Abstract
The cell nucleus is an organelle bounded by a double-membrane which undergoes drastic reorganization during major cellular events such as cell division and apoptosis. Maintenance of proper nuclear structure, function and dynamics is central to organelle vitality. Over recent years growing evidence has shown that parts of the nucleus can be specifically degraded by an autophagic process termed nucleophagy. The process is best described in the yeast, Saccharomyces cerevisiae, where piecemeal microautophagy of the nucleus or nucleophagy (micronucleophagy) requires direct interaction of the nuclear membrane with that of the vacuole (the yeast lytic compartment). Here, we review the process of nucleophagy in the context of nuclear membrane dynamics, and examine the evidence for autophagic degradation of the nucleus in mammalian cells. Finally, we discuss the importance of nucleophagy as a ‘housecleaning’ mechanism for the nucleus under both normal and disease conditions.
Key words: autophagy, mammalian cells, membrane, nuclear components, nuclear envelope, nucleophagy, nucleus, yeast
Introduction: The Need for Nuclear Turnover
The nucleus is a site of a number of essential metabolic activities relating to maintenance and expression of the genome. Such activities include: DNA replication, recombination and repair, gene transcription, RNA processing and ribosome subunit maturation and assembly.1,2 In order for these essential activities to be carried out in a correct and efficient manner the corresponding machinery must be maintained. Consequently, it is to be expected there are processes within cells which act to ‘repair’ nuclear damage through the coordinated removal of damaged non-functional components, or those surplus to requirement. A growing body of evidence indicates that parts of the nucleus can be specifically degraded by nucleophagy (micronucleophagy), an autophagic process involving localized changes in membrane organization and dynamics. Here, we discuss nucleophagy in this context as well as its role in ‘housecleaning’ during nutrient limitation and under various physiological and pathological conditions.
Nuclear Structure, Function and Compartmentalization
The nucleus is not only the largest membrane-enclosed organelle in eukaryotic cells, but arguably the most elaborate. Two main structures make up the metazoan (including mammalian) nucleus. The first of these is the nuclear envelope (NE) that acts as the interface between the nucleus and the rest of the cell enclosing the contents of the nucleus.3,4 The second is the nuclear lamina, a dense but fenestrated network composed of intermediate filaments made of lamins and lamin-associated (e.g., emerin) proteins underlying the inner surface of the NE. The NE is required for maintenance of nuclear shape, spacing of nuclear pore complexes (NPCs), organization of heterochromatin, DNA replication and regulation of transcription factors (Fig. 1).5–11 Certain organisms, such as fungi or plants lack lamins12 and consequently lack a nuclear lamina.13 Plants contain coiled-coil proteins, apparently unrelated to lamins, which serve as nucleoskeleton components.12,14,15 Coiled-coil proteins have been implicated also as nucleoskeleton components in non-metazoans, notably yeast and Trypanosoma brucei.12,16,17
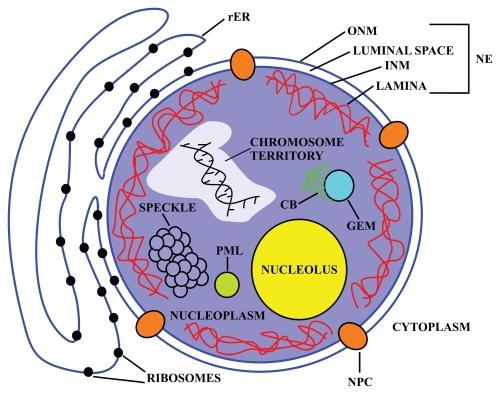
Figure 1.
Schematic representation of a cell nucleus showing the main structural components. The nucleoplasm is enclosed by the nuclear envelope (NE), which contains the inner nuclear membrane (INM), luminal space and the outer nuclear membrane (ONM), the nuclear pore complexes (NPC), and the nuclear lamina. The ribosome-studded ONM is contiguous with the rough endoplasmic reticulum (rER), whereas the ribosome free INM faces the nucleoplasm and lamina. The ONM and INM are joined at the NPC. The yeast NE also contains NPCs and is continuous with the rER, but a functional counterpart of the metazoan lamina has not yet been described in yeast. This diagram also depicts some of the subnuclear structures located in the nucleoplasm, namely, nucleolus, chromosome territory, the Cajal body (CB), the promyelocytic leukemia oncoprotein (PML), splicing speckles and gems. Although each of these subnuclear structures represents an independent entity, remarkably, they are highly dynamic both internally and as discrete entities within the nucleus.13,26
The NE is comprised of two concentric, closely opposed lipid bilayers, the outer and inner nuclear membranes (ONM and INM, respectively).4,18 The ONM is continuous with the rough endoplasmic reticulum (rER), studded with ribosomes and functions in secretion and lipid biosynthesis. The ribosome-free INM faces the viscous nucleoplasm, and contains a unique spectrum of integral and membrane-associated proteins that provide binding sites for the nuclear lamina and chromatin. The ONM and INM are separated by a luminal space but become contiguous at sites of high membrane curvature occupied by the NPCs (Fig. 1). NPCs exclusively mediate and regulate nucleocytoplasmic trafficking, the signal-dependent, bidirectional trafficking of macromolecules between the nucleus and cytoplasm which is crucial for both gene expression and chromosomal maintenance.7,18–20
The interior of the nucleus lacks membrane-bound subcompartments, but is organized into domains usually referred to as “subnuclear structures” (also often referred as “nuclear organelles” or “nuclear compartments”) with highly specialized functions and containing a unique complement of proteins and RNA molecules. This subnuclear organization is promoted and maintained through protein-protein and protein-nucleic acid interactions. The subnuclear structures [e.g., nucleolus, chromosome territory, the Cajal body (CB), promyelocytic leukemia oncoprotein (PML) nuclear bodies, splicing speckles and gems]20–25 (Fig. 1) are highly dynamic both internally and as discrete entities within the nucleus.13,26 Understanding in detail the functions of subnuclear structures and the mechanisms that regulate their occurrence is an active area of investigation.1,13,21,24–26
Dynamics of the Nucleus
In order to avoid the potential for inappropriate rapid exchange of the constituent molecules, subnuclear structures must contend with structural and functional challenges that arise from the lack of boundary membranes (conceptually different from the cell cytoplasm where many key structures are membrane-limited).2,27–30 In this context a number of highly dynamic processes must be accommodated, including nuclear import and export across the NE,31 constant localized chromatin remodelling, local and long-range movements and interactions of chromosomes, movement of subnuclear structures within the nucleus and rearrangement of the NE.29 The processes that lead to rearrangements of the NE and its components occur under a variety of physiological and pathological circumstances that occur during developmental, apoptotic and autophagic cell death pathways.
In recent years, a consensus model has been formulated that accounts for virtually all changes in NE structure and dynamics that occur during mitosis.32 In dividing metazoan cells, the interphase NE must undergo dramatic change. To successfully complete mitosis, the exclusively cytoplasmic microtubules of the spindle apparatus must contact the chromosomes normally enclosed within the nucleus and shielded by the NE. Mammalian cells disassemble their NE in prometaphase and undergo an ‘open mitosis’, effectively releasing the chromosomes into the cytoplasm allowing contact between the spindle apparatus and chromosome and facilitating their segregation following replication. The process of NE breakdown involves the disassembly and dispersal of all its structural units and their respective protein components (i.e., ONM, INM, NPCs and nuclear lamina). Once mitosis is completed, the dispersed NE components are reused to assemble new NE and nuclei in both mother and daughter cells. Functionally distinct classes of chromatin-interacting membrane proteins (e.g., lamin B, some nucleoporins) collaborate to rapidly re-establish the NE.33 NE breakdown and re-assembly require the coordinated action of many cellular activities such as mitotic phosphorylation/dephosphorylation (e.g., cyclins and cyclin-dependent kinases), nucleocytoplasmic transport, the action of microtubule motor-proteins and membrane fusion.8,34 Note that clustering of NPCs and disintegration of the NE and lamina also occur as part of the cellular changes associated with apoptotic cell death in mammalian cells (characterized by chromatin condensation, the NE breakage and the DNA fragmentation).8,35
In the yeast S. cerevisiae, by contrast, mitosis is ‘closed’ with the NE remaining intact. Nevertheless, the nucleus undergoes dramatic structural reorganization. Each mitotic spindle is confined within the intact NE (NE during mitotic entry restricts the attachment of spindle microtubules to only chromosomes within that nucleus) and components (e.g., tubulin) required for the process are shuttled across the NE through NPCs.36 The NE also remains intact during mating37 and meiotic ascospore formation.38
In addition to rearrangements of the nuclear membranes/nucleus during nuclear export/import, mitosis, mating and apoptosis accumulating evidence indicates the existence of a degradation process termed autophagy (specifically nucleophagy; see below), which is responsible for degradation of nuclear components in both yeast39–42 and mammalian cells.43 We now discuss the process of nucleophagy and its significance in cellular physiology and pathology, based mainly on the evidence from yeast studies and more recently from studies in mammalian systems.
Snapshot: Autophagy-Types, Mechanism(s), Function and Selectivity
Autophagy is a degradative process important for cellular homeostasis mediated via the vacuole (yeast cells) or the lysosome (mammalian cells).44–50 Three morphologically and mechanistically distinct modes of autophagy have been described: (1) chaperone-mediated autophagy (CMA) (described only in mammalian cells);46,51–53 (2) macroautophagy48,54 and (3) microautophagy.48,54 CMA targets cytosolic proteins bearing a KFERQ-like motif.55 Whether there are proteins which shuttle between the cytosol and the nucleus that could undergo CMA during their cytosolic “residency” has, to our knowledge, not been investigated. The latter two modes of autophagy have been described in both yeast and mammalian cells and can potentially serve as modes of nucleophagy.
Macroautophagy (often referred to simply as autophagy) is considered to be the major degradation route for turnover of cellular cytoplasmic constituents. Targets for autophagy include long-lived proteins, protein complexes (e.g., ribosomes) and aggregates, disease-related inclusions, exhausted, damaged or superfluous organelles (e.g., mitochondria, peroxisomes, ER), invading bacteria and viruses48,56–58 and most recently lipid droplets.59,60 During autophagy, portions of cytoplasm including macromolecules and whole organelles can be either non-selectively (bulk autophagy) or selectively (e.g., autophagic degradation of specific organelles such as mitochondria, peroxisomes) sequestered within double-membrane vesicles, termed autophagosomes (APs).48,54 Macroautophagy has been extensively characterized morphologically in both yeast and mammalian cells.49,61,62 Its mechanistic nature has been described, predominantly in yeast, as proceeding in seven sequential steps: (1) induction, (2) cargo selection and packaging, (3) AP nucleation, (4) AP expansion and completion, (5) retrieval of AP membrane components, (6) AP fusion with the vacuole (an example of point membrane fusion) and (7) breakdown of delivered cargo within the vacuole.46,48,49,54,63,64 The core (macro)autophagic machinery (autophagy-related ATG genes/Atg proteins) required for the process seems to be largely conserved in mammals65,66 and plants,66,67 as a number of homologues of yeast ATG genes have been identified.
Microautophagy (best described in yeast) is a related process with distinct membrane dynamics in which portions of cytoplasm including organelles (data exists for mitochondria, peroxisomes and the nucleus) are sequestered by direct invagination of the vacuole membrane.39,42,68–70 Until recently, the components of the core ATG machinery involved in microautophagic processes were relatively poorly defined except for autophagic degradation of the peroxisomes (micropexophagy).68,69 Descriptions of microautophagic degradation of mitochondria (micromitophagy)70 and of the nucleus (piecemeal microautophagy of the nucleus, PMN/micronucleophagy)39,42,71 have lead to the understanding that much of the core ATG machinery seems to be required for microautophagic processes.
Microautophagy was first described as a process in which the vacuolar membrane underwent localized invagination and ‘pinching off’ to form a vesicle within the vacuolar lumen. Components described as being required for this process, include some components of the homotypic vacuole fusion machinery and the vacuolar transporter chaperone-VTC complex (comprising Vtc1p-Vtc4p) which is present on the ER, vacuoles and at the cell periphery. On induction of autophagy by nutrient limitation the VTC complex is recruited to vacuoles becoming enriched on the membranes of autophagic tubes. Deletion of VTC genes blocks microautophagic uptake into vacuoles; although autophagic tubes still form the production of microautophagic vesicles from their tips is impaired.72–74 Additionally, the exit from rapamycin-induced growth (EGO) complex (composed of Ego1p, Gtr2p and Ego3p) has been implicated in the regulation of microautophagy.75 The EGO signalling complex acts in conjunction with target of rapamycin (TOR) signalling complex, to positively regulate microautophagy following treatment of cells with rapamycin (a pharmacological compound which markedly upregulates autophagy). The internalisation of vacuolar membrane through microautophagy is considered to counterbalance the massive membrane influx toward the vacuolar membrane resulting from rapamycin-induced macroautophagy,75 thereby maintaining vacuolar volume and membrane composition.73 The requirement for the VTC, EGO and TOR complexes in organelle specific microautophagy in yeast and for microautophagy in general for mammalian cells remains to be determined.
Autophagic Degradation of the Nucleus by PMN: Requirements and Membrane Rearrangements
In S. cerevisiae, PMN leads to the pinching off and degradation of non-essential nuclear components including portions of the NE and the granular nucleolus enriched in pre-ribosomes, but excludes chromosomal DNA, NPCs and spindle pole bodies (SPBs).39,40,48,58,76 More experiments are required to definitively determine what components of the nucleus are degraded by PMN under various conditions. With regard to the mechanistic basis of PMN, Thumm and colleagues42 showed that efficient PMN requires much of the core ATG machinery. Required components are: (1) the two ubiquitin-like conjugation systems, coupling Atg12 to Atg5 and Atg8 to phosphatidylethanolamine (PE) respectively; (2) the Atg9 cycling system (except for Atg23 and Atg27); (3) phosphatidylinositol (PtdIns) 3-kinase complex; (4) macroautophagy specific proteins (Atg17, Atg29 and Atg31); and (5) some cytoplasm-to-vacuole targeting (Cvt)-specific proteins (Atg11 and partially Atg21 and Atg24). On this basis they concluded that PMN represents a “true microautophagic process,” which can also be termed micronucleophagy (Scenario 1; Fig. 2A). Atg8p has been demonstrated to be involved in tethering between adjacent membranes and stimulating membrane hemifusion in vitro.77,78 Such functions, which mimic expansion of the autophagosomal membrane during macroautophagy, may contribute to membrane growth during microautophagy.
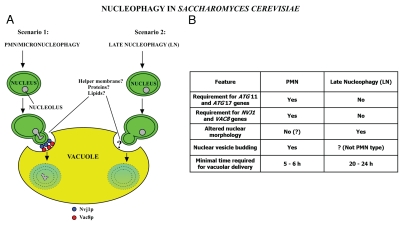
Figure 2.
Mechanistic nature of nucleophagy in yeast. (A) Autophagy-induction by nitrogen starvation (early periods of nitrogen starvation) induces the formation of NV junctions involving Nvj1p and Vac8p which initiates PMN. Then the nucleus bulges into invaginations of the vacuolar membrane, followed by fission of a nuclear membrane vesicle, and its release into the vacuolar lumen, where it is finally degraded (Scenario 1). Upon prolonged periods of nitrogen starvation (>20 h) delivery of nucleoplasm occurs by late nucleophagy (LN) (Scenario 2). Furthermore, it is possible that during both PMN and late nucleophagy additional factors such as nuclear and vacuolar proteins and lipids may be involved in the regulation of respective processes. (B) Major differences between PMN (Scenario 1) and LN (Scenario 2) are indicated.
PMN occurs in the context of nucleus-vacuole (NV) junctions between the nucleus and the vacuole generated by the interaction between two key proteins, a vacuolar membrane protein, Vac8p, and the ONM protein Nvj1p (Scenario 1; Fig. 2A).40,79 Upon starvation, these NV junctions bulge into invaginations of the vacuole, after which a portion of the nucleus (“micronucleus”) buds off and after fusion of the vacuolar extensions, a (PMN) vesicle limited by three membranes (the outermost vacuolar membrane, the two nuclear membrane; ONM and INM) is released into the vacuole and subsequently degraded (Scenario 1; Fig. 2A).39,40,42,58 Interestingly the vesicles produced in PMN can be very large, seemingly almost as large as the nucleus. It is not clear why these vesicles need to be of such size, or whether size is determined by the specific cargo being delivered to the vacuole. It has been reported that in addition to the presence of Vac8p and Nvj1p at NV junction sites, and the requirement of the core autophagic machinery (see above), lipid homeostasis (see below) is important also for the biogenesis of PMN vesicles.40,42,80,81 Thus, yeast mutants lacking certain proteins involved in lipid homeostasis exhibit defects in the maintenance of NE structure, accumulation of PMN intermediates, reduced size of PMN structures, perturbed intracellular sterol-lipid distributions and disrupted vacuole structure. However, a direct link between the factors involved in sterol-lipid homeostasis and the molecular mechanism of PMN has yet to be established.40
In addition to its involvement in the formation of NV junctions, Vac8p is a multi-purpose adaptor protein required for several other vacuolar processes in yeast such as the Cvt pathway, homotypic vacuole-vacuole fusion and the transfer and inheritance of vacuoles to budding daughter cells.82–85 In addition to Nvj1, other binding partners of Vac8p have been identified by yeast two hybrid analysis and include: Vac17p (a component of the actin-based vacuole inheritance apparatus),86 Tco89p (a vacuole membrane protein reported to be a member of the yeast TOR complex 1 and required for maintenance of cell wall integrity),87 Atg13p (a phosphorylated protein required for Cvt and autophagy),84 Vid21p (a component of the conserved NuA4 histone acetyltransferase complex),88 Vab2p (a vacuolar membrane protein that interacts with the vacuolar ATPase subunit Vma8p),79 Tao3p (involved in cell morphogenesis and proliferation)89 and two uncharacterized proteins Yel043w and Yfr035c.85 However, whether these Vac8p binding partners function in PMN has yet to be elucidated.
In addition to mediating PMN, Nvj1p also functions in sequestering two conserved proteins, namely, Osh1p and Tsc13p,40,90,91 with roles in lipid biosynthesis and trafficking to the NV junctions. Osh1p is thought to mediate sterol-dependent trafficking of the high-affinity tryptophan permease, Tat2p, that is routed to either the plasma membrane or the vacuole depending on the concentration of tryptophan in the medium.90–92 Targeting of Osh1p to NV junctions is specified by the ankyrin repeat region; Osh1p is diffusely distributed in strains lacking either Nvj1p or Vac8p alone, implying that binding requires both Nvj1p and Vac8p. Targeting would seem to require correct formation of the NV junction structure, such that Osh1p binds only to the Nvj1p/Vac8p complex, or to some other factor recruited to this complex.93 Interestingly, the human homologue of Osh1p, ORP1L, localises to perinuclear lysosomes and autophagic vacuoles (AVs) when overexpressed in HeLa cells,40,94 but the relationship between its localization and any function in mammalian nucleophagy is yet to be fully investigated.
The enoyl-CoA reductase Tsc13p is an essential ER membrane protein that catalyses the terminal step of very-long-chain fatty acid (VLCFA) biosynthesis, but which is also highly enriched in NV junctions.40,90,91,95,96 Whether Tsc13p requires Nvj1p or Vac8p for localization to NV junctions remains to be determined. VLCFAs are important constituents of several complex lipid species in yeast including ceramides, sphingolipids, inositolphospholipids and glycosylphosphatidylinositol anchors, and as such, play important roles in membrane and lipid raft biogenesis, membrane fluidity and thickness and cell signalling.40,90,91 The human homologue of Tsc13p, SC2, exhibits similar enoyl-CoA reductase activity during fatty acid elongation in vitro.40,97
Vesicles, highly enriched in Tsc13p, are observed to bud off the NE into the lumen of the vacuole (these vesicles markedly resemble PMN vesicles) when cells enter stationary phase in the presence of non-fermentable carbon sources, or in elo2 and elo3 mutants (both genes are required for VLCFA synthesis) indicating that nuclear-vacuolar interactions may provide a preferred site for NE recycling.95 In this context, one could ask what might be the physiological role of the dual localization of Tsc13p at ER and NV junctions, or of other components of the fatty acid elongation system which might be found at the sites of nuclear-vacuolar interaction.95
Why micronucleophagy requires such a complex apparatus to achieve membrane fusion between the nucleus and vacuole has been considered by Thumm and colleagues.42 They argue that fusion of the vacuolar and nuclear membranes at a site of invagination is unlikely to be able to occur at a single point based on observations of homotypic fusion of vacuoles. Vacuolar fusion does not take place at a single fusion point, which then expands radially, but at multiple vertex sites (microdomains at the apposed membranes).98 Presumably, several SNARE-complexes must form simultaneously over a larger surface area to generate enough force to achieve fusion. Krick et al.42 speculated that this problem could be overcome by using a membranous helper structure similar to that observed in micropexophagy in Pichia pastoris. Here, a cluster of peroxisomes is enclosed by vacuolar membrane protrusions and/or segmented vacuoles in concert with a newly formed membrane structure, the micropexophagy-specific membrane apparatus (MIPA), which mediates the enclosure of the vacuolar membrane,68,99 by heterotypical fusion with the surrounding vacuolar membrane (Scenario 1; Fig. 2A). At present there is no evidence to support the involvement of a MIPA-like apparatus in nucleophagy.
A Second Form of Autophagic Degradation of the Yeast Nucleus
We have recently obtained evidence for a second form of nucleophagy— late nucleophagy (LN)—in yeast that can be detected after prolonged periods of nitrogen starvation (Mijaljica et al., manuscript in preparation). Dual labelling of cells with Nvj1p-EYFP (an outer NE reporter of PMN)39 and NAB35-DsRed.T3 (nucleoplasm reporter) demonstrated that whereas induction of PMN can be detected as early as 3 hours of nitrogen starvation as reported previously,39 LN can be detected only after 20 hours. In addition to this clear temporal distinction between the two processes, our data suggest that they are also spatially separated, as we rarely observe dual-labelling of nuclear material (vesicles) taken up into the vacuole. LN does not require the same components of the core macroautophagic machinery, or even Vac8p or Nvj1, essential components of NV junctions in PMN. The inhibition of LN in some atg mutants is accompanied by gross alterations in the nuclear morphology (Scenario 2; Fig. 2A), again emphasising the differences between this form of nucleophagy and PMN. The differences between PMN and LN have been summarised in Figure 2B.
It is not clear (yet) whether these alterations are a consequence of LN being mechanistically deranged (and therefore the inability of the cell to remove material such as damaged components from the nucleus), or whether the conditions of cell growth (nitrogen starvation) under which the phenotype is viewed otherwise leads to the depletion or mis-localization of a key enzymatic activity in relation to maintenance of membrane morphology.
Previous findings have established that the amount and types of phospholipids are crucial for the biogenesis and homeostasis of the NE. Dramatic effects on yeast nuclear morphology have been reported in yeast strains perturbed in phospholipid homeostasis due to deletion of selected genes. For example, in cells null for expression of each of NEM1 or PAH1 display irregularly shaped nuclei.18,100 A striking nuclear membrane phenotype has been reported in cells of a temperature-conditional mtr7 mutant, defective in acetyl-CoA carboxylase, linking VLCFA synthesis to the structure and function of the nuclear membrane-pore complex.101
It is possible that alterations in nuclear morphology observed in cells null for autophagy genes required for efficient LN may relate to disturbance of phospholipid and/or VLCFA homeostasis. A change in lipid composition and distribution in the nuclear membranes may be a signal for nucleophagy events.102
Autophagic Degradation of the Nucleus and its Components in Mammalian Cells
The role of autophagy in nuclear turnover of mammalian cells has recently received increased attention. Early observations were disparate in nature and provided few details with the respect to the type and role of the particular autophagic processes described (Table 1). The degradation of the entire nucleus was reported to occur in cultured murine seminal vesicle epithelial cells through an autophagic process termed nucleophagy, but without any description of its mechanism (i.e., mechanistically uncharacterized nucleophagy).103 More recently it was reported that sequestration of mitotic (M-phase) chromosomes in APs of Chang liver cells could be induced by an OH· burst.104 These observations raised the intriguing question of how the normal mitotic spindle and chromosomes that are transiently resident in the cytosol compared to their normal sequestration within the NE during interphase, are protected from persistently robust autophagy that occurs during mitosis.105 A third study demonstrated that senescent keratinocytes seem to die through massive and specific autophagic degradation of their nuclei and mitochondria, although degradation of other cellular organelles was not examined.106 Other findings supported the role of autophagy in the death of senescent keratinocytes. The autophagy inhibitor 3-methyladenine can delay the death of senescent cells. Moreover, if the acidification of lysosomes, required for the final degradation of cell components, was blocked then AVs full of debris accumulated inside corpses. Finally, dying senescent cells acquired a particular intracellular organization, whereby a cytokeratin network developed and partitioned the cell into a cortical domain devoid of organelles and a central domain containing a high number of AVs, most of the mitochondria, and the nucleus. These observations gave rise to the assumption that due to their close proximity to AVs, nuclei and mitochondria could be degraded within such AVs. This notion was supported by the altered morphology of the nuclei and mitochondria within the central domain and by the level of DNA degradation observed.106
Table 1.
Autophagic degradation of the nucleus and its components in yeast and mammalian cells
| Evidence for autophagic process(s) involved in degradation | ||
| Nucleus/Nuclear component | Yeast cells | Mammalian cells |
| Entire nucleus | Not determined | Mechanistically uncharacterized nucleophagy103 |
| Nucleoplasm | Nucleophagy41,126,127 | Not determined |
| PMN42 | ||
| Inner Nuclear Membrane (INM) | PMN39 | INM and ONM participate in the formation of autophagosome-like membranes during viral infection of macrophages123,124 |
| Outer Nuclear Membrane (ONM) | PMN39 | |
| Nuclear Pore Complex (NPC) | Not degraded39,40 | Not determined |
| Nucleolus | PMN39 | Not determined |
| Nuclear Lamina | Yeast cells lack nuclear lamins | Mechanistically uncharacterized nucleophagy43 |
| Chromosomes/Chromosomal DNA | Not degraded39,40 | Mechanistically uncharacterized nucleophagy43,104 |
It is possible that the nuclei of senescent cells in general could be targeted for autophagy simply because their DNA is damaged, since inhibition of DNA-protein kinase, a nuclear kinase involved in DNA-break signaling, sensitizes to autophagy in malignant glioma cells,107 suggesting that persistence of damaged DNA can activate the autophagic process. Similarly, both an increase in Beclin-1 (mammalian homologue of Atg6p) expression and a concomitant increase in autophagic activity occur following treatment with the DNA-damaging agent etoposide.108 Furthermore, DNA damage has been shown to accumulate in cancer cells deficient in autophagy.109
Accumulating evidence suggests that reduced autophagy promotes DNA damage, gene amplification, chromosome instability, and aneuploidy, all of which are clinically associated with tumor progression and poor prognosis. Similar findings with allelic loss of beclin1 in immortalized breast epithelial cells and immortalized, autophagy-defective atg5−/− mouse embryo fibroblasts (MEFs) suggest that these observations are independent of tissue type. Analogous to the mammalian central nervous system where targeted deletion of either atg5 or atg7 revealed the protective role of constitutive autophagy through prevention of the accumulation of polyubiquitinated proteins and neurodegeneration, in tumor cells, preventing the genome damage through autophagy may be an essential cell-autonomous mechanism for tumor suppression.110 Furthermore, autophagy—defective beclin1+/− immortalized baby mouse kidney epithelial (iBMK) cells expressing anti-apoptotic Bcl-2 showed profound microtubule and centrosome abnormalities, including heterogeneity in cell and nuclear size and shape and an increase in the percentage of cells with centrosome abnormalities including increased centrosome number. Abnormalities in the microtubule framework can arise due to abnormally large cell size. Moreover, supernumerary centrosomes and large nuclei are defining traits of excess DNA content as well as genomic instability. These abnormalities in beclin1+/− iBMK cells suggested that autophagy and maintenance of metabolism might be crucial for limiting DNA damage and maintaining genome integrity.110
Most recently, it has been demonstrated that perinuclear APs/autolysosomes (sometimes bigger than the nucleus itself) contain extruded nuclear components in nuclear envelopathies/laminopathies caused by mutations in the genes encoding A-type lamins (LMNA) or emerin (EMD). These APs and autolysosomes were decorated with autophagy-related proteins (e.g., LC3) and the lysosomal marker protein, LAMP2. Moreover, APs appeared to contain the damaged portions of nuclei as demonstrated by γH2AX (a marker of DNA double-strand breaks) immunostaining.43
Nucleophagy: In Physiology and Pathophysiology
Focusing on S. cerevisiae.
In unicellular organisms, the primary role of autophagic processes (in general) is to regulate intracellular homeostasis (e.g., in response to starvation). Thus, PMN could function in yeast as a quality control mechanism (regulation of nuclear volume, quantity and quality) to dispose of damaged nuclear components or bulky nuclear aggregates that are resistant to attack by the proteasome.39,40 Strong evidence now indicates that proteasomes are active in both the cytoplasm (the ubiquitin-proteasome system-UPS) and the cell nucleus (nUPS).111 nUPS is active both in the nucleoplasm and distinct subnuclear structures (e.g., PML nuclear bodies, nuclear speckles) where it plays a major role in controlling the initial steps of gene expression and nuclear quality-control mechanisms including DNA repair.111,112 In mammalian cells, specific inhibition of proteasomal degradation by inhibitors such as lactacystin stabilises nuclear proteins such as histone H2A and PML protein. However, these proteins accumulate in the subnuclear structures in which they normally reside upon inhibition of proteasomes, findings that suggest that they represent substrates of the nUPS. By contrast, nucleolar and NE proteins such as fibrillarin and lamin A/C do not appear to represent proteasome substrates under normal conditions.111,112 In yeast cells, both the UPS and nUPS appear to play a more restricted role. Limitations in the use of proteasomal inhibitors (because only β-lactone can penetrate cells113), make it difficult to test if inhibition of nUPS enhances the level of PMN or late nucleophagy.
Focusing on mammals.
In higher eukaryotes autophagy is involved in a wide range of physiological and pathological processes, including stress responses, development, cell differentiation, antigen presentation, cancer, aging, neurodegeneration (e.g., Parkinson and Alzheimer diseases), various myopathies (e.g., Danon disease) and cell death.48,50,56,57,64,114,115 Recently, it has become evident that in several pathological situations where autophagy has a beneficial role, this degradation pathway is able to specifically and/or selectively eliminate unwanted structures including cellular organelles or bacterial and viral pathogens.64,116
It could be argued that autophagic degradation of the nucleus in mammalian cells is not generally necessary because, unlike yeast cells which have a closed nucleus during mitosis, they could utilise ‘standard’ autophagic degradation processes (either macro- or microautophagic) during S phase when the NE has been dis-assembled. As suggested by Kvam and Goldfarb40 specialized autophagy for turnover of the nucleus may be more important for long-lived amitotic cells such as neurons. Micronuclei are found in a variety of mammalian cells117 for example, micronuclei have been found in the cytoplasm of Bloom syndrome cells during S phase.118 While it is beyond the scope of this review to consider the production and fate of micronuclei, it is presently unknown whether micronuclei are turned over by autophagy. This could occur by a microautophagic process through the direct interaction of micronuclei membranes with lysosomes, or micronuclei could be sequestered in APs by non-selective macroautophagy. In both cases these processes could be considered as nucleophagy.
Links between human pathology and possible autophagy of the nucleus are now becoming apparent. Approximately 25% of cases of Diamond Blackfan Anemia (DBA), a severe hypoplastic anaemia, are linked to heterozygous mutations in the gene encoding ribosomal protein S19 and a haploinsufficiency for this protein.119 When amino acid substitutions from DBA patients known to induce defects in the processing of the pre-rRNA are introduced into yeast Rps19p, defects similar to those observed in cells under-expressing Rps19p are observed. A conspicuous feature in a majority of Rps19p-depleted yeast cells, as observed in electron microscopy sections, was what appeared to be the vacuole engulfing the nucleolus.120 These results demonstrate the effect of DBA-associated mutations on the function of Rps19p, strongly connecting the pathology to ribosome biogenesis and indicate a role for autophagy in the pathogenesis of disease.
The mutation of p.H222P in LMNA is one of several mutations causing muscular dystrophy in humans. A homozygous knock-in mouse model carrying the mutation, LmnaH222P/H222P reproduces the phenotype of human muscular dystrophy due to LMNA mutations.121 Recently, Nishino and colleagues43 demonstrated, using LmnaH222P/H222P MEFs, that where the NE interfaces with APs/autolysosomes there was an increase of NE proteins. The autophagic nature of such processes was further supported by the finding that LC3-II (a marker of autophagy that decorates APs) is upregulated in these cells. Furthermore, inhibition of autophagy in these cells led to the accumulation of nuclear abnormalities (irregular shape of the nucleus, the disruption of nuclear membrane) and reduced cell viability, strongly suggesting a beneficial role of nucleophagy. Interestingly evidence for the occurrence of nucleophagy was observed even in a small percentage of wild-type cells, suggesting that autophagic degradation of nuclear components is not confined to the disease condition. It was further suggested that autophagic degradation of particular nuclear components could contribute to the rapid repair of the nuclear membrane and prevention of cell death. However, the precise role of nucleophagy in LmnaH222P/H222P MEFs, remains to be fully elucidated.43
An intriguing observation suggests the NE and its interrelationship with an autophagic process may be crucial to the course of viral infection. In herpes simplex virus type 1 (HSV-1) infected macrophages, four-layered membrane structures (comprising the double AP membrane and NE membrane) were seen to emerge from the NE, disconnect from the nucleus and accumulate in the cytoplasm as a late response to infection (around 8 hours after infection). These four-layered structures were decorated with LC3 and subsequently fused with lysosomes. It seems that during this process, the nucleus is the source of the membrane that becomes the autophagosomal membrane. Thus the potential advantage of assembly of viral particles in the protected environment of the nucleus prior to movement through the NE, is counteracted by the nucleophagic reponse.122,123
Unanswered Questions
Yeast.
Many questions with regards to micronucleophagy remain unanswered. For example, what additional proteins (of either nuclear or vacuolar membranes) are required for establishing the exceptionally tight associations between Nvj1p and Vac8p? What is the influence of lipid composition of the respective membranes? Do the sites of vesicle formation represent sites used once, or for multiple sequential events? Do NV junctions form at specific (as yet unidentified) sub-domains of the nuclear and/or vacuolar membranes and what controls inclusion/exclusion of various proteins and/or lipids at those NV junctions? In this context it has been reported that NV junctions form at regions lacking NPCs.39,102 Another intriguing question is which nuclear components (target substrates) can be degraded by PMN and/or LN and whether the same nuclear components are degraded under different autophagy-induced conditions (e.g., nitrogen and/or carbon starvation, rapamycin treatment). Also, it is not clear if there is a temporal difference in the recruitment of different components of the nucleus into PMN vesicles or during LN.
Mammalian cells.
The findings of Park et al.43 indicate that the giant APs/autolysosomes observed appear to degrade the extruded nuclear components. Although such giant APs are quite unusual, there is precedent for large APs in mammalian cells. First the observations of Kovács et al.103 referred to above and second the APs reported to sequester infecting group A streptococcus in HeLa cells.124,125 The form of nucleophagy being observed in all these examples would appear to be macroautophagy. The involvement of microautophagic processes cannot yet be unequivocally ruled out. It may be that the particular conditions applying in cells influence the form of nucleophagy induced. Thus, in yeast PMN and LN are induced by nitrogen depletion or rapamycin induction, whereas in LmnaH222P/H222P cells it appears the NE structural defect relating to defective lamina structure contributes to the molecular mechanism. Interestingly, Park et al.43 noted evidence of nucleophagy in (apparently) wild-type cells, but at much lower frequency, implying that nucleophagy can occur as a result of other conditions that promote nuclear damage. Understanding what these conditions might be, the nature of the damage sustained when these conditions persist and the identity of the nuclear components subsequently degraded by nucleophagy, await elucidation.
Concluding Comments and Prospectus
The role of autophagy in the degradation of nuclear components is gaining wider significance, with reports of nucleophagy in mammalian cells, now building upon the foundation of observations in yeast. Even though there remains much to be determined regarding the mechanistic nature of the processes described to date, it is clear that nucleophagy involves largely yet to be revealed intricacies of membrane dynamics. The notion of pinching off parts (“pieces”) of the nucleus and eliminating them by nucleophagy (Fig. 3), in order to maintain the vitality of the nucleus and the cell is an attractive proposition. Presumably, nucleophagy (in whatever form) is sufficient to ‘repair’ damaged nuclei so that cells can maintain normal function and avoid cell death by apoptosis. Whether a similar mechanism apply across species (i.e., conservation across evolution) and under various environmental cues, has yet to be investigated in detail and awaits further studies in mammalian cells in particular, and may well be informed by studies in other genetically tractable metazoan species such as Drosophila melanogaster, Caenorhabditis elegans and Danio rerio (zebrafish). Thus, the next few years hold the promise of significant progress in our understanding of the mechanisms of nucleophagy and the accompanying intricacies of nuclear membrane rearrangements.
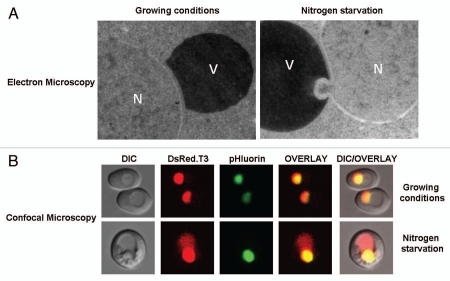
Figure 3.
Monitoring autophagic degradation of the nucleus in yeast. (A) Electron micrograph showing the contact between the outer membrane of the nucleus and the vacuolar membrane under both growing and nitrogen starvation conditions (24 h starvation). Note the partitioning of the nucleus (N) into the vacuole (V) under nitrogen starvation conditions where both the INM and ONM exvaginate in tandem into the vacuole (this could represent either a PMN or LN event). (B) Live-cell confocal imaging of delivery of the nucleus in the vacuole (LN) using NAB35-Rosella biosensor.41,126,127 Cells were either grown in the presence of both carbon and nitrogen (growing conditions) or subjected to nitrogen starvation for 24 h. This fluorescent reporter is targeted to the nucleoplasm, allowing nucleophagy to be monitored during the phase that represents uptake of material into the yeast vacuole for degradation. The reporter is a dual colour emission biosensor comprising a relatively pH-stable red fluorescent protein variant, DsRed.T3 and a pH-sensitive green fluorescent protein variant, pHluorin.126,127 NAB35-Rosella exhibits both red and green fluorescence in the nucleus (neutral pH environment) (growing conditions and nitrogen starvation), but loses green fluorescence and retains red fluorescence when within the vacuole (nitrogen starvation).
Acknowledgements
We appreciate critical feedback on the review from Prof. D.A. Jans. Work from the authors' laboratory was supported by Australian Research Council (ARC) Discovery Project funding for DP0986937 to R.J.D. We thank Sarah Ellis and Stephen Asquith (Peter MacCallum Cancer Centre, Melbourne) for the use of the EM facilities and acknowledge their help with EM sample preparation and analysis.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/11738
References
- 1.Dundr M, Misteli T. Functional architecture in the cell nucleus. Biochem J. 2001;356:297–310. doi: 10.1042/0264-6021:3560297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rippe K. Dynamic organization of the nucleus. Curr Opin Genet Dev. 2007;17:373–380. doi: 10.1016/j.gde.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hetzer MW, Walther TC, Mattaj IW. Pushing the envelope: structure, function and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- 5.Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly and interactions. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- 6.Foisner R. Inner nuclear membrane proteins and the nuclear lamina. J Cell Sci. 2001;114:3791–3792. doi: 10.1242/jcs.114.21.3791. [DOI] [PubMed] [Google Scholar]
- 7.Burke B, Stewart CL. Life at the edge: the nuclear envelope and human disease. Nat Rev Mol Cell Biol. 2002;3:575–585. doi: 10.1038/nrm879. [DOI] [PubMed] [Google Scholar]
- 8.Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16:533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- 9.Gruenbaum Y, Goldman RD, Meyuhas R, Mills E, Margalit A, Fridkin A, et al. The nuclear lamina and its functions in the nucleus. Int Rev Cytol. 2003;226:1–62. doi: 10.1016/s0074-7696(03)01001-5. [DOI] [PubMed] [Google Scholar]
- 10.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 11.Vlcek S, Foisner R. A-type lamin networks in light of laminopathic diseases. Biochim Biophys Acta. 2007;1773:661–674. doi: 10.1016/j.bbamcr.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 13.Huber MD, Gerace L. The size-wise nucleus: nuclear volume control in eukaryotes. J Cell Biol. 2007;179:583–584. doi: 10.1083/jcb.200710156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda K, Xu ZJ, Takahashi S, Ito A, Ono M, Nomura K, et al. Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp Cell Res. 1997;232:173–181. doi: 10.1006/excr.1997.3531. [DOI] [PubMed] [Google Scholar]
- 15.Gindullis F, Rose A, Patel S, Meier I. Four signature motifs define the first class of structurally related large coiled-coil proteins in plants. BMC Genomics. 2002;3:9. doi: 10.1186/1471-2164-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirzayan C, Copeland CS, Snyder M. The NUF1 gene encodes an essential coiled-coil related protein that is a potential component of the yeast nucleoskeleton. J Cell Biol. 1992;116:1319–1332. doi: 10.1083/jcb.116.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rout MP, Aitchison JD. The nuclear pore complex as a transport machine. J Biol Chem. 2001;276:16593–16596. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- 18.Siniossoglou S. Lipins, lipids and nuclear envelope structure. Traffic. 2009;10:1181–1187. doi: 10.1111/j.1600-0854.2009.00923.x. [DOI] [PubMed] [Google Scholar]
- 19.Holmer L, Worman HJ. Inner nuclear membrane proteins: functions and targeting. Cell Mol Life Sci. 2001;58:1741–1747. doi: 10.1007/PL00000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim RY, Fahrenkrog B. The nuclear pore complex up close. Curr Opin Cell Biol. 2006;18:342–347. doi: 10.1016/j.ceb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S. Chromosome territories-a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 24.Spector DL. Nuclear domains. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- 25.Zimber A, Nguyen QD, Gespach C. Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cell Signal. 2004;16:1085–1104. doi: 10.1016/j.cellsig.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 26.Spector DL, Gasser SM. A molecular dissection of nuclear function. Conference on the dynamic nucleus: questions and implications. EMBO Rep. 2003;4:18–23. doi: 10.1038/sj.embor.embor701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misteli T. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- 28.Pederson T. Dynamics and genome-centricity of interchromatin domains in the nucleus. Nat Cell Biol. 2002;4:287–291. doi: 10.1038/ncb1202-e287. [DOI] [PubMed] [Google Scholar]
- 29.Belmont A. Dynamics of chromatin, proteins and bodies within the cell nucleus. Curr Opin Cell Biol. 2003;15:304–310. doi: 10.1016/s0955-0674(03)00045-0. [DOI] [PubMed] [Google Scholar]
- 30.Lamond AI, Sleeman JE. Nuclear substructure and dynamics. Curr Biol. 2003;13:825–828. doi: 10.1016/j.cub.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Pemberton L, Paschal B. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 32.Trinkle-Mulcahy L, Lamond AI. Toward a high-resolution view of nuclear dynamics. Science. 2007;318:1402–1407. doi: 10.1126/science.1142033. [DOI] [PubMed] [Google Scholar]
- 33.Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW. Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J Cell Biol. 2009;186:183–191. doi: 10.1083/jcb.200901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burke B, Ellenberg J. Remodelling the walls of the nucleus. Nat Rev Mol Cell Biol. 2002;3:487–497. doi: 10.1038/nrm860. [DOI] [PubMed] [Google Scholar]
- 35.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Souza CP, Osmani SA. Mitosis, not just open or closed. Eukaryot Cell. 2007;6:1521–1527. doi: 10.1128/EC.00178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose MD. Nuclear fusion in the yeast Saccharomyces cerevisiae. Annu Rev Cell Dev Biol. 1996;12:663–695. doi: 10.1146/annurev.cellbio.12.1.663. [DOI] [PubMed] [Google Scholar]
- 38.Neiman AM. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kvam E, Goldfarb DS. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 2007;3:85–92. doi: 10.4161/auto.3586. [DOI] [PubMed] [Google Scholar]
- 41.Mijaljica D, Prescott M, Devenish RJ. Nibbling within the nucleus: turnover of the nuclear contents. Cell Mol Life Sci. 2007;64:581–588. doi: 10.1007/s00018-007-6395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krick R, Muehe Y, Prick T, Bremer S, Schlotterhose P, Eskelinen EL, et al. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell. 2008;19:4492–4505. doi: 10.1091/mbc.E08-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park YE, Hayashi YK, Bonne G, Arimura T, Noguchi S, Nonaka I, et al. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009;5:795–804. doi: 10.4161/auto.8901. [DOI] [PubMed] [Google Scholar]
- 44.Deretic V, Klionsky DJ. How cells clean house. Sci Am. 2008;298:74–81. doi: 10.1038/scientificamerican0508-74. [DOI] [PubMed] [Google Scholar]
- 45.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Todde V, Veenhuis M, van der Klei IJ. Autophagy: principles and significance in health and disease. Biochim Biophys Acta. 2009;1792:3–13. doi: 10.1016/j.bbadis.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 47.Cebollero E, Reggiori F. Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2009;1793:1413–1421. doi: 10.1016/j.bbamcr.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Kraft C, Reggiori F, Peter M. Selective types of autophagy in yeast. Biochim Biophys Acta. 2009;1793:1404–1412. doi: 10.1016/j.bbamcr.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 49.Nakatogawa H, Suzuki K, Kamada y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 50.Shoji-Kawata S, Levine B. Autophagy, antiviral immunity and viral countermeasures. Biochim Biophys Acta. 2009;1793:1478–1484. doi: 10.1016/j.bbamcr.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Cuervo AM. Autophagy: many paths to the same end. Mol Cell Biochem. 2004;263:55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- 53.Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 54.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 55.Kon M, Cuervo AM. Chaperone-mediated autophagy in health and disease. FEBS Lett. 2009 doi: 10.1016/j.febslet.2009.12.025. doi:10.1016/j.febslet.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farré JC, Krick R, Subramani S, Thumm M. Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol. 2009;21:522–530. doi: 10.1016/j.ceb.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Czaja MJ, Cuervo AM. Lipases in lysosomes, what for? Autophagy. 2009;5:866–867. doi: 10.4161/auto.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 62.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 2007;17:839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 64.van der Vaart A, Mari M, Reggiori F. A picky eater: exploring the mechanisms of selective autophagy in human pathologies. Traffic. 2008;9:281–289. doi: 10.1111/j.1600-0854.2007.00674.x. [DOI] [PubMed] [Google Scholar]
- 65.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 66.Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–116. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- 67.Bassham DC. Plant autophagy-more than a starvation response. Curr Opin Plant Biol. 2007;10:587–593. doi: 10.1016/j.pbi.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 68.Dunn WA, Jr, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, et al. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 69.Farré JC, Subramani S. Peroxisome turnover by micropexophagy: an autophagy-related process. Trends Cell Biol. 2004;14:515–523. doi: 10.1016/j.tcb.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 70.Kissová I, Salin B, Schaeffer J, Bhatia S, Manon S, Camougrand N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007;3:329–336. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- 71.Mijaljica D, Prescott M, Devenish RJ. Vacuolar turnover of the nucleus in yeast cells lacking ATG gene products. FEBS J. 2008;275:78. [Google Scholar]
- 72.Uttenweiler A, Schwarz H, Neumann H, Mayer A. The vacuolar trasporter chaperone (VTC) complex is required for microautophagy. Mol Biol Cell. 2007;18:166–175. doi: 10.1091/mbc.E06-08-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uttenweiler A, Mayer A. Microautophagy in the yeast Saccharomyces cerevisiae. Methods Mol Biol. 2008;445:245–259. doi: 10.1007/978-1-59745-157-4_16. [DOI] [PubMed] [Google Scholar]
- 74.Muller O, Neumann H, Bayer MJ, Mayer A. Role of the Vtc proteins in V-ATPase stability and membrane trafficking. J Cell Sci. 2003;116:1107–1115. doi: 10.1242/jcs.00328. [DOI] [PubMed] [Google Scholar]
- 75.Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 76.Millen JI, Krick R, Prick T, Thumm M, Goldfarb DS. Measuring piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae. Autophagy. 2009;5:75–81. doi: 10.4161/auto.5.1.7181. [DOI] [PubMed] [Google Scholar]
- 77.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 78.Subramani S, Farré JC. A ubiquitin-like protein involved in membrane fusion. Cell. 2007;130:18–20. doi: 10.1016/j.cell.2007.06.041. [DOI] [PubMed] [Google Scholar]
- 79.Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, et al. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p and Nvj1p. Mol Biol Cell. 2000;11:2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kvam E, Goldfarb DS. Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J Cell Sci. 2004;117:4959–4968. doi: 10.1242/jcs.01372. [DOI] [PubMed] [Google Scholar]
- 81.Kvam E, Gable K, Dunn TM, Goldfarb DS. Targeting of Tsc13p to nucleus-vacuole junctions: a role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles. Mol Biol Cell. 2005;16:3987–3998. doi: 10.1091/mbc.E05-04-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pan X, Goldfarb DS. YEB3/VAC8 encodes a myristylated armadillo protein of the Saccharomyces cerevisiae vacuolar membrane that functions in vacuole fusion and inheritance. J Cell Sci. 1998;111:2137–2147. doi: 10.1242/jcs.111.15.2137. [DOI] [PubMed] [Google Scholar]
- 83.Wang YX, Catlett NL, Weisman LS. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scott SV, Nice DC, 3rd, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 85.Tang F, Peng Y, Nau JJ, Kauffman EJ, Weisman LS. Vac8p, an armadillo repeat protein, coordinates vacuole inheritance with multiple vacuolar processes. Traffic. 2006;7:1368–1377. doi: 10.1111/j.1600-0854.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- 86.Tang F, Kauffman EJ, Novak JL, Nau JJ, Catlett NL, Weisman LS. Regulated degradation of a class V myosin receptor directs movement of the yeast vacuole. Nature. 2003;422:87–92. doi: 10.1038/nature01453. [DOI] [PubMed] [Google Scholar]
- 87.Reinke A, Anderson S, McCaffery JM, Yates J, 3rd, Aronova S, Chu S, et al. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- 88.Auger A, Galarneau L, Altaf M, Nourani A, Doyon Y, Utley RT, et al. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol Cell Biol. 2008;28:2257–2270. doi: 10.1128/MCB.01755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Du LL, Novick P. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:503–514. doi: 10.1091/mbc.01-07-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kvam E, Goldfarb DS. Nucleus-vacuole junctions in yeast: anatomy of a membrane contact site. Biochem Soc Trans. 2006;34:340–342. doi: 10.1042/BST0340340. [DOI] [PubMed] [Google Scholar]
- 91.Kvam E, Goldfarb DS. Structure and function of nucleus-vacuole junctions: outer-nuclear-membrane targeting of Nvj1p and a role in tryptophan uptake. J Cell Sci. 2006;119:3622–3633. doi: 10.1242/jcs.03093. [DOI] [PubMed] [Google Scholar]
- 92.Beck T, Schmidt A, Hall MN. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol. 1999;146:1227–1238. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levine TP, Munro S. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol Biol Cell. 2001;12:1633–1644. doi: 10.1091/mbc.12.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Johansson M, Lehto M, Tanhaunpaa K, Cover TL, Olkkonen VM. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol Biol Cell. 2005;16:5480–5492. doi: 10.1091/mbc.E05-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kohlwein SD, Eder S, Oh CS, Martin CE, Gable K, Bacikova D, et al. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:109–125. doi: 10.1128/MCB.21.1.109-125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rossler H, Rieck C, Delong T, Hoja U, Schweizer E. Functional differentiation and selective inactivation of multiple Saccharomyces cerevisiae genes involved in very-long-chain fatty acid synthesis. Mol Genet Genomics. 2003;269:290–298. doi: 10.1007/s00438-003-0836-0. [DOI] [PubMed] [Google Scholar]
- 97.Moon YA, Horton JD. Identification of two mammalian reductases involved in the two-carbon fatty acyl elongation cascade. J Biol Chem. 2003;278:7335–7343. doi: 10.1074/jbc.M211684200. [DOI] [PubMed] [Google Scholar]
- 98.Wang L, Seeley ES, Wickner W, Merz AJ. Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell. 2002;108:357–369. doi: 10.1016/s0092-8674(02)00632-3. [DOI] [PubMed] [Google Scholar]
- 99.Sakai Y, Oku M, van der Klei IJ, Kiel JA. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 100.Campbell JL, Lorenz A, Witkin KL, Hays T, Loidl J, Cohen-Fix O. Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol Biol Cell. 2006;17:1768–1778. doi: 10.1091/mbc.E05-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schneiter R, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD, Tartakoff AM. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Krick R, Mühe Y, Prick T, Bredschneider M, Bremer S, Wenzel D, et al. Piecemeal microautophagy of the nucleus: genetic and morphological traits. Autophagy. 2009;5:270–272. doi: 10.4161/auto.5.2.7639. [DOI] [PubMed] [Google Scholar]
- 103.Kovács AL, Réz G, Pálfia Z, Kovács J. Autophagy in the epithelial cells of murine seminal vesicle in vitro. Formation of large sheets of nascent isolation membranes, sequestration of the nucleus and inhibition by wortmannin and 3-ethyladenine. Cell Tissue Res. 2000;302:253–261. doi: 10.1007/s004410000275. [DOI] [PubMed] [Google Scholar]
- 104.Sit KH, Paramanantham R, Bay BH, Chan HL, Wong KP, Thong P, et al. Sequestration of mitotic (M-phase) chromosomes in autophagosomes: mitotic programmed cell death in human Chang liver cells induced by an OH* burst from vanadyl(4) Anat Rec. 1996;245:1–8. doi: 10.1002/(SICI)1097-0185(199605)245:1<1::AID-AR1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 105.Liu L, Xie R, Nguyen S, Ye M, McKeehan WL. Robust autophagy/mitophagy persists during mitosis. Cell Cycle. 2009;8:1616–1620. doi: 10.4161/cc.8.10.8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gosselin K, Deruy E, Martien S, Vercamer C, Bouali F, Dujardin T, et al. Senescent keratinocytes die by autophagic programmed cell death. Am J Pathol. 2009;174:423–435. doi: 10.2353/ajpath.2009.080332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Daido S, Yamamoto A, Fujiwara K, Sawaya R, Kondo S, Kondo Y. Inhibition of the DNA-dependent protein kinase catalytic subunit radiosensitizes malignant glioma cells by inducing autophagy. Cancer Res. 2005;65:4368–4375. doi: 10.1158/0008-5472.CAN-04-4202. [DOI] [PubMed] [Google Scholar]
- 108.Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 109.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.von Mikecz A. The nuclear ubiquitin-proteasome system. J Cell Sci. 2006;119:1977–1984. doi: 10.1242/jcs.03008. [DOI] [PubMed] [Google Scholar]
- 112.Rockel DT, von Mikecz A. Proteasome-dependent processing of nuclear proteins is correlated with their subnuclear localization. J Struct Biol. 2002;140:189–199. doi: 10.1016/s1047-8477(02)00527-0. [DOI] [PubMed] [Google Scholar]
- 113.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 114.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 115.Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta. 2009;1793:664–673. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 116.Tolkovsky AM. Mitophagy. Biochim Biophys Acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 117.Mateuca R, Lombaert N, Aka PV, Decordier I, Kirsch-Volders M. Chromosomal changes: induction, detection methods and applicability in human biomonitoring. Biochimie. 2006;88:1515–1531. doi: 10.1016/j.biochi.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 118.Yankiwski V, Marciniak RA, Guarente L, Neff NF. Nuclear structure in normal and Bloom syndrome cells. Proc Natl Acad Sci USA. 2000;97:5214–5219. doi: 10.1073/pnas.090525897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Flygare J, Karlsson S. Diamond-Blackfan anemia: erythropoiesis lost in translation. Blood. 2007;109:3152–3154. doi: 10.1182/blood-2006-09-001222. [DOI] [PubMed] [Google Scholar]
- 120.Léger-Silvestre I, Caffrey JM, Dawaliby R, Alvarez-Arias DA, Gas N, Bertolone SJ, et al. Specific role for yeast homologs of the Diamond Blackfan Anemia-associated Rps19 protein in ribosome synthesis. J Biol Chem. 2005;280:38177–38185. doi: 10.1074/jbc.M506916200. [DOI] [PubMed] [Google Scholar]
- 121.Arimura T, Helbling-Leclerc A, Massart C, Varnous S, Niel F, Lacène E, et al. Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Human Mol Genet. 2005;14:155–169. doi: 10.1093/hmg/ddi017. [DOI] [PubMed] [Google Scholar]
- 122.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.English L, Chemali M, Desjardins M. Nuclear membrane-derived autophagy, a novel process that participates in the presentation of endogenous viral antigens during HSV-1 infection. Autophagy. 2009;5:1026–1029. doi: 10.4161/auto.5.7.9163. [DOI] [PubMed] [Google Scholar]
- 124.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 125.Yoshimori T. Autophagy vs. Group A Streptococcus. Autophagy. 2006;2:154–155. doi: 10.4161/auto.2822. [DOI] [PubMed] [Google Scholar]
- 126.Devenish RJ, Prescott M, Turcic K, Mijaljica D. Monitoring organelle turnover in yeast using fluorescent protein tags. Methods Enzymol. 2008;451:109–131. doi: 10.1016/S0076-6879(08)03209-6. [DOI] [PubMed] [Google Scholar]
- 127.Rosado CJ, Mijaljica D, Hatzinisiriou I, Prescott M, Devenish RJ. Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy. 2008;4:205–213. doi: 10.4161/auto.5331. [DOI] [PubMed] [Google Scholar]