope at the Core Cell Imaging Facility, Cross Cancer institute.
Flow Cytometric Detection of IL-22RA1 MCL Cell Lines
MCL cells were fixed in the CytoFix Buffer from Becton Dickinson Biosciences (Franklin Lakes, NJ) washed in cold PBS, centrifuged, and resuspended in the FACS staining buffer purchased from Becton Dickinson. Cells were incubated with primary antibodies for 60 minutes at 4°C in the dark and washed twice using cold buffer between incubations. The following antibodies were used: unconjugated mouse IgG1 as the isotype control (10 µg/ml; Santa Cruz Biotechnology), unconjugated mouse anti-human IL-22RA1 (IgG1, 10 µg/ml; R&D Systems), and phycoerythrin-conjugated rat anti-mouse antibody (IgG1, 5 µg/ml; Becton Dickinson). Flow cytometry was performed using the FACScan (Becton Dickinson, Toronto, Canada), and data were analyzed using the accompanying CELLQuest software as per manufacturer's guidelines.
Reverse Transcription-Polymerase Chain Reaction
Total cellular RNA was extracted from 1 x 106 cells from Mino, SP53, Jeko-1, purified human B cells, and HepG2 cell lines using the TRIzol extraction method (Invitrogen). Reverse transcription (RT) was performed using 500 ng of total RNA in a first-strand complementary DNA synthesis reaction with SuperScript Reverse Transcriptase as recommended by the manufacturer (Invitrogen). Primer pairs were designed to detect IL-22, IL-22RA1, and IL-10R2. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. Polymerase chain reaction (PCR) was performed by adding 5 µl of RT product into a 25-µl volume reaction containing 1x buffer, 200 µM of each dNTPs, oligonucleotide primer, and 0.2 U of AmpliTaq polymerase. The primer sequences and PCR cycles are shown in Table 1. For DNA amplification, complementary DNA was denatured at 94°C for 1 minute, subjected to primer annealing at 60°C for 1 minute, and then subjected to DNA extension at 72°C for 1minute for 35 cycles in a thermal cycler (Applied Biosystems, Foster City, CA). Amplified products were analyzed by DNA gel electrophoresis in 1% agarose and visualized by the Alpha Imager 3400 (Alpha Innotech, San Leandro, CA).
Table 1.
Primer Sequence.
| Sense Primers | Antisense Primers | Product Size (bp) | |
| IL-22 | TTCTCTTGGCCCTCTTGGTA | TTCTCCCCAATGAGACGAAC | 169 |
| IL-22RA1 | TGCTGACCATCTTGACTGTG | TCCCTCTCTCCGTACGTCTTAT | 181 |
| IL-22BP | GGAACTCAGTCAACGCATGA | TTGGCTTCTGGTGAGAGCTT | 178 |
| IL-10Rb | TACCACCTCCCGAAAATGTC | CAAATTCAGCCCTGACTCTCA | 219 |
| GAPDH | AAGGTCATCCCTGAGCTGAA | CCCTGTTGCTGTAGCCAAAT | 316 |
Cell Growth Assay
For the MTS assay (Promega, Madison, WI), 5000 cells were seeded in 96-well culture plates and treated daily with recombinant IL-22 up to 6 days. Cell proliferation was then measured colorimetrically at 450 nm using a Microplate reader (BioRad, Hercules, CA), and absorbance values were normalized using the Microplate Manager 5.2.1 (BioRad).
MCL Tumors and Immunohistochemistry
All MCL primary tumors were diagnosed at the Cross Cancer Institute, and the diagnostic criteria were based on those described in the World Health Organization Classification Scheme [1]. All cases were confirmed to express cyclin D1 by immunohistochemistry. The use of these tissues has been approved by our institutional ethics committee. Immunohistochemical staining was performed using standard techniques. Briefly, formalin-fixed, paraffin-embedded tissue sections of 4-µm thickness were deparaffinized and hydrated. Heat-induced epitope retrieval was performed using Tris buffer (pH 9.9; Dako) and a rapid microwave histoprocessor (RHS; Milestone, Bergamo, Italy). After incubating at 100°C for 10 minutes, slides were washed in running tap water for 5 minutes, and endogenous peroxidase was blocked using 10% H2O2 and methanol followed by washing in running tap water. Tissue sections were then incubated with anti-IL-22RA1 (1:500; Capralogics, Hardwick, MA) overnight in a humidified chamber at 4°C. After three washes with PBS, tissue sections were incubated with anti-rabbit IgG (EnVision, Dako) for 30 minutes at room temperature. The tissue sections were incubated with 3,3′-diaminobenzidine/H2O2 (Dako) for color development, using hematoxylin as a counterstain. For IL-22RA1, liver tissues were used as a positive control, and benign tonsils were used as a negative control.
Statistical Analysis
All the experiments were performed in triplicate. Student's t test was used. P < .05 was considered to be statistically significant.
Results
Expression and Subcellular Localization of IL-22RA1 in MCL Cell Lines and Tumors
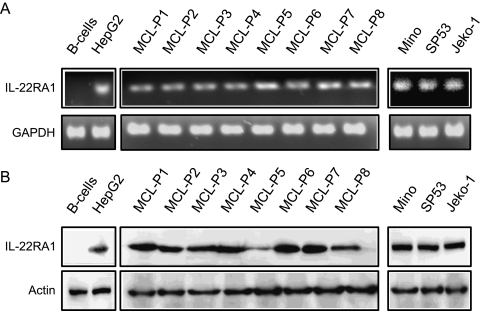
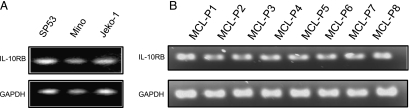
We first demonstrated the expression of IL-22RA1 using RT-PCR. RNA extracted from eight MCL frozen tumors and three cell lines were used, and the results are illustrated in Figure 1A. Amplifiable IL-22RA1 signal was detectable in all eight tumors (labeled MCL-P1 to MCL-P8) and in all three cell lines examined (Mino, SP53, and Jeko-1). HepG2 (a hepatocarcinoma cell line) and benign B cells served as the positive and negative controls, respectively. The PCR products were sequenced and confirmed to be full-length IL-22RA1 (not shown).
Figure 1.
The expression of IL-22RA1 in MCL cell lines and patient tumors. RT-PCR studies demonstrated the presence of IL-22RA1 messenger RNA (mRNA) in all three MCL cell lines and in the eight MCL patient samples (A). The HepG2 cell line was included as a positive control for IL-22RA1, and normal B cells was included as a negative control. GAPDH was included as an internal control. Western blot analysis confirmed the protein expression of IL-22RA1 in all three MCL cell lines and the eight MCL patient samples (B). HepG2 cells were used as a positive control for IL-22RA1, and normal human B cells were used as a negative control (B).
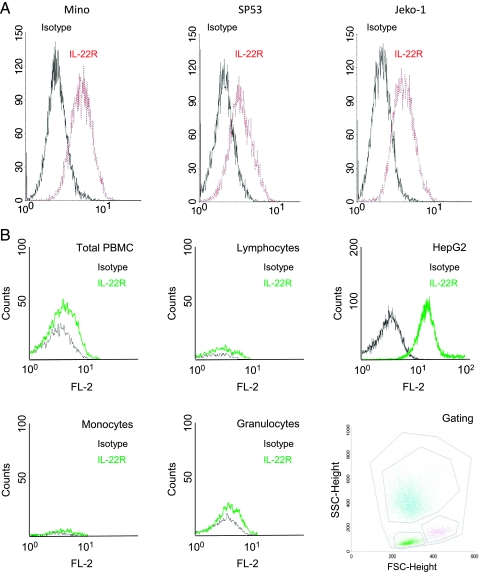
We then detected the protein expression of IL-22RA1 in MCL cells using Western blot. As shown in Figure 1B, IL-22RA1 protein was detectable in the same eight MCL tumors and three MCL cell lines used for Figure 1A. HepG2 and benign peripheral blood B cells served as the positive and negative controls, respectively. As shown in Figure 2A, the expression of IL-22RA1 on the surface of MCL cells was detectable by flow cytometry; specifically, all three MCL cell lines showed a significant shift to the right compared with the isotype controls. Triplicate experiments were performed, and results from a representative experiment are shown. In contrast, peripheral blood mononuclear cells, as well as the gated lymphocytes, monocytes, and granulocytes, revealed no appreciable shift compared with the isotype controls (Figure 2B). HepG2 cells served as the positive controls in these experiments.
Figure 2.
Flow cytometric detection of IL-22RA1 in MCL cell lines. (A) Three MCL cell lines were subjected to flow cytometry analysis with a specific anti-IL-22RA1 antibody. All three cell lines were shown to express IL-22RA1. The black lines correspond to the isotype control and the red lines illustrate the IL-22RA1 reactivity. (B) Flow cytometry analysis was also performed using the peripheral blood mononuclear cells from a healthy donor. The black lines correspond to the isotype control, and the green lines illustrate IL-22RA1 reactivity. Normal peripheral blood mononuclear cells (total) as well as specific subsets of cells (including lymphocytes, monocytes, and granulocytes) were negative for IL-22RA1. The positive control (HepG2 cell line) showed a strong signal. The gating strategy is illustrated in the right lower panel.
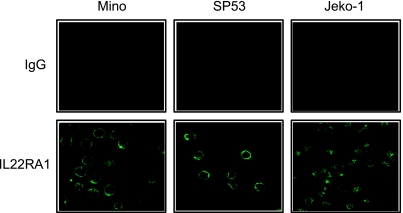
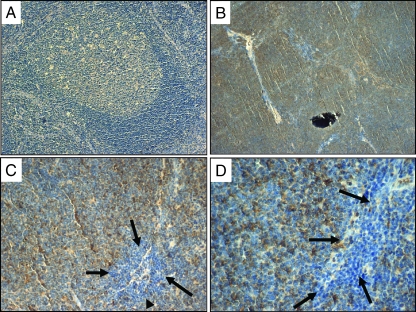
We then assessed the subcellular localization of IL-22RA1 using immunofluorescence staining and confocal microscopy. As shown in Figure 3, IL-22RA1 was found localized to the cell membrane of MCL cells. The isotype control revealed no detectable signal. Using immunohistochemistry applied to paraffin-embedded MCL tumors (n = 10), we were able to demonstrate membranous staining of IL-22RA1 in all cases (Figure 4, B–D). In contrast, reactive tonsilar lymphoid tissues including the mantle zone were negative (Figure 4A).
Figure 3.
Subcellular localization of IL-22RA1 in MCL cell lines. Immunofluorescence staining using antibodies directed against IL-22RA1 (lower panel) or control IgG (upper panel) was assessed by confocal microscopy. The results revealed a membranous staining pattern for IL-22RA1 in the three MCL cell lines, Mino, SP53, and Jeko-1.
Figure 4.
Detection of IL-22RA1 by immunohistochemistry in MCL tumors. Immunohistochemical staining of IL-22RA1 using paraffin-embedded tissue sections. (A) Benign lymphoid tissues in reactive tonsil showed no reactivity in the mantle zones and a faint background staining in the germinal centers (magnification, x200). (B) Neoplastic lymphoid nodules in a case of MCL showed definitive reactivity with anti-IL-22RA1 (magnification, x40). (C and D) The neoplastic nodules in a case of MCL showed reactivity with anti-IL-22RA1, whereas the benign infiltrating lymphocytes adjacent to the neoplastic nodules (as highlighted by the black arrows) were negative (magnifications, x200 and x400, respectively).
Expression of IL-22RB in MCL Cell Lines and Tumors
Using RT-PCR, we then assessed the expression of IL-10RB, the other subunit of the IL-22 receptor, in MCL cell lines and patient samples. As shown in Figure 5, amplifiable IL-10RB signals were detected in all cell lines and patient samples examined. The PCR products were sequenced and confirmed to be full-length IL-10RB.
Figure 5.
IL-10RB expression in MCL cell lines and tumors. RT-PCR to detect the expression of IL-10RB was performed using a specific set of primer (Table 1). Amplifiable IL-10RB signals were detectable in all three MCL cell lines (A) and five MCL tumor samples (B).
IL-22 Expression in MCL Cell Lines and Tumors
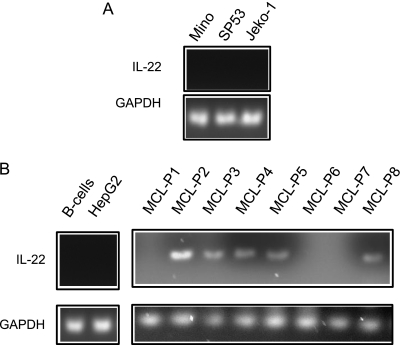
Using RT-PCR, we assessed if IL-22 is produced by MCL cells. As shown in Figure 6A, no IL-22 signal was detectable in all three MCL cell lines. In contrast, of the eight MCL tumor samples, amplifiable IL-22 signal was detectable in five (63%) cases. Both benign peripheral blood B cells and HepG2 were negative (Figure 6B).
Figure 6.
IL-22 expression in MCL. RT-PCR studies demonstrated the presence of IL-22 mRNA in MCL tumors. HepG2 cells were included as a negative control and GAPDH was included as an internal control. MCL cell lines are negative for IL-22 (A). In eight MCL tumor samples, five are positive for IL-22 mRNA (B). In contrast, HepG2 cells and normal B cells were negative.
IL-22 Promotes Cell Growth and Activates STAT3 in MCL Cells
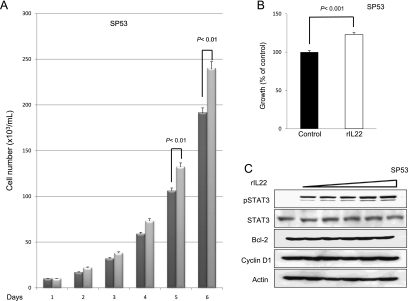
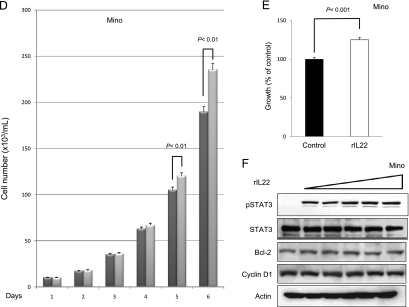
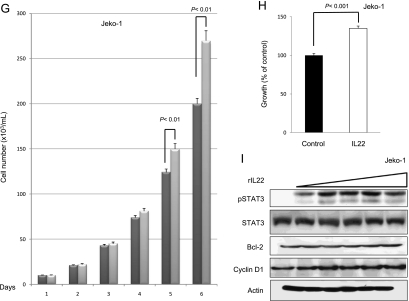
To assess if the IL-22 signaling pathway is functionally intact and if it exerts any significant biological effects in MCL cells, we added rIL-22 to the cell culture of all three MCL cell lines (i.e., with 20 ng/ml daily for up to 6 days). Daily cell counts were performed using Trypan blue exclusion test. A significant increase in the number of viable cells was observed on days 5 and 6 in all three cell lines (Figure 7, A, D, and G). To further support this finding, we performed MTS assay under the same experimental condition; cells treated with rIL-22 had a significant higher viable cell number than those without rIL-22 on day 5 (Figure 7, B, E, and H).
Figure 7.
Addition of rIL-22 increased the number of viable cells. Treatment of three MCL cell lines with rIL-22 (20 ng/ml daily) induced a significant increase in the number of viable cells, as revealed by the Trypan blue exclusion assay (A, D, and G). Four independent experiments were performed. Error bars, SDs with the indicated P value. Using the MTS assay, we found that the rIL-22 treatment of MCL cells induced a significant increase in the number of viable cells (B, E, and H). Six independent experiments were performed. Western blot studies revealed that treatment with rIL-22 (0, 5, 10, 20, 40, and 80 ng/ml) for 30 minutes increased the level of pSTAT3 in all three MCL cell lines (C, F, and I).
Because rIL-22 has been reported to activate various cell signaling pathways, including that of STAT3, in a number of nonhematologic cell lines [18,28,44,45], we investigated if rIL-22 also contributes to STAT3 activation in MCL cells. We found that MCL cells treated with increasing concentrations (from 5 to 80 ng/ml) of rIL-22 for 30 minutes showed a dose-dependent increase in the level of pSTAT3. The expression levels of cyclin D1 and Bcl-2 were unchanged after 30 minutes of incubation with rIL-22.
IL-22RA1 Expression Can Be Partly Attributed to NF-κB Activation in MCL
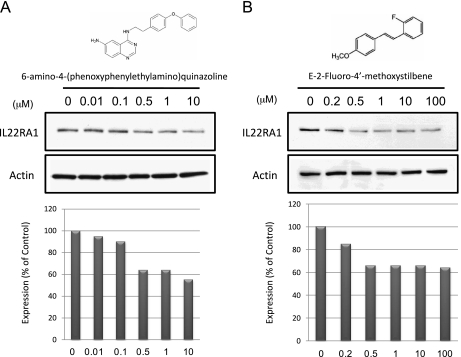
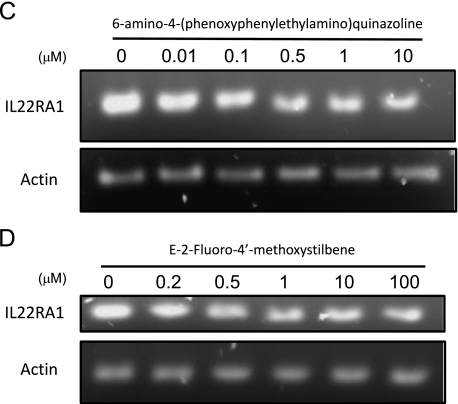
To investigate the mechanism underlying the aberrant expression of IL-22RA1 in MCL, we analyzed the gene promoter of the IL-22RA1 gene. Gene sequence analysis revealed that the IL-22RA1 gene promoter contains three consensus binding sequences for NF-κB (illustrated in Table 2). Because NF-κB has been shown to be biologically important for MCL [46,47], we hypothesized that NF-κB may contribute to the aberrant expression of IL-22RA1 in this cell type. Thus, we treated MCL cells lines with two highly specific inhibitors of NF-κB, including 6-amino-4-(phenoxyphenylethylamino) quinazoline [48–52] and E-2-fluoro-4′-methoxystilbene [53]. As shown in Figure 8, treatment of Mino cells with these two NF-κB inhibitors for 24 hours induced a dose-dependent decrease in the protein expression of IL-22RA1. In this period, no evidence of cell death, as assessed by Trypan blue exclusion test, was noted.
Table 2.
IL-22RA1 Human Promoter.
 |
▭ NF-κB consensus site.
Figure 8.
IL-22RA1 expression is dependent on NF-κB activation. MCL Mino cells were treated with two potent NF-κB inhibitors, including 6-amino-4-(phenoxyphenylethylamino)quinazoline (A and C) and E-2-fluoro-4′-methoxystilbene (B and D). Inhibition of NF-κB activation induced a dose-dependent decrease in the level of IL-22RA1 protein expression (A and B) and transcript expression (C and D). Quantification of the IL-22RA1 protein expression is summarized in the lower panel for each inhibitor.
Discussion
Recent studies have provided insights into the biology of MCL. In addition to the overexpression of cyclin D1, a large number of biochemical and genetic abnormalities have been identified in these neoplasms, such as the constitutive activation of various signaling pathways including those of STAT3 [17], Wnt [16], and NF-κB [46,47]. Our results have provided evidence that IL-22RA1 is consistently expressed in MCL cell lines and tumors. This conclusion is based on the results of three different techniques, including RT-PCR, Western blots, and confocal imaging. By contrast, IL-22RA1 is not detectable in benign B cells. To our knowledge, this represents the first report in which the aberrant expression of IL-22RA1 in a B-cell lymphoma is described.
We found that the IL-22 signaling is functional and biologically significant in MCL cells. Specifically, the addition of rIL-22 activates STAT3 and significantly promotes cell growth in all three MCL cell lines. Our observations that rIL-22 promotes cell growth in MCL correlates well with the previous reports that IL-22 promotes cell proliferation and antiapoptotic effects in epithelial cell lines [39,54]. Our finding that rIL-22 activates STAT3 in MCL cells also correlates well with the findings in various epithelial cell lines [41,45].
Regarding the source of IL-22, we found that most (63%) of our cohort of MCL frozen tumors had detectable IL-22 expression, whereas all three MCL cell lines did not produce detectable IL-22. This finding indicates that IL-22 stimulation of MCL cells exists in vivo in a subset of cases. Because IL-22 is normally produced by T cells, endothelial cells, and histiocytes [23,24,26], it is highly possible that these same cell types may also release IL-22 in the tumor microenvironment and promote the cell growth of MCL cells in vivo. Moreover, IL-22 also has been shown to be present in the peripheral blood of healthy people [37,55,56], and thus stimulation of MCL cell can be maintained while they are in the circulation.
We also investigated the mechanism by which IL-22RA1 is aberrantly expressed in MCL cells. In this study, we found that IL-22RA1 expression can be partly attributed to NF-κB signaling because inhibition of this pathway using two structurally different pharmacologic agents resulted in a dramatic reduction of IL-22RA1 protein expression. The specificity of these two compounds has been previously documented [48,53]. Of note, we previously investigated how IL-22RA1 is aberrantly expressed in another type of malignant lymphoma, namely ALK-positive anaplastic large cell lymphoma; we found that IL-22RA expression in these cells was related to the oncogenic fusion protein NPM-ALK that is characteristic of this type of lymphoma [42]. Taken together, the mechanisms leading to IL-22RA1 expression in malignant lymphoid cells are likely to be variable and/or multifactorial.
In summary, we have demonstrated for the first time that IL-22RA1 is aberrantly expressed in a B-cell lymphoma. Our data also has provided evidence that IL-22 stimulation is biologically important, by promoting the growth of MCL cells and activating the signaling of STAT3, a protein known to be oncogenic when inappropriately activated. Targeting IL-22RA1 and/or blocking the endogenous production of IL-22 may be a useful therapeutic strategy for MCL.
Footnotes
This study is supported by research grants from the Alberta Cancer Foundation and the Canadian Cancer Research Institute awarded to RL. PG and ZZ are research fellows supported by the Alberta Cancer Foundation. MA is a clinical research fellow supported by the Canadian Cancer Society Research Institute.
References
- 1.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. Classification of Tumours of Haematopoietic and Lymphoid Tissues, Fourth Edition. WHO Classification of Tumours, Volume 2. Geneva, Switzerland: WHO; 2008. pp. 229–232. [Google Scholar]
- 2.Ghielmini M, Zucca E. How I treat mantle cell lymphoma. Blood. 2009;114:1469–1476. doi: 10.1182/blood-2009-02-179739. [DOI] [PubMed] [Google Scholar]
- 3.Gill S, Ritchie D. Therapeutic options in mantle cell lymphoma. Leuk Lymphoma. 2008;49(3):398–409. doi: 10.1080/10428190701851364. [DOI] [PubMed] [Google Scholar]
- 4.Herrmann A, Hoster E, Zwingers T, Brittinger G, Engelhard M, Meusers P, Reiser M, Forstpointner R, Metzner B, Peter N, et al. Improvement of overall survival in advanced stage mantle cell lymphoma. J Clin Oncol. 2009;27(4):511–518. doi: 10.1200/JCO.2008.16.8435. [DOI] [PubMed] [Google Scholar]
- 5.Smith MR. Mantle cell lymphoma: advances in biology and therapy. Curr Opin Hematol. 2008;15(4):415–421. doi: 10.1097/MOH.0b013e328302c9c5. [DOI] [PubMed] [Google Scholar]
- 6.Gladden AB, Diehl JA. Location, location, location: the role of cyclin D1 nuclear localization in cancer. J Cell Biochem. 2005;96(5):906–913. doi: 10.1002/jcb.20613. [DOI] [PubMed] [Google Scholar]
- 7.Klier M, Anastasov N, Hermann A, Meindl T, Angermeier D, Raffeld M, Fend F, Quintanilla-Martinez L. Specific lentiviral shRNA-mediated knockdown of cyclin D1 in mantle cell lymphoma has minimal effects on cell survival and reveals a regulatory circuit with cyclin D2. Leukemia. 2008;22(11):2097–2105. doi: 10.1038/leu.2008.213. [DOI] [PubMed] [Google Scholar]
- 8.Vater I, Wagner F, Kreuz M, Berger H, Martin-Subero JI, Pott C, Martinez-Climent JA, Klapper W, Krause K, Dyer MJ, et al. GeneChip analyses point to novel pathogenetic mechanisms in mantle cell lymphoma. Br J Haematol. 2009;144(3):317–331. doi: 10.1111/j.1365-2141.2008.07443.x. [DOI] [PubMed] [Google Scholar]
- 9.Kawamata N, Ogawa S, Gueller S, Ross SH, Huynh T, Chen J, Chang A, Nabavi-Nouis S, Megrabian N, Siebert R, et al. Identified hidden genomic changes in mantle cell lymphoma using high-resolution single nucleotide polymorphism genomic array. Exp Hematol. 2009;37(8):937–946. doi: 10.1016/j.exphem.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schraders M, Jares P, Bea S, Schoenmakers EF, van Krieken JH, Campo E, Groenen PJ. Integrated genomic and expression profiling in mantle cell lymphoma: identification of gene-dosage regulated candidate genes. Br J Haematol. 2008;143(2):210–221. doi: 10.1111/j.1365-2141.2008.07334.x. [DOI] [PubMed] [Google Scholar]
- 11.Cecconi D, Zamo A, Bianchi E, Parisi A, Barbi S, Milli A, Rinalducci S, Rosenwald A, Hartmann E, Zolla L, et al. Signal transduction pathways of mantle cell lymphoma: a phosphoproteome-based study. Proteomics. 2008;8(21):4495–4506. doi: 10.1002/pmic.200800080. [DOI] [PubMed] [Google Scholar]
- 12.Henrickson SE, Hartmann EM, Ott G, Rosenwald A. Gene expression profiling in malignant lymphomas. Adv Exp Med Biol. 2007;593:134–146. doi: 10.1007/978-0-387-39978-2_13. [DOI] [PubMed] [Google Scholar]
- 13.Camps J, Salaverria I, Garcia MJ, Prat E, Bea S, Pole JC, Hernandez L, Del Rey J, Cigudosa JC, Bernues M, et al. Genomic imbalances and patterns of karyotypic variability in mantle-cell lymphoma cell lines. Leuk Res. 2006;30(8):923–934. doi: 10.1016/j.leukres.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Rizzatti EG, Falcao RP, Panepucci RA, Proto-Siqueira R, Anselmo-Lima WT, Okamoto OK, Zago MA. Gene expression profiling of mantle cell lymphoma cells reveals aberrant expression of genes from the PI3K-AKT, WNT and TGFbeta signalling pathways. Br J Haematol. 2005;130(4):516–526. doi: 10.1111/j.1365-2141.2005.05630.x. [DOI] [PubMed] [Google Scholar]
- 15.Ghobrial IM, McCormick DJ, Kaufmann SH, Leontovich AA, Loegering DA, Dai NT, Krajnik KL, Stenson MJ, Melhem MF, Novak AJ, et al. Proteomic analysis of mantle-cell lymphoma by protein microarray. Blood. 2005;105(9):3722–3730. doi: 10.1182/blood-2004-10-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelebart P, Anand M, Armanious H, Peters AC, Dien Bard J, Amin HM, Lai R. Constitutive activation of the Wnt canonical pathway in mantle cell lymphoma. Blood. 2008;112(13):5171–5179. doi: 10.1182/blood-2008-02-139212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai R, Rassidakis GZ, Medeiros LJ, Leventaki V, Keating M, McDonnell TJ. Expression of STAT3 and its phosphorylated forms in mantle cell lymphoma cell lines and tumours. J Pathol. 2003;199(1):84–89. doi: 10.1002/path.1253. [DOI] [PubMed] [Google Scholar]
- 18.Dumoutier L, Louahed J, Renauld JC. Cloning and characterization of IL-10-related T cell-derived inducible factor (IL-TIF), a novel cytokine structurally related to IL-10 and inducible by IL-9. J Immunol. 2000;164(4):1814–1819. doi: 10.4049/jimmunol.164.4.1814. [DOI] [PubMed] [Google Scholar]
- 19.Dumoutier L, Van Roost E, Ameye G, Michaux L, Renauld JC. IL-TIF/IL-22: genomic organization and mapping of the human and mouse genes. Genes Immun. 2000;1(8):488–494. doi: 10.1038/sj.gene.6363716. [DOI] [PubMed] [Google Scholar]
- 20.Commins S, Steinke JW, Borish L. The extended IL-10 superfamily: IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, IL-28, and IL-29. J Allergy Clin Immunol. 2008;121(5):1108–1111. doi: 10.1016/j.jaci.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 21.Chung Y, Yang X, Chang SH, Ma L, Tian Q, Dong C. Expression and regulation of IL-22 in the IL-17-producing CD4+ T lymphocytes. Cell Res. 2006;16(11):902–907. doi: 10.1038/sj.cr.7310106. [DOI] [PubMed] [Google Scholar]
- 22.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203(10):2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, Doherty JM, Mills JC, Colonna M. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457(7230):722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nograles KE, Zaba LC, Shemer A, Fuentes-Duculan J, Cardinale I, Kikuchi T, Ramon M, Bergman R, Krueger JG, Guttman-Yassky E. IL-22-producing “T22” T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J Allergy Clin Immunol. 2009;123(6):1244–1252. doi: 10.1016/j.jaci.2009.03.041. e1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricciardi L, Minciullo PL, Saitta S, Trombetta D, Saija A, Gangemi S. Increased serum levels of IL-22 in patients with nickel contact dermatitis. Contact Dermatitis. 2009;60(1):57–58. doi: 10.1111/j.1600-0536.2008.01454.x. [DOI] [PubMed] [Google Scholar]
- 26.Vivier E, Spits H, Cupedo T. Interleukin-22-producing innate immune cells: new players in mucosal immunity and tissue repair? Nat Rev Immunol. 2009;9(4):229–234. doi: 10.1038/nri2522. [DOI] [PubMed] [Google Scholar]
- 27.Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat Immunol. 2009;10(8):864–871. doi: 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- 28.Xie MH, Aggarwal S, Ho WH, Foster J, Zhang Z, Stinson J, Wood WI, Goddard AD, Gurney AL. Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2-4 and IL-22R. J Biol Chem. 2000;275(40):31335–31339. doi: 10.1074/jbc.M005304200. [DOI] [PubMed] [Google Scholar]
- 29.Bleicher L, de Moura PR, Watanabe L, Colau D, Dumoutier L, Renauld JC, Polikarpov I. Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism. FEBS Lett. 2008;582(20):2985–2992. doi: 10.1016/j.febslet.2008.07.046. [DOI] [PubMed] [Google Scholar]
- 30.de Oliveira Neto M, Ferreira JR, Jr, Colau D, Fischer H, Nascimento AS, Craievich AF, Dumoutier L, Renauld JC, Polikarpov I. Interleukin-22 forms dimers that are recognized by two interleukin-22R1 receptor chains. Biophys J. 2008;94(5):1754–1765. doi: 10.1529/biophysj.107.112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones BC, Logsdon NJ, Walter MR. Structure of IL-22 bound to its high-affinity IL-22R1 chain. Structure. 2008;16(9):1333–1344. doi: 10.1016/j.str.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shioya M, Andoh A, Kakinoki S, Nishida A, Fujiyama Y. Interleukin 22 receptor 1 expression in pancreas islets. Pancreas. 2008;36(2):197–199. doi: 10.1097/MPA.0b013e3181594258. [DOI] [PubMed] [Google Scholar]
- 33.Ramanathan M, Jr, Spannhake EW, Lane AP. Chronic rhinosinusitis with nasal polyps is associated with decreased expression of mucosal interleukin 22 receptor. Laryngoscope. 2007;117(10):1839–1843. doi: 10.1097/MLG.0b013e31811edd4f. [DOI] [PubMed] [Google Scholar]
- 34.Brand S, Dambacher J, Beigel F, Zitzmann K, Heeg MH, Weiss TS, Prufer T, Olszak T, Steib CJ, Storr M, et al. IL-22-mediated liver cell regeneration is abrogated by SOCS-1/3 overexpression in vitro. Am J Physiol Gastrointest Liver Physiol. 2007;292(4):G1019–G1028. doi: 10.1152/ajpgi.00239.2006. [DOI] [PubMed] [Google Scholar]
- 35.Nagalakshmi ML, Rascle A, Zurawski S, Menon S, de Waal Malefyt R. Interleukin-22 activates STAT3 and induces IL-10 by colon epithelial cells. Int Immunopharmacol. 2004;4(5):679–691. doi: 10.1016/j.intimp.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Lecart S, Morel F, Noraz N, Pene J, Garcia M, Boniface K, Lecron JC, Yssel H. IL-22, in contrast to IL-10, does not induce Ig production, due to absence of a functional IL-22 receptor on activated human B cells. Int Immunol. 2002;14(11):1351–1356. doi: 10.1093/intimm/dxf096. [DOI] [PubMed] [Google Scholar]
- 37.Ratsep R, Kingo K, Karelson M, Reimann E, Raud K, Silm H, Vasar E, Koks S. Gene expression study of IL10 family genes in vitiligo skin biopsies, peripheral blood mononuclear cells and sera. Br J Dermatol. 2008;159(6):1275–1281. doi: 10.1111/j.1365-2133.2008.08785.x. [DOI] [PubMed] [Google Scholar]
- 38.Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206(7):1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Chen Y, Wei H, Zheng C, Sun R, Zhang J, Tian Z. Antiapoptotic activity of autocrine interleukin-22 and therapeutic effects of interleukin-22-small interfering RNA on human lung cancer xenografts. Clin Cancer Res. 2008;14(20):6432–6439. doi: 10.1158/1078-0432.CCR-07-4401. [DOI] [PubMed] [Google Scholar]
- 40.Ziesche E, Bachmann M, Kleinert H, Pfeilschifter J, Muhl H. The interleukin-22/STAT3 pathway potentiates expression of inducible nitric-oxide synthase in human colon carcinoma cells. J Biol Chem. 2007;282(22):16006–16015. doi: 10.1074/jbc.M611040200. [DOI] [PubMed] [Google Scholar]
- 41.Weber GF, Gaertner FC, Erl W, Janssen KP, Blechert B, Holzmann B, Weighardt H, Essler M. IL-22-mediated tumor growth reduction correlates with inhibition of ERK1/2 and AKT phosphorylation and induction of cell cycle arrest in the G2-M phase. J Immunol. 2006;177(11):8266–8272. doi: 10.4049/jimmunol.177.11.8266. [DOI] [PubMed] [Google Scholar]
- 42.Bard JD, Gelebart P, Anand M, Amin HM, Lai R. Aberrant expression of IL-22 receptor 1 and autocrine IL-22 stimulation contribute to tumorigenecity in ALK+ anaplastic large cell lymphoma. Leukemia. 2008;22:1595–1603. doi: 10.1038/leu.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amin HM, McDonnell TJ, Medeiros LJ, Rassidakis GZ, Leventaki V, O'Connor SL, Keating MJ, Lai R. Characterization of 4 mantle cell lymphoma cell lines. Arch Pathol Lab Med. 2003;127(4):424–431. doi: 10.5858/2003-127-0424-COMCLC. [DOI] [PubMed] [Google Scholar]
- 44.Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheets H, Donnelly RP, Pestka S. Identification of the functional interleukin-22 (IL-22) receptor complex: the IL-10R2 chain (IL-10Rbeta) is a common chain of both the IL-10 and IL-22 (IL-10-related T cell-derived inducible factor, IL-TIF) receptor complexes. J Biol Chem. 2001;276(4):2725–2732. doi: 10.1074/jbc.M007837200. [DOI] [PubMed] [Google Scholar]
- 45.Lejeune D, Dumoutier L, Constantinescu S, Kruijer W, Schuringa JJ, Renauld JC. Interleukin-22 (IL-22) activates the JAK/STAT, ERK, JNK, and p38 MAP kinase pathways in a rat hepatoma cell line. Pathways that are shared with and distinct from IL-10. J Biol Chem. 2002;277(37):33676–33682. doi: 10.1074/jbc.M204204200. [DOI] [PubMed] [Google Scholar]
- 46.Shishodia S, Amin HM, Lai R, Aggarwal BB. Curcumin (diferuloylmethane) inhibits constitutive NF-kappaB activation, induces G1/S arrest, suppresses proliferation, and induces apoptosis in mantle cell lymphoma. Biochem Pharmacol. 2005;70(5):700–713. doi: 10.1016/j.bcp.2005.04.043. [DOI] [PubMed] [Google Scholar]
- 47.Roue G, Perez-Galan P, Lopez-Guerra M, Villamor N, Campo E, Colomer D. Selective inhibition of IkappaB kinase sensitizes mantle cell lymphoma B cells to TRAIL by decreasing cellular FLIP level. J Immunol. 2007;178(3):1923–1930. doi: 10.4049/jimmunol.178.3.1923. [DOI] [PubMed] [Google Scholar]
- 48.Choi S, Kim JH, Roh EJ, Ko MJ, Jung JE, Kim HJ. Nuclear factor-kappaB activated by capacitative Ca2+ entry enhances muscarinic receptor-mediated soluble amyloid precursor protein (sAPPalpha) release in SH-SY5Y cells. J Biol Chem. 2006;281(18):12722–12728. doi: 10.1074/jbc.M601018200. [DOI] [PubMed] [Google Scholar]
- 49.Curry CL, Reed LL, Broude E, Golde TE, Miele L, Foreman KE. Notch inhibition in Kaposi's sarcoma tumor cells leads to mitotic catastrophe through nuclear factor-kappaB signaling. Mol Cancer Ther. 2007;6(7):1983–1992. doi: 10.1158/1535-7163.MCT-07-0093. [DOI] [PubMed] [Google Scholar]
- 50.Tobe M, Isobe Y, Tomizawa H, Nagasaki T, Takahashi H, Hayashi H. A novel structural class of potent inhibitors of NF-kappa B activation: structure-activity relationships and biological effects of 6-aminoquinazoline derivatives. Bioorg Med Chem. 2003;11(18):3869–3878. doi: 10.1016/s0968-0896(03)00438-3. [DOI] [PubMed] [Google Scholar]
- 51.Yang RH, Strong JA, Zhang JM. NF-kappaB mediated enhancement of potassium currents by the chemokine CXCL1/growth related oncogene in small diameter rat sensory neurons. Mol Pain. 2009;5(26) doi: 10.1186/1744-8069-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armstrong SJ, Wiberg M, Terenghi G, Kingham PJ. Laminin activates NF-kappaB in Schwann cells to enhance neurite outgrowth. Neurosci Lett. 2008;439(1):42–46. doi: 10.1016/j.neulet.2008.04.091. [DOI] [PubMed] [Google Scholar]
- 53.Heynekamp JJ, Weber WM, Hunsaker LA, Gonzales AM, Orlando RA, Deck LM, Jagt DL. Substituted trans-stilbenes, including analogues of the natural product resveratrol, inhibit the human tumor necrosis factor alpha-induced activation of transcription factor nuclear factor kappaB. J Med Chem. 2006;49(24):7182–7189. doi: 10.1021/jm060630x. [DOI] [PubMed] [Google Scholar]
- 54.Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, Diepolder H, Marquardt A, Jagla W, Popp A, et al. IL-22 is increased in active Crohn's disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am J Physiol Gastrointest Liver Physiol. 2006;290(4):G827–G838. doi: 10.1152/ajpgi.00513.2005. [DOI] [PubMed] [Google Scholar]
- 55.Pan HF, Zhao XF, Yuan H, Zhang WH, Li XP, Wang GH, Wu GC, Tang XW, Li WX, Li LH, et al. Decreased serum IL-22 levels in patients with systemic lupus erythematosus. Clin Chim Acta. 2009;401(1–2):179–180. doi: 10.1016/j.cca.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 56.Cheng F, Guo Z, Xu H, Yan D, Li Q. Decreased plasma IL22 levels, but not increased IL17 and IL23 levels, correlate with disease activity in patients with systemic lupus erythematosus. Ann Rheum Dis. 2009;68(4):604–606. doi: 10.1136/ard.2008.097089. [DOI] [PubMed] [Google Scholar]