Abstract
Peroxisome proliferator-activated receptorδ (PPARδ) regulates a multiplicity of physiological processes associated with glucose and lipid metabolism, inflammation, and proliferation. One or more of these processes likely create risk factors associated with the ability of PPARδ agonists to promote tumorigenesis in some organs. In the present study, we describe a new gastric tumor mouse model that is dependent on the potent and highly selective PPARδ agonist GW501516 following carcinogen administration. The progression of gastric tumorigenesis was rapid as determined by magnetic resonance imaging and resulted in highly metastatic squamous cell carcinomas of the forestomach within two months. Tumorigenesis was associated with gene expression signatures indicative of cell adhesion, invasion, inflammation, and metabolism. Increased PPARδ expression in tumors correlated with increased PDK1, Akt, β-catenin, and S100A9 expression. The rapid development of metastatic gastric tumors in this model will be useful for evaluating preventive and therapeutic interventions in this disease.
1. Introduction
Gastric cancer is the second leading cause of cancer-related death globally, particularly in developing countries, due to its metastatic nature at the time of diagnosis [1, 2]. Among the many risk factors for gastric cancer are diet, smoking, and inflammation associated with H. pylori infection [3–5] that are likely exacerbated in patients with proinflammatory gene polymorphisms [6].
PPARs are ligand-dependent nuclear receptors that regulate expression of a multiplicity of genes associated with metabolic disorders, such as type II diabetes and lipodystrophies [7, 8]. PPARs consist of the α, γ and δ isotypes that regulate not only glucose and lipid metabolism, but also proliferation, inflammation, and angiogenesis [9–13]. PPARδ expression is increased in breast, colon, and head and neck cancers [9, 14–17] and is associated with a more aggressive phenotype in breast cancer cells [18]. The selective PPARδ agonist GW501516 acts as a tumor promoter in mammary carcinogenesis [19] and colon tumorigenesis [15, 20, 21], whereas disruption of PPARδ expression blocks mammary [22] and colon tumorigenesis [23, 24]. PPARδ is regulated by risk factors implicated in cancer progression, including K-Ras [25], Wnt [26], and Pges/Cox2 [22, 27], and is associated with activation of angiogenesis [20, 28–30]. PPARδ regulates proinflammatory signaling through NFκB and IL-1β [31], and is activated by metabolites of arachidonic acid metabolism that serve as PPAR ligands [14, 29, 32]. Notwithstanding these results, there are several reports showing that azoxymethane-induced colon carcinogenesis is increased in PPARδ nullizygous mice [33, 34] and by the selective PPARδ ligand GW0742 [35] (reviewed in [36]). Differences between the various nullizygous models that may account for some of these disparities have been discussed [9, 21].
Here we demonstrate that activation of PPARδ by a selective agonist, GW501516, rapidly induces highly metastatic gastric tumors following carcinogen administration, which expressed a markedly increased inflammatory gene expression signature. These findings suggest a crucial role for PPARδ in the progression of this disease.
2. Materials and Methods
2.1. Materials
GW501516 was synthesized as previously described [37] and was provided by the Chemoprevention Branch, National Cancer Institute, NIH, Bethesda, MD.
2.2. Carcinogenesis
DMBA (Sigma-Aldrich, St Louis, MO) was dissolved in cottonseed oil at a concentration of 10 mg/ml, and six week-old female FVB mice were administered 4 weekly doses of 1 mg DMBA by gavage. Animals were fed a standard diet or a diet supplemented with 0.005% (w/w) GW501516 beginning one day after the last dose of DMBA as previously described [19]. Mice were euthanized when gastric tumors reached ≥2cm3 as determined by MRI imaging or if mice became moribund. All protocols were approved by the Georgetown University Animal Care and Use Committee.
2.3. Magnetic Resonance Imaging
MRI was performed on a 7.0 Tesla Bruker horizontal spectrometer/imager with a 20 cm bore equipped with 100 gauss/cm microimaging gradients and run by Paravision 4.0 software in the Preclinical Imaging Research Laboratory, Lombardi Comprehensive Cancer Center. Mice were anesthetized using 1.5% isoflurane and 30% nitrous oxide, positioned in a custom-made stereotaxic animal holder with temperature and respiration control, and imaged in a 72 mm birdcage radiofrequency volume coil as previously described [38]. The imaging protocol used was a T2-weighted RARE (rapid acquisition with refocused echos) two-dimensional sequence with the following parameters: Matrix: 256, TR: 5822 msec, TE: 36 msec, number of averages: 4, RARE factor: 8, FOV: 4.0 cm, number of slices: 4, slice thickness: 0.5 mm, and with respiratory gating to account for breathing movement.
2.4. Histopathology and Immunohistochemistry (IHC)
Stomach and tumors were excised, and formalin-fixed, paraffin-embedded sections were prepared for H&E staining and IHC. Antigen retrieval was carried out by incubation of tissue sections in 10 mM sodium citrate buffer (pH 6.0) for 20 min at a subboiling temperature in an electric steamer as previously described [19, 39]. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide for 10 min and incubated for 30 min with blocking solution (10% goat serum in Tris-buffered saline), followed by incubation overnight at 4°C with the appropriate primary antibody diluted in blocking solution. Biotin-conjugated secondary antibodies were diluted in TBS containing 0.1% Tween-20 and incubated for 30 min at room temperature using the ABC Vectastain (Vector Laboratories) detection system and diaminobenzidine (Pierce), and slides were counterstained with Harris-modified hematoxylin (Fisher Scientific) and dehydrated and mounted in Permount (Fisher Scientific). The following antibodies and their dilutions were used: anti-CK14 (1 : 200, ms-115-P1, Neomarkers), anti-CK18 (1 : 200, sc-7197, Santa Cruz Biotechnology), PDK1 (1 : 50, sc-17765, Santa Cruz Biotechnology), anti-pS473 Akt (1 : 200, 40515, Cell Signaling Biotechnology), anti-β-catenin (1 : 50, sc-7963, Santa-Cruz Biotechnology) and anti-S100a9 (1 : 50, sc-65580, Santa Cruz Biotechnology).
2.5. Gene Microarray Analysis
Three groups of tissues were analyzed: (1) gastric tumors, (2) forestomach (nonglandular) from GW501516-treated mice, and (3) forestomach from DMBA-treated mice. Tissue was excised, washed in phosphate-buffered saline, and stored in RNAlater (Ambion) at ‒20°C until RNA extraction. Tissue was snap-frozen in liquid nitrogen, pulverized in a mortar and pestle, and RNA extracted using an RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. RNA purity was assessed by an A260/A280 ratio of ≥1.9, and by the integrity of 18S and 28S rRNA using an Agilent microfluidic chip. Array analysis was carried out on cRNA prepared from equal amounts of RNA (1 μg) pooled from 5 mice per group as previously described [19, 40]. Biotin-labeled cRNA was fragmented at 94°C for 35 min and hybridized overnight to an Affymetrix mouse 430A 2.0 GeneChip representing approximately 14,000 annotated mouse genes. GeneChips were scanned with an Agilent Gene Array scanner, and grid alignment and raw data generation with the Affymetrix GeneChip Operating software 1.1. A noise value (Q) based on the variance of low-intensity probe cells was used to calculate a minimum threshold for each GeneChip. Samples were averaged and data refined by eliminating genes with signal intensities <300 in both comparison groups, and heat maps were generated from ≥3-fold changes in gene expression normalized to untreated forestomach using unsupervised hierarchical cluster analysis as previously described [41]. Gene ontology analysis utilized Ariadne Pathway Studio version 7.1
2.6. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted as described above, and equal amounts of RNA (1 μg) were pooled from each group (five samples per group) and reverse transcribed with the Omniscript RT kit (Qiagen) in a total volume of 20 μL as previously described [19, 39]. PCR was performed in triplicate using an ABI-Prism 7700 (Applied Biosystems, Foster City, CA) with SYBRGreen I detection (Qiagen) according to the manufacturer's protocol. Amplification using the appropriate primers (see Table S1 in supplementary material available at doi:10.1155/2010/571783) was confirmed by ethidium bromide staining of the PCR products on an agarose gel. The expression of each target gene was normalized to GAPDH and is presented as the ratio of the target gene to GADPH expression calculated using the formula, 2−ΔCt, where ΔCt = CtTarget − Ct18s.
3. Results
3.1. PPARδ Agonist GW501516 Rapidly Promotes Gastric Tumorigenesis
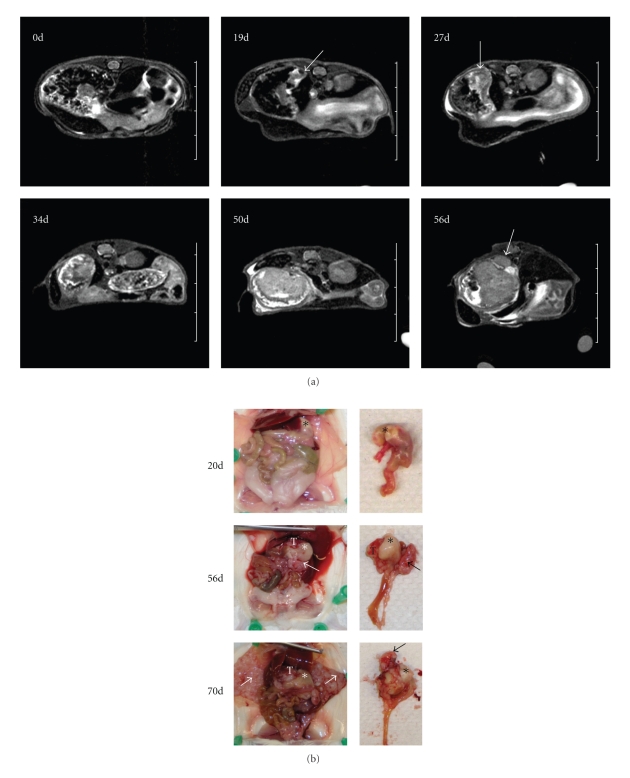
Mice maintained on a diet supplemented with PPARδ agonist GW501516 following carcinogen administration resulted in the rapid development of gastric tumors in 12/15 animals, whereas treatment with either GW501516 or DMBA alone was not tumorigenic (Table 1). To follow the onset and progression of tumorigenesis more precisely, five mice were monitored by MRI (Figure 1(a)). Tumors were visible as early as 19 days after beginning the GW501516 diet and appeared to initiate in the forestomach (Figure 1(a)). By 50 days, tumor had filled the stomach lumen, and by 56 days it had extravasated through the gastric wall (Figure 1(a)). Gross inspection of the stomach confirmed that the tumor was confined within the stomach at day 20 but had invaded through the stomach wall forming local metastases by day 56 (Figure 1(b)). Mice showed signs of morbidity between days 63 and 70 (mean survival 67 days), where metastases were present throughout the mesentery and adjacent serosal organ surfaces including the abdominal wall (Figure 1(b)).
Table 1.
Gastric tumor occurrence after treatment with DMBA and GW501516.
Figure 1.
PPARδ agonist GW501516 induces gastric tumorigenesis after DMBA treatment. (a) In vivo axial T2-weighted abdominal MR Images showing gastric tumor progression at the indicated time points; d, number of days animals were administered the GW501516-supplemented diet. Tumor growth initiated in the forestomach (arrow) at 19 days, rapid tumor growth between 27 and 50 days, and tumor invasion through the stomach (arrow) at 56 days. (b) Gross morphology of tumors at indicated time points. Forestomach (*), gastric tumors (T) and metastases (arrows) are indicated. Tumor is confined within the stomach at 20 days. Invasion of primary tumor through the stomach (T) and metastases (arrow) were evident at 56 days, and intraperitoneal metastases along the abdominal wall (white arrows) occurred at 70 days.
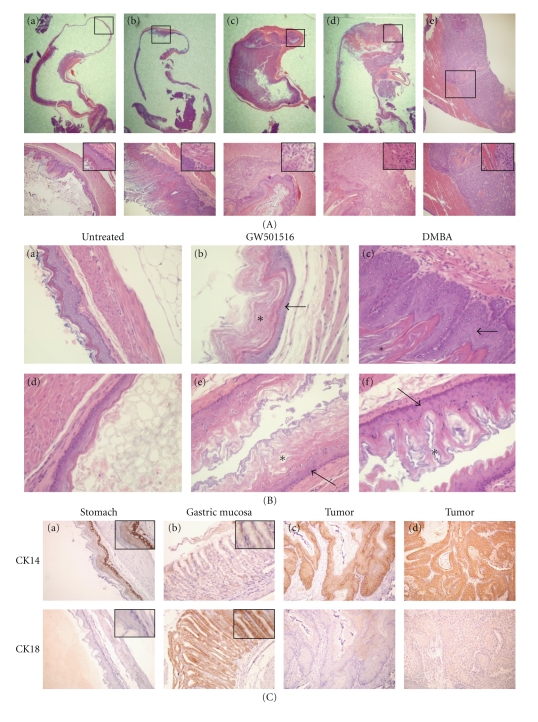
Primary tumors and metastases were uniformly squamous cell carcinomas with varying degrees of keratinization (Figure 2(A)). Animals fed the GW501516 diet for six months without prior DMBA treatment did not exhibit hyperplasia or dysplasia (Figure 2(B)), and DMBA treatment alone produced squamous cell hyperplasia of the forestomach without signs of dysplasia (Figures 2(A) and 2(B)). No changes occurred in the gastric mucosa resulting from DMBA and GW501516 treatment alone (data not shown), and esophageal squamous epithelium was unaffected by DMBA treatment (Figure 2(B)). Gastric tumors were positive for the squamous basal cell marker CK14, and negative for the columnar epithelial cell marker CK18 [42] (Figure 2(C)).
Figure 2.
Pathophysiology of gastric tumorigenesis. (A) Histological changes in the forestomach after GW501516 and/or DMBA treatment. Upper panel: Stereoscopic images of H&E stained sections of the stomach in (a) untreated, (b) DMBA-treated, (c, d) DMBA plus GW501516-treated mice, and (e) metastases of the abdominal wall. Lower panel: higher magnification (200x) of the boxes in areas in the upper panel. Insets, magnification 400x. (a) Morphology of the normal forestomach wall. (b) Squamous epithelial hyperplasia, where the basal membrane is well defined. (c) Squamous cell carcinoma. (d) Invasive squamous cell carcinoma showing disruption of the basement membrane. (e) Metastatic squamous carcinoma showing invasion into the abdominal wall. (B) Histological changes in the stomach and esophagus after treatment with either GW501516 or DMBA. H&E sections of the forestomach wall (a, b, c) and esophagus (d, e, f) six months after administration of GW501516 or five months after treatment with DMBA. Both GW501516 and DMBA were associated with increased keratinization (*) of squamous epithelium (arrow). Magnification 400x. (C) Cytokeratin 14 (CK14) and cytokeratin18 (CK18) expression in gastric tissue and tumors. Squamous epithelium of the forestomach (a), gastric mucosa (b), and gastric tumors (c, d). Tumors are CK14+/CK18− indicating a squamous epithelial origin. Magnification 200x, insets, 400x.
3.2. Differential Gene Expression in Gastric Tumors and Stomach
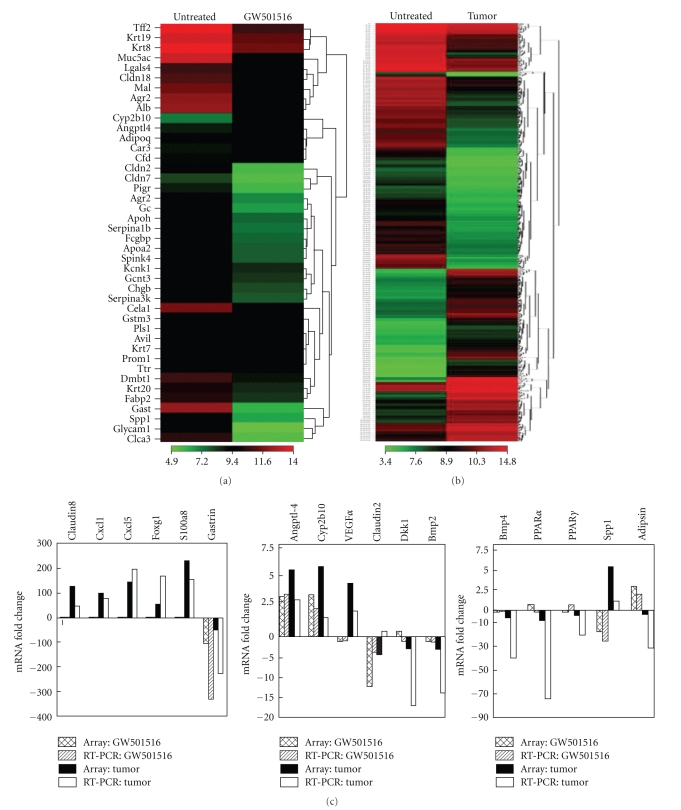
Tumors manifested a marked inflammatory phenotype denoted by increased expression of chemokines Cxcl-1,-2,-5,-9,-14 and Ccl-2,-3,-8, S100a8, S100a9, and S100a3 and interleukins IL-1β, IL-6, and IL-24 (Table 2, Figure 3(b), and Table S2). In addition to these changes, increased expression of prostaglandin metabolism genes Ptgs2/Cox2 and Ptges and reduced expression of PPARγ and PPARα were noted. To determine if these changes were tumor specific, gene expression was assessed in stomach tissue after treatment with either GW501516 for seven days (Figure 3(a), Table S3) or DMBA for four weeks (Table S4). GW501516 increased expression of only five genes ≥3-fold, Angptl4, Cyp2b10, Cfd/Adipsin, Adipoq and Chi3l4 and markedly reduced expression of Gast, Ccla3, Glycam1, Spp1, Serpina1a, Cela1, Cldn2, and Fabp2 (Table S3). DMBA increased expression of S100a8, S100a9, and Ccl8 4–10-fold and reduced expression of the same subset of genes as GW501516 (Table S4). Thus, changes in Gast, Ccla3, Glycam1, Spp1, Serpina1a, Cela1, Cldn2, and Fabp2 are a result of GW501516 treatment and are not tumor specific. On the other hand, DMBA produced an inflammatory response denoted by increase S100A8, Ccl8, and S100A9 although it was an order of magnitude less than in tumors. The increase in Krt6a by DMBA is consistent with increased keratinization in the stomach (Figure 2(B)) but was less pronounced than in tumors. Real-time qRT-PCR analysis generally confirmed the changes in expression of several genes, including Cldn8, Cxcl1, Cxcl5, Foxg1, S100a8, Angptl4, Cyp2b10, Vegfα and Spp1, Gast, Dkk1, Bmp3, Bmp4, PPARα, and PPARγ (Figure 3(c)).
Table 2.
Differentially expressed genes in gastric tumors.
| Gene Symbol | Gene Name | Fold Change |
|---|---|---|
| S100a8 | S100 calcium-binding protein A8 (calgranulin A) | 229.0 |
| S100a9 | S100 calcium-binding protein A9 (calgranulin B) | 74.6 |
| S100a3 | chemokine (C-X-C motif) ligand 2 | 17.7 |
| Cxcl2 | chemokine (C-X-C motif) ligand 2 | 109.2 |
| Cxcl5 | chemokine (C-X-C motif) ligand 5 | 145.6 |
| Cxcl1 | chemokine (C-X-C motif) ligand 1 | 100.0 |
| Cxcl9 | chemokine (C-X-C motif) ligand 9 | 13.3 |
| Ccl2 | chemokine (C-C motif) ligand 2 | 12.8 |
| Ccl3 | chemokine (C-C motif) ligand 3 | 28.7 |
| Ccl8 | chemokine (C-C motif) ligand 8 | 13.4 |
| Il1b | interleukin 1 beta | 20.9 |
| Il24 | interleukin 24 | 23.7 |
| Il6 | interleukin 6 | 39.4 |
| Mmp10 | matrix metallopeptidase 10 | 15.8 |
| Mmp12 | matrix metallopeptidase 12 | 104.1 |
| Mmp13 | matrix metallopeptidase 13 | 62.5 |
| Mmp3 | matrix metallopeptidase 3 | 9.3 |
| Mmp9 | matrix metallopeptidase 9 | 6.0 |
| Ppara | peroxisome proliferator activated receptor alpha | −8.6 |
| Pparg | peroxisome proliferator activated receptor gamma | −4.3 |
| Ptges | prostaglandin E synthase | 11.4 |
| Ptgs2 | prostaglandin-endoperoxide synthase 2 | 14.1 |
| Krt16 | keratin 16 | 116.0 |
| Krt6a | keratin 6A | 86.2 |
Figure 3.
Differential gene expression in GW501516-treated stomach and gastric tumors. Gene microarray analysis was carried out with pooled RNA samples prepared from either five tissue samples of forestomach from untreated and GW501516 treated mice or six gastric tumors. Heat maps represent unsupervised hierarchical clustering of ≥3-fold changes in signal intensity normalized to untreated forestomach. (a) Heatmap of GW501516-treated versus stomach representing 42 genes. (b) Tumor versus stomach representing 811 genes. (c) qRT-PCR analysis of the relative changes in gene expression in the stomach after GW501516 treatment (GW501516), and in tumors (Tumor). GW501516 treated: array, cross-hatch; qRT-PCR, diagonal; Tumors: array, black; qRT-PCR, white.
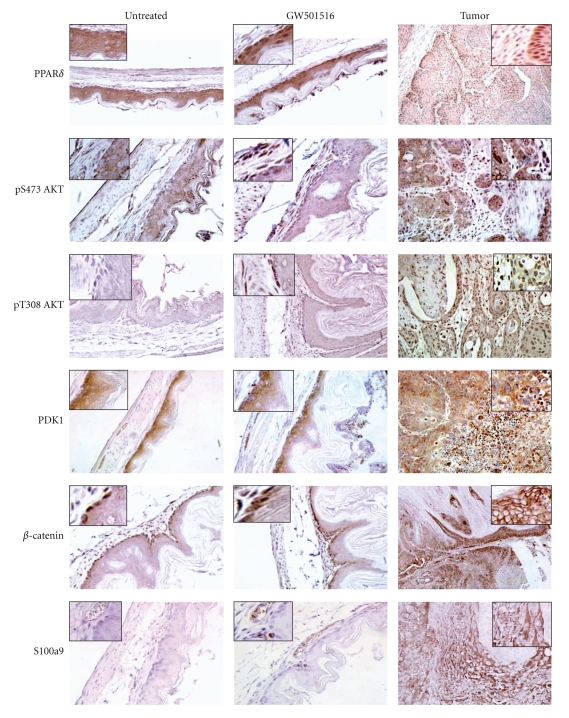
The expression of PPARδ and factors known to be associated with its signaling were assessed in tumors and forestomach after GW501516 treatment (Figure 4). GW501516 increased nuclear localization of PPARδ in gastric squamous epithelium and tumors, in contrast to its diffuse cytoplasmic staining in untreated gastric tissue. GW501516 also elicited strong pS473Akt and pT308Akt staining in basal cells and in the submucosal layer, as well as in tumor and stromal tissue, which correlated with more intense PDK1 expression. β-Catenin was expressed in the nuclei of basal squamous epithelial cells and was not altered by GW501516 treatment, whereas tumors expressed increased β-catenin at cellular junctions. S100a9 was absent in untreated gastric epithelium but was expressed in endothelial and epithelial cells from GW501516-treated mice. Tumors expressed S100a9 in a diffuse pattern, with strong expression in blood vessels and adjacent epithelial cells.
Figure 4.
Immunohistochemical analysis of forestomach after GW501516 treatment and gastric tumors. IHC detection of PPARδ, pS473Akt, pT308 Akt, PDK1, β-catenin, and S100a9. Magnification: untreated and GW501516 treated, 400x; tumor, PPARδ, PDK1, and β-catenin, 200x; pT308Akt, pS473Akt, S100a9, and 400x. Inset, 2x original magnification. PPARδ shows diffuse reactivity in untreated gastric squamous epithelium, increased nuclear localization after GW501516 treatment, and strong nuclear expression in tumor and stromal cells (inset). pS473Akt and pT308Akt expressed weak diffuse reactivity in untreated gastric tissue, increased staining in basal cells and the submucosal cell layer after GW501516 treatment, and strong reactivity in tumor and stromal cells. PDK1 exhibited diffuse cytoplasmic localization throughout the untreated squamous epithelium and was unchanged after GW501516 treatment, whereas PDK1 was increased in tumors similarly to pS473Akt and pT308Akt. Nuclear β-catenin was present in basal cells of untreated squamous epithelium and was unchanged after GW501516 treatment, whereas tumors expressed increased β-catenin at cellular junctions (inset). S100a9 was undetectable in untreated tissue and was increased in blood vessels and squamous epithelium (inset), whereas tumors exhibited increased diffuse cytoplasmic staining.
4. Discussion
The present study describes a new model of metastatic gastric cancer that is dependent on the tumor promoting activity of PPARδ agonist GW510516 following carcinogen administration. In contrast to a previous study reporting a low percentage of squamous cell carcinomas of the forestomach by DMBA [43], our DMBA regimen produced only forestomach hyperplasia without signs of dysplasia up to five months after treatment (Figure 2(B)). This suggests a high sensitivity of mouse forestomach squamous epithelium to dysplasia, and the predilection of GW501516 to promote tumors of this histotype [19]. This model differs from N-methyl-N-nitrosourea-induced gastric tumors in wild-type and APCMin transgenic mice [44, 45] in that it is dependent on both DMBA-induced mutagenesis and the tumor-promoting effects of GW501516. A feature of this model is its short latency of approximately three weeks in comparison to 10 to 20 weeks for NMU-treated wild-type and APCMin mice. An important histopathological distinction, and perhaps disadvantage of the GW501516 tumor model, is that it produces squamous cell carcinomas from the nonglandular forestomach rather than adenocarcinomas from the glandular tissue that comprises the majority of human gastric cancer [46]. Since this model was dependent on the selective PPARδ agonist GW501516 [47], it is important to note that the dose of GW501516 used in the present and previous studies [19] is equivalent to daily oral doses of 3–10 mg/kg that were previously shown to specifically enhance PPARδ-dependent fatty acid oxidation in mice [48]. In addition, PPARδ agonist GW7042, which is almost identical to GW501516 in structure, potency, and specificity, was inactive in inducing gene expression in PPARδ knockout mice [49], suggesting that the tumor promoting effects of GW501516 and GW7042 are not due to off-target effects.
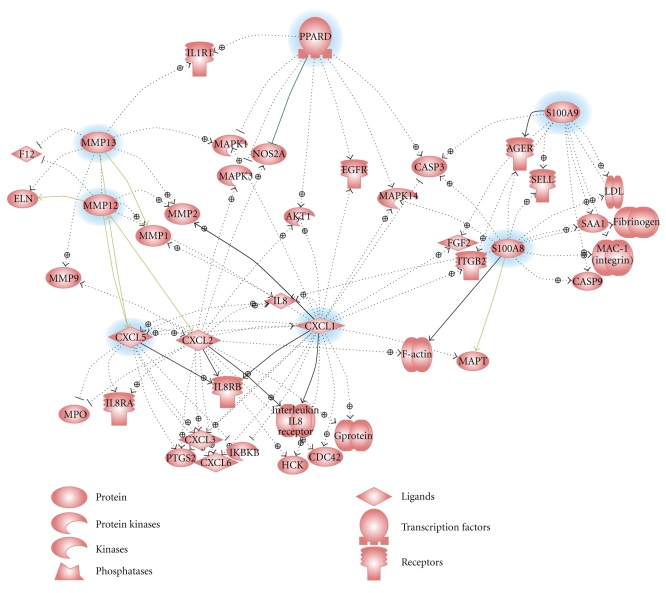
Tumors induced by GW501516 exhibited a distinct inflammatory gene expression signature comprised predominantly of chemokine, MMP, and S100 genes (Table S2). This was unexpected in view of the lack of a similar signature after treatment with GW501516 (Table S3), and the fact that GW501516 induces an anti-inflammatory response in macrophages [50] and protects the heart against oxidative stress [51, 52]. Gene ontology analysis of gene expression in the gastric tumors indicates that PPARδ, MMP12, MMP13, Cxcl1, Cxcl5, S100A8, and S100A9 share both common and disparate pathway interactions that likely contributed to the tumorigenic phenotype (Figure 5). PPARδ is associated with activation of genes related to proliferation (EGFR, Akt1) and adhesion (Itgb2), whereas S100A9 is associated with angiogenesis (Fgf2) and inflammation (Ager). Cxcl1 activates proliferation (Mapk3, Mapk14, and Akt1), angiogenesis (Fgf2), and invasion (MMP2, MMP9), and Cxcl5 activates other chemokines (Cxcl1, Cxcl2, and Cxcl3) and Ptgs2/Cox2. This scheme reiterates the ability of S100A8 and S100A9 to act as ligands for Ager (advanced glycation end-product receptor), which mediates acute and chronic inflammation, tumor development, and metastasis [53, 54]. This paradigm is also consistent with GW501516-induced activation of Ptges and Ptgs2/Cox-2 expression [55, 56], which initiate the production of prostacyclins [14] and arachidonic acid metabolites [29] that serve as PPARδ ligands [57]. Since Cox-2 inhibitors reduce inflammation-related gastrointestinal carcinogenesis [58], and overexpression or deletion of Ptgs2 increases or suppresses tumorigenesis, respectively [59, 60]; this suggests cooperativity between PPARδ and inflammatory signaling pathways in gastric tumorigenesis. The ability of PPARδ to have an anti-inflammatory effect in normal cells [51, 52] and a proinflammatory effect in tumors is reminiscent of the dual roles of TGF-β in tumor cells [61, 62]. TGFβ can function as a proinflammatory cytokine to activate S100A8 and S100A9 expression in the presence of activated Ras [63] but acts as a repressor of inflammation-induced PPARδ expression in normal cells [64, 65]. The increased expression of the PPARδ target gene, Agptl4, the TGFβ-activated genes Runx1 and Runx2, and S100A8 and S100A9 in the gastric tumors indeed suggests a duality of function of both PPARδ and TGFβ signaling in gastric tumorigenesis.
Figure 5.
Signaling networks associated with PPARδ and inflammatory and invasive gene expression. Common signaling pathways were analyzed for six genes with the greatest changes in expression in gastric tumors (highlighted in blue) versus forestomach using Ariadne Pathway Studio 7.1.
PPARδ is ubiquitously expressed in gastrointestinal tissue and gastric tumors [27], and GW501516 elicited increased PPARδ nuclear staining and elevated pAkt in gastric epithelium and tumors. PPARδ-dependant activation of Akt is required for the growth-promoting and antiapoptotic effects of PPARδ [66–68], as shown by the delayed wound-healing response of PPARδ-deficient keratinocytes [57, 69]. Enhanced Krt6a and Krt16 expression in tumors further suggests that PPARδ plays an important role in gastric squamous cell differentiation and tissue renewal.
Tumors also exhibited reduced PPARγ and PPARα expression that may have resulted, in part, from the negative regulation of PPARγ by PPARδ [70]. PPARγ suppresses the growth and invasion of human colon [71] and gastric [72, 73] and esophageal carcinoma cells [74], and both PPARγ [75] and PPARα have anti-inflammatory actions [76]. Thus, reduction of PPARα and PPARγ expression may be an additional mechanism for facilitating the proinflammatory and tumor-promoting effects of GW501516.
In summary, we describe a rapidly developing metastatic gastric cancer model dependent on the tumor-promoting effects of GW501516 following carcinogen treatment, which suggests a proinflammatory switch in PPARδ function. This animal model will therefore be useful to delineate the role of PPARδ in tumor initiation and progression and as a possible target for early intervention.
Supplementary Material
Supplementary material contains qRT-PCR (Table S1) primers and differential gene expression data in gastric tumors (Table S2), and in the stomach of GW501516-treated (Table S3) and DMBA-treated (Table S4) animals.
Acknowledgments
This work was supported by Grant no. R01 CA111482 and Contract no. N01 CN43309 from the National Institutes of Health, Bethesda, MD. This investigation was conducted using the Animal Research, Histopathology and Tissue, Genomics and Epigenomics, and Preclinical Imaging Shared Resources supported by Research Facilities Improvement Grant no. C06 RR14567 from the National Center for Research Facilities, and by Cancer Center Support Grant no. 1P30-CA-51008 from the National Cancer Institute. The authors thank Yi Chien Lee for his help with the MRI.
References
- 1.Lochhead P, El-Omar EM. Gastric cancer. British Medical Bulletin. 2008;85(1):87–100. doi: 10.1093/bmb/ldn007. [DOI] [PubMed] [Google Scholar]
- 2.Forman D, Burley VJ. Gastric cancer: global pattern of the disease and an overview of environmental risk factors. Best Practice and Research: Clinical Gastroenterology. 2006;20(4):633–649. doi: 10.1016/j.bpg.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 3.van Loon AJM, Botterweck AAM, Goldbohm RA, Brants HAM, van Klaveren JD, van den Brandt PA. Intake of nitrate and nitrite and the risk of gastric cancer: a prospective cohort study. British Journal of Cancer. 1998;78(1):129–135. doi: 10.1038/bjc.1998.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishino Y, Inoue M, Tsuji I, et al. Tobacco smoking and gastric cancer risk: an evaluation based on a systematic review of epidemiologic evidence among the Japanese population. Japanese Journal of Clinical Oncology. 2006;36(12):800–807. doi: 10.1093/jjco/hyl112. [DOI] [PubMed] [Google Scholar]
- 5.Eslick GD. Helicobacter pylori infection causes gastric cancer? A review of the epidemiological, meta-analytic, and experimental evidence. World Journal of Gastroenterology. 2006;12(18):2991–2999. doi: 10.3748/wjg.v12.i19.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Omar EM. Role of host genes in sporadic gastric cancer. Best Practice and Research: Clinical Gastroenterology. 2006;20(4):675–686. doi: 10.1016/j.bpg.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Barish GD, Narkar VA, Evans RM. PPARδ: a dagger in the heart of the metabolic syndrome. Journal of Clinical Investigation. 2006;116(3):590–597. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger JP, Akiyama TE, Meinke PT. PPARs: therapeutic targets for metabolic disease. Trends in Pharmacological Sciences. 2005;26(5):244–251. doi: 10.1016/j.tips.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Glazer RI, Yuan H, Xie Z, Yin Y. PPARγ and PPARδ as modulators of neoplasia and cell fate. PPAR Research. 2008;2008:8 pages. doi: 10.1155/2008/247379. Article ID 247379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zang C, Wächter M, Liu H, et al. Ligands for PPARγ and RAR cause induction of growth inhibition and apoptosis in human glioblastomas. Journal of Neuro-Oncology. 2003;65(2):107–118. doi: 10.1023/b:neon.0000003728.80052.a8. [DOI] [PubMed] [Google Scholar]
- 11.Kopelovich L, Fay JR, Glazer RI, Crowell JA. Peroxisome proliferator-activated receptor modulators as potential chemopreventive agents. Molecular Cancer Therapeutics. 2002;1(5):357–363. [PubMed] [Google Scholar]
- 12.Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers: complex stories. Nature Reviews Cancer. 2004;4(1):61–70. doi: 10.1038/nrc1254. [DOI] [PubMed] [Google Scholar]
- 13.Stephen RL, Gustafsson MCU, Jarvis M, et al. Activation of peroxisome proliferator-activated receptor δ stimulates the proliferation of human breast and prostate cancer cell lines. Cancer Research. 2004;64(9):3162–3170. doi: 10.1158/0008-5472.can-03-2760. [DOI] [PubMed] [Google Scholar]
- 14.Gupta RA, Tan J, Krause WF, et al. Prostacyclin-mediated activation of peroxisome proliferator-activated receptor δ in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(24):13275–13280. doi: 10.1073/pnas.97.24.13275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta RA, Wang D, Katkuri S, Wang H, Dey SK, DuBois RN. Activation of nuclear hormone receptor peroxisome proliferator-activated receptor-δ accelerates intestinal adenoma growth. Nature Medicine. 2004;10(3):245–247. doi: 10.1038/nm993. [DOI] [PubMed] [Google Scholar]
- 16.Knutsen HK, Ølstørn HB, Paulsen JE, et al. Increased levels of PPARβ/δ and cyclin D1 in flat dysplastic ACF and adenomas in Apc Min/+ mice. Anticancer Research. 2005;25(6B):3781–3789. [PubMed] [Google Scholar]
- 17.Jaeckel EC, Raja S, Tan J, et al. Correlation of expression of cyclooxygenase-2, vascular endothelial growth factor, and peroxisome proliferator-activated receptor δ with head and neck squamous cell carcinoma. Archives of Otolaryngology—Head and Neck Surgery. 2001;127(10):1253–1259. doi: 10.1001/archotol.127.10.1253. [DOI] [PubMed] [Google Scholar]
- 18.Suchanek KM, May FJ, Lee WJ, Holman NA, Roberts-Thomson SJ. Peroxisome proliferator-activated receptor β expression in human breast epithelial cell lines of tumorigenic and non-tumorigenic origin. International Journal of Biochemistry and Cell Biology. 2002;34(9):1051–1058. doi: 10.1016/s1357-2725(02)00025-0. [DOI] [PubMed] [Google Scholar]
- 19.Yin Y, Russell RG, Dettin LE, et al. Peroxisome proliferator-activated receptorδ and γ agonists differentially alter tumor differentiation and progression during mammary carcinogenesis. Cancer Research. 2005;65(9):3950–3957. doi: 10.1158/0008-5472.CAN-04-3990. [DOI] [PubMed] [Google Scholar]
- 20.Wang D, Wang H, Guo Y, et al. Crosstalk between peroxisome proliferator-activated receptorδ and VEGF stimulates cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(50):19069–19074. doi: 10.1073/pnas.0607948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuo X, Peng Z, Moussalli MJ, et al. Targeted genetic disruption of peroxisome proliferator-activated receptor-δ and colonic tumorigenesis. Journal of the National Cancer Institute. 2009;101(10):762–767. doi: 10.1093/jnci/djp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh M, Ai Y, Narko K, Wang Z, Peters JM, Hla T. PPARδ is pro-tumorigenic in a mouse model of COX-2-induced mammary cancer. Prostaglandins and Other Lipid Mediators. 2009;88(3-4):97–100. doi: 10.1016/j.prostaglandins.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park BH, Vogelstein B, Kinzler KW. Genetic disruption of PPARδ decreases the tumorigenicity of human colon cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(5):2598–2603. doi: 10.1073/pnas.051630998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwak H, Hwang I, Kim JH, Kim MY, Yang JS, Jeong S. Modulation of transcription by the peroxisome proliferator-activated receptor δ-binding RNA aptamer in colon cancer cells. Molecular Cancer Therapeutics. 2009;8(9):2664–2673. doi: 10.1158/1535-7163.MCT-09-0214. [DOI] [PubMed] [Google Scholar]
- 25.Shao J, Sheng H, DuBois RN. Peroxisome proliferator-activated receptors modulate K-Ras-mediated transformation of intestinal epithelial cells. Cancer Research. 2002;62(11):3282–3288. [PubMed] [Google Scholar]
- 26.He TC, Chan TA, Vogelstein B, Kinzler KW. PPARδ is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99(3):335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu J, Leung WK, Chen J, Ebert MPA, Malfertheiner P, Sung JJY. Expression of peroxisome proliferator-activated receptor δ in human gastric cancer and its response to specific COX-2 inhibitor. Cancer Letters. 2005;223(1):11–17. doi: 10.1016/j.canlet.2004.09.052. [DOI] [PubMed] [Google Scholar]
- 28.Yoshinaga M, Kitamura Y, Chaen T, et al. The simultaneous expression of peroxisome proliferator-activated receptor delta and cyclooxygenase-2 may enhance angiogenesis and tumor venous invasion in tissues of colorectal cancers. Digestive Diseases and Sciences. 2009;54(5):1108–1114. doi: 10.1007/s10620-008-0465-x. [DOI] [PubMed] [Google Scholar]
- 29.Naruhn S, Meissner W, Adhikary T, et al. 15-hydroxyeicosatetraenoic acid is a preferential peroxisome proliferator-activated receptor β/δ agonist. Molecular Pharmacology. 2010;77(2):171–184. doi: 10.1124/mol.109.060541. [DOI] [PubMed] [Google Scholar]
- 30.Piqueras L, Reynolds AR, Hodivala-Dilke KM, et al. Activation of PPARβ/δ induces endothelial cell proliferation and angiogenesis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2007;27(1):63–69. doi: 10.1161/01.ATV.0000250972.83623.61. [DOI] [PubMed] [Google Scholar]
- 31.Wu CW, Yu J, Sung JJY. Peroxisome proliferator-activated receptor δ and gastric cancer (review) Oncology Reports. 2009;22(3):451–457. [PubMed] [Google Scholar]
- 32.Xu HE, Lambert MH, Montana VG, et al. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Molecular Cell. 1999;3(3):397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 33.Harman FS, Nicol CJ, Marin HE, Ward JM, Gonzalez FJ, Peters JM. Peroxisome proliferator-activated receptor-δ attenuates colon carcinogenesis. Nature Medicine. 2004;10(5):481–483. doi: 10.1038/nm1026. [DOI] [PubMed] [Google Scholar]
- 34.Hollingshead HE, Borland MG, Billin AN, Willson TM, Gonzalez FJ, Peters JM. Ligand activation of peroxisome proliferator-activated receptor-β/ δ (PPARβ/δ) and inhibition of cyclooxygenase 2 (COX2) attenuate colon carcinogenesis through independent signaling mechanisms. Carcinogenesis. 2008;29(1):169–176. doi: 10.1093/carcin/bgm209. [DOI] [PubMed] [Google Scholar]
- 35.Marin HE, Peraza MA, Billin AN, et al. Ligand activation of peroxisome proliferator-activated receptor β inhibits colon carcinogenesis. Cancer Research. 2006;66(8):4394–4401. doi: 10.1158/0008-5472.CAN-05-4277. [DOI] [PubMed] [Google Scholar]
- 36.Peters JM, Hollingshead HE, Gonzalez FJ. Role of peroxisome-proliferator-activated receptor β/δ (PPARβ/δ) in gastrointestinal tract function and disease. Clinical Science. 2008;115(3-4):107–127. doi: 10.1042/CS20080022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei ZL, Kozikowski AP. A short and efficient synthesis of the pharmacological research tool GW501516 for the peroxisome proliferator-activated receptor δ . Journal of Organic Chemistry. 2003;68(23):9116–9118. doi: 10.1021/jo035140g. [DOI] [PubMed] [Google Scholar]
- 38.Fricke ST, Rodriguez O, VanMeter J, et al. In vivo magnetic resonance volumetric and spectroscopic analysis of mouse prostate cancer models. Prostate. 2006;66(7):708–717. doi: 10.1002/pros.20392. [DOI] [PubMed] [Google Scholar]
- 39.Yin Y, Yuan H, Zeng X, Kopelovich L, Glazer RI. Inhibition of peroxisome proliferator-activated receptor γ increases estrogen receptor-dependent tumor specification. Cancer Research. 2009;69(2):687–694. doi: 10.1158/0008-5472.CAN-08-2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin Y, Bai R, Russell RG, et al. Characterization of medroxyprogesterone and DMBA-induced multilineage mammary tumors by gene expression profiling. Molecular Carcinogenesis. 2005;44(1):42–50. doi: 10.1002/mc.20119. [DOI] [PubMed] [Google Scholar]
- 41.Herschkowitz JI, Simin K, Weigman VJ, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biology. 2007;8(5, article R76) doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi H, Shikata N, Senzaki H, Shintaku M, Tsubura A. Immunohistochemical staining patterns of keratins in normal oesophageal epithelium and carcinoma of the oesophagus. Histopathology. 1995;26(1):45–50. doi: 10.1111/j.1365-2559.1995.tb00619.x. [DOI] [PubMed] [Google Scholar]
- 43.Chandra SA, Nolan MW, Malarkey DE. Chemical carcinogenesis of the gastrointestinal tract in rodents: an overview with emphasis on NTP carcinogenesis bioassays. Toxicologic Pathology. 2010;38(1):188–197. doi: 10.1177/0192623309356452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomita H, Yamada Y, Oyama T, et al. Development of gastric tumors in Apc Min/+ mice by the activation of the β-catenin/Tcf signaling pathway. Cancer Research. 2007;67(9):4079–4087. doi: 10.1158/0008-5472.CAN-06-4025. [DOI] [PubMed] [Google Scholar]
- 45.Yamachika T, Nakanishi H, Inada KI, et al. N-methyl-N-nitrosourea concentration-dependent, rather than total intake-dependent, induction of adenocarcinomas in the glandular stomach of BALB/c mice. Japanese Journal of Cancer Research. 1998;89(4):385–391. doi: 10.1111/j.1349-7006.1998.tb00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vauhkonen M, Vauhkonen H, Sipponen P. Pathology and molecular biology of gastric cancer. Best Practice and Research: Clinical Gastroenterology. 2006;20(4):651–674. doi: 10.1016/j.bpg.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 47.Oliver WR, Shenk JL, Snaith MR, et al. A selective peroxisome proliferator-activated receptor δ agonist promotes reverse cholesterol transport. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5306–5311. doi: 10.1073/pnas.091021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka T, Yamamoto J, Iwasaki S, et al. Activation of peroxisome proliferator-activated receptorδ induces fatty acid β-oxidation in skeletal muscle and attenuates metabolic syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palkar PS, Borland MG, Naruhn S, et al. Cellular and pharmacological selectivity of the peroxisome proliferator-activated receptor-β/δ antagonist GSK3787. Molecular Pharmacology. 2010;78(3):419–430. doi: 10.1124/mol.110.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee CH, Chawla A, Urbiztondo N, Liao D, Boisvert WA, Evans RM. Transcriptional repression of atherogenic inflammation: modulation by PPARδ . Science. 2003;302(5644):453–457. doi: 10.1126/science.1087344. [DOI] [PubMed] [Google Scholar]
- 51.Ali F, Ali NS, Bauer A, et al. PPARσ and PGC1α act cooperatively to induce haem oxygenase-1 and enhance vascular endothelial cell resistance to stress. Cardiovascular Research. 2010;85(4):701–710. doi: 10.1093/cvr/cvp365. [DOI] [PubMed] [Google Scholar]
- 52.Pesant M, Sueur S, Dutartre P, et al. Peroxisome proliferator-activated receptor δ (PPARδ) activation protects H9c2 cardiomyoblasts from oxidative stress-induced apoptosis. Cardiovascular Research. 2006;69(2):440–449. doi: 10.1016/j.cardiores.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 53.Gebhardt C, Németh J, Angel P, Hess J. S100A8 and S100A9 in inflammation and cancer. Biochemical Pharmacology. 2006;72(11):1622–1631. doi: 10.1016/j.bcp.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 54.Ghavami S, Chitayat S, Hashemi M, et al. S100A8/A9: a Janus-faced molecule in cancer therapy and tumorgenesis. European Journal of Pharmacology. 2009;625(1–3):73–83. doi: 10.1016/j.ejphar.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 55.Glinghammar B, Skogsberg J, Hamsten A, Ehrenborg E. PPARδ activation induces COX-2 gene expression and cell proliferation in human hepatocellular carcinoma cells. Biochemical and Biophysical Research Communications. 2003;308(2):361–368. doi: 10.1016/s0006-291x(03)01384-6. [DOI] [PubMed] [Google Scholar]
- 56.Han S, Ritzenthaler JD, Wingerd B, Roman J. Activation of peroxisome proliferator-activated receptor α/δ (PPARβ/δ) increases the expression of prostaglandin E receptor subtype EP4: the roles of phosphatidylinositol 3-kinase and CCAAT/enhancer-binding protein β . Journal of Biological Chemistry. 2005;280(39):33240–33249. doi: 10.1074/jbc.M507617200. [DOI] [PubMed] [Google Scholar]
- 57.Tan NS, Michalik L, Noy N, et al. Critical roles of PPARβ/δ in keratinocyte response to inflammation. Genes and Development. 2001;15(24):3263–3277. doi: 10.1101/gad.207501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oba M, Miwa K, Fujimura T, et al. A selective cyclooxygenase-2 inhibitor prevents inflammation-related squamous cell carcinogenesis of the forestomach via duodenogastric reflux in rats. Cancer. 2009;115(2):454–464. doi: 10.1002/cncr.23990. [DOI] [PubMed] [Google Scholar]
- 59.Liu CH, Chang SH, Narko K, et al. Overexpression of cyclooxygenase-2 is sufficient to induce tumorigenesis in transgenic mice. Journal of Biological Chemistry. 2001;276(21):18563–18569. doi: 10.1074/jbc.M010787200. [DOI] [PubMed] [Google Scholar]
- 60.Oshima M, Dinchuk JE, Kargman SL, et al. Suppression of intestinal polyposis in Apc(Δ716) knockout mice by inhibition of cyclooxygenase 2 (COX-2) Cell. 1996;87(5):803–809. doi: 10.1016/s0092-8674(00)81988-1. [DOI] [PubMed] [Google Scholar]
- 61.Bierie B, Moses HL. Tumour microenvironment: TGFβ: the molecular Jekyll and Hyde of cancer. Nature Reviews Cancer. 2006;6(7):506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 62.Xu J, Lamouille S, Derynck R. TGF-β-induced epithelial to mesenchymal transition. Cell Research. 2009;19(2):156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pérez-Lorenzo R, Markell LM, Hogan KA, Yuspa SH, Glick AB. Transforming growth factor β1 enhances tumor promotion in mouse skin carcinogenesis. Carcinogenesis. 2010;31(6):1116–1123. doi: 10.1093/carcin/bgq041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan NS, Michalik L, Desvergne B, Wahli W. Genetic- or transforming growth factor-β1-induced changes in epidermal peroxisome proliferator-activated receptor β/δ expression dictate wound repair kinetics. Journal of Biological Chemistry. 2005;280(18):18163–18170. doi: 10.1074/jbc.M412829200. [DOI] [PubMed] [Google Scholar]
- 65.Tan NS, Michalik L, Di-Poï N, et al. Essential role of Smad3 in the inhibition of inflammation-induced PPARβ/δ expression. The EMBO Journal. 2004;23(21):4211–4221. doi: 10.1038/sj.emboj.7600437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han S, Ritzenthaler JD, Zheng Y, Roman J. PPARβ/δ agonist stimulates human lung carcinoma cell growth through inhibition of PTEN expression: the involvement of PI3K and NF-κB signals. American Journal of Physiology. 2008;294(6):L1238–L1249. doi: 10.1152/ajplung.00017.2008. [DOI] [PubMed] [Google Scholar]
- 67.Di-Po N, Tan NS, Michalik L, Wahli W, Desvergne B. Antiapoptotic role of PPARβ in keratinocytes via transcriptional control of the Akt1 signaling pathway. Molecular Cell. 2002;10(4):721–733. doi: 10.1016/s1097-2765(02)00646-9. [DOI] [PubMed] [Google Scholar]
- 68.Pedchenko TV, Gonzalez AL, Wang D, DuBois RN, Massion PP. Peroxisome proliferator-activated receptor β/δ expression and activation in lung cancer. American Journal of Respiratory Cell and Molecular Biology. 2008;39(6):689–696. doi: 10.1165/rcmb.2007-0426OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Michalik L, Desvergne B, Tan NS, et al. Impaired skin wound healing in peroxisome proliferator-activated receptor (PPAR)α and PPARβ mutant mice. Journal of Cell Biology. 2001;154(4):799–814. doi: 10.1083/jcb.200011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi Y, Hon M, Evans RM. The peroxisome proliferator-activated receptor δ, an integrator of transcriptional repression and nuclear receptor signaling. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(5):2613–2618. doi: 10.1073/pnas.052707099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sarraf P, Mueller E, Jones D, et al. Differentiation and reversal of malignant changes in colon cancer through PPARγ . Nature Medicine. 1998;4(9):1046–1052. doi: 10.1038/2030. [DOI] [PubMed] [Google Scholar]
- 72.Cheon CW, Kim DH, Kim DH, Cho YH, Kim JH. Effects of ciglitazone and troglitazone on the proliferation of human stomach cancer cells. World Journal of Gastroenterology. 2009;15(3):310–320. doi: 10.3748/wjg.15.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He Q, Pang R, Song X, et al. Rosiglitazone suppresses the growth and invasiveness of SGC-7901 gastric cancer cells and angiogenesis in vitro via PPAR γ dependent and independent mechanisms. PPAR Research. 2008;2008:9 pages. doi: 10.1155/2008/649808. Article ID 649808. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Takashima T, Fujiwara Y, Hamaguchi M, et al. Relationship between peroxisome proliferator-activated receptor-gamma expression and differentiation of human esophageal squamous cell carcinoma. Oncology Reports. 2005;13(4):601–606. [PubMed] [Google Scholar]
- 75.Slomiany BL, Piotrowski J, Slomiany A. Gastric mucosal inflammatory responses to Helicobacter pylori lipopolysaccharide: suppression of caspase-3 and nitric oxide synthase-2 by omeprazole and sucralfate. Inflammopharmacology. 1999;7(2):163–177. doi: 10.1007/BF02918388. [DOI] [PubMed] [Google Scholar]
- 76.Delerive P, De Bosscher K, Besnard S, et al. Peroxisome proliferator-activated receptor α negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-κB and AP-1. Journal of Biological Chemistry. 1999;274(45):32048–32054. doi: 10.1074/jbc.274.45.32048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material contains qRT-PCR (Table S1) primers and differential gene expression data in gastric tumors (Table S2), and in the stomach of GW501516-treated (Table S3) and DMBA-treated (Table S4) animals.