Abstract
The ability of small interfering RNA (siRNA) to efficiently silence the expression of specific genes provides the basis for exciting new therapies based on RNA interference (RNAi). The efficient intracellular delivery of siRNA from cell uptake through the endosomal trafficking pathways into the cytoplasm remains a significant challenge. Previously we described the synthesis of a new family of diblock copolymer siRNA carriers using controlled reversible addition fragmentation chain transfer (RAFT) polymerization. The carriers were composed of a positively-charged block of dimethylaminoethyl methacrylate (DMAEMA) to mediate siRNA binding and a second pH-responsive endosome releasing block composed of DMAEMA and propylacrylic acid (PAA) in roughly equimolar ratios, and butyl methacylate (BMA). Here we describe the development of a new generation of siRNA delivery polymers based on this design that exhibit enhanced transfection efficiency and low cytotoxicity. This design incorporates a longer endosomolytic block with increased hydrophobic content to induce micelle formation. These polymers spontaneously form spherical micelles in the size range of 40 nm with CMC (critical micelle concentration) values of approximately 2 μg/ml based on dynamic light scattering (DLS), 1H-NMR, electron microscopy, and selective partitioning of the small molecule pyrene into the hydrophobic micelle core. The siRNA binding to the cationic shell block did not perturb micelle stability or significantly increase particle size. The self-assembly of the diblock copolymers into particles was shown to provide a significant enhancement in mRNA knockdown at siRNA concentrations as low as 12.5 nM. Under these conditions the micelle--based systems showed an 89 % reduction in GAPDH mRNA levels as compared to only 23 % (10 nM siRNA) for the non-micelle system. The reduction in mRNA levels becomes nearly quantitative as the siRNA concentration is increased to 25 nM and higher. Flow cytometry analysis of fluorescent-labeled siRNA showed uptake in 90% of cells and a 3-fold increase in siRNA per cell compared to a standard lipid transfection agent. These results demonstrate the potential utility of this carrier design for siRNA drug delivery.
Keywords: micelle, pH responsive polymer, nanoparticle, siRNA, RNA interference, gene silencing, RAFT polymerization, propylacryilic acid, dimethyaminoethyl methacrylate, endosome release
Introduction
The discovery that short interfering RNA (siRNA) can efficiently silence specific genes and inhibit protein expression by selective degradation of mRNA could provide the basis for revolutionary new treatments based on RNA inference (RNAi). 1,2 Effective intracellular delivery of siRNA, however, remains a significant obstacle which must be overcome before clinical therapeutics based on RNAi can be realized. 3–6 Cellular uptake of siRNA typically occurs by passive or receptor-mediated endocytosis where the predominant fate of internalized siRNA is endosomal localization and ultimately enzymatic degradation in the lysosome or extracellular clearance. A number of natural and synthetic strategies have been adopted to mediate intracellular nucleic acid delivery. For example, viral vectors have been employed to deliver plasmid DNA through the endosomal pathway into the cellular cytoplasm. 7 These vectors mediate endosomal escape by incorporating fusogenic proteins on their viral coat that undergo a pH-induced conformational change from hydrophilic at physiological pH to hydrophobic in response to the acidic endosomal environment.8–10 Synthetic peptides that mimic the behavior of fusogenic proteins have also been shown to enhance cytoplasmic delivery.11 Despite the endosomolytic activity of such peptides, their potential toxicity and immunogenicity has thus far limited their clinical utility. Synthetic lipids and polycations such as PEI and polylysine have been employed to facilitate the intracellular delivery of bound therapeutics. 12–18
In order to overcome these delivery challenges, we have focused on the design and synthesis of pH-responsive, membrane-destabilizing polymers. 19,20 These polymers respond to changes in pH by transitioning from an ionized, hydrophilic structure at physiologic pH (~7.4) to a hydrophobic, membrane-destabilizing conformation at endosomal pH values (<6.6). Poly(propylacrylic acid) (PPAA) in particular has been shown to be effective at endosomal release resulting in the delivery of different types of biological molecules to the cytoplasmic compartment.20–22 We have shown that conjugation of PPAA to model antibody targeted proteins significantly enhances cytoplasmic delivery in Jurkat T-cells.18 Subsequent studies have examined the effect of copolymerizing hydrophobic monomers with PAA.22 In these studies a large increase in the concentration dependent hemolysis was observed by incorporating 20 % butyl acrylate into copolymers of PAA. Polymer synthesis was carried out using reversible addition-fragmentation chain transfer (RAFT) polymerization,23,24 which provided good control over molecular weight distribution and composition. Multiple applications to siRNA delivery have been described. 25–30
Recently we have shown that polymers containing a mixture of positive, negative, and hydrophobic residues exhibit significant pH-responsive hemolysis.29 This system, which was designed to facilitate siRNA delivery, is a diblock copolymer consisting of a cationic poly(N,N-dimethylaminoethyl methacrylate) (polyDMAEMA) block to mediate siRNA binding and a second pH-responsive membrane disruptive block. This hydrophobic block is a mixture of positively charged DMAEMA residues, negatively charged proplylacrylic acid (PAA) residues, and hydrophobic butyl methacrylate (BMA) residues. This mixture of positive and negative charged groups throughout the second block reduces the overall net charge of the polymer at physiological pH. Upon acidification of endosomal compartments protonation of the carboxyl residues results in a more hydrophobic block with a net positive charge. This shift allows the polymer to interact with endosomal membranes and has been shown to efficiently delivery siRNA to the cellular cytoplasm. These polymers become strongly hemolytic at endosomal pHs (5.8–6.6), with increasing hemolytic activity as the percentage of BMA in the second block is increased. As demonstrated in this report, these diblock copolymers can be induced to spontaneously form micelle-like particles by modification of the endosomolytic block size and composition. These particles show significantly enhanced in vitro mRNA knockdown and low cytotoxicity in the presence of serum.
1. Materials and Methods
2.1 Materials
Chemicals and all materials were supplied by Sigma-Aldrich unless otherwise specified. 2,2′-Azobis(4-methoxy-2.4-dimethyl valeronitrile) (V70) was obtained from Wako chemicals. PD10 desalting columns were obtained from GE life sciences. siRNA against glyceraldehyde phosphate dehydrogenase (GAPDH, #4624) and a negative control (#4611) were obtained from Ambion. SYBR Green II gel stain was purchased from Molecular Probes/Invitrogen. The colorimetric Cytotoxicity Detection Kit was acquired from Roche Applied Science. Gene specific primers against GAPDH, HPRT1 and RPL13A were ordered from Integrated DNA Technologies (IDT, Coralville, IA). Dimethylaminoethyl methacrylate (DMAEMA), butyl methacrylate (BMA), and propylacrylic acid (PAA) were distilled prior to use. HeLa cells, human cervical carcinoma cells (ATTC), were maintained in minimum essential media (MEM) containing L-glutamine (Gibco), 1% penicillin-streptomycin (Gibco), and 10% fetal bovine serum (FBS, Invitrogen) at 37 °C and 5% CO2.
2.2 Methods
Synthesis of poly(dimethylaminoethyl methacrylate) (polyDMAEMA) macro chain transfer agent (macroCTA). PolyDMAEMA was synthesized as previously described.29 Briefly DMAEMA (2.5 g, 15.9 mmol), 4-Cyano-4-(ethylsulfanylthiocarbonyl) sulfanylvpentanoic acid (ECT) (27.9 g, 0.106 mmol), and V70 (3.27 mg, 0.01 mmol) were dissolved in N,N-dimethylformamide (DMF) (5 g). The solution was purged with nitrogen for 30 minutes and then allowed to react at 30 °C for 18 h. The resultant polymer was isolated by repeated precipitation from ether into a 50x excess of pentane:ether (3:1 v/v). The final molecular weight and polydispersities were determined via SEC.
Synthesis of poly[(DMAEMA)-b-(BMA)-co-(DMAEMA)-co-(PAA)]. Poly[(DMAEMA)-b-(BMA)-co-(DMAEMA)-co-(PAA)] was prepared by adding the poly(DMAEMA) macroCTA (9,100 g/mol) to a solution of BMA, DMAEMA and PAA (40:30:30 mol %) in DMF such that the final solvent concentration was 60 % by weight. The initial macroCTA to initiator ratio ([macroCTA]o/[I]o) and initial monomer to macroCTA ([M]o/[macroCTA]o) was 5:1 and 250:1 respectively. The polymerization solution was purged with nitrogen for 30 minutes before being allowed to react at 30 °C for 24 hours. The final polymers were isolated by precipitation from ether into a 50x excess of pentane:ether (3:1 v/v). The solutions resultant oils were then redissolved in ether and precipitated into neat pentane (x5). Following precipitation, the polymers were dissolved in deionized water and further purified by passing them through PD10 desalting columns. The final dry polymers were obtained by lyophilization.
2.2.1 Size Exclusion Chromatography (SEC)
Absolute molecular weights and polydispersities (PDI) were determined via SEC laser light scattering (LLS) using a Viscotek GPCmax VE2001 (Viscotek, Houston, TX) equipped with light scattering, refractive index (VE3580), UV, and viscosity detectors connected in series to Tosoh TSK-GEL R-3000 and R-4000 columns (Tosoh Bioscience, Montgomeryville, PA). HPLC-grade DMF containing 0.1 wt.% LiBr at 60 °C was used as the mobile phase at a flow rate of 1 mL/min.
2.2.2 NMR spectroscopy
1H NMR spectra were recorded on a Bruker AV301 in deuterated chloroform (CDCl3) and deuterated water (D2O) at 25°C. A deuterium lock (CDCl3, D2O) was used and chemical shifts were determined in ppm from tetramethylsilane (for CDCl3) and 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid, sodium salt (for D2O). Polymer concentration was 6 mg/mL.
2.2.3 Dynamic light scattering
Particle sizes of polymer alone or polymer/siRNA complexes were measured by dynamic light scattering (DLS) using a Malvern Zetasizer Nano ZS. Lyophilized polymer was dissolved in 100 % ethanol at 10–50 mg/mL, then diluted 10-fold into phosphate buffer, pH 7.4. Polymers were analyzed in phosphate buffered saline, pH 7.4 (PBS) at 1 mg/mL for polymer alone or at 0.7 mg/mL polymer complexes to 1 μM GAPDH-specific 21 mer-siRNA with the indicated theoretical charge ratio (positive charges on polymer: negative charges on siRNA).
2.2.4. Gel Shift Assay
Gel retardation assays were conducted in order to determine the charge ratio (+/−) at which complete complexation of the polymer to the siRNA occurs. The charge ratio is the molar ratio of amines on the DMAEMA (50% are assumed charged) to phosphorus (in the form of negatively charged phosphate groups from the nucleic acid). A 2% agarose gel was loaded with each lane containing 100 ng of siRNA premixed with varying quantities of diblock copolymer. The gels were run at 60 volts for one hour. They were then stained with SYBR Safe dye diluted 1:5000 for 30 minutes before UV visualization.
2.2.5. Flow Cytometry
Intracellular uptake of siRNA/polymer complexes was measured using flow cytometry (Becton Dickinson LSR benchtop analyzer). HeLa cells were seeded at 15,000 cells/cm2 (6-well plates) and allowed to adhere overnight. FAM-labeled siRNA (Ambion) was complexed with polymer for 30 min at room temperature in PBS buffer and then added to the plated HeLa cells at a final siRNA concentration of 25 nM (1000 μL volume). After incubation with the complexes for 4 h, the cells were trypsinized and resuspended in PBS with 0.5% BSA and 0.01% trypan blue. Trypan blue was utilized as previously described for quenching of extracellular fluorescence and discrimination of complexes that have been endocytosed by cells.31 10,000 cells were analyzed per sample and fluorescence gating was determined using samples receiving no treatment and treated with polymer alone.
2.2.6. Cytotoxicity Measurements
Cytotoxicity of diblock copolymer and siRNA/polymer complex was determined by assaying for cell metabolic activity. HeLa cells were seeded in 96-well plates at a density of 8000 cells/cm2 and allowed to adhere over night. Complexes were formed by the addition of polymer (0.1 mg/mL stock solutions) to GAPDH siRNA (50 μM) to attain a concentration of 25 nM siRNA/well (100 μL volume). Complexes were added to wells in triplicate. After cells had been incubated for 24 h with the polymer/complexes, 10ul of Cell Titer Blue (Promega, Madison WI) reagent was added to each well. The cells were incubated at 37C for 120 min for color formation, the fluorescence (560/590nm) was determined according to the manufacturer’s instructions. Percent viability is expressed as function of 1% Triton X-100 treated cells (control for 0% viability). Figure S-1 (Supplementary Information), shows no loss in cell viability at the polymer-siRNA concentrations used in this study.
2.2.7 Transmission electron microscopy (TEM)
A 0.5 mg/ml solution of poly[(DMAEMA)-b-(BMA)-co-(DMAEMA)-co-(PAA)] in PBS was applied to a carbon coated copper grid for 30 minutes. The grid was fixed in Karnovsky’s solution and washed in cacodylate buffer once and then in water 8 times. The grid was stained with a 6% solution of uranyl acetate for 15 minutes and then dried until analysis. Transmission electron microscopy was carried out on a JEOL 1230 microscope, run at 80 KV.
2.2.8. Measurement of siRNA knock-down activity
Knock-down (KD) activity of siRNA/poly[(DMAEMA)-b-(BMA)-co-(DMAEMA)-co-(PAA)] complexes was assayed in 96-well format by measuring specific gene expression after 24 hours of treatment with polymer:siRNA complexes. Polymer and GAPDH targeting siRNA or negative control siRNA (Ambion) were mixed in 25 μL to obtain various charge ratios and concentrations at 5-fold over final transfection concentration and allowed to complex for 30 minutes before addition to HeLa cells in 100 μL normal media containing 10% FBS. Final siRNA concentrations were evaluated at 100, 50, 25, and 12.5 nM. Polymer was added either at 4:1, 2:1 or 1:1 charge ratios, or at fixed polymer concentrations of 18, 9, 4.5, and 2.2 μg/ml in PBS buffer to determine what conditions result in highest mRNA knockdown. For different charge ratios, the complexes were prepared at higher concentrations, incubated for 30 minutes, and then serial diluted at 5-fold over concentration shown on graphs just prior to addition to cells. For fixed polymer concentration, the siRNA and polymer were complexed at 5-fold over concentrations shown on the graph, incubated for 30 minutes then added to cells for final concentrations. Polymer-siRNA complexes were incubated overnight until assayed for knock down activity. Total RNA was isolated 24 hours post treatment and GAPDH expression was measured relative to 2 internal normalizer genes, RPL13A and HPRT, by quantitative PCR.
2.2.9. Red blood cell hemolysis assay
pH responsive membrane destabilizing activity was assayed by titrating polymer alone or polymer/siRNA complexes into preparations of human red blood cells (RBC) and determining membrane-lytic activity by hemoglobin release (determined by measuring absorbance at 540 nm) under three different pH conditions. Human red blood cells (RBC) were isolated by centrifugation from whole blood collected in vaccutainers containing EDTA. RBC were washed 3 times in normal saline, and brought to a final concentration of 2% RBC in PBS at a specific pH (5.8, 6.6 or 7.4). Polymer alone or polymer/siRNA complex was tested at concentrations just above and below the critical micelle concentration (CMC). For polymer/siRNA complex, 25 nM siRNA was added to polymer at 1:1, 2:1, 4:1 and 8:1 charge ratios (same polymer concentrations for polymer alone). Solutions of polymer alone or polymer/siRNA complexes were formed at 20 times final assayed concentration for 30 minutes and diluted into each RBC preparation. RBC with polymer alone or polymer/siRNA complex were incubated at 37°C for 60 minutes and centrifuged to remove intact RBC. Supernatants were transferred to cuvettes and absorbance determined at 540 nm. Percent hemolysis isexpressed as A540 sample/A540 of 1% Triton X-100 treated RBC (control for 100% lysis).
2.2.10 Determination of critical micelle concentration (CMC). Two complimentary methods were used
Pyrene binding assay
The formation of polymer micelles with or without siRNA was confirmed by a fluorescence probe technique using pyrene (C16H10, MW = 202), in which the partitioning of pyrene into the micelle core was determined using the ratio of 2 emission maxima of the pyrene spectrum. The fluorescence emission spectrum of pyrene in the polymer micelle solution was measured from 300 to 360 nm using a fixed excitation wavelength of 395 nm with a constant pyrene concentration of 6 × 10−7 M. The polymer was varied from 0.001% to 20% (w/w) with or without 100 nM siRNA. The spectral data were acquired using a Varian fluorescence spectrophotometer. All fluorescence experiments are carried out at 25°C. The critical micelle concentration (CMC) was determined by plotting the intensity ratio I336/I333 as a function of polymer concentration. CMC values were calculated from the low concentration break point to be approximately 2 ug/ml (see Figure S-1 in Supporting Information).
Dynamic light scattering assay
The stability of polymer micelles to incremental dilution was measured by dynamic light scattering (DLS). Particle size was measured by DLS over a 5-fold range of serial dilutions with PBS buffer from 1 mg/ml to 1.6 ug/ml. Particle sizes of about 45 nm were stable down to a concentration of about 10ug/ml with instability below about 5 ug/ml (the CMC) where individual polymer chains appear to form higher MW polydisperse aggregates and lower particle sizes including monomer chains. (see Figure S-2 in Supporting Information).
3. Results
Recently we described the development of a new class of diblock copolymers that are able to deliver siRNA from the endosomal pathway into the cellular cytoplasm.29 These polymers contain a poly(DMAEMA) cationic block for siRNA binding and a hydrophobic ampholyte block capable of mediating endosomal escape in response to endosomal acidification (Figure 1). This membrane interactive block contains PAA and DMAEMA, which are designed to change from an inactive conformation at physiological pH to a more hydrophobic membrane-interactive conformation in response to the endosomal pH drop. The addition of hydrophobic BMA residues increases the hydrophobicity of the copolymer, increasing the pKa of the PAA carboxylate residues, thus raising the pH at which protonation occurs. The optimal incorporation of hydrophobic BMA residues was found to be between 40 and 50 mol %. In all cases the molecular weight of both blocks was held constant at approximately 10 kDa. At polymer concentrations of between 0.1 and 10 mg/ml at pH=7.4, these materials did not form micelles or any higher order structures according to light scattering data. However, upon the addition of siRNA, particles with sizes between 85 and 236 nm were observed depending on the +/− charge ratio. Here we prepared a diblock copolymer with a hydrophobic endosomolytic second block approximately 2.5 times larger than the hydrophilic first block (Figure 1) in order to investigate the structural and biological impact of such a modification. The polyDMAEMA macroCTA was first prepared by polymerizing DMAEMA in the presence of the RAFT CTA and a radical initiator. The resultant polyDMAEMA block (Mn = 9,100 g/mol; PDI =1.19) was then used to prepare the poly[DMAEMA-b-(BMA-co-DMAEMA-co-PAA)] diblock copolymer (Mn = 31,000 g/mol; PDI = 1.57). Copolymer composition of the second block was determined to be 52 % BMA, 26 % DMAEMA, and 22 % PAA via a combination of 1H NMR and size exclusion chromatography.
Figure 1.
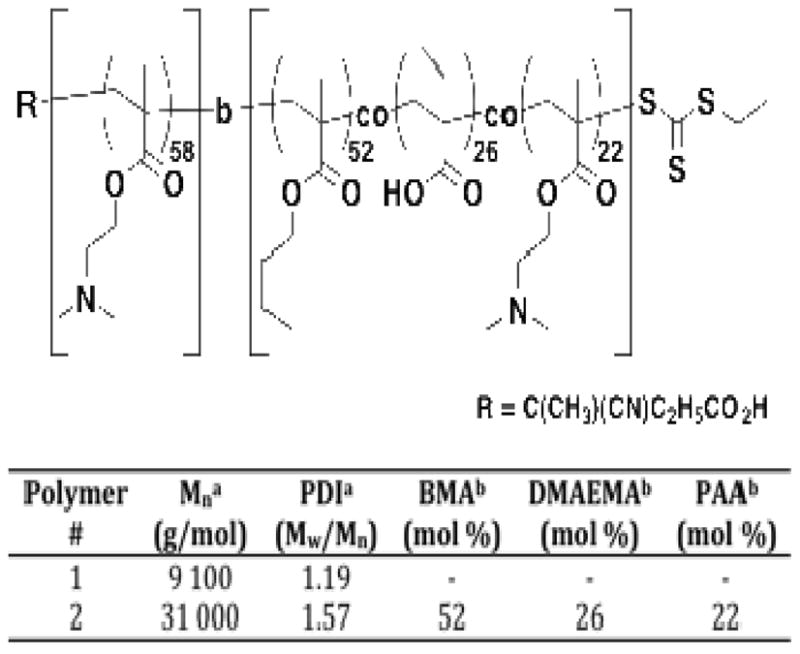
Synthetic structure, molecular weight, and chemical composition of the diblock copolymer siRNA carrier employed in these studies.
a. As determined by SEC Tosoh TSK-GEL R-3000 and R-4000 columns (Tosoh Bioscience, Montgomeryville, PA) connected in series to a Viscotek GPCmax VE2001 and refractometer.
VE3580 (Viscotek, Houston, TX). HPLC-grade DMF containing 0.1 wt.% LiBr was used as the mobile phase. The molecular weights of the synthesized copolymers were determined using a series of poly(methyl methacrylate) standards.
b. As determined by 1H NMR spectroscopy (3 wt.% in D2O or CDCl3; Bruker DRX 499).
3.1 Aqueous particle sizes and solution morphology of the diblock copolymer
Particle sizes for the diblock copolymer alone and in the presence of siRNA were investigated at various charge ratios using DLS. Lyophilized polymer was first dissolved in ethanol at a concentration of 10–50 mg/mL. The ethanolic solutions were then diluted 10-fold into phosphate buffer at pH of 7.4. Following dissolution, the diblock copolymers at a final aqueous concentration of 1 mg/mL spontaneously self assembled to form particles with average particle sizes of 45 nm. For comparison, no particles were observed for any of our previous diblock copolymer systems even at similar comonomer compositions. This result suggests that increasing the second block size and the ratio of the hydrophilic DMAEMA block to the endosome pH responsive block is sufficient to induce the formation of micelles. The addition of free siRNA to the micelle solutions did not significantly change the particle sizes over a range of +/− charge ratios. For example, at a +/− charge ratio of 4:1 the complexed particles showed hydrodynamic diameters of 47 nm as compared to 45 nm for the free polymer. It should be noted that complete siRNA binding to the polymer was observed at theoretical +/− charge ratios of 4:1 and higher. For both the free diblock copolymer as well as the polymer/siRNA complexes, near uniform distributions (PDI < 0.1) were observed. This result strongly suggests that the addition of the negatively charged siRNA molecules is not causing bridging between the positively charged micelles.
Particle morphology of micelles prepared from aqueous solutions of the diblock copolymer was also evaluated by electron microscopy (Figure 2). From the electron micrograph the average diameter of the particles, which appear spherical, was determined to be 27.5 nm ± 3.9 nm (n =45). The surface-exposed shell of the particles appears to be collapsed on the core as indicated by an electron dense region surrounding the particle cores. It is likely that the hydrophilic poly(DMAEMA) block, which stabilizes the particles under aqueous conditions, is collapsed under the anhydrous conditions of the experiment.
Figure 2.
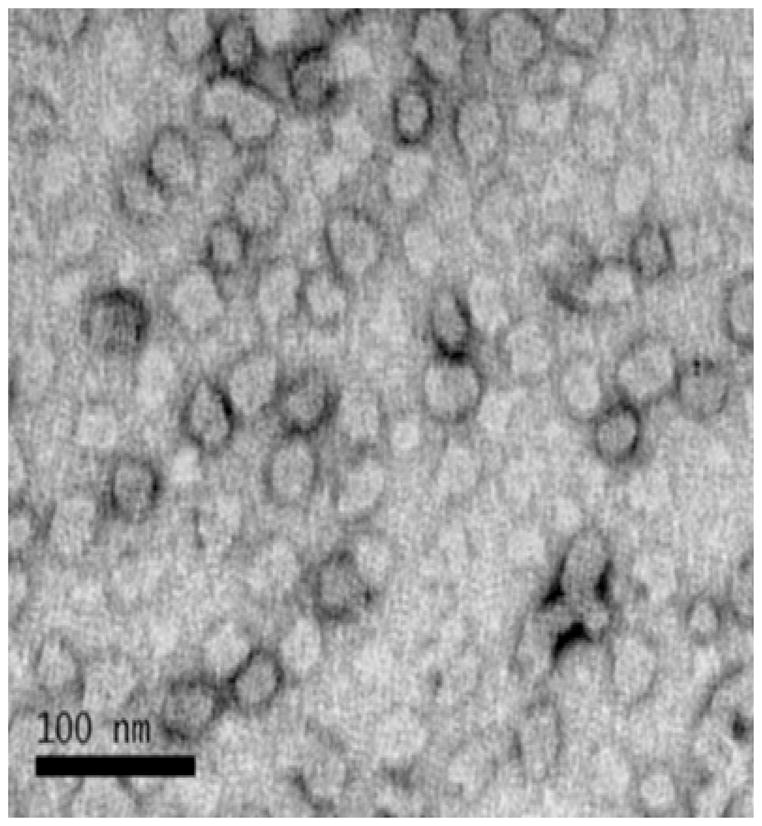
Transmission electron microscopy image of micelles formed from an aqueous solution of the diblock copolymer. A 0.5 mg/ml solution of the diblock copolymer in PBS was applied to a carbon coated copper grid for 30 minutes. The grid was fixed in Karnovsky’s solution and washed in cacodylate buffer once and then in water 8 times. The grid was stained with a 6% solution of uranyl acetate for 15 minutes and then dried until analysis. Transmission electron microscopy was carried out on a JEOL 1230 microscope.
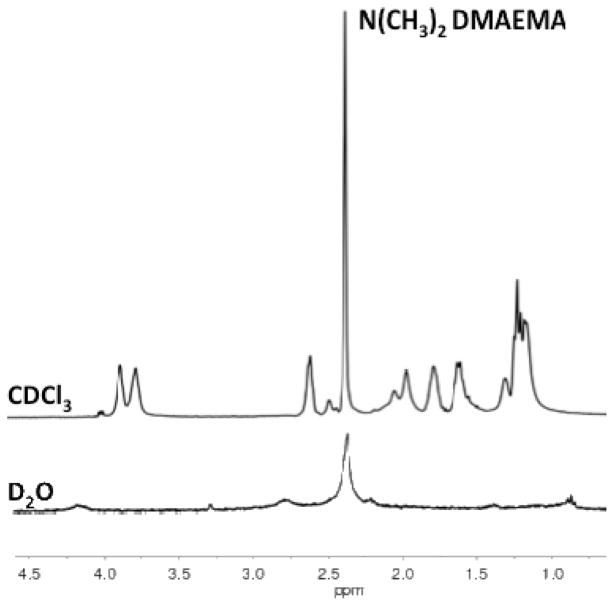
Additional evidence that the diblock copolymers exist as particles stabilized by a hydrophilic poly(DMAEMA) block was provided by 1H NMR spectroscopy. Shown in Figure 3 are 1H NMR spectra for diblock copolymer in CDCl3 and D2O respectively. The 1H NMR spectrum of the diblock copolymer in CDCl3 shows resonances associated with both the poly(DMAEMA) block (e.g. (CH3)2N at 2.38 ppm and OCH2CH2N at 4.1 ppm) as well as those associated with the pH responsive block (e.g., BMA OCH2CH2C at 3.95 ppm). Under these conditions both blocks are solvated and show free segmental motion, which is consistent with a molecularly dissolved unimeric polymer. In contrast the 1H NMR spectrum of diblock copolymer in D2O (Figure 3) shows significant peak suppression and broadening of the resonances associated with the comonomers present in the hydrophobic block (i.e., BMA, PAA, and DMAEMA). Resonances associated with the DMAEMA residues remain visible in D2O, however, the signal is significantly attenuated. For example, the signal associated with the dimethyl groups (δ = 2.28 ppm) is reduced from 5600 in CDCl3 to 2800 in D2O.
Figure 3.
1H NMR spectra of the diblock copolymer in deuterated chloroform (CDCl3) and deuterated water (D2O) at 25ºC. In CDCl3 resonances associated with both the poly(DMAEMA) block as well as the endosomolytic block (poly(BMA-co-DMAEMA-co-PAA) are visible. In comparison, under aqueous conditions (D2O) only resonances associate with the poly(DMAEMA) block are visible. These resonances are also suppressed suggesting the endosomolytic block is sequestered from the aqueous phase.
These findings provide direct spectroscopic evidence for the formation of a core-shell structure under aqueous conditions with a hydrated poly(DMAEMA) corona stabilizing a hydrophobic core composed of hydrophobic BMA units and electrostatically stabilizing units of opposite charge PAA and DMAEMA).
3.2 Effect of pH on polymer structure and CMC
CMC values of particles formed from the diblock copolymer were also measured by a DLS-based dilution method. Hydrodynamic diameters of the particles in pH 7.4 PBS buffer at a concentration of 1 mg/mL were measured by dynamic light scattering over a 5-fold range of serial dilutions from 1 mg/mL to 1.6 μg/mL. Using this method the particles were observed to remain stable at 45 nm with a low PDI down to a polymer concentration of about 10 μg/mL (> 100-fold dilution). CMC values at pH 7.4 were determined to be 2 ug/ml (pyrene assay) and 4 ug/ml (DLS dilution assay). As the diblock copolymer concentration was further reduced below approximately 5 μg/mL the particles become increasingly unstable suggesting a CMC value in this concentration range. This result is in good agreement with the pyrene assay, where individual polymer chains appear to dissociate and form non-specific aggregates (data not shown). A significant pH dependence was observed for CMC values as determined by DLS. Under physiological pH conditions (i.e. pH 7.4) the diblock copolymer micelles remain stable at concentrations as low as 5 μg/ml. In contrast, CMC values determined at pH 4.7 show a large increase in the concentration at which a significant population shows sizes of 1–8 nm, which is consistent with unimeric polymer chains. The CMC value at pH 4.7 was estimated to be approximately100 ug/ml (DLS dilution assay). This finding is likely a result of increased ionization of DMAEMA residues from both the corona and core forming segments. The increase in net positive charge would serve to increase the overall hydrophilicity of the diblock copolymer enhancing the micellar exchange rate. These conditions of low pH reflect the natural pH gradient found within endosomal compartments and could provide the basis by which the polymer mediates cytoplasmic delivery of siRNA.
3.3 pH responsive membrane destabilizing activity of polymeric micelles and their siRNA complexes
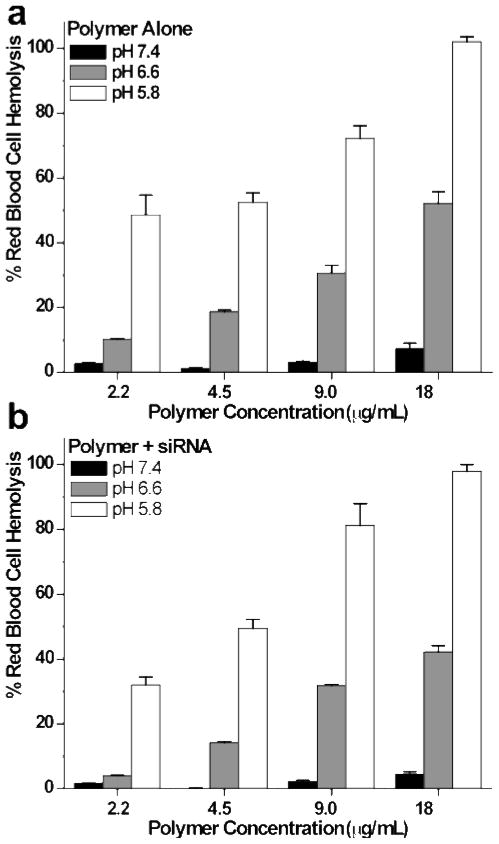
The pH-responsive membrane destabilizing activity of diblock copolymer was assayed using a red blood cell hemolysis assay. Three different pH conditions were used to mimic endosomal pH environments: extracellular pH = 7.4, early endosome pH = 6.6, late endosome pH = 5.8 (Figure 4). The dependence of hemolytic properties on the morphology of the diblock copolymer was evaluated by incubating red blood cells with concentrations above and below the polymer CMC. Intact red blood cells were then removed by centrifugation and the amount of hemoglobin in solution was determined by measuring the absorbance of the supernatant at 540 nm. In order to establish that complexation of the diblock copolymer micelle to a hydrophilic nucleic acid does not interfere with the intrinsic membrane disruptive properties of the polymer, hemolysis experiments were conducted on both the polymer alone as well as the polymer/siRNA complexes. Polymer/siRNA complexes at theoretical +/− charge ratios of 8:1, 4:1, 2:1 and 1:1 were prepared by adding increasing quantities of 25 nM siRNA to a fixed concentration of the diblock copolymer. The concentrated polymer and polymer/siRNA stocks were then added to the red blood cell suspensions to prepare solutions with a final polymer concentration between 2.2 and 18 μg/mL. No significant hemolytic activity was observed at pH 7.4 for polymer and polymer/siRNA complexes at any of the concentrations evaluated. Significant increases in red blood cell lysis were observed as the pH was reduced to 6.6. Under these conditions very similar hemolytic properties were observed between the polymer and polymer/siRNA complexes at a given pH and polymer concentration. For example, at a diblock copolymer concentration of 18 μg/mL the polymer alone showed 52 % red blood cell lysis at pH 6.6 which increased to 102 % at pH 5.8. (Figure 4a). In comparison, the polymer/siRNA complexes showed 42 % red blood cell lysis at pH 6.6 with a similar increase to 98 % at pH 5.8 (Figure 4b). Taken together, there is less than a 23 % and 4 % difference between the polymer and the polymer/siRNA at pH 6.6 and 5.8 respectively. Given these trends it was concluded that the complexation of siRNA to the diblock copolymer micelles does not significantly compromise the hemolytic activity.
Figure 4.
Hemolysis of the (a) diblock copolymer as a function of pH at concentrations of 2.2, 4.5, 9.0, and 18 μg/mL and (b) diblock copolymer/siRNA complexes at theoretical charge ratios of 1:1, 2:1, 4:1 and 8:1 (25 nM siRNA). Hemolytic activity is normalized relative to a positive control, 1 % v/v Triton X-100, and the data represent a single experiment conducted in triplicate ± standard deviation.
3.4 Knockdown activity and toxicity of siRNA-polymer complexes in cultured mammalian cells
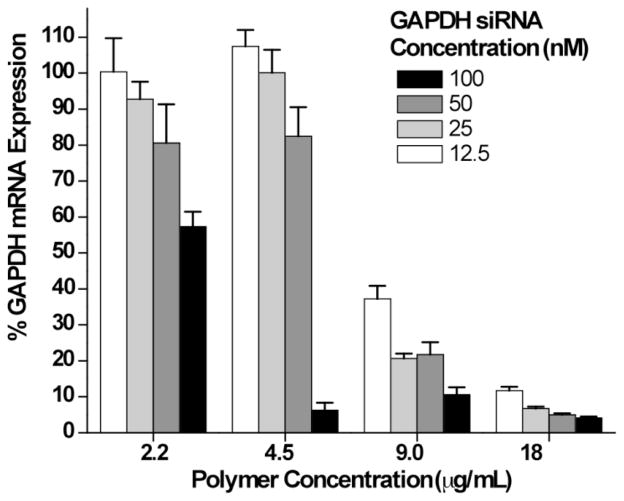
The ability of the polymeric micelles to deliver siRNA to the cellular cytoplasm in an active form was evaluated by conducting mRNA knockdown experiments in HeLa cells (Figure 5). Solutions of the diblock copolymer were mixed with siRNA targeting GAPDH as well as a scrambled sequence to serve as a negative control. These solutions were then allowed to incubate for 30 minutes after which time they were added directly to HeLa cells in media containing 10 % FBS. Final siRNA concentrations were evaluated at 12.5, 25, 50, and 100 nM at polymer concentrations such that the theoretical +/− charge ratios were 8:1, 4:1, 2:1, and 1:1 to determine what conditions result in highest knockdown activity. It should be noted that at these polymer concentrations LDH cytotoxicity experiments showed complete HeLa cell viability (data not shown). Specific gene knockdown activity was then measured 24 hours post transfection via real-time quantitative PCR. At a theoretical +/− charge ratio of 4/1, high mRNA knockdown was observed at all four siRNA concentrations. The observed knockdown at this charge ratio varies between greater than 90% at 25, 50, and 100 nM to around 75 % at 12.5 nM. In contrast to the 4/1 charge ratio, a precipitous decline in mRNA knockdown as a function of siRNA concentration was observed at +/− charge ratios of 2/1 and 1/1. Under these conditions high mRNA knockdown efficiency was retained at 100 nM for both charge ratios, however, a dramatic loss in mRNA knockdown was seen as the siRNA concentration is reduced to 25 and 50 nM for the 2/1 and 1/1 charge ratios respectively. These trends are also associated with differences in polymer concentration which are necessary in order to prepare different charge ratios at a given siRNA concentration. Under these conditions (i.e., [polymer] < 5 μg/mL) the particles are below the CMC of the polymer. In this concentration region destabilization of the micelle core results in almost complete loss of knockdown activity for all but the highest siRNA concentrations where siRNA binding may stabilize the micelle. These results suggest self-assembly of the diblock copolymer into stable micelles results in a significant enhancement of siRNA mediated mRNA knockdown. This knockdown is greater at higher +/− ratios and siRNA concentrations but significant mRNA knockdown levels remain high (~90 %) even at siRNA concentrations as low as 12.5 nM.
Figure 5.
GAPDH knockdown in HeLa cells was measured using real time RT-PCR following a 24 hour incubation period with the diblock copolymer/siRNA complexes. Final siRNA concentrations were evaluated at 12.5, 25, 50, and 100 nM at polymer concentrations such that the theoretical +/− charge ratios were 8:1, 4:1, 2:1, and 1:1. Scrambled siRNA and a commercially available transfection reagent, Lipofectamine, were used as negative and positive controls, respectively.
3.5 Cell uptake properties and cellular distribution of diblock copolymer/siRNA complexes
The cellular internalization of the diblock copolymer-siRNA complexes at a 4:1 charge ratio was examined using flow cytometry with untreated cells and the commercial transfection reagent Lipofectamine (L2k) serving as the negative and positive controls respectively (Figure 6). Measurements were taken with and without the addition of the fluorescence quenching agent Trypan blue in order to control for surface bound noninternalized fluorescence. As expected, untreated cells showed negligible fluorescence while the L2k control showed moderate fluorescence with approximately 39 % cellular uptake of the FAM-labeled siRNA (Figure 6a). By comparison, very high cellular uptake of the labeled siRNA was observed for cells treated with the diblock copolymer complexes. Indeed, near quantitative siRNA delivery (91 %) was observed for the diblock copolymer at a charge ratio of 4:1 following a 24 hour incubation period. The mean fluorescent intensity for the diblock copolymer and positive controls was also evaluated via flow cytometry (Figure 6b). From these experiments a dramatic increase in the fluorescent intensity is observed for cells treated with the diblock copolymer/siRNA complexes as compared to L2k/siRNA samples.
Figure 6.
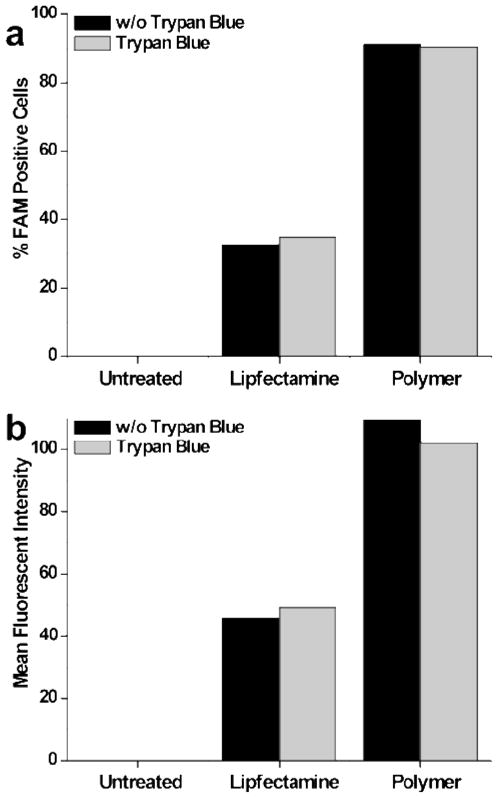
(a) HeLa cell internalization of 25 nM FAM-labeled siRNA, Lipofecatmine/siRNA, and polymer/siRNA complexes formed with the polymers at a theoretical charge ratios of 4:1 (after 4 h). Trypan blue was utilized for quenching of extracellular fluorescence and discrimination of complexes that have been endocytosed by cells. 10,000 cells were analyzed per sample and fluorescence gating was determined using samples receiving no treatment and treated with polymer alone.
In order to better understand the cellular distribution of the polymer/siRNA complexes fluorescent microscopy experiments were performed (Figure 7). Shown in Figure 7a is a fluorescent microscopy image of L2K mediated delivery of FAM labeled siRNA to HeLa cells. DAPI staining, shown in blue, was employed in order to visualize the nucleus. While some diffuse fluorescence (green) is observed for the L2k based system, the majority of the fluorescence is located in discrete punctuate spots. This observation suggests that most of the FAM-labeled siRNA is confined to endosomal compartments. Because siRNA must reach the cytoplasm in order to become active this is clearly not optimal. In comparison, the diblock copolymer micelle mediated delivery of siRNA shows diffuse fluorescence throughout the cellular cytoplasm (Figure 7b). Taken with the high mRNA knockdown levels these results suggest that the diblock copolymer is able to transition to a membrane disruptive conformation in response to endosomal pH changes, resulting in endosomal release of the siRNA.
Figure 7.
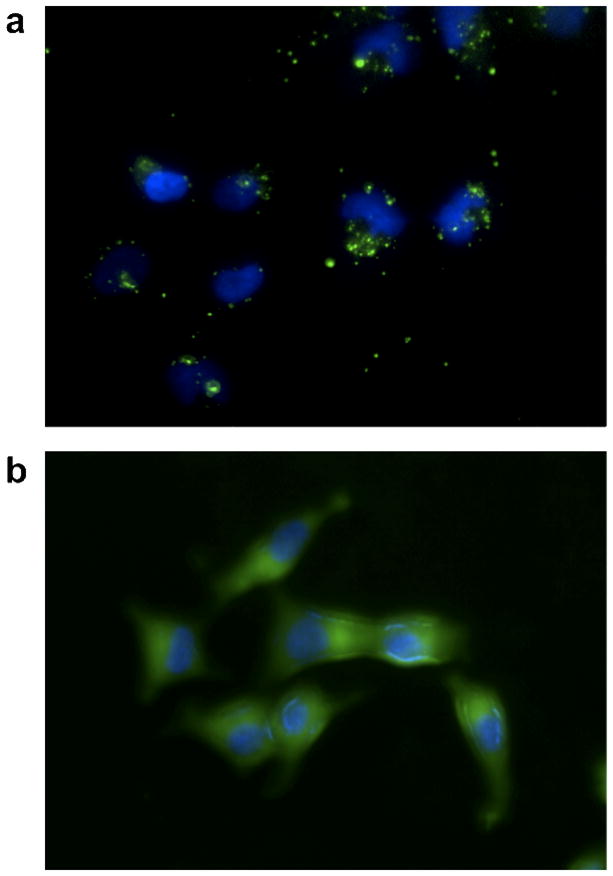
Polymer enhanced intracellular delivery of FAM labeled siRNA. Representative images illustrating (a) punctate staining (green) in the samples treated with lipofectamine/siRNA complexes alone and (b) dispersed fluorescence within the cytosol following delivery of diblock copolymer/siRNA complexes. Samples were treated for 15 min with 25 nM FAM-siRNA and prepared for microscopic examination following DAPI nuclear staining (blue).
4. Discussion
In these studies we detail the development of a new generation of siRNA delivery polymers that exhibits enhanced transfection efficiency and low cytotoxicity compared to previous polymer-based systems. Zhu et al. have previously described DMAEMA containing copolymers containing varying amounts of primary and tertiary amine groups for enhanced gene transfection (ref 1) and that form cationic micelles for the combinatorial delivery of siRNA and paclitaxol (ref 2). Our design retains the overall diblock copolymer structure with a poly(DMAEMA) block for siRNA complexation and a second block that becomes membrane disruptive under endosomal conditions (i.e. pH ≤ 6.6). Using this basic design the second block was lengthened in an attempt to induce the formation of well-defined micelles with sizes less than 100 nm. Dynamic light scattering measurements conducted in PBS buffer at a pH of 7.4 show that this change in diblock copolymer architecture does indeed induce spontaneous self-assembly of the polymer over a wide range of concentrations. These particles show hydrodynamic diameters of approximately 45 nm and low polydispersity. DLS experiments show a progressive loss in particle stability as evidenced by the appearance of a low size population (2–10 nm) when the polymer concentration is reduced below 10 μg/mL. This critical concentration threshold increases when the pH is decreased from 7.4 to 5.8 suggesting that increased protonation of DMAEMA residues from both the core and corona segments enhances the hydrophilicity of the polymer destabilizing the micelle. No significant increase in particle size or heterogeneity was observed when negatively charged siRNA was added at theoretical +/− charge ratios of 4:1 and 8:1. Transmission electron microscopy images of these particles formed from aqueous solution show a spherical morphology with a thin electron rich ring surrounding an inner core. 1H NMR analysis of the diblock copolymer in aqueous (D2O) and organic (CDCl3) solvents showed a significant suppression of the resonances associated with the pH-responsive endosomolytic block under aqueous conditions while no attenuation was observed for solutions of the polymer in deuterated chloroform. These results combined with the DLS and TEM findings support the conclusion that the increase in block size does induce the formation of particles with sizes and shapes that are consistent with micelles. This result contrasts with our previous finding where no micelle-like particles were observed for the diblock copolymer in the absence of siRNA. The morphology of these micelles, under aqueous conditions, is likely a hydrophilic corona of poly(DMAEMA) stabilizing a more hydrophobic core composed of the BMA containing second block.
In order to determine if self-assembly of the diblock copolymers into micelles disrupts the pH-responsive membrane disruptive properties, red blood cell hemolysis experiments were conducted. These experiments were performed at polymer concentrations above and below the CMC. Hemolysis experiments were also conducted in the presence of siRNA to evaluate if charge-charge interactions would suppress hemolysis. No significant hemolytic activity was observed at pH 7.4 for polymer and polymer/siRNA complexes at any of the concentrations evaluated. Upon reduction of the pH to 6.6, however, a significant increase in red blood cell hemolysis was observed. Indeed the hemolysis profiles as a function of pH are very similar for both the polymer as well as the polymer/siRNA complexes. As expected red blood cell hemolysis increased with increasing polymer concentration and the diblock copolymer displayed high levels of hemolysis at polymer concentrations below the CMC. This result supports the conclusion that self-assembly of the diblock copolymer does not reduce the pH-selective membrane disruptive properties of the polymer.
The ability of these polymers to deliver siRNA through the endosomal pathway into the cytoplasm was investigated by conducting mRNA knockdown experiments against GAPDH. Transfection experiments in HeLa cells were conducted at siRNA concentrations between 12.5 and 100 nM under high serum conditions. At theoretical +/− charge ratio of 4/1 and greater, high mRNA knockdown was observed at all four siRNA concentrations. The observed knockdown at this charge ratio varied between greater than 90% at 25, 50, and 100 nM to around 75 % at 12.5 nM. Increasing the +/− charge ratio to 8:1 resulted in near quantitative mRNA knockdown even at the lowest siRNA concentration. In contrast to the 4/1 charge ratio a dramatic reduction in mRNA knockdown was observed for all but the highest siRNA concentrations when the polymer concentration was reduced to 4.5 μg/mL and below. Under these conditions the diblock copolymers are believed to be below their CMC. From these results, it is clear that self-assembly of the diblock copolymer into micelles is necessary to retain high mRNA knockdown levels (~90 %) at the lower siRNA concentrations. Since hemolytic activity is high below the CMC, loss of KD activity suggests that siRNA binding is lost below the CMC where micelle structure is disrupted. Therefore, micelle structure appears to be important for RNA binding. As the micelle approaches its CMC and below, the polymer chains begin to form less ordered aggregates and dissociates which is expected to reduce cationic charge density and therefore RNA binding. We have also directly observed reduced RNA binding to polymer at concentrations near the CMC by gel shift assay (unpublished results)
These findings represent a significant advance over our previous system in terms of knockdown efficiency. Likely explanations for the enhancement in knockdown potential of the micellar system were provided by flow cytometry analysis of cell uptake properties of labeled siRNA. These experiments showed that greater than 90 % of cells transfected with the diblock copolymer contained FAM labeled siRNA as compared to a maximum of 25 % FAM positive cells in our previous studies. Fluorescence micrographs showed that the labeled siRNA is dispersed throughout the cellular cytoplasm in contrast to the L2K commercial reagent that was primarily punctate. These results clearly demonstrate the ability of the new micellar carriers to mediate endosomal escape of siRNA in a pH-dependent fashion.
5. Conclusion
The applications of RNAi based therapeutics have the potential to revolutionize the treatment of serious diseases such as cancer. Here we have described the development of a new generation of polymeric siRNA carriers based on our previously described technology. Physical-chemical characterization of the resultant diblock copolymer shows the formation of stable micelle-like particles under aqueous conditions with sizes less than 50 nm and low polydispersity. These results contrast with our previous work with short endosomolytic block segments where micelles were not observed until siRNA was added. The high serum stability of the new micelle-based delivery system combined with its low in vitro cytotoxicity and near quantitative mRNA knockdown makes these polymers promising candidates for in vivo characterization.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the National Institutes of Health (EB2991) and grant #2496490 from the Washington State Life Sciences Discovery Fund to the Center for Intracellular Drug Delivery.
Footnotes
Supporting Information Available: Figure S-1. Cell viability as a function of polymer concentration and charge ratio. Figure S-2. Critical micelle concentration (CMC) determination via pyrene fluorescence. Figure S-3. pH dependence of the critical micelle concentration (CMC) determined by DLS dilution method. Figure S-4. 1H NMR Analysis of pDMAEMA and diblock copolymers. Figure S-5. Size Exclusion Chromatography (SEC) for the pol(DMAEMA) macrocCTA and the corresponding diblock copolymer. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hamilton A, Baulcombe D. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 2.Elbashi S, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.White PJ. Clin Exp Pharmacol Physiol. 2008;35:1371–1376. doi: 10.1111/j.1440-1681.2008.04992.x. [DOI] [PubMed] [Google Scholar]
- 4.Walchli S, Sioud M. Front Biosci. 2008;13:3488–3493. doi: 10.2741/2943. [DOI] [PubMed] [Google Scholar]
- 5.Ramon AL, Bertrand JR, Malvy C. Tumorigenesis. 2008;94:254–263. doi: 10.1177/030089160809400218. [DOI] [PubMed] [Google Scholar]
- 6.Bahadori M. Arch Iran Med. 2008;11:435–443. [PubMed] [Google Scholar]
- 7.Grimm D, Kay MA. Curr Gene Ther. 2003;3:281–304. doi: 10.2174/1566523034578285. [DOI] [PubMed] [Google Scholar]
- 8.Wiley DC, Skehel JJ. Ann Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 9.Hughson FM. Curr Biol. 1995;5:265–274. doi: 10.1016/s0960-9822(95)00057-1. [DOI] [PubMed] [Google Scholar]
- 10.Ren J, Sharpe JC, Collier RJ, London E. Biochemistry. 1999;38:976–984. doi: 10.1021/bi981576s. [DOI] [PubMed] [Google Scholar]
- 11.Plank C, Oberhauser B, Mechtler K, Koch C, Wagner E. J Biol Chem. 1994;269:12918–12924. [PubMed] [Google Scholar]
- 12.Jiang T, Chang JB, Wang C, Ding Z, Chen J, Zhang J, Kang ET. Biomacromolecules. 2007;8:1951–1957. doi: 10.1021/bm0700486. [DOI] [PubMed] [Google Scholar]
- 13.Veron L, Ganee A, Ladaviere C, Delair T. Macromol Biosci. 2006;6:540–554. doi: 10.1002/mabi.200600071. [DOI] [PubMed] [Google Scholar]
- 14.You YZ, Manickam DS, Zhou QH, Oupicky D. J Controlled Release. 2007;122:217–225. doi: 10.1016/j.jconrel.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aral C, Akbuga J. J Pharm Sci. 2003;6:321–326. [PubMed] [Google Scholar]
- 16.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DWY, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin RJ, Qin J, Lam K, Kallanthottathil RG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Qingmin C, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 17.Geisbert TW, Lee ACH, Robbins M, Geisbert JB, Honko AN, Sood V, Johnson JC, de Jong S, Tavakoli I, Judge AJ, Hensley LE, MacLachlan I. Lancet. 2010;375:1896–1905. doi: 10.1016/S0140-6736(10)60357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kathryn A, Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discovery. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murthy N, Chang I, Stayton PS, Hoffman AS. Macromol Symp. 2001;172:49–55. [Google Scholar]
- 20.Murthy N, Robichaud JR, Tirrell DT, Stayton PS, Hoffman AS. J Controlled Release. 1999;61:137–143. doi: 10.1016/s0168-3659(99)00114-5. [DOI] [PubMed] [Google Scholar]
- 21.Lackey CA, Press OW, Hoffman AS, Stayton PS. Bioconj Chem. 2002;13:996–1001. doi: 10.1021/bc010053l. [DOI] [PubMed] [Google Scholar]
- 22.Mohamed EH, Hoffman AS, Stayton PS. J Controlled Release. 2005;101:47–58. doi: 10.1016/j.jconrel.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 23.McCormick CL, Lowe AB. Acc Chem Res. 2004;37:312–325. doi: 10.1021/ar0302484. [DOI] [PubMed] [Google Scholar]
- 24.Thang SH, Chiefari J, Mayadunne RTA, Moad G, Rizzardo E. Macromolecules. 1998;31:5559–5562. [Google Scholar]
- 25.Lenz RW, Marchessault RH. Biomacromolecules. 2005;6:1–8. doi: 10.1021/bm049700c. [DOI] [PubMed] [Google Scholar]
- 26.Heredia KL, Nguyen TH, Chang CW, Bulmus V, Davis TP, Maynard HD. Chem Commun. 2008;28:3245–3247. doi: 10.1039/b804812f. [DOI] [PubMed] [Google Scholar]
- 27.Cyrille B, Priyanto P, Thomas DP, Dakrong P, Bulmus V, Kavallaris M, Teoh WY, Amal R, Carroll M, Woodward R, St Pierre T. J Mat Chem. 2010;202:255–265. [Google Scholar]
- 28.York AW, Zhang Y, Holley AC, Guo Y, Huang F, McCormick CL. Biomacromolecules. 2009;10:936–943. doi: 10.1021/bm8014768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Convertine AJ, Benoit DSW, Duvall CL, Hoffman AS, Stayton PS. J Controlled Release. 2009;133:221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.York AW, Huang F, McCormick CL. Biomacromolecules. 2010;2:505–514. doi: 10.1021/bm901249n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyer C, Priyanto P, Davis TP, Pissuwan D, Bulmus V, Kavallaris M, Teoh WY, Amal R, Carroll M, Woodward R. J Mat Chem. 2010;20:255–265. pH-Responsive Polymeric Micelle Carriers for siRNA Drugs. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.