Abstract
Several human diseases are associated with the formation of amyloid aggregates, but experimental characterization of these amyloid fibrils and their oligomeric precursors has remained challenging. Experimental and computational analysis of simpler model systems has therefore been necessary, for instance on the peptide fragment GNNQQNY7-13 of yeast prion protein Sup35p. Expanding on a previous publication, we report here a detailed structural characterization of GNNQQNY fibrils using magic angle spinning (MAS) NMR. Based on additional chemical shift assignments we confirm the coexistence of three distinct peptide conformations within the fibrillar samples, as reflected in substantial chemical shift differences. Backbone torsion angle measurements indicate that the basic structure of these co-existing conformers is an extended β-sheet. We structurally characterize a previously identified localized distortion of the β-strand backbone specific to one of the conformers. Intermolecular contacts are consistent with each of the conformers being present in its own parallel and in-register sheet. Overall the MAS NMR data indicate a substantial difference between the structure of the fibrillar and crystalline forms of these peptides, with a clear increased complexity in the GNNQQNY fibril structure. These experimental data can provide guidance for future work, both experimental and theoretical, and provide insights into the distinction between fibril growth and crystal formation.
There are more than 20 human diseases, include Alzheimer’s, Parkinson’s and Huntington’s diseases(1), that are associated with the formation of amyloid fibrils, through the misfolding and aggregation of different amyloidogenic proteins. To further our understanding of these diseases, which affect increasingly large fractions of the population, it is essential to understand the misfolding process, in terms of the mechanism of fibril formation as well as the structural features of the resulting aggregates. Unfortunately, it has proven challenging to obtain structural information on the highly stable but insoluble and non-crystalline amyloid or amyloid-like fibrils, because they do not diffract to high resolution and are insoluble. Thus, the two major techniques used in structural biology, X-ray diffraction and solution NMR, are not applicable to these systems. The steps that constitute the misfolding process itself are even more difficult to examine experimentally. This includes the structure determination of oligomers that are thought to form precursors to the fibrillar form, and are increasingly suspected to be toxic species in amyloid-related disorders. Obtaining structural information on these systems often relies on a combination of indirect methods(2). Furthermore, the paucity of experimental data has stimulated a significant interest in exploring theoretical or computational studies of fibril formation and the intermediates formed along the pathway to mature fibrils(3).
Many computational studies aim to understand amyloid formation by focusing on relatively simple model systems that should represent the essential features of amyloid fibril structure. Recently, experimental atomic level data on an amyloid core structure were obtained via X-ray crystallography on a crystalline (i.e. non-fibrillar) form of a fibril-forming peptide, GNNQQNY(4, 5). This peptide is one of various peptide fragments from the yeast prion protein Sup35p that have been found to form amyloid-like fibrils, but this is one that also forms microcrystals(6). With newly developed micro-diffractometers, these crystals yielded a high resolution structure, and the short length of the peptide and the relative simplicity of the cross-β spine motif present in the crystals have made it an attractive model amyloid fibril system. As such it has stimulated numerous in silico studies of the characteristics and formation of model amyloid fibrils (7-27). The purpose of such simulations is to produce insights into the common features of the fibril formation process. This specifically includes information on unstable intermediates, thereby complementing experimental data that is most easily obtained on the more stable states along the fibril formation pathway. Note that such intermediates are of particular interest due to their potential role as toxic agents in various amyloid disorders(28).
However, it is important to keep in mind that the widely used structural data on GNNQQNY are based on the crystalline form of the peptide, and that the same peptide also forms amyloid fibrils under very similar conditions(6, 29, 30). In electron micrographs these latter moieties have a striated, flat ribbon-like appearance(29, 30), similar to fibrils formed by other peptides and proteins (31-36). Experimental studies have indicated significant structural differences between the crystalline and fibrillar structures(30, 37). Given the significant interest in GNNQQNY as a model system of fibril formation, it is important to characterize the molecular structure of these fibrils and compare them to the crystal structure, an experimental challenge for which few techniques are available. Magic angle spinning (MAS) NMR has proved uniquely valuable in the structural characterization of amyloid fibrils(38, 39), due to its ability to provide site-specific structural constraints on biological solids, without requiring crystalline samples.
Accordingly, we have previously used MAS NMR to obtain initial qualitative data comparing the GNNQQNY crystals and fibrils, which revealed significant differences in chemical shifts between the two forms, suggestive of differences in atomic structure.(30) In addition, the fibrils featured three distinct peptide conformations, which differ among themselves in terms of their chemical shifts. However,, the data was semi-quantitative and limited to the few N-terminal residues which were labeled in the fibrils. Within those sites the MAS NMR results suggested a localized helix-like deviation from β-sheet structure for one of these conformers, in contrast to more typical amyloid β-sheet structures (as were found in the other two fibril forms). As such the nature and structure of these conformers has remained a surprising and poorly understood aspect of these a priori (and based on the X-ray data) seemingly simple peptide fibrils.
To clarify this situation and provide further details on the structure of the GNNQQNY fibrils, we performed additional MAS experiments on more extensively labeled fibril samples. Thus, we present here more complete chemical shift assignments and structural NMR measurements. Our experiments reveal that all three conformers basically have a β-sheet conformation, based on both chemical shift analysis and backbone torsion angle measurements that relate the relative orientations of 13C-15N dipolar interaction vectors (40). The previously observed chemical shift deviations for the second fibril form reflect a localized backbone distortion rather than a completely non-β monomer structure. We also examine the arrangement of monomers within these β-sheets by measuring intermolecular 13C-13C and 13C-15N distance measurements. 13C-13C double quantum techniques(41) can generate coherences between singly 13C-labeled carbonyl sites in neighboring peptide monomers, and measure the 13C=O—13C=O distance to help constrain the strand-strand arrangement within amyloid fibrils(42-46). Rotational resonance width (R2W) measurements(47, 48) are used to determine additional intermolecular 13C-13C distances(49-51) in an approach recently used in other peptides and proteins(52-54). A mixture of singly 15N- and 13C-labeled peptides was used to determine 13C-15N distances via 1D REDOR(55) and 2D TEDOR(56, 57) experiments. Note that analogous 13C-13C and 13C-15N experiments have been used for the identification of inter-strand contacts in other amyloids(58-60). These intermolecular distance measurements consistently show that the Gly-7 residue of different monomers is in close proximity, and in a site-specific fashion show that G7-G7 intermolecular interactions only occur between identical fibril conformers (i.e. not between different conformers). 13C-13C proton-assisted-recoupling (PAR) experiments(61) reveal similar same-to-same contacts elsewhere along the sequence. Taken together this data this points to the different fibril conformers each adopting an in-register parallel (IP) structure, somewhat analogous to the crystals. Nonetheless, our data also show that significant chemical shift differences are present between the crystals and fibrils. Some of these correlate clearly to differences in backbone conformation between the fibril forms, in contrast to the two crystalline structures that feature virtually identical backbone (and side chain) conformations. We examine these differences with the crystals, and discuss the implications for past and future computational studies on this model system and its formation of amyloid fibrils.
Experimental Procedures
Sample Preparation
GNNQQNY crystalline and fibrillar samples were prepared as described previously (30). The peptides were obtained by solid phase peptide synthesis from CS Bio Inc. (Menlo Park, CA) and New England Peptide (Gardner, MA), and were isotopically labeled with 13C and/or 15N labeled amino acids from Cambridge Isotope Laboratories (Andover, MA). Various differently labeled peptides were prepared, including segmentally and site-specifically labeled versions. Crystal and fibril formation was accomplished by rapid dissolution of the lyophilized peptide in water, resulting in an acidic peptide solution. Largely as a function of concentration, the peptides form monoclinic or orthorhombic crystals and/or fibrils(29, 30). After centrifugation and removal of excess water, the peptide aggregates were packed into MAS rotors (Revolution NMR, Fort Collins, CO). The samples were maintained in a hydrated state at all times and were stored at 4°C.
NMR methods
Assignment methods
NMR measurements were performed using custom-made spectrometers (designed by D. J. Ruben, Francis Bitter Magnet Laboratory, MIT) operating at 1H Larmor frequency ωH0/2π = 500 MHz (11.7T), and ωH0/2π = 700 MHz (16.4T). Experiments at 500 MHz utilized a Varian triple-resonance HCN probe equipped with 4-mm stator, whereas experiments at 700 MHz used Varian triple-resonance HCN probes with a 3.2 mm MAS stator outfitted with either a solenoid or scroll coil (Varian Inc, Palo Alto, CA).(62) Spinning was regulated with Bruker MAS I spinning frequency controllers (Bruker BioSpin, Billerica MA). Assignment experiments used methods similar to those described previously (30). 13C-13C 2D assignment spectra were obtained using DARR/RAD mixing (63, 64) or PAR recoupling(65). 15N–13C correlations were obtained using double CP-based measurements(66), to give NCA, NCO, NCOCX, and NCACX 2D and 3D spectra(67-69). The NCOCX and NCACX(70) pulse sequences included a 10 to 20 ms DARR/RAD period to establish the intra-residue 13C-13C correlations. TPPM 1H decoupling(71) was applied during acquisition and t1 evolution. 13C chemical shifts were referenced to aqueous DSS using external referencing via the published 13C chemical shifts of adamantane.(72) 15N chemical shifts were referenced to liquid ammonia, via indirect referencing using the suggested IUPAC frequency ratios (13C/1H) of aqueous DSS and liquid NH3 (15N/1H).(73, 74) NMR data processing and assignment were done with the aid of the NMRPipe(75), Sparky(76), and CCPNMR/Analysis (77) software packages.
Torsion angle measurements
Chemical shift based backbone torsion angle analysis was performed with the TALOS program (version 2003.027.13.05)(78), using the default database of 78 proteins. The results were examined by hand to evaluate individual cases. To resolve inconclusive cases, and to provide more precise results, complementary measurements of the Ψ angles were done via NCCN torsion angle experiments (40, 79), which applied REDOR dephasing of 13C double quantum coherence obtained with the SPC5 pulse sequence (80). Data fitting was done using the numerical simulation software SPINEVOLUTION(81), employing secondary constraints (bond distances and bond angles) as determined from the crystal structure of the monoclinic crystals of GNNQQNY(4). Measurements on 100% [U-13C,15N-GNNQ]QNY and 100% GNN[U-13C,15N-QQNY] monoclinic crystals were done at ω0H/2π = 500MHz, and measurements on 100% [U-13C,15N-GNNQ]QNY and 100% GNN[U-13C,15N-QQN]Y fibrils were done at ω0H/2π = 700 MHz. Note that the higher magnetic field was essential to allow the distinct conformers within the fibrils to be resolved(30). The supporting information (SI) contains pulse sequence schematics and additional experimental details for these as well as subsequent MAS NMR experiments.
Intermolecular distance measurements
DQ DRAWS (dipolar recoupling with a windowless sequence) experiments (41, 44) (45, 46) were aimed at establishing strand-strand contacts characteristic of IP sheets by detecting the buildup of DQ-filtered coherence buildup in GNNQQNY fibrils and crystals containing a single 13C-carbonyl labeled residue, [13C’-Gly]. The experiments were performed at ω0H/2π = 500MHz using a 4mm Varian HCN probe. The resulting buildup curves were analyzed by fitting with the numerical simulation package SPINEVOLUTION (81), employing a linear spin-system consisting of seven 13C sites. These CPU-intensive calculations were performed on the 156-core Beowulf cluster at the Department of Structural Biology (University of Pittsburgh).
Fibril samples were prepared from 50/50 mixtures of specifically labeled [13C’-Gly]-NNQQNY and [15N-Gly]-NNQQNY. REDOR was used to measure intermolecular distances between these labels, using 3.2 mm Varian MAS probes at ω0H/2π = 700 MHz. 15N and 13C observed measurements were done using a Varian solenoid HCN probe at a MAS rate ωr/2π = 8 kHz and employing sample cooling with −8 °C cooling gas. We used REDOR mixing period of up to 25 ms with a 4s recycle delay, 91 kHz TPPM 1H decoupling(71) during mixing, evolution and acquisition, and 50 kHz REDOR pulses (on 13C when observing 15N and reversed). Extended mixing time (up to 50ms) REDOR measurements were performed using a scroll-coil BioMAS probe (Varian Inc., Palo Alto, CA), observing 13C. Data fitting and simulation of these REDOR curves was accomplished with SPINEVOLUTION (81), by simulating the normalized S/S0 data (i.e. in the absence of relaxation), where S0 data were obtained experimentally with the same mixing time but without the application of REDOR pulses. To supplement these 1D experiments, a single 2D z-filtered TEDOR (57) spectrum with 16 ms mixing time was recorded.
To measure the intermolecular 13C-13C distance in 50/50 mixed [13C’-Gly]-NNQQNY and [13Cα-Gly]-NNQQNY fibril samples, we used rotational resonance (R2) width measurements (47), implemented as described previously (52). These spectra were recorded at ω0H/2π = 700 MHz using a Varian solenoid HCN probe with a 3.2 mm spinner. A series of 2D spectra was recorded at different MAS rates using a constant 30 ms R2 mixing time, with 95 kHz TPPM 1H decoupling (71). These measurements were done near ωr/2π = ~11 kHz to match the n=2 R2 conditions between the Gly C’ and Gly Cα sites. At each MAS rate the peak intensities for C’-Cα cross peaks were measured and scaled relative to C’ intensities of reference data with 0 ms R2 mixing time. The resulting R2W profiles were fit with a multipole-multimode Floquet theory based program (52) to determine the distances.
Fibrils were also prepared from approximately equal amounts of GN[U-13C,15N-N]QQNY and GNN[U-13C,15N-Q]QNY fibrils which were studied at ω0H/2π = 700 MHz, with a 2D 13C-13C PAR experiment with τmix=14ms (65), and a 2D 13C-13C DARR correlation experiment with τmix=10 ms, both at ωr/2π=9.5 kHz.
Results
Solid-state NMR assignments
We have previously described the partial chemical shift assignments of the three coexisting fibril forms of GNNQQNY based on several samples where the first four residues (GNNQ) were uniformly 13C,15N-labeled. This allowed for some initial insight into the fibril dynamics and structure. We now present additional results on a peptide that has a subsequent segment 13C,15N-labeled (GNN[U-13C,15N-QQN]Y). We used standard 2D and 3D homo- and heteronuclear methods to obtain complete assignments on the labeled residues, the results of which are summarized in Table 1, together our previously published data.(30) We relied on 3D NCACX and NCOCX experiments to resolve various heavily overlapped resonances. Slices from these 3Ds, along with further experimental details, are included in the supporting information. Figure 1 contains 2D NCO spectra obtained for the crystalline and fibrillar forms of the peptide. The figure demonstrates that the shifts of the fibrillar peptides vary substantially from their crystalline counterparts and shows the multiplicity that is due to the coexistence of three distinct peptide conformers within the fibrillar sample. The intensity variations between the fibril resonances, which can be correlated to differences in their population and local dynamical variations, agree with our earlier observations on the other residues.
Table 1.
Assignment of fibril form resonances for the three predominant forms found in GNNQQNY fibrils. These assignments are based on various SSNMR assignments measurements performed at ω0H/2π = 700 MHz. The uncertainty in the chemical shifts is estimated to be ~ 0.25 ppm
| 13C chemical shift (ppm) | 15N chem. shift (ppm) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| C’ | Cα | Cβ | Cγ | Cδ | N | Nδ | Nε | ||
|
fibril
form 1 |
Gly7 | 171.2 | 43.5 | 27.6 | |||||
| Asn8 | 172.8 | 53.0 | 39.7 | 177.3 | 121.1 | 114.0 | |||
| Asn9 | 173.5 | 52.7 | 40.4 | 176.4 | 123.0 | 114.5 | |||
| Gln10 | 176.5 | 55.0 | 34.0 | 34.7 | 180.8 | 123.3 | 114.3 | ||
| Gln11 | 174.8 | 55.2 | 34.4 | 34.5 | 179.2 | 121.9 | 114.2 | ||
| Asn12 | 172.5 | 53.3 | 41.4 | 173.9 | 124.2 | 115.4 | |||
|
fibril
form 2 |
Gly7 | 170.6 | 44.1 | 27.4 | |||||
| Asn8 | 173.8 | 56.0 | 37.3 | 178.5 | 113.2 | 112.0 | |||
| Asn9 | 176.0 | 53.3 | 39.2 | 176.2 | 116.8 | 113.5 | |||
| Gln10 | 174.8 | 54.0 | 33.4 | 33.3 | 178.2 | 117.5 | 112.0 | ||
| Gln11 | 174.1 | 54.6 | 32.4 | 34.1 | 179.9 | 121.8 | 111.3 | ||
| Asn12 | 173.1 | 49.8 | 43.1 | 180.8 | 127.1 | 110.2 | |||
|
fibril
form 3 |
Gly7 | 170.1 | 43.4 | 27.5 | |||||
| Asn8 | 174.2 | 52.6 | 40.1 | 177.0 | 118.6 | 115.2 | |||
| Asn9 | 173.3 | 53.7 | 42.6 | 176.2 | 121.3 | 115.5 | |||
| Gln10 | 174.8 | 54.7 | 32.5 | 34.2 | 180.8 | 127.1 | 114.5 | ||
| Gln11 | 175.0 | 53.8 | 36.2 | 35.2 | 180.0 | 118.2 | 116.9 | ||
| Asn12 | 173.6 | 52.6 | 40.6 | 174.3 | 123.3 | 114.6 | |||
Figure 1.

MAS NMR 15N13CO spectra on (a,b) [U-13C,15N-GNNQ]QNY monoclinic and orthorhombic crystals and (c) fibrils, as well as (d,e) GNN[U-13C,15N-QQNY] monoclinic and orthorhombic crystals and (f) GNN[U-13C,15N-QQN]Y fibrils. All data were acquired at ω0H/2π = 700 MHz except (a) and (d) which were acquired at 500MHz. Backbone cross-peaks connecting neighboring residues are highlighted in black and connected between panels. The chemical shifts of the two crystalline forms were found to be similar, but the fibrils exhibit significantly different chemical shifts and contain three forms (#1-3), which vary in intensity as well as chemical shift.
TALOS torsion angle analysis
The observed chemical shifts can be used to determine the secondary structure and backbone torsion angles. Chemical shift indexing (CSI) (82, 83) provides a qualitative analysis of the secondary structure, and indicated a predominantly β-sheet conformation (see Table 2 and the SI). The assignment data are also used to determine the backbone torsion angles in a more quantitative manner, employing the widely-used TALOS software package (78) that compares observed chemical shifts with those for known protein structures. Despite being based on solution NMR shifts of mostly globular proteins, this approach proved reasonably reliable for the monoclinic crystals (30). It also seems to work well for most, but not for all, residues in the fibril conformers. In particular for residue N9 it proved problematic for the TALOS analysis to provide a clear answer (Figure S5). While we can identify a single constrained backbone conformation for forms #1 and #3, no unequivocal solution is found for conformer #2. The TALOS results are tabulated in Table 2, and further details are included in the SI. Analogous to the CSI data, conformers 1 and 3 are consistently β-sheet, while conformer 2 may feature an ill-defined distortion from such an extended β-sheet conformation.
Table 2.
CSI and TALOS-based torsion angle data for GNNQQNY fibrils. Some of these data were previously reported (30). Asterisks indicate data at the termini of the peptide that are unsuitable for TALOS analysis, since it relies on triplets of residues. The data for Asn-9 in fibril form #2 were inconclusive (marked with X; see also Figure S5). More CSI data analysis is included in the SI (fig. S4).
| residue | Fibril form 1 | Fibril form 2 | Fibril form 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| TALOS | CSI | TALOS | CSI | TALOS | CSI | ||||
| ϕ | ψ | ϕ | ψ | ϕ | ψ | ||||
| G7 | * | * | β | * | * | β | * | * | β |
| N8 | −108 ±20 | 127 ±10 | β | 54 ±6 | 40 ±13 | α | −119 ±18 | 131 ±12 | β |
| N9 | −101 ±11 | 131 ± 13 | β | −88 ±22 | X | α | −116 ±19 | 128 ± 8 | β |
| Q10 | −131 ±14 | 151 ±13 | β | −142 ±13 | 156 ±10 | β | −146 ±15 | 155 ±15 | β |
| Q11 | −150 ±12 | 149 ±12 | β | −129 ±22 | 132 ±11 | β | −145 ±12 | 153 ±9 | β |
| N12 | * | * | β | * | * | β | * | * | β |
NCCN torsion angle measurements
To verify the TALOS analysis and determine the backbone conformation of additional residues (e.g. the N-terminal Gly that are not suitable for TALOS analysis), we also performed NCCN torsion angle measurements. These rely on the generation of DQ coherence between the backbone carbons of residue i, which is then dephased under the influence of the 13C-15N dipolar interactions involving the 15N sites of the (i) and (i+1) residues(40). As reference data, we first performed these measurements on monoclinic crystals prepared from segmentally labeled GNNQQNY peptides. Figure 2(a) shows a representative 2D spectrum, illustrating the assignments of the recoupled sites. Note that here the only sites of specific interest for the ψ torsion angle experiments are those involving the backbone carbons (indicated with dashed rectangles). The intensity of these cross peaks was monitored as a function of the REDOR time, resulting in dephasing curves (e.g. Figure 2b). Numerical simulations of these experimental data were done with the SPINEVOLUTION program(81), in order to determine the ψ backbone torsion angles. The resulting ψ values are listed in Table 3, alongside the results from the TALOS analysis(30) and the X-ray crystallographic data(4). Note that the best-fit values are consistent with the TALOS results, but are significantly closer to the X-ray values, suggesting an improved accuracy (see also Figure S7 in the SI and the discussion below).
Figure 2.

Representative NCCN torsion angle data on GNNQQNY monoclinic crystals, obtained at ω0H/2π = 500 MHz. (a) 2D reference spectrum on 100% [U-13C,15N-GNNQ]QNY crystals, without REDOR dephasing, showing the assignments of the signals. Boxed cross peaks are due to backbone sites, which were used for ψ torsion angle measurements. Other signals reflect Asn and Gln side chains. (b) Experimental dephasing curves as a function of the REDOR mixing time, for three residues (G7, N9, and N12). The solid lines indicate best-fit simulations of the experimental data, obtained with SPINEVOLUTION (81).
Table 3.
Backbone torsion angle ψ for monoclinic crystalline GNNQQNY. X-ray and TALOS data are literature values (4, 30). Y13 is omitted since it has no defined ψ angle, as the C-terminal residue. Multiple minima are listed where applicable and uncertainty ranges are included in parentheses. In these cases the global minimum (‘best fit’) is listed first
| residue | X-ray ψ/° |
TALOS ψ/° |
NCCN |ψ|/° |
|---|---|---|---|
| G7 | −162.2 | - | 162 (154-168) |
| N8 | 141.2 | 131 ±10 | 151 (137-158) |
| N9 | 125.2 | 136 ± 9 | 122 (<146) |
| Q10 | 112.8 | 126 ±14 | 116 (87-136); 18 (<64) |
| Q11 | 126.8 | 142 ±15 | 117; 23 (<139) |
| N12 | 97.7 | 119 ±13 | 105; 44 (<137) |
Analogous NCCN measurements were performed on GNNQQNY fibril samples with overlapping segmental labeling: [U-13C,15N-GNNQ]QNY] and GNN[13C,15N-QQN]Y (Figure 3). Due to the multiple forms being present, we observe significant crosspeak overlap. Peak integrations were only done on cross-peaks that could be resolved or at least de-convoluted reliably. This meant that several sites were missing, but none of these involved data that were ambiguous or missing in the TALOS analysis (see SI). In addition, the fibril data had fewer mixing times and a generally poorer signal-to-noise than the crystal data (in part due to the multiple conformations and broader linewidths). Examplary results are illustrated in Figure 4 for the N9 residue, where the TALOS data were ambiguous (ref. Figure S5). Consistent with large differences in the chemical shift (and the CSI in figure S3 of the SI), the NCCN dephasing curve for conformer #2 is significantly different from the other two fibril forms, corresponding to different ψ values (Figure 4a,b). In this case, both the NCCN and the TALOS results are somewhat ambiguous. However, the NCCN data clearly exclude several of the TALOS matches (panel e). The TALOS data had a large uncertainty in ψ, which we can now resolve, but was reasonably consistent in terms of the φ angle. Similar observations apply to N8, where fibril form 1 and the monoclinic crystals show similar dephasing, but the curve is quite different for conformer 2 (Figure S8). For form #2, the best solution occurs at the non-β torsion angles, which would explain the substantial chemical shift differences seen in this conformer’s N-terminus (Figure 1) and the associated non-β CSI values(30). Although less consistent with all the available data (including the CSI and TALOS results), there is also a secondary and less likely solution that is closer to β-sheet torsion angles at (φ, ψ) < (−80°, 117°).
Figure 3.
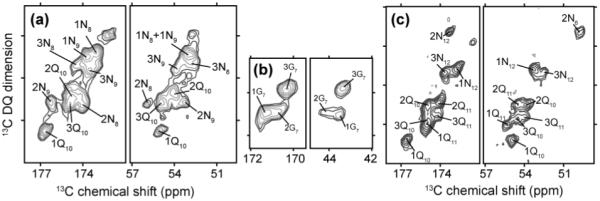
Assignments of fibril NCCN SQ-DQ 2D spectra. These are the reference data with 0 ms REDOR mixing time, for (a,b) 100% [U-13C,15N-GNNQ]QNY and (c) GNN[U-13C,15N-QQN]Y fibrils. Panel (b) shows the spectral region with the G7 cross peaks. Measurements were done at ω0H/2π = 700 MHz. Note the intensity variations and the significant overlap of various signals.
Figure 4.

Complementary data from NCCN and TALOS torsion angle analyses. (a) Experimental and simulated NCCN dephasing data for N9 in fibril forms #1 (circles, dashed line), #2 (triangles, black line), and #3 (diamonds, gray line). The simulations of the experimental data result in fits that are consistent with multiple torsion angles: N9 in (b) form #1, (c) form #2, and (d) form #3. These data exclude many of the ambiguous TALOS data form #2, as illustrated in (e) for the NCCN fit (left) and the TALOS-based Ramachandran plot (right) of N9 in form #2. The dashed line indicates the solution most consistent with both data sets (shown as lines in panel a).
By combining the NCCN results with the TALOS data we obtained the unified backbone torsion angles summarized in Table 4 (see also SI). Note that many of these values (and their errors) basically reflect the TALOS results, and that application of TALOS to the crystalline peptides suggests that the TALOS errors are likely to underestimate the true uncertainty (see SI and discussion).
Table 4.
Summary of MAS NMR-based torsion angles in GNNQQNY fibrils. The data include data determined from TALOS chemical shift analysis (Table 2), chemical shift index analysis and Ψ torsion angles determined via NCCN torsion angle measurements
| residue | Fibril conformer 1 | Fibril conformer 2 | Fibril conformer 3 | |||
|---|---|---|---|---|---|---|
| ϕ | ψ | ϕ | ψ | ϕ | ψ | |
| G7 | n/a | 166 ±10 | n/a | 166 ±20 | n/a | 180 ± 13 |
| N8 | −108 ±20 | 145 ±20 | 54 ±6a) | 40 ±13a) | −119 ±18 | 131 ±12 |
| N9 | −101 ±11 | 136 ± 18 | −88 ±22 | 150 ±13 | −116 ±19 | 130 ± 12 |
| Q10 | −131 ±14 | 130 ±15 | −142 ±13 | 135 ±15 | −146 ±15 | 121 ±20 |
| Q11 | −150 ±12 | 136 ±15 | −129 ±22 | 132 ±11 | −145 ±12 | 153 ±9 |
| N12 b) | β | β | β | β | β | β |
While both TALOS and NCCN analysis favor the listed torsion angles, there appears to be a secondary minimum with β-sheet-like torsion angles (ϕ~ −80°; ψ~ 117°) that may also be consistent with our experimental data (ref. Figure S9 in the SI and main text below).
Results for N12 reflect the secondary structure identified by CSI analysis (figure S4 in SI).
Intermolecular G7-G7 backbone contacts
We also performed experiments providing initial constraints for the supramolecular structure of the fibrils, initially targeting characteristic distances between the β-strands in an IP β-sheet. For the IP β-sheets found within the GNNQQNY crystals, the X-ray distance between subsequent peptide monomers is 4.9 Å (4, 5), as typical for amyloid fibrils. As reference data, we used DQ-filtered DRAWS (41, 44-46) to measure the DQ-buildup in monoclinic crystals prepared from GNNQQNY in which a single carbonyl site is labeled (Gly-13C’). Figure 5a shows the DQ signal (as a fraction of the cross polarization signal) as a function of the DQ DRAWS mixing time. The presence of any significant intensity suggests that the 13C’ sites must be in close proximity. Numerical simulation of the experimental data suggested a distance of 4.4 Å, which is in reasonably good agreement with the X-ray distance.
Figure 5.
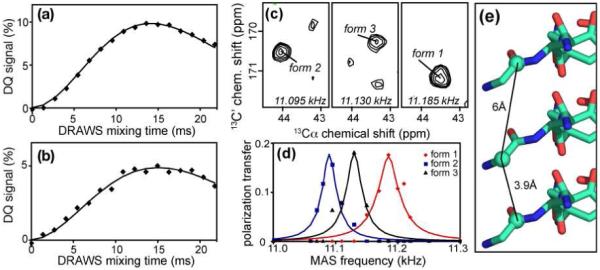
Intermolecular 13C-13C distance measurements. (a) DQ coherence buildup of G7-13C’ labeled GNNQQNY monoclinic crystals, characteristic of their IP β-sheet structure. Diamonds indicate experimental values, and the solid line shows simulations for a 4.4Å distance with 28ms T2 relaxation. (b) DQ buildup for G7-13C’ labeled fibrils, showing the combined intensity of all three fibril forms, along with simulated data for a 4.9 Å C’-C’ distance and 16 ms T2 relaxation. Both DRAWS experiments were done at ω0H/2π = 500 MHz with a MAS rate of 5.88 kHz. (c) 2D panels from R2W experiments on 51/49 mixed G7-13C’ and G7-13Cα labeled GNNQQNY fibrils, at ω0H/2π = 700 MHz and 11.095, 11.130, and 11.185 kHz MAS frequencies. Cross-peaks reflect an intermolecular contact between the G7-C’ and G7-Cα and occur consistently within the same conformer. (d) Intermolecular R2W profiles, where red diamonds, blue squares and black triangles indicate experimental Cα intensities, normalized to the corresponding carbonyl peak volume. Solid lines indicate simulated R2W profiles for distances of 4.5 - 4.9 (±1.0) Å. (e) Schematic showing the arrangement of the labeled sites in the IP β-sheet of the monoclinic crystal structure.
Identical measurements were performed on analogously labeled amyloid fibrils, as shown in Figure 5b. Since we were unable to reliably resolve the three individual fibril conformers at the 11.74 T field used in these experiments, the combined intensity was used for the fibril data analysis. A simple model that assumes an identical distance and relaxation for all three fibril forms results in a 13C’-13C’ distance of 4.9 Å (included in Table 5), possibly consistent with IP β-sheets as in the crystals. If the simple model were not correct, but one or more of the peptides were not sufficiently close and thus not contributing to the DQ buildup, we would expect the 13C-13C distance in the remaining conformers to be even shorter. For instance simulations for two IP fibrillar peptide forms, and one that is anti-parallel (with its 13C’ sites too far apart to participate), yield an apparent distance of 4.7 Å for the parallel peptides (data not shown). Note that these measurements did not allow us to confirm that the intermolecular interactions involve the same or different conformers, but do clearly show that the N-termini of at least some peptides must be close together. We address the uncertainties with additional intermolecular experiments.
Table 5.
Overview of intermolecular distance measurements. Where applicable, distances between non-identical sites (i.e. C’-Cα and C’-N contacts) only reflect the shorter distance of the two in-equivalent pairings that characterize intra-sheet contacts (see e.g. Fig 6e). X-ray crystallographic distances are based on PDB files 1YJP and 2OMM, respectively (4, 5). Note that the systematic errors listed reflect the assumptions made in the analysis, most notably assuming a lack of (undetermined) dynamical averaging and sometimes does not account for all multispin interactions (see text and SI for specific details). The presence of signification molecular motion would imply shorter actual distances than listed below.
| Fibrils by MAS NMR | X-ray crystallography | |||||
|---|---|---|---|---|---|---|
| intermolecular distance |
MAS NMR Experiment |
conformer # 1 |
conformer # 2 |
conformer # 3 |
monoclinic crystals |
orthorhombic crystals |
| G7C’-G7C’ | DRAWS1) | 4.9±1.0Å | 4.9±1.0Å | 4.9±1.0Å | 4.9 | 4.9 |
| G7C’-G7Cα | R2W | 4.5±1.0Å | 4.85±1.0Å | 4.7±1.0Å | 3.9 | 3.9 |
| G7C’-G7N | REDOR2) | 4.5±0.8Å | 4.5±0.6Å | 5.0±0.8Å | 3.9 | 4.0 |
Analysis assumed equivalent distance for all three fibril forms.
Results based on simple model with single 13C-15N coupling.
We performed additional 13C-13C distance measurements via R2 width (R2W) experiments (47, 52) on samples consisting of 50/50 (by mass) mixed 13C’-Gly/13Cα-Gly GNNQQNY fibril samples. In the mixed fibrils we measure intermolecular distances between labeled sites in neighboring peptides. For any peptide there is a 50% chance that its neighbor has the appropriate (i.e. different) labeling, in which case these intermolecular interactions would include two different closest distances (to peptides above and below) within the same β-sheet (see Figure 5(e) and SI). Figure 5(c) shows representative 2D panels from the R2W experiment, near the polarization transfer maxima (where the MAS rate matches half the frequency difference between the selectively recoupled nuclei). The observed cross peaks show that intermolecular polarization transfer occurs only between peptides of the same conformation (not between different conformers). Integration of cross peak volumes as a function of the MAS rate yields R2W profiles as shown in panel (d). The maximum transfer occurs at different MAS rates (due to differences in chemical shift), but the maximum transfer is similar among the three fibril forms. Fits of these profiles yield 13C-13C distances of 4.5 to 4.9 (±1.0) Å for each of these fibril forms (see Table 5). These simulations assumed a single closest distance dominating the interaction as is the case in the monoclinic and orthorhombic crystal structures (Figure 5e). Note that the observed distance measurements are actually consistent with the crystalline values, within the margin of error.
Figure 6 includes the results of REDOR-based intermolecular N-C distance measurements on a 57/43 (molar ratio) mixed 15N-Gly/13C’-Gly GNNQQNY fibril sample, showing significant dephasing due to the 15N-13C interaction. To check for any potential background dephasing, the experiment was also performed on 13C’-Gly / 13Cα-Gly GNNQQNY fibrils (as used for the R2W experiments). These data confirmed that none of the three fibril forms showed any significant background dephasing and thus that the observed REDOR dephasing is predominantly due to the labeled 15N sites. These 1D REDOR data do not reveal which 13C was dephased by which 15N site. To examine this aspect we recorded a 2D ZF-TEDOR spectrum, with a 16 ms mixing time, as shown in Figure 6(c). It confirmed that each of the 13C’sites interacts (only) with the 15N site of the same fibril form, consistent with the R2W data.
Figure 6.

(a) 1D 13C spectrum of 13C’-Gly-labeled GNNQQNY fibrils at ωH0/2π = 700MHz and ωr/2π = 10 kHz, showing the overlapping 13C’ signals from the three distinct conformers; (b) Experimental (triangles) and simulated (lines) REDOR dephasing curves from a 57/43 15N-Gly/13C’-Gly mixed GNNQQNY fibril sample. These simulations correspond to 15N-13C distances of 4.6, 4.7, and 5.5 Å, respectively (see text for details). (c) 2D TEDOR spectrum showing form-specific cross-peaks within each fibril form (16 ms TEDOR mixing time).
For a more quantitative analysis, the REDOR curves were simulated with SPINEVOLUTION. Initially a simple simulation was performed where the interaction was described by a single 13C-15N coupling that accounts for the experimentally determined 15N/13C ratio. These distance fits suggest distances of 4.5±0.8Å, 4.5±0.6Å, and 5.0±0.8Å for the fibril conformers 1, 2, and 3, respectively (Table 5). Note that in such a simulation we ignore the fact that there are multiple distance contributions to the interaction. To examine this aspect, we also tried a geometrical model that assumes an IP geometry (more details in the SI). The data do match this model, with the best fits shown in Figure 6(b) corresponding to 15N-13C distances of 4.6, 4.7, and 5.5 Å, respectively (with longer secondary distances of 6.2, 6.2, and 5.5 Å). We do note that under the constraints of this model, the fit of the data achieved for the third conformer is not perfect. Since the N-termini could be less rigid, and we have seen some indications of increased motion (not shown), we tried simulations that included a limited amount of isotropic motional averaging (as a scaling of the dipolar interaction). These did yield improved fits to these form #3 data (data not shown). Generally, dynamics would imply actual distances that are shorter than our ‘rigid lattice’ simulations suggest.
Intermolecular N9-Q10 backbone contacts
Additional data regarding intermolecular backbone contacts were obtained from a fibril sample prepared from a 50%/50% mixture of GN[U-13C,15N-N]-QQNY and GNN[U-13C,15N-Q]QNY. Here, any interresidue contacts should reflect intermolecular interactions. Such inter-molecular contacts were indeed observed, including a number of backbone-backbone contacts that are of particular interest here. Figure 7(a) shows the Cα-Cα region of a 13C-13C PAR spectrum with τmix=14 ms obtained from this sample. Several interresidue, off-diagonal cross-peaks can be identified, based on our assignments and by comparison with shorter mixing-time DARR data that show only the diagonal peaks (panel b). The internuclear distances associated with these contacts are harder to estimate than in the more quantitative measurements discussed earlier, but should reflect distances of less than 7 Å (61). The figure also includes one inter-form cross-peak, which reflects the fact that we do actually observe inter-form contacts in this sample. However, most of these involve side chain resonances and thus appear consistent with steric-zipper-like interactions. However, due to the inherent ambiguity and signal overlap, their detailed analysis is beyond the scope of this publication. It necessitates additional complementary experimental data and is the topic of ongoing research, but does suggest the possibility of inter-form polarization exchange, which implies an additional level of complexity in the GNNQQNY fibril assembly.
Figure 7.

Cα-Cα segments of 13C-13C 2D spectra on a mixed GN[U-13C,15N-N]QQNY / GNN[U-13C,15N-Q]QNY fibril sample. (a) Experiment employing 14 ms 13C-13C PAR mixing at 9.5 kHz MAS rate and ωH0/2π = 700 MHz. Several intermolecular cross-peaks can be observed, between the backbone Cα’s of N9 and Q10. Most are reflect ‘intra-form’ contacts, similar to the results on G7. One inter-form (between conformer #1 and #2) contact is indicated in grey. (b) Same section from a 2D 13C-13C DARR experiment with 10 ms DARR mixing, showing the diagonal peaks along with their assignments (ωr/2π = 9.5 kHz at ωH0/2π = 700MHz).
Discussion
Fibril shifts
The newly determined chemical shift assignments in the C-terminal half of the peptide fibrils confirm a number of observations that we reported previously, and also clarify some puzzling aspects of the results. Clearly, the shifts of the fibrils are different from the crystalline peptides throughout the length of the peptide (e.g. Figure 1 and Figure S3). Also, the conformational heterogeneity extends into these newly labeled residues, again showing three different sets of shifts. In this context, it is worth pointing out that these additional samples once again give the same resonances and intensity ratios compared to the fibril forms reported previously. Furthermore, we excluded the possibility that a pre-existing seeding aggregate drives the polymorphism of these samples (which are sourced from different peptide syntheses from different companies) by filtering the freshly dissolved peptide through a 3 kDa cutoff centrifugal filter, and again obtained identical spectra with the same fibril forms (data not shown). One of those (conformer #2) previously diverged most strongly from the others and its shifts appeared indicative of local helix-like backbone structure (30). However, based on the shifts themselves, the remaining residues in fibril form #2 appear to be β-sheet indicating that the ‘helicity’ is very localized. It was previously noted that there are apparent differences in the Tyr behavior in the crystals and fibrils, both from our NMR data and reported by others (37). Unfortunately, we did not label the Tyr residue (due to earlier synthetic difficulties). We can deduce some information on the Tyr shifts by comparison with limited spectral data from before, although in a non-form-specific manner (see Figure S2 and SI). This meant that the Tyr shifts could not be included in the conformer-specific backbone analysis. A more detailed picture may be obtained from future NMR investigations.
Fibril backbone structure
We have used our MAS NMR data to determine the backbone structure of each of the three conformers present in the GNNQQNY amyloid fibrils. This was primarily motivated by the fact that the data indicated that one of the three co-existing fibril conformers might contain significant non-β structure (30). This was noteworthy since this is in contrast to the uniform β-sheet conformation of both GNNQQNY crystal structures and seemed counter-intuitive in a short amyloid-forming peptide. Using more extensive labeling, extended chemical shift assignments, and explicit torsion angle measurements we now provide a more complete and quantitative analysis of the backbone structure.
To examine any concerns regarding the applicability of TALOS and NCCN-based MAS NMR approaches to an amyloid fibril system (of such an unusually Q/N rich sequence), we took advantage of the availability of the crystal structures of known conformation. In agreement with our previous measurements(30, 84), we find the TALOS program generates results in good agreement with the known torsion angles (Figure 8). We do note that the errors reported by TALOS appear to underestimate the actual deviations from the X-ray based structures. There could be a number of factors that negatively affect the performance of TALOS in the samples at hand. One concern could be that the chemical shifts are affected by non-local contributions (e.g. the effect of nearby aromatic rings like the Tyr side chain, on neighboring 13C shifts (85)). In this context comparison of the chemical shifts of the two crystalline forms is of interest, since the monomers (in the X-ray structures) are virtually identical in structure, yet display significant differences in chemical shift. Based on the X-ray data, the heavy atom RMSD between the two crystal forms is 0.35 Å (0.26 Å for the backbone), as determined by the SUPERPOSE function in the CCP4 suite(86, 87). The difference in chemical shift thus appears largely due to the difference in supramolecular packing of the monomers in the two crystal lattices. Another consideration in this particular comparison is that the X-ray data were obtained at 100 K, whereas the NMR chemical shifts are all obtained near room temperature, which may affect the structural and/or dynamical nature of the sample (88, 89). Finally, the TALOS approach relies on the availability of segments of similar sequence and structure in the reference database, which may not be valid for these peptides.
Figure 8.

Torsion angle measurements. (a) Column graph comparing the TALOS and NCCN torsion angle results for the monoclinic GNNQQNY crystals to the angles obtained by X-ray crystallography(4). (b) Ramachandran plots of the GNNQQNY crystals, based on the X-ray results. The crystalline backbone angles fall within the β-sheet region (N12 is marked as open symbols). Here each point represents a particular amino acid. (c) Torsion angles determined for the GNNQQNY fibrils (for residues N8 to Q11). The fibril secondary structure shows a different distribution, with a significant deviation for the N-terminal residues of form #2. Overall, we see a structure composed primarily of β-sheet in the fibrils, based on our new MAS NMR data.
Despite these potential concerns, the TALOS data from the crystals match the X-ray data with average deviations of 14-21° (see SI). We then examined the NCCN experiment and found the results to be quite accurate (see Figure 8(a)). While the NCCN torsion angles are consistent with the TALOS results, they do tend to represent an improvement in accuracy as the best-fit NCCN results match the X-ray data very well (see also figure S7 in the SI). However, its precision can be limited and it suffers from degeneracy in the fit solutions. In some cases the NCCN precision appears comparable to or inferior to the errors reported by TALOS. Recall however that the latter are likely to be underestimates (see table S3 in the SI), whether due to features specific to these (solid) samples or in general (90).
For the fibrils we also applied such a combination of the NCCN and TALOS approaches, as summarized in Table 4. TALOS was unable to provide data for a number of residues (e.g. the N-terminal Gly and Asn-9 of conformer 2), and the NCCN torsion angle data provide insights into those residues. For a visual comparison, the results are plotted in panels (b) and (c) of Figure 8, in the form of Ramachandran plots (for the crystal residues 8-12 or residues 8-11 for the fibril conformers). The crystalline peptides have a uniform β-sheet structure, a feature that is reproduced in the conformers 1 and 3. Fibril conformer 2, which previously showed some non-β structure (30), now is found to be largely β-sheet in terms of its backbone structure. A very notable distortion occurs its N-terminus (Figure 8c), in a conformation that is somewhat reminiscent of a β-bulge conformation in terms of the backbone torsion angles (see Figure S14 in the SI for a schematic representation). Intriguingly, β-bulges have previously been suggested to prevent the lateral association of β-sheets (91, 92). However, here the fibril appears to be able to accommodate this distorted backbone conformation within the β-sheets, perhaps analogous to amyloid-like crystals formed by the peptide MVGGVV derived from Aβ reported by Eisenberg and colleagues(5), which also feature a turn in the peptide backbone.
Intermolecular arrangements
One of the key characteristics of the GNNQQNY crystal structures is the IP intra-sheet alignment of the monomers. As pointed out by Sawaya, et al.(5), both structures represent one out of many possible β-sheet configurations. We employed several independent measurements that showed intermolecular contacts between the N-terminal glycines, consistently only between peptides having the same conformation. In addition, we see intra-form backbone-backbone contacts for each of the three fibril conformers in the long-mixing 13C-13C PAR experiments. Taken together, these data are consistent with, and even suggestive of, an IP alignment analogous to the crystalline aggregates. Especially with such short peptides in amyloid fibrils manifesting a multiplicity of conformations it is difficult to completely exclude all non-parallel and/or out-of-register sheet configurations, based on the current data. We note that the N-terminus is positively charged, as expected given the acidic conditions and confirmed by the 15N chemical shift. This makes a very close proximity less desirable, but this tendency can be overcome by the extensive stabilizing interactions characteristic of the highly stable amyloid fibrils. This is indeed seen in several of the amyloid-like peptide crystal systems examined by Eisenberg and colleagues (5), as well as simulation studies on the GNNQQNY IP structure(9, 22). Given the across-the-board intra-form intermolecular interactions, the IP nature of both crystal forms (which form under very similar conditions and have even been reported to form from GNNQQNY fibrils(37)), the appearance that steric zipper features may be preserved in the fibrils(30) and the fact that various simulations find it to be the most stable alignment for GNNQQNY (7, 13, 16, 18, 22), we are inclined to consider the MAS NMR results to strongly suggest a parallel in-register conformation. Note that seeing the intermolecular contacts only within a single conformer is specifically supportive of an IP β-sheet. Anti-parallel β-sheets, as observed in crystals observed for other short peptides(5) tend to display structurally different monomers within the β-sheet, instead of the internally structurally identical, parallel β-sheets seen in both GNNQQNY crystal forms. Solid state NMR studies of fibrils prepared from amylin fragments were found to be antiparallel within the β-sheets, as indicated by the detected of multiple sets of resonances of equal intensities(93). The latter observation also contrasts with the different intensities that characterize the different conformers detected in the GNNQQNY fibrils. Any anti-parallel arrangement involving two of the fibril forms would necessitate equal populations for those conformers. Note that in other experiments we have actually observed certain inter-form contacts (such as the one indicated in Figure 7), but these commonly involve side-chains and are more consistent with steric-zipper-like sheet-to-sheet contacts. However, those data on apparent sheet-to-sheet contacts necessitate a more detailed analysis and discussion that go beyond this publication.
If we compare the MAS NMR distance measurements to the equivalent distances in the crystal structures (Table 4), then we see that the measured distances in the fibrils tend to be slightly longer. However, these differences are within the margin of error of the experiments. There are several contributions that increase the uncertainty in these distances. First, the presence of multiple, partially overlapping conformers in all spectra makes it difficult to de-convolute the peaks. A second complication is related to the fact that the G7-labeled sites are near the peptide’s N-terminus. This was done for practical reasons and related to the availability (and cost) of various singly-labeled glycine variants (in contrast to appropriately backbone- and sidechain-protected Gln, Asn and Tyr). Since the Gly is near the N-terminus, increased dynamics may be expected (relative to the rest of the peptide). This is reflected in the temperature factors of the X-ray crystal structures(4, 5) and simulations on GNNQQNY aggregates(11, 22). Ongoing MAS NMR relaxation measurements suggest only moderate dynamics in the N-terminus and are the focus of a forthcoming publication. In any case, dynamics would effectively reduce the dipolar coupling and thus lead to overestimate of the distances. This would mean that the true distances might be shorter, therefore not changing the essence of our observed interactions, or how they reflect on the supramolecular arrangement.
Experimentally, the observation of an IP β-sheet is reminiscent not only of the structure of the crystalline forms of GNNQQNY (4, 5), but has also been observed in MAS NMR experiments on the parent protein Sup35p, Rnq1p and fragments of Ure2p (94-97). All of these are Gln- and Asn-rich prion proteins found in yeast, but this pattern of IP β-sheet formation is quite common in amyloid fibrils in general and appears to facilitate amyloid formation in proteins of non-symmetric (and purposely scrambled) primary sequences(96, 98).
Conclusion
The GNNQQNY crystal structures have stimulated a great deal of interest as an atomic-level perspective of the core structure of amyloid fibrils. It also has spawned considerable research using in silico methods to further our understanding of the amyloid fibril formation and stability. The structural and mechanistic correlation between the crystalline and fibrillar aggregates has remained enigmatic however, especially in the context of recent experimental data suggesting a transition between the fibrillar and crystalline conformations(37). We previously identified the presence of multiple conformations within the fibrils, which prominently included one with apparent non-β structure. We then also highlighted a number of indicators suggesting structural differences between the crystals and fibrils. Here we have presented new experimental data that explicitly address some of the key features of the crystal structures and show a number of basic similarities between the fibrils and crystals. Our data indicate that the conformers are actually all three predominantly β-sheet in structure, but with a highly localized distortion in one. In addition, we have presented experimental data supporting a parallel, in-register assembly into form-specific β-sheets. These observations should serve as valuable experimental reference data to allow the continued development of GNNQQNY as an model amyloid system in theoretical studies. Conversely, given their independently known crystal structures, GNNQQNY (and other amyloid-like crystalline peptides) proved to be useful as an experimental amyloid test system for the development and demonstration of MAS NMR and other experimental techniques. Here we have used the crystalline form of the peptide as a reference system to examine the effectiveness of the TALOS analysis, NCCN torsion angle measurements and 13C-13C distance measurements in a realistic amyloid-like β-sheet context, of an independently known conformation.
Questions do remain regarding the role of these conformers within the fibrils. Such issues are the target of continued MAS NMR studies, but will also benefit from experimentally-informed in silico studies. For instance, it will be interesting to see whether a twisting or distortion of the peptide backbone is observed in fibrils assembled in silico. In the context of our results, theoretical studies may also be able to address the question of how the localized backbone twist (of conformer 2) may be accommodated into the fibrils and their seemingly IP β-sheet arrangement. As it is unclear whether this has been observed in the current computational studies, it may be important to more closely approximate the experimental conditions. In this regard, we point out that the concentration and pH appear to play critical roles that have not necessarily been sufficiently examined in the computational studies. Overall, our data point to an increased complexity in the behavior of the fibrillar form of GNNQQNY, even though it does share a variety of features with its crystals that continue to serve as a canonical model of the cross-β spine.
Supplementary Material
Acknowledgment
We want to thank Dr. Marc Caporini, Dr. Vikram Bajaj and Marvin Bayro for their help and feedback, and gratefully acknowledge Dr. Mikhail Veshtort for providing the SPINEVOLUTION software. Molecular schematics were generated using Pymol (99).
Footnotes
This research was supported by the National Institutes of Health through grants EB-003151 and EB-002026.
Supporting Information Available: Assignment spectra, Tyr chemical shifts, and chemical shift comparisons to the crystals; CSI and TALOS data for the fibrils; additional experimental details on the MAS NMR experiments; additional details on the torsion angle data and analyses; schematics of the inter-strand, intra-sheet contacts in in-register parallel β-sheets; backbone schematics for the fibril conformers. These supplemental materials may be accessed free of charge online at http://pubs.acs.org.
- CSI
- chemical shift indexing
- DQ
- double-quantum
- DRAWS
- dipolar recoupling with a windowless sequence
- IP
- in-register parallel
- MAS
- magic angle spinning
- R2
- rotational resonance
- R2W
- rotational resonance width
- REDOR
- rotational echo double resonance
- SQ
- single-quantum
- TEDOR
- transferred echo double resonance
- TPPM
- two-pulse phase-modulation
References
- 1.Chiti F, Dobson CM. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Langkilde AE, Vestergaard B. Methods for structural characterization of prefibrillar intermediates and amyloid fibrils. FEBS Lett. 2009;583:2600–2609. doi: 10.1016/j.febslet.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 3.Ma B, Nussinov R. Simulations as analytical tools to understand protein aggregation and predict amyloid conformation. Curr. Opin. Chem. Biol. 2006;10:445–452. doi: 10.1016/j.cbpa.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 4.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 6.Balbirnie M, Grothe R, Eisenberg DS. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated beta-sheet structure for amyloid. Proc. Natl. Acad. Sci. USA. 2001;98:2375–2380. doi: 10.1073/pnas.041617698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gsponer J, Haberthür U, Caflisch A. The role of side-chain interactions in the early steps of aggregation: Molecular dynamics simulations of an amyloid-forming peptide from the yeast prion Sup35. Proc. Natl. Acad. Sci. 2003;100:5154–5159. doi: 10.1073/pnas.0835307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cecchini M, Rao F, Seeber M, Caflisch A. Replica exchange molecular dynamics simulations of amyloid peptide aggregation. J. Chem. Phys. 2004;121:10748–10756. doi: 10.1063/1.1809588. [DOI] [PubMed] [Google Scholar]
- 9.Lipfert J, Franklin J, Wu F, Doniach S. Protein Misfolding and Amyloid Formation for the Peptide GNNQQNY from Yeast Prion Protein Sup35: Simulation by Reaction Path Annealing. J. Mol. Biol. 2005;349:648–658. doi: 10.1016/j.jmb.2005.03.083. [DOI] [PubMed] [Google Scholar]
- 10.Fernández A. What factor drives the fibrillogenic association of beta-sheets? FEBS Lett. 2005;579:6635–6640. doi: 10.1016/j.febslet.2005.10.058. [DOI] [PubMed] [Google Scholar]
- 11.Zheng J, Ma B, Tsai C-J, Nussinov R. Structural Stability and Dynamics of an Amyloid-Forming Peptide GNNQQNY from the Yeast Prion Sup-35. Biophys. J. 2006;91:824–833. doi: 10.1529/biophysj.106.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esposito L, Pedone C, Vitagliano L. Molecular dynamics analyses of cross-beta-spine steric zipper models: Beta-Sheet twisting and aggregation. Proc. Natl. Acad. Sci. 2006;103:11533–11538. doi: 10.1073/pnas.0602345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Chen H, Bai H, Lai L. Molecular Dynamics Simulations on the Oligomer-Formation Process of the GNNQQNY Peptide from Yeast Prion Protein Sup35. Biophys. J. 2007;93:1484–1492. doi: 10.1529/biophysj.106.100537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C, Wang Z, Lei H, Zhang W, Duan Y. Dual Binding Modes of Congo Red to Amyloid Protofibril Surface Observed in Molecular Dynamics Simulations. J. Am. Chem. Soc. 2007;129:1225–1232. doi: 10.1021/ja0662772. [DOI] [PubMed] [Google Scholar]
- 15.Tsemekhman K, Goldschmidt L, Eisenberg D, Baker D. Cooperative hydrogen bonding in amyloid formation. Protein Sci. 2007;16:761–764. doi: 10.1110/ps.062609607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strodel B, Whittleston CS, Wales DJ. Thermodynamics and Kinetics of Aggregation for the GNNQQNY Peptide. J. Am. Chem. Soc. 2007;129:16005–16014. doi: 10.1021/ja075346p. [DOI] [PubMed] [Google Scholar]
- 17.Knowles TP, Fitzpatrick AW, Meehan S, Mott HR, Vendruscolo M, Dobson CM, Welland ME. Role of Intermolecular Forces in Defining Material Properties of Protein Nanofibrils. Science. 2007;318:1900–1903. doi: 10.1126/science.1150057. [DOI] [PubMed] [Google Scholar]
- 18.Meli M, Morra G, Colombo G. Investigating the Mechanism of Peptide Aggregation: Insights from Mixed Monte Carlo-Molecular Dynamics Simulations. Biophys. J. 2008;94:4414–4426. doi: 10.1529/biophysj.107.121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Simone A, Esposito L, Pedone C, Vitagliano L. Insights into Stability and Toxicity of Amyloid-Like Oligomers by Replica Exchange Molecular Dynamics Analyses. Biophys. J. 2008;95:1965–1973. doi: 10.1529/biophysj.108.129213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Tan C, Chen H-F, Luo R. All-Atom Computer Simulations of Amyloid Fibrils Disaggregation. Biophys. J. 2008;95:5037–5047. doi: 10.1529/biophysj.108.131672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esposito L, Paladino A, Pedone C, Vitagliano L. Insights into Structure, Stability, and Toxicity of Monomeric and Aggregated Polyglutamine Models from Molecular Dynamics Simulations. Biophys. J. 2008;94:4031–4040. doi: 10.1529/biophysj.107.118935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vitagliano L, Esposito L, Pedone C, De Simone A. Stability of single sheet GNNQQNY aggregates analyzed by replica exchange molecular dynamics: Antiparallel versus parallel association. Biochem. Biophys. Res. Commun. 2008;377:1036–1041. doi: 10.1016/j.bbrc.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 23.Periole X, Rampioni A, Vendruscolo M, Mark AE. Factors That Affect the Degree of Twist in Beta-Sheet Structures: A Molecular Dynamics Simulation Study of a Cross-Beta Filament of the GNNQQNY Peptide. The Journal of Physical Chemistry B. 2009;113:1728–1737. doi: 10.1021/jp8078259. [DOI] [PubMed] [Google Scholar]
- 24.Berryman JT, Radford SE, Harris SA. Thermodynamic Description of Polymorphism in Q- and N-Rich Peptide Aggregates Revealed by Atomistic Simulation. Biophys. J. 2009;97:1–11. doi: 10.1016/j.bpj.2009.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy G, Straub JE, Thirumalai D. Dynamics of locking of peptides onto growing amyloid fibrils. Proc. Natl. Acad. Sci. 2009;106:11948–11953. doi: 10.1073/pnas.0902473106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brovchenko I, Singh G, Winter R. Aggregation of Amyloidogenic Peptides near Hydrophobic and Hydrophilic Surfaces. Langmuir. 2009;25:8111–8116. doi: 10.1021/la9006058. [DOI] [PubMed] [Google Scholar]
- 27.Park J, Kahng B, Hwang W. Thermodynamic Selection of Steric Zipper Patterns in the Amyloid Cross-β Spine. PLoS Comput Biol. 2009;5:e1000492. doi: 10.1371/journal.pcbi.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glabe CG. Structural Classification of Toxic Amyloid Oligomers. J. Biol. Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diaz-Avalos R, Long C, Fontano E, Balbirnie M, Grothe R, Eisenberg D, Caspar DLD. Cross-beta order and diversity in nanocrystals of an amyloid-forming peptide. J. Mol. Biol. 2003;330:1165–1175. doi: 10.1016/s0022-2836(03)00659-4. [DOI] [PubMed] [Google Scholar]
- 30.Van der Wel PCA, Lewandowski JR, Griffin RG. Solid state NMR study of amyloid nanocrystals and fibrils formed by the peptide GNNQQNY from yeast prion protein Sup35p. J. Am. Chem. Soc. 2007;129:5117–5130. doi: 10.1021/ja068633m. [DOI] [PubMed] [Google Scholar]
- 31.Jimenez JL, Guijarro JI, Orlova E, Zurdo J, Dobson CM, Sunde M, Saibil HR. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J. 1999;18:815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jimenez JL, Nettleton EJ, Bouchard M, Robinson CV, Dobson CM, Saibil HR. The protofilament structure of insulin amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferguson N, Berriman J, Petrovich M, Sharpe TD, Finch JT, Fersht AR. Rapid amyloid fiber formation from the fast-folding WW domain FBP28. Proc. Natl. Acad. Sci. 2003;100:9814–9819. doi: 10.1073/pnas.1333907100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saiki M, Honda S, Kawasaki K, Zhou D, Kaito A, Konakahara T, Morii H. Higher-order Molecular Packing in Amyloid-like Fibrils Constructed with Linear Arrangements of Hydrophobic and Hydrogen-bonding Side-chains. J. Mol. Biol. 2005;348:983–998. doi: 10.1016/j.jmb.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 35.Petkova AT, Leapman RD, Guo Z, Yau W-M, Mattson MP, Tycko R. Self-Propagating, Molecular-Level Polymorphism in Alzheimer’s β-Amyloid Fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 36.Luca S, Yau WM, Leapman R, Tycko R. Peptide Conformation and Supramolecular Organization in Amylin Fibrils: Constraints from Solid-State NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall KE, Hicks MR, Williams TL, Hoffmann SV, Rodger A, Dafforn TR, Serpell LC. Characterizing the Assembly of the Sup35 Yeast Prion Fragment, GNNQQNY: Structural Changes Accompany a Fiber-to-Crystal Switch. Biophys. J. 2010;98:330–338. doi: 10.1016/j.bpj.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 39.Heise H. Solid-State NMR Spectroscopy of Amyloid Proteins. ChemBioChem. 2008;9:179–189. doi: 10.1002/cbic.200700630. [DOI] [PubMed] [Google Scholar]
- 40.Costa PR, Gross JD, Hong M, Griffin RG. Solid-state NMR Measurement of ψ in Peptides: a NCCN 2Q-Heteronuclear Local Field Experiment. Chem. Phys. Lett. 1997;280:95–103. [Google Scholar]
- 41.Gregory DM, Mitchell DJ, Stringer JA, Kiihne S, Shiels JC, Callahan J, Mehta MA, Drobny GP. Windowless dipolar recoupling: the detection of weak dipolar couplings between spin 1/2 nuclei with large chemical shift anisotropies. Chem. Phys. Lett. 1995;246:654–663. [Google Scholar]
- 42.Gregory DM, Benzinger TLS, Burkoth TS, Miller-Auer H, Lynn DG, Meredith SC, Botto RE. Dipolar recoupling NMR of biomolecular self-assemblies: determining inter- and intrastrand distances in fibrilized Alzheimer’s β-amyloid peptide. Solid State Nucl. Magn. Reson. 1998;13:149–166. doi: 10.1016/s0926-2040(98)00086-1. [DOI] [PubMed] [Google Scholar]
- 43.Benzinger TLS, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, Botto RE, Meredith SC. Propagating structure of Alzheimer’s β-amyloid(10-35) is parallel β-sheet with residues in exact register. Proc. Natl. Acad. Sci. USA. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporini M, Bajaj VS, Griffin RG. High Resolution Determination of Interstrand Distances in Amyloid Fibrils. 2010 doi: 10.1021/jp106675h. in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caporini MA. Ph.D. Thesis. Massachusetts Institute of Technology; Cambridge, MA: 2008. Structural Studies of Amyloid Fibrils Using Solid-State NMR. [Google Scholar]
- 46.Bajaj VS. Ph.D. Thesis. Massachusetts Institute of Technology; Cambridge, MA: 2007. Dynamic Nuclear Polarization in Biomolecular Solid State NMR: Methods and Applications in Peptides and Membrane Proteins. [Google Scholar]
- 47.Costa PR, Sun B, Griffin RG. Rotational resonance NMR: separation of dipolar coupling and zero quantum relaxation. J. Magn. Reson. 2003;164:92–103. doi: 10.1016/s1090-7807(03)00083-1. [DOI] [PubMed] [Google Scholar]
- 48.Ramachandran R, Ladizhansky V, Bajaj VS, Griffin RG. 13C-13C Rotational Resonance Width Distance Measurements in Uniformly 13C-Labeled Peptides. J. Am. Chem. Soc. 2003;125:15623–15629. doi: 10.1021/ja037761x. [DOI] [PubMed] [Google Scholar]
- 49.Raleigh DP, Levitt MH, Griffin RG. Rotational resonance in solid state NMR. Chem. Phys. Lett. 1988;146:71–76. [Google Scholar]
- 50.Colombo MG, Meier BH, Ernst RR. Rotor-driven spin diffusion in natural-abundance C-13 spin systems. Chem. Phys. Lett. 1988;146:189. [Google Scholar]
- 51.Levitt MH, Raleigh DP, Creuzet F, Griffin RG. Theory and simulations of homonuclear spin pair systems in rotating solids. J. Chem. Phys. 1990;92:6347–6364. [Google Scholar]
- 52.Ramachandran R, Lewandowski JR, van der Wel PCA, Griffin RG. Multipole multimode Floquet theory of rotational resonance width experiments: 13C-13C distance measurements in uniformly labeled solids. J. Chem. Phys. 2006;124:214107. doi: 10.1063/1.2194905. [DOI] [PubMed] [Google Scholar]
- 53.van der Wel PCA, Eddy MT, Ramachandran R, Griffin RG. Targeted 13C-13C distance measurements in a microcrystalline protein via J-decoupled rotational resonance width measurements. ChemPhysChem. 2009;10:1566–1663. doi: 10.1002/cphc.200900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barnes AB, Andreas LB, Huber M, Ramachandran R, van der Wel PCA, Veshtort M, Griffin RG, Mehta MA. High-resolution solid-state NMR structure of Alanyl-Prolyl-Glycine. J. Magn. Reson. 2009;200:95–100. doi: 10.1016/j.jmr.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gullion T, Schaefer J. Rotational-echo double-resonance NMR. J. Magn. Reson. 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 56.Hing AW, Vega S, Schaefer J. Transferred-echo double-resonance NMR. J. Magn. Reson. 1992;96:205–209. [Google Scholar]
- 57.Jaroniec CP, Filip C, Griffin RG. 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly 13C, 15N-labeled solids. J. Am. Chem. Soc. 2002;124:10728–10742. doi: 10.1021/ja026385y. [DOI] [PubMed] [Google Scholar]
- 58.Madine J, Jack E, Stockley PG, Radford SE, Serpell LC, Middleton DA. Structural Insights into the Polymorphism of Amyloid-Like Fibrils Formed by Region 20–29 of Amylin Revealed by Solid-State NMR and X-ray Fiber Diffraction. J. Am. Chem. Soc. 2008;130:14990–15001. doi: 10.1021/ja802483d. [DOI] [PubMed] [Google Scholar]
- 59.Gordon DJ, Balbach JJ, Tycko R, Meredith SC. Increasing the Amphiphilicity of an Amyloidogenic Peptide Changes the [beta]-Sheet Structure in the Fibrils from Antiparallel to Parallel. Biophys. J. 2004;86:428–434. doi: 10.1016/S0006-3495(04)74119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verel R, Tomka IT, Bertozzi C, Cadalbert R, Kammerer RA, Steinmetz MO, Meier BH. Polymorphism in an Amyloid-Like Fibril-Forming Model Peptide13. Angew. Chem. Intl. Ed. 2008;47:5842–5845. doi: 10.1002/anie.200800021. [DOI] [PubMed] [Google Scholar]
- 61.De Paepe G, Lewandowski JR, Loquet A, Bockmann A, Griffin RG. Proton assisted recoupling and protein structure determination. J. Chem. Phys. 2008;129:245101. doi: 10.1063/1.3036928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stringer JA, Bronnimann CE, Mullen CG, Zhou DH, Stellfox SA, Li Y, Williams EH, Rienstra CM. Reduction of RF-induced sample heating with a scroll coil resonator structure for solid-state NMR probes. J Magn Reson. 2005;173:40–48. doi: 10.1016/j.jmr.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 63.Takegoshi K, Nakamura S, Terao T. C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 64.Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J. Am. Chem. Soc. 2004;126:7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- 65.Lewandowski J, De Paëpe G, Griffin RG. Proton Assisted Insensitive Nuclei Cross Polarization. J. Am. Chem. Soc. 2007;129:728–729. doi: 10.1021/ja0650394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schaefer J, Mckay RA, Stejskal EO. Double-Cross-Polarization NMR of Solids. J. Magn. Reson. 1979;34:443–447. [Google Scholar]
- 67.Baldus M, Petkova AT, Herzfeld J, Griffin RG. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol. Phys. 1998;95:1197–1207. [Google Scholar]
- 68.Petkova AT, Baldus M, Belenky M, Hong M, Griffin RG, Herzfeld J. Backbone and side chain assignment strategies for multiply labeled membrane peptides and proteins in the solid state. J. Magn. Reson. 2003;160:1–12. doi: 10.1016/s1090-7807(02)00137-4. [DOI] [PubMed] [Google Scholar]
- 69.Egorova-Zachernyuk TA, Hollander J, Fraser N, Gast P, Hoff AJ, Cogdell J, de Groot HJM, Baldus M. Heteronuclear 2D-correlations in a uniformly [13C, 15N] labeled membrane-protein complex at ultra-high magnetic fields. J. Biomol. NMR. 2001;19:243–253. doi: 10.1023/a:1011235417465. [DOI] [PubMed] [Google Scholar]
- 70.Pauli J, Baldus M, van Rossum B-J, de Groot HJM, Oschkinat H. Backbone and side-chain 13C and 15N signal assignments of the α-Spectrin SH3 Domain by magic angle spinning solid-state NMR at 17.6 Tesla. CHEMBIOCHEM. 2001;2:272–281. doi: 10.1002/1439-7633(20010401)2:4<272::AID-CBIC272>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 71.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J. Chem. Phys. 1995;103:6951–6957. [Google Scholar]
- 72.Morcombe CR, Zilm KW. Chemical shift referencing in MAS solid state NMR. J. Mag. Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- 73.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. Pure & Appl. Chem. 1998;70:117–142. doi: 10.1046/j.1432-1327.1998.2560001.x. [DOI] [PubMed] [Google Scholar]
- 74.Harris RK, Becker ED, Cabral de Menezes SM, Goodfellow R, Granger P. NMR Nomenclature: Nuclear Spin Properties and Conventions for Chemical Shifts. Solid State Nuc. Mag. Reson. 2002;22:458–483. doi: 10.1006/snmr.2002.0063. [DOI] [PubMed] [Google Scholar]
- 75.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 76.Goddard TD, Kneller DG. SPARKY 3. SPARKY 3 ed. University of California; San Francisco: [Google Scholar]
- 77.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins: Structure, Function, and Bioinformatics. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 78.Cornilescu G, Delaglio F, Bax A. Protein Backbone Angle Restraints from Searching a Database for Chemical Shift and Sequence Homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 79.Feng X, Lee YK, Sandström D, Eden M, Maisel H, Sebald A, Levitt MH. Direct determination of a molecular torsional angle by solid-state NMR. Chem. Phys. Lett. 1996;257:314–320. [Google Scholar]
- 80.Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG. Fivefold symmetric homonuclear dipolar recoupling in rotating solids: Application to double quantum spectroscopy. J. Chem. Phys. 1999;110:7983–7992. [Google Scholar]
- 81.Veshtort M, Griffin RG. SPINEVOLUTION: A powerful tool for the simulation of solid and liquid state NMR experiments. J. Magn. Reson. 2006;178:248–282. doi: 10.1016/j.jmr.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 82.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 83.Zhang H, Neal S, Wishart DS. RefDB: A database of uniformly referenced protein chemical shifts. J. Biomol. NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 84.Jaroniec CP, MacPhee CE, Astrof NS, Dobson CM, Griffin RG. Molecular conformation of a peptide fragment of transthyretin in an amyloid fibril. Proc. Natl. Acad. Sci. 2002;99:16748–16753. doi: 10.1073/pnas.252625999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ma Z, Halling MD, Solum MS, Harper JK, Orendt AM, Facelli JC, Pugmire RJ, Grant DM, Amick AW, Scott LT. Ring Current Effects in Crystals. Evidence from 13C Chemical Shift Tensors for Intermolecular Shielding in 4,7-Di-t-butylacenaphthene versus 4,7-Di-t-butylacenaphthylene. J. Phys. Chem. A. 2007;111:2020–2027. doi: 10.1021/jp068400h. [DOI] [PubMed] [Google Scholar]
- 86.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Cryst. D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 87.Collaborative Computational Project Number 4 The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. D. 1994;50 doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 88.Bajaj VS, van der Wel PCA, Griffin RG. Observation of a Low-Temperature, Dynamically Driven Structural Transition in a Polypeptide by Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2008;131:118–128. doi: 10.1021/ja8045926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Debelouchina GT, Bayro MJ, Van Der Wel PCA, Caporini MA, Barnes AB, Rosay M, Maas WE, Griffin RG. Dynamic nuclear polarization-enhanced solid-state NMR spectroscopy of GNNQQNY nanocrystals and amyloid fibrils. Phys Chem Chem Phys. 2010:1–11. doi: 10.1039/c003661g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheung M-S, Maguire ML, Stevens TJ, Broadhurst RW. DANGLE: A Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J. Magn. Reson. 202:223–233. doi: 10.1016/j.jmr.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 91.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Simone A, Dodson GG, Fraternali F, Zagari A. Water molecules as structural determinants among prions of low sequence identity. FEBS Lett. 2006;580:2488–2494. doi: 10.1016/j.febslet.2006.02.083. [DOI] [PubMed] [Google Scholar]
- 93.Nielsen JT, Bjerring M, Jeppesen MD, Pedersen RO, Pedersen JM, Hein Kim L., Vosegaard T, Skrydstrup T, Otzen DE, Nielsen NC. Unique Identification of Supramolecular Structures in Amyloid Fibrils by Solid-State NMR Spectroscopy. Angew. Chem. Intl. Ed. 2009;48:2118–2121. doi: 10.1002/anie.200804198. [DOI] [PubMed] [Google Scholar]
- 94.Shewmaker F, Kryndushkin D, Chen B, Tycko R, Wickner RB. Two prion variants of Sup35p have in-register parallel beta-sheet structures, independent of hydration. Biochemistry. 2009;48:5074–5082. doi: 10.1021/bi900345q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register beta-sheet structure. Proc Natl Acad Sci USA. 2008;105:2403–2408. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baxa U, Wickner RB, Steven AC, Anderson DE, Marekov LN, Yau W-M, Tycko R. Characterization of beta-sheet structure in Ure2p1-89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 97.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ross ED, Minton A, Wickner RB. Prion domains: sequences, structures and interactions. Nat Cell Biol. 2005;7:1039–1044. doi: 10.1038/ncb1105-1039. [DOI] [PubMed] [Google Scholar]
- 99.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


