Abstract
Previous studies of Brugia malayi promoters have suggested that they are unusual in that they lack the CAAT or TATAA boxes that are often emblematic of eucaryotic core promoter domains. Instead, the region surrounding the spliced leader (SL) addition site appears to function as the core promoter domain in B. malayi. To test the hypothesis that polymorphisms in this SL addition domain are important determinants of promoter activity, a series of domain swap mutants were prepared replacing the SL addition domain of the B. malayi 13 kDa large subunit ribosomal protein (BmRPL13) with those of other ribosomal protein (RP) promoters exhibiting a wide range of activities. These constructs were then tested for promoter activity in a homologous transient transfection system. On average, polymorphisms in the SL addition domain were found to be responsible for 80% of the variation in promoter activity exhibited by the RP promoters tested. Essentially all of this effect could be attributable to polymorphisms in the 10nt located directly upstream of the SL addition site. A comparison of the sequence of this domain to the promoter activity exhibited by the domain swap mutants suggested that promoter activity was related to the number of T residues present in the coding strand of the upstream domain. Confirming this, mutation of the upstream domain of the promoter of the BmRPS4 gene to a homogeneous stretch of 10 T residues resulted in a significant increase in promoter activity.
Keywords: filariasis, promoter, spliced leader, transfection, ribosomal promoter
1. Introduction
Lymphatic filariasis represents an important public health problem in the developing world, particularly in Southeast Asia and Africa. It is estimated that 120 million individuals worldwide are infected with the two major causative agents of this disease, Wuchereria bancrofti and Brugia malayi [1]. Recently, substantial progress has been made in controlling filariasis, using a strategy based upon the mass distribution of drugs to at-risk populations [2]. However, a need still exists for the development of new drugs to support these efforts. This need to develop novel chemotherapeutic agents was a major force behind the successful effort to determine the sequence of the B. malayi genome [3]. The completion of a draft B. malayi genome sequence has also opened up new avenues of research into how these parasites regulate their gene expression. This question will be central to understanding how this parasite has adapted to life in two very different host environments (the vertebrate host and insect vector) and how it survives in the face of an active attack by the host’s immune system.
Previous studies have shown that Brugia malayi isolated embryos can be transiently transfected by biolistic methods [4]. Studies using this method to explore promoter structure have suggested that promoter structure in this parasite is rather unusual. Detailed mapping studies of the promoter of the B. malayi 70 kDa heat shock protein (BmHSP70) and the 12 kDa small subunit ribosomal protein (BmRPS12) demonstrated that the essential domains of both of these promoters do not contain sequences normally emblematic of the core domain of a typical eucaryotic promoter, such as CAAT or TATAA boxes [5, 6]. Instead, both promoters contain essential domains which flank the splice leader (SL) splice acceptor site (the SL addition domain). The promoter activity of these domains is not related to SL addition, as transgenic transcripts produced from B. malayi transfected with constructs containing either promoter alone were not trans-spliced [7, 8]. These data suggest the hypothesis that the region flanking the SL addition domain of B. malayi genes might form at least part of the core promoter domain in these organisms. This hypothesis was subsequently tested in a study of the putative promoters of 12 additional genes encoding ribosomal proteins (RPs), all of which are trans-spliced and thus contain the SL addition domain found to be essential for activity of the BmHSP70 and BmRPS12 promoters [9]. Deletion and substitution mutagenesis of six of these promoters demonstrated that the SL addition domain was essential for activity of all of these promoters [9]. Furthermore, as was the case with the BmHSP70 and BmRPS12 promoters, this activity was independent of the trans-splicing process [9]. Interestingly, the activity of the RP promoters examined varied over an 80 fold range [9]. It was possible that some of this variation was due to polymorphisms among the SL addition domains. However, from studies of both the BmHSP70 and BmRPS12 promoters, it is clear that sequences located upstream of the SL addition domain also play an important role in determining the overall activity of a given promoter [5, 6]. Thus, in these experiments it was impossible to determine how much of the variation in promoter activity seen among the RP promoters was due to polymorphisms in the SL addition domain and how much was due to differences in the less well characterized regulatory elements located outside of the SL addition domain. In the experiments described below, we constructed a series of domain switch mutants in which the SL addition domains from a number of different RP promoters were inserted in place of the SL addition domain present in the BmRPL13 promoter. By placing these SL addition domains in identical genomic contexts, it has been possible to directly evaluate the role that polymorphisms in the SL addition domain play in promoter activity.
2. Materials and Methods
2.1 Preparation of mutants
The constructs used in this study were derived from clones of 12 ribosomal protein promoters developed in a previous study [9]. The locus tags and corresponding accession numbers of the genes included in this study are given in the supplementary material (Table S1). Domain swap mutants were prepared using the BmRPL13 promoter cloned into the vector pGL3 basic (BmRPL13/luc) as a backbone employing the GeneTailor in vitro mutagenesis system (Invitrogen). In constructing the domain swap mutants, the 10nt located upstream of the SL addition site of BmRPL13/luc were first mutated to the sequence of the SL addition domain to be swapped. A total of 9 such upstream domain swap mutants were prepared, replacing the 10nt upstream of the SL addition site in BmRPL13 with the corresponding sequences from the BmRPS4, BmRPL9, BmRPL11.1, BmRPS12, BmRPS18, BmRPL21, BmRPL23, BmRPL24.e and BmRPL30 promoters. The DNA sequence of a number of clones resulting from this in vitro mutagenesis reaction were determined. One clone with the correct mutated sequence was then chosen and used as a template in a second in vitro mutagenesis to replace the downstream portion of the BmRPL13 SL addition domain with the corresponding domain derived from the SL addition domain to be swapped, following the procedure summarized above. In vitro mutagenesis reactions replacing the downstream sequences of BmRPS4, BmRPL9 and BmRPL23 were unsuccessful, leaving a total of six swap mutants in which the entire 22nt SL addition domain was replaced.
2.2 Transient transfection of B. malayi embryos
Isolated B. malayi embryos were transfected and promoter activity assayed by luciferase activity as previously described [7]. In brief, embryos were isolated from gravid female parasites and transfected with the experimental DNA driving the expression of firefly luciferase mixed with a constant amount of an internal standard, consisting of the BmHSP70 promoter fragment driving the expression of renilla luciferase (construct BmHSP70(−659 to −1)/ren) [7]. Transfected embryos were maintained in culture for 48 hours before being assayed for transgene activity. Firefly luciferase activity was normalized to the amount of renilla luciferase activity in each sample to control for variations in transfection efficiency. Firefly/renilla luciferase activity ratios for each sample were further normalized to the activity ratio found in embryos transfected in parallel with the parental construct BmRPL13/luc. This permitted comparisons of data collected in experiments carried out on different days. Each construct was tested in two independent experiments, with each experiment containing triplicate transfections of each construct to be analyzed. The statistical significance of the differences in activity of the native constructs and the paired experimental constructs was assessed based on the population marginal means (least squares means) using a Dunnett’s test. A multiple comparison analysis was performed using the Tukey method and the Tukey-Kramer adjustments to determine significant differences between the mutant promoter activities and their corresponding wild type promoters. All calculations were performed using the PROC GLM procedure in the SAS system, version 9.
3. Results
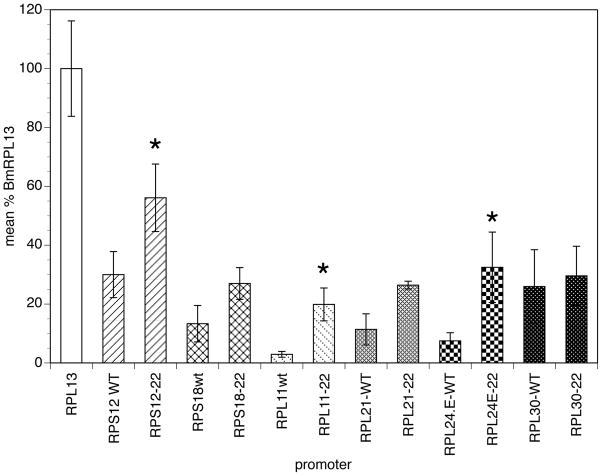
In order to evaluate the role that polymorphisms in the SL addition domain play in determining promoter activity, it was necessary to evaluate the activity of the different SL addition domains in a context in which all other motifs affecting promoter activity were identical. To accomplish this, a series of domain swap mutants were prepared replacing the SL addition domain of the BmRPL13 promoter with the SL addition domains from six other RP promoters. The BmRPL13 promoter was chosen as the backbone in the domain swap experiments because it was the RP promoter with the highest level of activity in the transient transfection assay [9]. This provided the highest signal to noise ratio, permitting the most accurate comparison of the activity differences among the different RP SL addition domains. Transient transfection assays were carried out to compare the activity of the domain swap mutants to that of BmRPL13 and to their corresponding native promoters. We predicted that if the polymorphisms in the SL addition domain were exclusively responsible for the differences in promoter activity exhibited by the RP promoters, the activity of the domain swap mutants would not be significantly different from that of their corresponding native promoters. In contrast, if the SL polymorphisms played no role in promoter activity, the domain swap mutants should all exhibit activities that were not significantly different from that of the parental BmRPL13 promoter. When the activity of the six domain swap mutants were compared to that of BmRPL13/luc and to their corresponding native promoter constructs, all were found to exhibit activities which were significantly different from that produced by BmRPL13/luc (Figure 1; p < 0.001). In contrast, the activity exhibited by three domain swap mutants was not significantly different from their corresponding native promoter constructs (BmRPS18, BmRPL21 and BmRPL30; Figure 1).
Figure 1. Activity of 22nt SL addition domain swap mutants prepared in the BmRPL13 backbone.
Replacement mutants were prepared and tested for promoter activity in the transfection assay as described in Section 2. Columns represent means, and error bars standard deviations, of six independent transfections. Columns with identical patterns represent activities produced by RP wild type promoters and their corresponding 22nt replacement mutants. Columns highlighted with asterisks indicate replacement mutants whose activity differed significantly from that of the corresponding wild type promoter (p < 0.05).
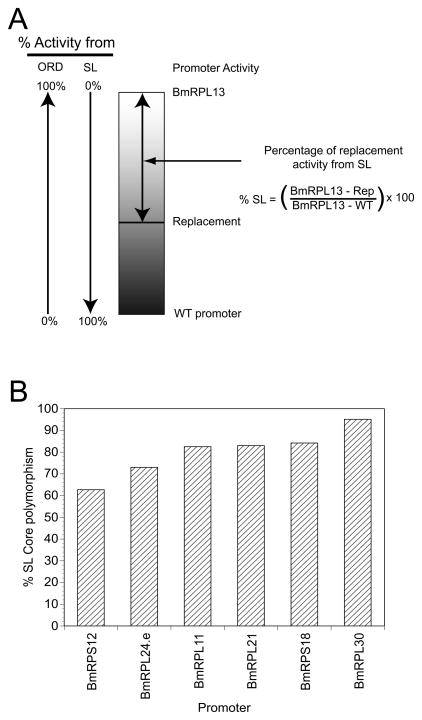
All of the replacement constructs tested exhibited activity levels that were greater than those produced by their corresponding wild type promoters. This might have been predicted, given the fact that BmRPL13, the promoter used as the backbone in the replacement experiments, was the most active RP promoter tested. The fact that all of the replacement mutants exhibited higher activities than those produced by their wild type counterparts offered the opportunity of estimating the relative contribution of polymorphisms in the SL addition domains and other regulatory domains to the overall activity exhibited by each promoter. To accomplish this, we first recognized that the activity of each wild type promoter (and therefore its activity level when compared to the BmRPL13 promoter) was a sum of the effect of polymorphisms in both the SL addition domain and the other uncharacterized regulatory domains. By extension, the difference in activity seen between each wild type promoter and the BmRPL13 promoter was a reflection of the effect of the polymorphisms in both regulatory elements. However, as noted above, the substitution mutants were constructed so that all of the SL addition domains were placed in the context of an identical backbone sequence. Furthermore, if the activity difference between a given wild type promoter and BmRPL13 was solely attributable to polymorphisms in the SL addition domain, the replacement mutant prepared from this promoter should exhibit an activity which was identical to that of the corresponding wild type promoter, and the difference between the activity exhibited by the replacement and the corresponding wild type promoter would therefore be zero (Figure 2, Panel A). Conversely, if all of the activity difference between a given wild type promoter and BmRPL13 was attributable to polymorphisms in other elements, the activity exhibited by the replacement mutant would be the same as that exhibited by the BmRPL13 promoter (Figure 2, Panel A). Thus, the difference between the activity exhibited by the BmRPL13 promoter and that of a given replacement mutant, when normalized for the difference of the activity between the BmRPL13 promoter and the corresponding wild type promoter would reflect the proportion of the activity difference due to polymorphisms in the SL addition domain (Figure 2, Panel A). When these normalized activity differences were calculated, the proportion of the activity attributable to polymorphisms in the SL addition domain varied between 63% and 95% (Figure 2, Panel B). On average, 80% of the activity of the six promoters examined was attributable to polymorphisms in the SL addition domain.
Figure 2. Estimation of what proportion of the activity exhibited by each promoter was attributable to polymorphisms in the SL addition domain.
Panel A: Rationale for calculation of the proportion of activity derived from the SL addition domain and other regulatory domains. The rectangle represents the possible activity range exhibited by the 22nt replacement mutants, ranging from that of the corresponding wild type promoter to that of the BmRPL13 promoter. Vertical arrows indicate the proportion of activity exhibited by a given replacement mutant that can be attributed to either the SL addition domain (SL) or the other regulatory domains (ORD). For the SL addition domain, as described in Section 3, this contribution would range from 0% for a replacement mutant with an activity identical to that of BmRPL13, to 100% for a mutant with an activity identical to that of the corresponding wild type promoter. Conversely, for the ORDs, the contribution would range from 0% for a replacement mutant with an activity identical to the corresponding wild type promoter to 100% for a replacement mutant exhibiting an activity identical to BmRPL13. The double headed arrow highlights the proportion of the overall activity difference that can be attributed to polymorphisms in the SL addition domain. Panel B: The relative contribution of polymorphisms in the SL addition domain to RP promoter activity, calculated using the method outlined schematically in Panel A.
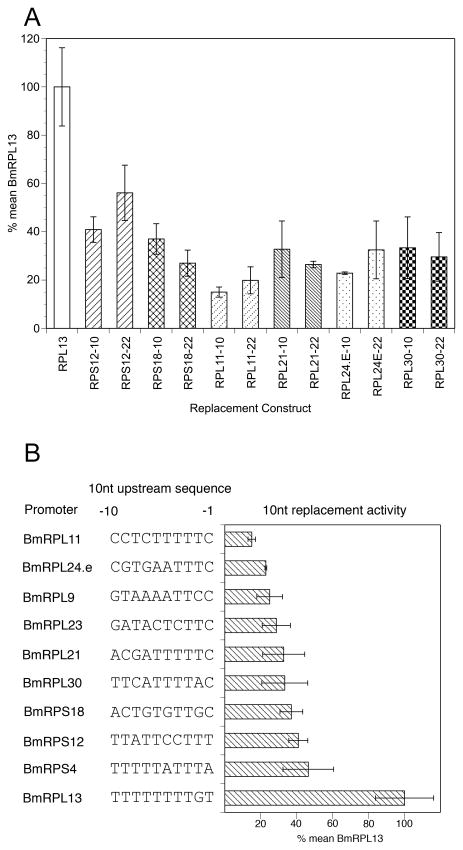
Sequence alignments of the SL addition domains of the RP genes revealed that the 10nt upstream of the invariant AG representing the SL addition site itself were more highly conserved than the corresponding region located downstream of the SL addition site [9]. In particular, the 10nt upstream of the addition site contained a conserved polypyrimidine tract that was previously shown to be essential for activity [9]. This finding suggested that the polymorphisms in the upstream 10 nt of the SL addition domain might prove to be the more important in determining promoter activity than polymorphisms region downstream of the SL addition site. To test this hypothesis, the activity of the six 22nt domain swap mutants were compared to a set of domain swap mutants in which only the 10nt upstream of the SL addition site were swapped. The activities exhibited by all of the 10nt upstream domain swap mutants were not significantly different than that exhibited by the mutants in which the entire 22nt SL addition domain had been swapped into the BmRPL13/luc backbone (Figure 3, Panel A; p > 0.1). This result confirmed that the polymorphisms in the 10nt upstream domain were the most important in determining promoter activity, while polymorphisms in the downstream portion of the domain had little or no effect on activity.
Figure 3. Activity of 22nt SL addition domain swap mutants and the corresponding upstream 10nt swap mutants.
Panel A: Comparison of activity of 22nt and 10nt domain swap mutants. Replacement mutants were prepared and tested for promoter activity in the transfection assay as described in Section 2. Columns represent means, and error bars standard deviations, of six independent transfections. Columns with identical patterns represent activities produced by paired 22nt and 10nt swap mutants. None of the activities exhibited by the 10nt swap mutants were significantly different from those exhibited by the corresponding 22nt swap mutant ( p > 0.1). Panel B: Activity and sequence analysis of the 10nt swap mutants: Columns represent means, and error bars standard deviations, of six independent transfections.
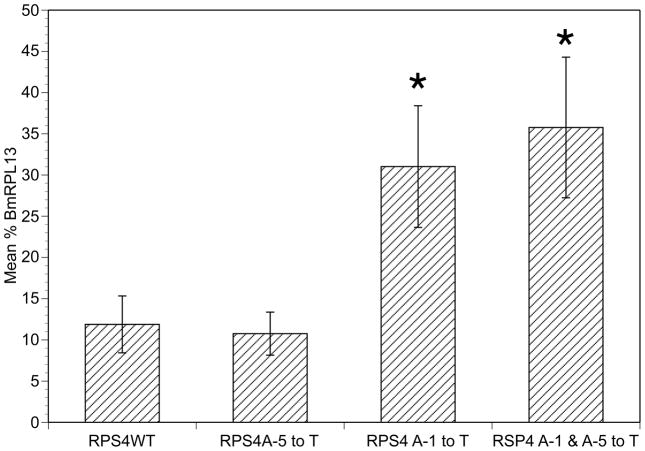
A total of nine swap mutants replacing the 10nt upstream portion of the SL addition domain were prepared as described in Section 2.1 and their activities determined. The results of these experiments are summarized in Figure 3, Panel B. Comparing the relative activity of each of these mutants to their corresponding sequences revealed that those domains containing larger numbers of T residues in the coding strand in general seemed to be more active than those with lesser numbers of T residues. For example, BmRPL13, the most active RP promoter, contained 9 T residues in its 10nt upstream domain, with only a single G present at position -2. Similarly, the most active 10nt domain swap mutant, BmRPS4, contained 8 T residues, with A residues at positions -1 and -5 (Figure 3, Panel B). Conversely, the least active 10nt domains, BmRPL24.e and BmRPL11, contained only 4–6 T residues (Figure 3, Panel B). These results suggested that an increased proportion of T residues in the 10nt upstream domain was associated with an increased promoter activity, and therefore mutation of either or both of the A residues in the upstream domain of BmRPS4 (the latter resulting in an upstream domain consisting entirely of T residues) might result in an increase in BmRPS4 promoter activity. To test this hypothesis, a mutant was prepared changing these two A residues to T residues. Mutation of both A residues in the BmRPS4 promoter resulted in a significant increase in promoter activity (Figure 4). When the A residues were mutated individually, changing the A residue at position -5 from A to T resulted in no significant change in activity (Figure 4). Changing the A residue at position -1 to T resulted in an increased activity over the wild type promoter which did not differ significantly from the activity produced by the construct in which both A residues had been changed to T (Figure 4; p > 0.5).
Figure 4. Activity of single nucleotide replacement mutants of the SL addition domain of the BmRPS4 promoter.
Columns represent means, and error bars standard deviations, of six independent transfections. Asterisks highlight mutants whose activity was significantly different from that exhibited by the BmRPS4 wild type promoter (p < 0.05).
4. Discussion
Previous studies have shown that the SL addition domain of the RP promoters is essential for transgenic reporter gene activity in transiently transfected B. malayi embryos [9]. This effect is not associated with trans-splicing per se, as none of the transgenic mRNAs derived from embryos transfected with constructs containing the RP promoters alone are trans-spliced [9]. Trans-splicing of such transgenic mRNAs requires the presence of a trans-splicing domain, a motif encoded downstream of the promoter domain [8]. These studies also revealed that the RP promoters exhibited a wide range of activities in the transient transfection assay [9]. The overall goal of the experiments described above was to test the hypothesis that the difference seen in the activity of these RP promoters was at least in part due to the observed polymorphisms in the SL addition domain. To address this hypothesis, we evaluated the activity of the different SL addition domains when placed in the same genomic context, that of the BmRLP13 promoter. If the polymorphisms played no role in determining promoter activity, we hypothesized that the activity of all of these mutants would be the same as that of the BmRPL13 promoter used as the backbone for these experiments. In fact, the activity of all of the replacement mutants was significantly different than that exhibited by the BmRPL13 promoter. Thus, these data strongly support the hypothesis that polymorphisms in the SL addition domain do play a role in determining the level of activity exhibited by the RP promoters.
Overall, the analysis presented above suggested that between 5% and 37% of the activity exhibited by the RP promoters tested could be attributed to polymorphisms in regulatory elements located outside of the SL addition domain. Interestingly, the promoter in which these other domains appeared to play the greatest role in promoter activity was BmRPS12. Previous detailed mapping studies of this promoter have demonstrated that the region located at positions -318 to -65 relative to the start codon consists of 5 3/4 almost exact 44nt tandem repeats, which together act as an enhancer of promoter activity [6]. None of the other RP promoters tested here contain a similar repeat domain. Further experiments will be necessary to define the other regulatory elements of these promoters.
The data presented above indicate that replacement mutants encompassing the 10nt upstream of the SL addition site produced promoter activities that were not significantly different from those produced by mutants in which the entire 22nt SL addition domain was replaced. These data support the hypothesis that polymorphisms present in the 10nt upstream of the SL addition domain play a much greater role in determining the level of promoter activity than do polymorphisms located downstream of the SL addition site. This finding is in concordance with the fact that the 10nt upstream of the SL addition site are more highly conserved than the 10nt located downstream of the SL addition site. Furthermore, previous studies have demonstrated that the upstream domain consists of a pyrimidine rich tract that is essential for promoter activity [9]. In particular, a T residue present at position -3 relative to the SL addition site has been shown to be essential for promoter activity [9]. The results presented above support the finding of the importance of the polypyrimidine tract in promoter activity, and further suggest that an increased proportion of T residues in the upstream domain is usually associated with increased promoter activity. Thus, mutation of the only two A residues in the BmRPS4 promoter (resulting in an upstream domain consisting entirely of T residues) resulted in a significantly increased level of promoter activity. Interestingly however, this increase in activity was associated solely with the mutation of the A residue in position -1, while mutation of the A residue at position -5 had no effect on promoter activity. These data, when taken together, suggest that the relative importance of the residues encoded in the upstream domain varies considerably. In particular, the T residues encoded in positions -1 and -3 in the coding strand relative to the SL addition site appear to play important roles in promoter activity.
Overall, the promoter activity in the 10 wild type RP promoters examined varied over an 80 fold range [9]. This is surprising, in that one might expect that the stable levels of the ribosomal proteins might be expected to be roughly equivalent, given the fact that they are present in equimolar amounts in the ribosome [10]. There are several possible explanations for this difference. First, it is possible that despite their presence in the ribosome in equimolar amounts, the actual cellular pool of these proteins varies in B. malayi, reflecting the differences in their promoter strengths. Second, it is possible the promoters examined here contain additional regulatory elements that were not included in the constructs studied here. Finally, it is possible that substantial post-transcriptional regulation of RP protein expression exists in B. malayi, occurring perhaps at the level of mRNA stability or translational efficiency. In support of this latter hypothesis, translational regulation of RP mRNAs has been demonstrated in many other eukaryotic organisms, a process that involves specific sequestration of the RP mRNAs in translationally inactive ribonuclear particles [11]. More experiments will be needed to determine if a similar or related mechanism is present in B. malayi.
The data presented above, together with our previous studies [5, 6, 9] support the hypothesis that the 10nt located upstream of the SL addition site likely represents the core promoter domain of B. malayi genes. The use of the SL addition domain as a core promoter suggests that the B. malayi homologues of the proteins that form the pre-initiation complex of RNA polymerase (e.g. the TATA box binding protein and the TATA box associated factor TAF9) may exhibit novel sequence binding properties in B. malayi. Alternatively, B. malayi may contain unique transcription factors that interact with the SL addition domain. In either case, these data suggest that the transcriptional machinery of B. malayi may be distinct from that of its human host, and may therefore represent a potentially important chemotherapeutic target. In this regard, it is interesting to note that transcription has proven to be a very fruitful drug target, representing the site of action of many bacterial antibiotics (reviewed in [12]).
Supplementary Material
Acknowledgments
We thank Dr. Charles R. Katholi for assistance in carrying out the statistical analyses and Dr. Naomi Lang-Unnasch for critically reading the manuscript. Parasite material was provided by the Filariasis Research Reagent Resource Center (FR3) at the University of Georgia, under a contract from the National Institute of Allergy and Infectious Diseases. This work was supported by a grant from the National Institutes of Health to TRU (project #R01AI048562).
Abbreviations
- nt
nucleotides
- ORD
Other regulatory domains
- RP
ribosomal protein
- RPL
large ribosomal subunit protein
- RPS
small ribosomal subunit protein
- SL
spliced leader
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization. Lymphatic filariasis - epidemiology. 2010 Available from http://www.who.int/lymphatic_filariasis/epidemiology/en/
- 2.Hooper PJ, Bradley MH, Biswas G, et al. Ann Trop Med Parasitol. 2009;103 (Suppl 1):S17–21. doi: 10.1179/000349809X12502035776513. [DOI] [PubMed] [Google Scholar]
- 3.Ghedin E, Wang S, Spiro D, et al. Science. 2007;317:1756–60. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higazi TB, Merriweather A, Shu L, et al. Brugia malayi: Transient transfection by microinjection and particle bombardment. Exp Parasitol. 2002;100:95–102. doi: 10.1016/S0014-4894(02)00004-8. [DOI] [PubMed] [Google Scholar]
- 5.Higazi TB, DeOliveira A, Katholi CR, et al. J Mol Biol. 2005;353:1–13. doi: 10.1016/j.jmb.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 6.de Oliveira AD, Katholi CR, Unnasch TR. Characterization of the promoter of the Brugia malayi 12kDa small subunit ribosomal protein (RPS 12) gene. Int J Parasitol. 2008;38:1111–19. doi: 10.1016/j.ijpara.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shu L, Katholi C, Higazi T, et al. Mol Biochem Parasitol. 2003;128:67–75. doi: 10.1016/s0166-6851(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, de Oliveira A, Higazi TB, et al. Mol Biochem Parasitol. 2007;156:62–73. doi: 10.1016/j.molbiopara.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Chauhan C, Katholi CR, et al. Mol Biochem Parasitol. 2009;166:15–21. doi: 10.1016/j.molbiopara.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wool IG, Chan Y-L, Gluck A. Mammalian ribosomes: The structure and evolution of the proteins. In: Hershey JWB, Mathews MB, Sononberg N, editors. Translational control. Vol. 30. Cold Spring Harbor: Cold Spring Harbor Press; 1996. pp. 685–732. [Google Scholar]
- 11.Meyuhas O, Avni D, Shama S. Translational control of ribosomal protein mRNAs in eukaryotes. In: Hershey JWB, Mathews MB, Sononberg N, editors. Translational control. Vol. 30. Cold Spring Harbor: Cold Spring Harbor Press; 1996. pp. 363–88. [Google Scholar]
- 12.Villain-Guillot P, Bastide L, Gualtieri M, et al. Drug Discov Today. 2007;12:200–8. doi: 10.1016/j.drudis.2007.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.