Abstract
Nuclear intermediate filament networks formed by A- and B-type lamins are major components of the nucleoskeleton. Lamins have growing links to human physiology and disease including Emery-Dreifuss muscular dystrophy (EDMD), lipodystrophy, cardiomyopathy, neuropathy, cerebellar disorders and segmental accelerated ‘aging’ syndromes. How lamins interact with other nucleoskeletal components, and even the identities of these other components, are open questions. Previous studies suggested lamins might bind actin. We report that the recombinant C-terminal tail domain of human A- and B-type lamins binds directly to purified actin in high-speed pelleting assays. This interaction maps to a conserved Actin Binding site (AB-1) comprising lamin A residues 461–536 in the Ig-fold domain, which are 54% identical in lamin B1. Two EDMD-causing missense mutations (R527P and L530P) in lamin A that are predicted to disrupt the Ig-fold, each reduced F-actin binding by ∼66%, whereas the surface-exposed lipodystrophy-causing R482Q mutation had no significant effect. The lamin A tail was unique among lamins in having a second actin-binding site (AB-2). This second site was mapped to lamin A tail residues 564–608, based on actin-binding results for the lamin C tail and internal deletions in the lamin A tail that cause Hutchinson-Gilford Progeria Syndrome (Δ35, Δ50) or restrictive dermopathy (Δ90). Supporting the presence of two actin-binding sites, recombinant precursor (unmodified) and mature lamin A tails (not C or B1 tails) each bundled F-actin in vitro: furthermore F-actin bundling was reduced 25–40% by the R527P, L530P, Δ35 and Δ50 mutations, and was abolished by Δ90. Unexpectedly, the mature lamin A tail bound F-actin significantly more efficiently than did the prelamin A tail; this suggested unmodified residues 647–664, unique to prelamin A, might auto-inhibit binding to actin (and potentially other partners). These biochemical results suggest direct mechanisms by which lamins, particularly lamin A, might impact the concentration of free actin in the nucleus or pathways including transcription, nuclear export, chromatin remodeling, chromatin movement and nuclear assembly that require nuclear myosin 1c and polymerizable actin.
Key words: lamin, nuclear actin, nuclear envelope, laminopathy, Emery-Dreifuss muscular dystrophy, Hutchinson-Gilford progeria syndrome, atypical Werner syndrome
Introduction
Lamins polymerize to form nuclear intermediate filaments, which are major components of the nucleoskeleton.1 Two mammalian genes (LMNB1, LMNB2) encode somatic B-type lamins (B1 and B2, respectively), which are essential for cell viability.2,3 A single mammalian gene, LMNA, is alternatively spliced to generate A-type lamins (lamins A, C, AΔ10 and C2), which are nonessential at the cellular level. Collectively lamins and lamin-binding proteins form ‘lamina networks’ that tether the genome and support most nuclear activities including transcription, DNA replication and genome repair; these networks also support mechanotransduction, signaling and development.4,5
The mechanisms by which the genome depends on lamins are open questions with growing relevance to human physiology and disease. For example, LMNB1 gene duplication can cause leukodystrophy6 or leucoencephalopathy,7 and mutations in LMNA cause a range of “laminopathies” in non-neuronal tissues.1,5,8 Missense mutations in LMNA can cause dominant syndromes including Emery-Dreifuss muscular dystrophy (EDMD; e.g., R527P or L530P mutation), Dunnigan-type familial partial lipodystrophy (e.g., R482Q mutation) or the accelerated aging disease, atypical Werner syndrome (e.g., E578V mutation). Other mutations in LMNA that activate cryptic mRNA splice sites can result in the deletion of 35 or 50 residues within the lamin A tail domain (mutations Δ35 and Δ50, respectively); these deletions block proteolytic processing of the farnesylated lamin A precursor protein (prelamin A) and cause Hutchinson-Gilford Progeria Syndrome (HGPS).9–11 In nuclei from HGPS patient cells, all lamins aberrantly accumulate near the nuclear inner membrane; these nuclei have unusual mechanical properties (increased stiffness) when challenged by mechanical force,12 consistent with other evidence that A-type lamins have major mechanical roles in the nucleus.13 A third mutation that deletes 90 residues within lamin A (Δ90) causes a postnatally lethal disorder, restrictive dermopathy.14
The mechanisms that link specific lamin mutations to specific diseases are open questions, to which current evidence suggests there will be multiple correct answers. For example A-type lamins confer ‘stiffness’ properties to the nucleoskeleton that are important not only for the structural integrity of the nucleus, but also for force-induced changes in gene regulation (mechanotransduction) and the structural integrity of the cytoskeleton.13 Indeed, the inner nuclear membrane protein emerin, which binds lamins directly,15 is also a pointed-end F-actin capping protein,16 binds nuclear myosin 1c (NM1) directly,17 and is required for mechanotransduction.18
Previous studies19,20 suggested two regions of human lamin A might associate with actin. All isoforms of actin can actively enter and exit nuclei, and the two Nuclear Export Sequences are identical in all six human isoforms.21,22 Actin functions in many nuclear pathways including chromatin remodeling, transcription and RNA export.21,22 Within the nucleus, actin does not assemble conventional F-actin filaments; instead ∼80% is free and ∼20% adopts alternative polymeric states to which phalloidin cannot bind.21,23,24 One potential actin-binding region of lamin A, here designated Actin-Binding site 1 (AB-1; residues 461–536) was identified in a yeast two-hybrid screen.19 The second proposed site, here designated AB-2 (residues 563–646) was sufficient as an exogenous lamin fragment to co-immunoprecipitate actin from human lung carcinoma A549 cells.20 Whether actin and lamin A associated directly or indirectly was not determined.19,20
We hypothesized lamin A might bind actin directly. To test this hypothesis we studied purified recombinant lamin tail-domain polypeptides and conventionally assembled F-actin, since biochemical studies of nuclear actin polymers are not yet feasible. We report that F-actin binds directly to both B- and A-type lamins, via AB-1. Under these in vitro conditions mature lamin A tails bound almost twice as much actin, at steady state, than did unmodified prelamin A tails, suggesting potential auto-inhibition. We further report that AB-2, which is unique to lamin A, confers the ability to bundle or crosslink actin filaments in vitro.
Results
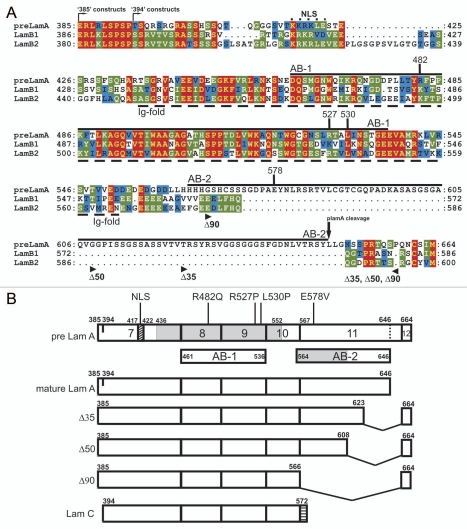
Lamin polypeptides have three major domains: a small N-terminal globular ‘head’, long coiled-coil ‘rod’ and large C-terminal globular ‘tail’.4 Within the tail is one known structural feature: a conserved lamin-specific Ig-fold domain comprising residues 436–545 in human lamin A and residues 438–545 in human lamin B1.25,26 The Ig-fold appears to mediate many specific protein-protein interactions.27 For example, lamin A tail residues 461–536 within the Ig-fold (Fig. 1A; AB-1) were sufficient to recover actin (and other proteins, including nuclear titin) in a two-hybrid study.19 The AB-1 region of lamin A is 68 and 75% conserved in lamins B1 and B2, respectively. By contrast AB-2 was unique to lamin A; this region, encoded by exon 11 of LMNA, is missing not only from B-type lamins (Fig. 1A) but also from lamin C (Fig. 1B).
Figure 1.
Conservation of AB-1 but not AB-2 in A- and B-type lamin tails; constructs used in this work. (A) Amino acid sequences of the tail domains of human prelamin A, lamin B1 and lamin B2. Predicted actin-binding sites AB-1 and AB-2 are indicated by solid lines. The nuclear localization signal (NLS)4 and Ig-fold domain are indicated by dashed lines, and residues R482, R527, L530 and E578 are marked. Arrowheads mark the left and right borders of the Δ35, Δ50 and Δ90 deletions in lamin A. (B) Schematic diagram showing the positions of AB-1 and AB-2 in the precursor and mature lamin A tails (pre Lam A and mature Lam A, respectively), and showing the missense and deletion mutations studied in this work. Shaded region in prelamin A indicates the Ig-fold. Shaded region in AB-2 indicates the minimal actin-binding site deduced in this work.
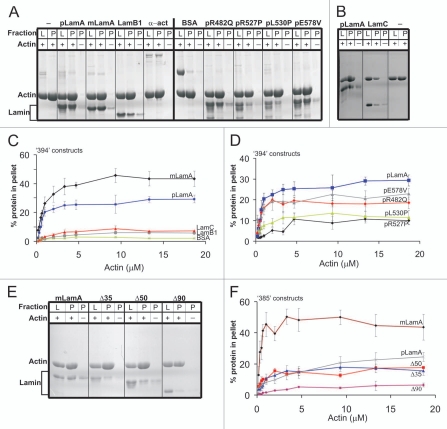
Potential direct binding to F-actin was tested in vitro using a high-speed pelleting assay16 with the purified recombinant tail domains (only) of either prelamin A (pLamA; residues 394–664), mature lamin A (mLamA; residues 394–646), lamin B1 (LamB1; residues 395–586) or lamin C (LamC; residues 394–572; depicted schematically in Fig. 1B). We also tested prelamin A tails that began at residue T394 as wildtype or bearing a disease-causing missense mutation (R482Q, R527P, L530P or E578V). Purified skeletal muscle G-actin (7.9 µM) was polymerized, incubated 1 hour with each tested tail polypeptide (6.7 µM) at a molar ratio of ∼1:1 (actin monomer to tail monomer), and then centrifuged at 100,000 g to pellet F-actin, with the actin-binding protein α-actinin28 as positive control and BSA as negative control. Inputs and pellets were then resolved by SDS-PAGE and stained with GelCode Blue (Fig. 2A and B). Qualitatively, all four wildtype lamin tails (pLamA, mLamA, LamB1 and LamC) pelleted with F-actin above background, suggesting each bound F-actin directly (Fig. 2A and B). The lamin B1 and C tails appeared to bind F-actin less well than lamin A tails (Fig. 2A and B). Qualitatively normal binding was seen for the R482Q-mutated and E578V-mutated prelamin A tails (Fig. 2A); residue R482 is surface-exposed on the Ig-fold25,26 whereas E578 is located in predicted AB-2. By contrast, the R527P-and L530P-mutated tails had qualitatively reduced binding to F-actin (Fig. 2A); both are located in AB-1 and predicted to disrupt the Ig-fold.25,26 We concluded that F-actin binding is a conserved property of lamin A and B1 tails that might be disrupted by two EDMD-causing missense mutations in AB-1.
Figure 2.
High speed pelleting assay to detect and quantify lamin tail binding to F-actin. G-actin (7.9 µM) was polymerized and incubated 1 hour with each tested protein (final concentration 6.7 µM lamin tails or BSA, 1.33 µM α-actinin), then centrifuged 30 minutes at 100,000 g. To control for background pelleting of lamins alone, actin was omitted from reactions marked (−). Gels were stained with GelCode Blue. L, load (10%) and P, pellet (20%). Gels shown are representative of three independent repeats. (A and B) Proteins tested for binding to F-actin were mature lamin A tail (mLamA), prelamin A tail (pLamA), lamin B1 tail (LamB1), α-actinin, BSA and prelamin A tail mutants R482Q, R527P, L530P, E578V and lamin C tail (LamC). (C and D) Independent binding assays in which F-actin (0–18.9 µM G-actin) was incubated 1 hour with a constant concentration (13.3 µM) of each tested protein; pellets were resolved and stained with GelCode Blue as in (A), and results were quantified by densitometry and plotted in (C and D) (n ≥ 3; bars indicate SEM). (E) Same type of assay as (A), testing the tails of mature lamin A (mLamA), prelamin A (pLamA) and corresponding deletion mutants Δ35, Δ50 and Δ90 for binding to F-actin. (F) Quantification of independent binding assays in which F-actin (0–18.9 µM G-actin) was incubated 1 hour with a constant concentration (13.3 µM) of either mature lamin A tail (mLamA), prelamin A tail (pLamA), or deletion mutants Δ35, Δ50, Δ90. (n ≥ 3, bars indicate SEM).
To independently test and quantify binding to F-actin, we incubated a constant amount (13.3 µM) of each recombinant lamin tail for 1 hour with different concentrations of actin (0–18.9 µM G-actin), then pelleted F-actin at 100,000 g, resolved by SDS-PAGE, stained with GelCode Blue and graphed the percentage of each tested protein that bound F-actin, after subtracting the amount of each tested protein that pelleted in the absence of actin (Fig. 2C and D). Under these conditions the initial rates of binding at low (<2 µM) actin concentrations were indistinguishable for mature and prelamin A tails (Fig. 2C). However, at high (>5 µM) actin concentrations, approximately 43% of wildtype mature lamin A tails pelleted but surprisingly, only ∼30% of prelamin A tails pelleted under the same conditions (Fig. 2C). These results suggested residues 647–664, unique to prelamin A, reduced binding to F-actin at steady-state. Lamin B1 and C tails bound F-actin significantly less well (5–7% bound) than the mature and precursor lamin A tails at high (>5 µM) actin concentrations (Fig. 2C).
Only ∼11% of the R527P and L530P mutants (in prelamin A tail) pelleted at high actin concentrations (Fig. 2D), ∼66% less than the prelamin A control. In other words, lamin tails with AB-1 only (e.g., lamins B1 and C) bound actin to a similar extent as tails with AB-2 plus Ig-fold-disrupting mutations in AB-1 (e.g., lamin A mutants R527P or L530P; maximum ∼5–11%). This suggested AB-1 and AB-2 each contribute to binding actin. Interestingly, the R482Q and E578V mutations (also in the prelamin A tail) had apparently normal rates of binding at low (<2 µM) actin concentrations but then plateaued at 18 and 22%, respectively; ∼30% less than their prelamin A control, suggesting mild but detectable impairment at steady state (Fig. 2D).
The contribution of AB-2 to F-actin binding was independently evaluated by testing internal deletions Δ35, Δ50 and Δ90 in polypeptides that initiated at residue E385 (see Fig. 1B). Interestingly compared to the mature control, this prelamin A tail showed a substantially lower initial rate of binding to F-actin, suggesting residues 385–393 contribute to auto-inhibition. F-actin binding of Δ35 and Δ50 plateaued at ∼18% (Fig. 2E, quantified in F); 60% less than the mature A tail, but only 25% less than the control prelamin A tail (Fig. 2F). This value (maximum ∼18% bound) was substantially higher than the maximum binding of either the Δ90 tail (∼7% bound; Fig. 2F), or the B1 and C tails that initiated at residue 394 (∼5–7% bound; Fig. 2C), which have AB-1 but completely lack AB-2. We concluded that Δ35 and Δ50 both retain a minimally functional AB-2 domain, comprising residues 564–608.
The lamin A tail, but not lamin B1 or C tails, can bundle F-actin.
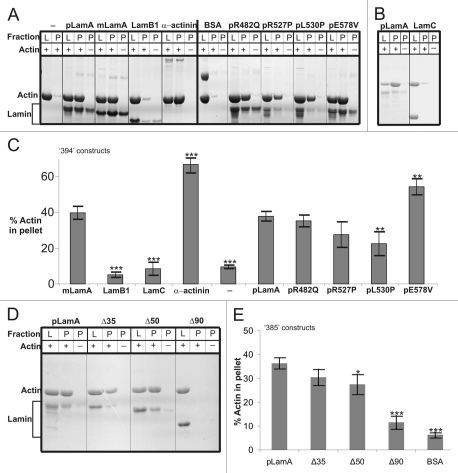
Other proteins with two actin-binding sites (e.g., filamins) can ‘bundle’ F-actin filaments.29 Bundling can be assayed by low speed sedimentation since cross-linked bundles of F-actin readily pellet, whereas single actin filaments remain in solution.30 To determine if lamin A tails bundled F-actin, we polymerized purified G-actin (7.9 µM), then incubated 1 hour with each tested tail polypeptide (6.7 µM) at a molar ratio of ∼1:1 (actin monomer to tail monomer), and centrifuged at 17,000 g, resolved pellets by SDS-PAGE, stained with GelCode Blue (Fig. 3A, B and D), and quantified by densitometry (Fig. 3C and E). Controls verified that F-actin alone and F-actin plus BSA remained soluble, and that F-actin was pelleted efficiently (∼70%) by α-actinin, the positive control31 (Fig. 3A and C). The lamin B1 and C tails showed only background levels of bundling, consistent with having a single actin-binding site (AB-1; Fig. 3A–C). The mature and precursor wildtype lamin A tails pelleted F-actin with similar efficiency (∼38%) (Fig. 3A and C), suggesting similar F-actin bundling activity. This result was initially puzzling, since the prelamin A tail showed (at a higher concentration, 13.3 µM) reduced steady-state binding to F-actin (Fig. 2), but can be explained by the more ‘permissive’ nature of the bundling assay (F-actin can be bundled by a few, or many, lamin molecules).
Figure 3.
Lamin A tails can bundle F-actin. Low speed pelleting assays. G-actin (7.9 µM) was polymerized and incubated 1 hour with each tested protein (final concentration 6.7 µM lamin tails or BSA; 1.33 µM α-actinin), then centrifuged 30 minutes at 17,000 g. To control for background pelleting of lamins, actin was omitted from reactions marked (−). (A and B) Proteins tested for binding to F-actin were prelamin A tail (pLamA), mature lamin A tail (mLamA), lamin B1 tail (LamB1), α-actinin (positive control), BSA (negative control), prelamin A mutants R482Q, R527P, L530P, E578V, or lamin C tail (LamC). L, load (10%) and P, pellet (20%). (C) Quantification by densitometry of bundling assays in (A and B) (n ≥ 3, bars indicate SEM). Differences were significant as determined by Student's t-test: *p < 0.05, **p < 0.01, ***p < 0.005. (D and E) Same low speed pelleting assays as (A) for F-actin, plus or minus prelamin A tail (pLamA) or deletion mutants Δ35, Δ50 or Δ90. L, load (10%) and P, pellet (20%). Results from (D) were quantified by densitometry in (E) (n ≥ 3, bars indicate SEM). Differences were significant as determined by Student's t-test: *p < 0.05, ***p < 0.005.
Supporting the idea that the R482Q mutant has two functional actin-binding sites, R482Q bundled F-actin nearly as well as wildtype lamin A tails (Fig. 3A and C). The R527P and L530P mutations (in AB-1) decreased F-actin bundling by 27 and 40% respectively (Fig. 3A and C), but this reduction was statistically significant only for L530P (p = 0.01, n = 4). Both mutants bundled significantly more F-actin than did the wild-type lamin B1 or C tails (Fig. 3C). These bundling results suggested the R527P and L530P mutations in AB-1 reduced, but did not abolish, binding to F-actin (see Discussion).
The effects of the Δ35, Δ50 and Δ90 deletions on F-actin bundling were studied using prelamin A tail polypeptides that started at residue E385. The Δ90 deletion failed to bundle actin filaments above the BSA negative control (Fig. 3D and E). This result was both significant (p = 10−6, n = 6) and consistent with the absence of AB-2. F-actin bundling by the Δ35 mutant did not differ significantly from the wildtype prelamin A control (Fig. 3D and E) suggesting AB-2 residues 624–646 were dispensable. The Δ50 tail bundled 24% less F-actin (Fig. 3D and E), a statistically significant result (p = 0.04, n = 6) that suggested residues 609–623 contribute to AB-2 activity, either by contributing to F-actin affinity or, we speculate, by helping overcome the proposed auto-inhibition by residues 647–664. Based on these results we suggest the ‘minimal’ AB-2 site comprises residues 564–608. Interestingly the E578V mutation, located within the proposed minimal AB-2 site, caused 50% more F-actin to pellet in this assay (Fig. 3A and C). This increase was significant (p = 0.008, n = 3) and suggests excessive or enhanced lamin A binding to actin might contribute to atypical Werner syndrome.
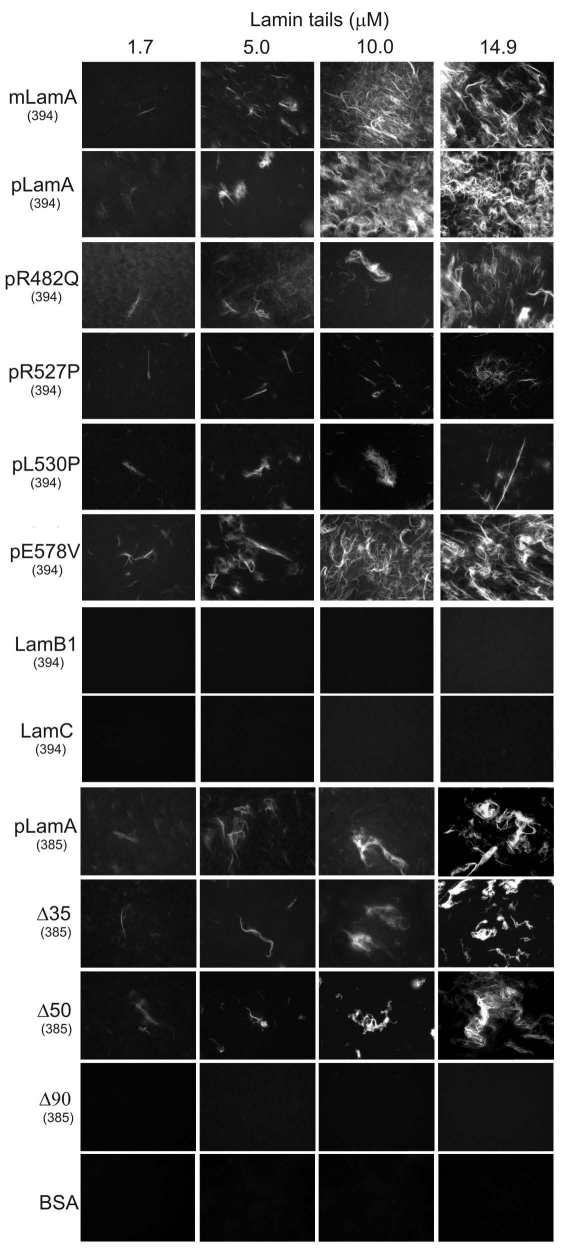
We independently assayed F-actin bundling using a fluorescence-based method that did not involve centrifugation. Purified G-actin (1.7 µM) was polymerized, then incubated 1 hour with increasing (1.7–14.9 µM) concentrations of each lamin tail to allow bundling; F-actin bundles were then stained with rhodamine-phalloidin and directly visualized by fluorescence microscopy (Fig. 4). Fluorescent bundles were detected, and increased qualitatively as a function of lamin tail concentration in reactions containing mature and prelamin A tails, but not in reactions with BSA alone, lamin B1, lamin C or Δ90 tails (Fig. 4), consistent with the low-speed bundling results (Fig. 3). Also consistent with centrifugation-based assays, the R527P and L530P mutations reduced bundling in the rhodamine labeling assay, consistent with impaired function of AB-1. The Δ35 and Δ50 tails also bundled F-actin in the rhodamine labeling assay, confirming the presence of a minimal AB-2 site. The E578V mutation, which increased F-actin bundling 50% in the low-speed pelleting assay, was indistinguishable from wildtype in the qualitative rhodamine assay (Fig. 4). These findings prompted three conclusions: (a) wildtype mature and prelamin A tails can bind at least two actin filaments simultaneously in vitro, (b) disease-associated lamin A mutations can impair bundling and (c) the lamin B1, lamin C and Δ90 tails fail to bundle and therefore have only one functional site, AB-1.
Figure 4.
Rhodamine-phalloidin assay for lamin-bundled F-actin. G-actin (1.7 µM) was polymerized and incubated 1 hour with the indicated concentration of each lamin tail, then stained with rhodamine-labeled phalloidin and directly visualized by fluorescence microscopy. Tails tested were: prelamin A (pLamA), mature lamin A (mLamA), lamin B1 (LamB1), lamin C (LamC), prelamin A mutants R482Q, R527P, L530P, E578V, deletion mutants Δ35, Δ50 or Δ90, or BSA as negative control. (394) and (385) indicate the first residue of each tested polypeptide.
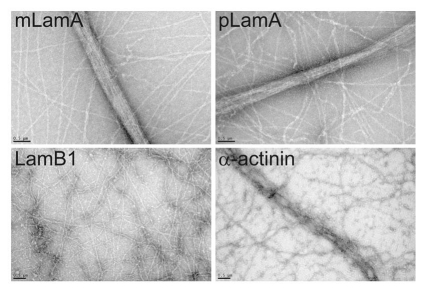
We visualized actin filaments bundled by either mature lamin A tails, prelamin A tails, lamin B1 tails or α-actinin by transmission electron microscopy (TEM). Control bundles formed by α-actinin were somewhat loose (Fig. 5) as expected.31,32 Wildtype mature and precursor lamin A tails produced relatively tight F-actin bundles (Fig. 5) consistent with the smaller molecular dimensions of lamin A tail monomers (mature 26.9 kD; precursor 28.8 kD) compared to the antiparallel dimers formed by α-actinin (116 kD).31,32 The lamin B1 tail, which showed no bundling activity in previous assays, was also negative by TEM (Fig. 5).
Figure 5.
Transmission electron microscopy of lamin-bundled F-actin. Transmission electron micrographs of negatively-stained lamin-bundled F-actin in vitro. Actin was polymerized and incubated with either the wildtype mature lamin A tail (mLamA), pre-lamin A tail (pLamA), lamin B1 tail (LamB1), or α-actinin as positive control. Scale bars, 0.5 µm.
Discussion
Both A- and B-type lamin tail domains are shown here to bind actin filaments directly, extending previous studies.19,20 Our results support two non-exclusive models. The first model is that native lamin filaments, especially those formed by lamin A, can directly interact with actin polymers in the nucleus. Since lamins are abundant, the second model is that lamins, and particularly lamin A, function to sequester G-actin or otherwise regulate actin polymers, as a mechanism to avoid spontaneous formation of F-actin in the nucleus.21,22,33 Whether lamins bind G-actin has not yet been determined. However, actin ‘suppression’ models are strongly supported by the formation of phalloidin-stainable fibers in Drosophila larval body wall muscle nuclei that lack the A-type lamin (lamin C).34 Our biochemical studies used purified rabbit skeletal muscle actin, which is predominantly skeletal α-actin and in humans differs from non-muscle β-actin and γ-actin by only 18 residues.35 Further work is needed to determine which actin residues contact lamins.
We discovered that lamin A tails are uniquely capable of bundling actin filaments in vitro, consistent with the presence of two independent actin binding sites. The first site, AB-1, is shared and conserved in all lamins whereas the second site, AB-2, is unique to lamin A. F-actin bundles were detected by rhodamine-phalloidin labeling and by TEM imaging, suggesting lamin-mediated bundling was not an artifact of centrifugation in the high- and low-speed pelleting assays. These findings suggest potential concerted mechanisms for pathways that are known to require both polymerizable actin and A-type lamins, such as mRNA export,36–39 intranuclear chromatin movement40,41 and transcription.5,42–44
Although nuclei lack phalloidin-stainable F-actin, approximately 20% of actin in the nucleus is found in alternative polymeric state(s).21,23,24 We speculate that short ‘unconventional’ actin filaments, anchored by emerin16 and potentially bundled by lamin A near the inner nuclear membrane, might help space and reinforce lamin filaments and contribute to dynamic changes in the mechanical properties of the nucleoskeleton at the nuclear envelope, and nuclear interior. Furthermore, lamin A expression, an early mark of differentiation,45,46 might contribute to development in part by supporting diverse actin-dependent pathways.
Implications for lamin A tail structure: proposed Ig-fold ‘stabilizing’ and ‘auto-inhibitory’ domains.
AB-1 comprises most of the Ig-fold domain of the lamin tail. The Ig-fold is necessary for lamin A tails to bind F-actin efficiently, since binding was reduced by the R527P and L530P mutations. However, these mutations did not eliminate binding as expected from the Ig-fold structure.25,26 R527 is a surface exposed residue predicted to form hydrogen bonds26 and a salt bridge25 that stabilize the Ig-fold. L530 is an interior (buried) residue predicted to be important for the overall structure of the fold.25,26 Based on this milder-than-expected disruption of binding, and the weak binding of lamin C and Δ90 tails we suggest the Ig-fold may be stabilized by interactions with another region of the tail, specifically residues 564–608 in AB-2. Inefficient actin binding and bundling by the lamin A Δ50 protein (‘progerin’) might contribute to the structural collapse of the lamina network in HGPS patient nuclei12 and might also impair dynamic rearrangements of nuclear actin polymers in response to mechanical stress.12,47
Based on these findings, we propose that prelamin A residues 647–664, which are normally cleaved, carboxymethylated, farnesylated and then proteolytically removed during lamin A maturation in vivo, might act in concert with residues 385–39352 to auto-inhibit prelamin A binding to actin and potentially other partners that bind the Ig-fold region,27 including nuclear titin,19 Sun1,48 Sun2,49 emerin15 and nesprins.50,51 We hypothesize that lamin head-to-tail polymerization, proposed to involve lamin A residues 385–393,52 is also subject to auto-inhibition in prelamin A. These models, and the potential impact of carboxymethylation and farnesylation will be important to test in the future.
Materials and Methods
Lamin tail constructs.
Human lamin tail constructs in pET23b (Novagen) which places a His6-tag at the C-terminus and a T7 tag at the N-terminus, were described previously19 for the mature lamin A tail (residues 394–646), prelamin A tail (residues 394–664, either wildtype or bearing single missense mutations R482Q, R527P, L530P, E578V), and lamin B1 tail (residues 395–586). Expression constructs for the slightly longer mature lamin A tail (residues 385–646) and the wildtype or deleted (Δ35, Δ50, Δ90) prelamin A tail residues 385–664 were generated by PCR amplification of full length lamin A cDNAs and then subcloned into pET23b. For prelamin A tail residues 385–664 the 5′ and 3′ primers used were 5′-GGA TCC AGA GAG GCT ACG CCT GTC-3′ and 5′-CTC GAG CAT GAT GCT GCA GTT CTG-3, respectively. For mature lamin A tail (residues 385–646) the 5′ primer was the same as above and the 3′ primer was 5′-CTC GAG GTA GGA GCC CGT GAC-3′. The 5′ and 3′ primers for the lamin C tail (residues 394–572) were 5′-GGA TCC TAC CTC GCA GCG CAG CCG TGG CCG-3′, and 5′-CTC GAG GCG GCG GCT ACC ACT CTC-3′ respectively. Recombinant proteins were expressed in E. coli BL-21, purified using nickel NTA-agarose per manufacturer instructions (Qiagen) and stored in 50 mM NaHPO4 pH 8.0, 300 mM NaCl, 100 mM Imidazole, 0.5 mM PMSF at −80°C. Purified mature and prelamin A tails eluted as monomers in sizing chromatography (data not shown).
High speed F-actin pelleting assay.
One milligram of ≥95% purified rabbit skeletal muscle actin (Cytoskeleton) was resuspended in G-buffer (5 mM Hepes pH 7.5, 0.2 mM CaCl2, 0.2 mM ATP pH 8.0) and then polymerized by adding one-tenth volume of 10X KMEI buffer (500 mM KCl, 10 mM MgCl2, 10 mM EGTA, 100 mM Imidazole, pH 7.0, 10 mM DTT, 2 mM ATP pH 8.0) to yield F-actin at a final concentration of 1 mg/ml. Purified recombinant lamin proteins were first pre-cleared by centrifugation at 100,000 g for 30 minutes at 4°C prior to determining protein concentration by SDS-PAGE relative to a BSA standard curve. Reactions containing 7.9 µM F-actin plus 6.7 µM lamin tail (or 1.3 µM α-actinin) in 1X KMEI, were supplemented with 1X KMEI to achieve a final volume of 75 µl, then rotated (1 hour, 22–25°C), pelleted (30 minutes, 100,000 g, 22°C), resuspended in SDS-sample buffer, resolved by 12% SDS-PAGE and stained with GelCode Blue Stain (Pierce Chemical Company) to visualize proteins. To generate binding curves the high speed F-actin pelleting assays were repeated holding the lamin tail concentration constant (13.3 µM), with increasing concentrations of F-actin (0–18.9 µM) in a final reaction volume of 75 µL. Gels were scanned and analyzed using Quantity One software (BioRad).
Low speed F-actin bundling assay.
Actin bundling was assayed as described.30 In brief, F-actin, assembled as described above was incubated 1 hour at 22–25°C with recombinant purified lamins, BSA, or α-actinin, then pelleted (17,000 g, 30 minutes, 22°C), resuspended in SDS-sample buffer, resolved by 12% SDS-PAGE, visualized by Coomassie brilliant blue and analyzed using Quantity One software (BioRad).
Rhodamine-phalloidin bundling assay.
Actin filaments (1.7 µM) were incubated 1 hour at 22–25°C with each indicated concentration of recombinant lamin tails or BSA, then supplemented with tetramethyl rhodamine-labeled phalloidin (Molecular Probes) to achieve a 1:1 molar ratio (phalloidin:G-actin) for 10 more minutes, spotted on coverslips and visualized using a Nikon Eclipse E600 equipped with a Nikon Plan APO 60X X.A. 1.40 oil objective. Images were acquired with a Q Imagine Retiga Exi 12 bit digital camera using IP Lab software from Scanalytics, Inc.
Transmission electron microscopy.
F-actin, made as above, was incubated 1 hour at at 22–25°C with recombinant purified lamin tails or α-actinin. Each reaction contained 7.9 µM F-actin plus 6.7 µM of the indicated lamin tail protein or 1.3 µM α-actinin in 1X KMEI, and was supplemented with 1X KMEI to a final volume of 150 µl. Carbon coated grids were then placed over each reaction for 2 minutes, then washed 1 minute in 1X KMEI, fixed 2 minutes (1% glutaraldehyde, 0.1 M sodium cacodylate pH 7Δ35), and washed in 0.1 M sodium cacodylate pH 7Δ35. Grids were stained in 1% uranyl acetate, 0.04% Tylose for 1 minute and imaged using a Philips BioTwin CM-120 Transmission Electron Microscope.
Acknowledgements
We thank Michael Delannoy and Carol Cooke at the Johns Hopkins University School of Medicine Microscopy Facility for help with TEM imaging, Robert Goldman (Northwestern University) for full length wildtype lamin A and lamin C cDNAs, and Susan Michaelis (Johns Hopkins University School of Medicine) for full-length Δ35, Δ50 and Δ90 cDNAs. We thank James Holaska, Kathryn Tifft, Rocio Montes de Oca, Jason Berk and Ueli Aebi for spirited discussions. This research was funded by the National Institutes of Health (GM48646 to K.L.W.).
Abbreviations
- AB-1
actin-binding site 1
- AB-2
actin-binding site 2
- EDMD
Emery-Dreifuss muscular dystrophy
- HGPS
Hutchinson-Gilford progeria syndrome
- TEM
transmission electron microscopy
- SEM
standard error of the mean
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/11799
References
- 1.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 2.Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 3.Stewart CL, Roux KJ, Burke B. Blurring the boundary: the nuclear envelope extends its reach. Science. 2007;318:1408–1412. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- 4.Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parnaik VK. Role of nuclear lamins in nuclear organization, cellular signaling, and inherited diseases. Int Rev Cell Mol Biol. 2008;266:157–206. doi: 10.1016/S1937-6448(07)66004-3. [DOI] [PubMed] [Google Scholar]
- 6.Padiath QS, Saigoh K, Schiffmann R, Asahara H, Yamada T, Koeppen A, et al. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat Genet. 2006;38:1114–1123. doi: 10.1038/ng1872. [DOI] [PubMed] [Google Scholar]
- 7.Brussino A, Vaula G, Cagnoli C, Mauro A, Pradotto L, Daniele D, et al. A novel family with Lamin B1 duplication associated with adult-onset leucoencephalopathy. J Neurol Neurosurg Psychiatry. 2009;80:237–240. doi: 10.1136/jnnp.2008.147330. [DOI] [PubMed] [Google Scholar]
- 8.Somech R, Shaklai S, Amariglio N, Rechavi G, Simon AJ. Nuclear envelopathies—raising the nuclear veil. Pediatr Res. 2005;57:8–15. doi: 10.1203/01.PDR.0000159566.54287.6C. [DOI] [PubMed] [Google Scholar]
- 9.De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 10.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuchi K, Katsuya T, Sugimoto K, Kuremura M, Kim HD, Li L, et al. LMNA mutation in a 45 year old Japanese subject with Hutchinson-Gilford progeria syndrome. J Med Genet. 2004;41:67. doi: 10.1136/jmg.2003.014688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahl KN, Scaffidi P, Islam MF, Yodh AG, Wilson KL, Misteli T. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2006;103:10271–10276. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape, mechanics and mechanotransduction. Circ Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro CL, De Sandre-Giovannoli A, Bernard R, Boccaccio I, Boyer A, Genevieve D, et al. Lamin A and ZMPSTE24 (FACE-1) defects cause nuclear disorganization and identify restrictive dermopathy as a lethal neonatal laminopathy. Hum Mol Genet. 2004;13:2493–2503. doi: 10.1093/hmg/ddh265. [DOI] [PubMed] [Google Scholar]
- 15.Clements L, Manilal S, Love DR, Morris GE. Direct interaction between emerin and lamin A. Biochem Biophys Res Commun. 2000;267:709–714. doi: 10.1006/bbrc.1999.2023. [DOI] [PubMed] [Google Scholar]
- 16.Holaska JM, Kowalski AK, Wilson KL. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2004;2:231. doi: 10.1371/journal.pbio.0020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holaska JM, Wilson KL. An emerin “proteome”: purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing and nuclear architecture. Biochemistry. 2007;46:8897–8908. doi: 10.1021/bi602636m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J Cell Biol. 2005;170:781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zastrow MS, Flaherty DB, Benian GM, Wilson KL. Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J Cell Sci. 2006;119:239–249. doi: 10.1242/jcs.02728. [DOI] [PubMed] [Google Scholar]
- 20.Sasseville AM, Langelier Y. In vitro interaction of the carboxy-terminal domain of lamin A with actin. FEBS Lett. 1998;425:485–489. doi: 10.1016/s0014-5793(98)00294-4. [DOI] [PubMed] [Google Scholar]
- 21.Gieni RS, Hendzel MJ. Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem Cell Biol. 2009;87:283–306. doi: 10.1139/O08-133. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann WA. Cell and molecular biology of nuclear actin. Int Rev Cell Mol Biol. 2009;273:219–263. doi: 10.1016/S1937-6448(08)01806-6. [DOI] [PubMed] [Google Scholar]
- 23.McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172:541–552. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoenenberger CA, Buchmeier S, Boerries M, Sutterlin R, Aebi U, Jockusch BM. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J Struct Biol. 2005;152:157–168. doi: 10.1016/j.jsb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Dhe-Paganon S, Werner ED, Chi YI, Shoelson SE. Structure of the globular tail of nuclear lamin. J Biol Chem. 2002;277:17381–17384. doi: 10.1074/jbc.C200038200. [DOI] [PubMed] [Google Scholar]
- 26.Krimm I, Ostlund C, Gilquin B, Couprie J, Hossenlopp P, Mornon JP, et al. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy and partial lipodystrophy. Structure. 2002;10:811–823. doi: 10.1016/s0969-2126(02)00777-3. [DOI] [PubMed] [Google Scholar]
- 27.Zastrow MS, Vlcek S, Wilson KL. Proteins that bind A-type lamins: integrating isolated clues. J Cell Sci. 2004;117:979–987. doi: 10.1242/jcs.01102. [DOI] [PubMed] [Google Scholar]
- 28.Sjoblom B, Salmazo A, Djinovic-Carugo K. Alpha-actinin structure and regulation. Cell Mol Life Sci. 2008;65:2688–2701. doi: 10.1007/s00018-008-8080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popowicz GM, Schleicher M, Noegel AA, Holak TA. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006;31:411–419. doi: 10.1016/j.tibs.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Vitavska O, Merzendorfer H, Wieczorek H. The V-ATPase subunit C binds to polymeric F-actin as well as to monomeric G-actin and induces cross-linking of actin filaments. J Biol Chem. 2005;280:1070–1076. doi: 10.1074/jbc.M406797200. [DOI] [PubMed] [Google Scholar]
- 31.Meyer RK, Aebi U. Bundling of actin filaments by alpha-actinin depends on its molecular length. J Cell Biol. 1990;110:2013–2024. doi: 10.1083/jcb.110.6.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wachsstock DH, Schwartz WH, Pollard TD. Affinity of alpha-actinin for actin determines the structure and mechanical properties of actin filament gels. Biophys J. 1993;65:205–214. doi: 10.1016/S0006-3495(93)81059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuven T, Hartmann E, Gorlich D. Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 2003;22:5928–5940. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulze SR, Curio-Penny B, Speese S, Dialynas G, Cryderman DE, McDonough CW, et al. A comparative study of Drosophila and human A-type lamins. PLoS One. 2009;4:7564. doi: 10.1371/journal.pone.0007564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tondeleir D, Vandamme D, Vandekerckhove J, Ampe C, Lambrechts A. Actin isoform expression patterns during mammalian development and in pathology: insights from mouse models. Cell Motil Cytoskeleton. 2009;66:798–815. doi: 10.1002/cm.20350. [DOI] [PubMed] [Google Scholar]
- 36.Parfenov VN, Davis DS, Pochukalina GN, Sample CE, Bugaeva EA, Murti KG. Nuclear actin filaments and their topological changes in frog oocytes. Exp Cell Res. 1995;217:385–394. doi: 10.1006/excr.1995.1101. [DOI] [PubMed] [Google Scholar]
- 37.Kimura T, Hashimoto I, Yamamoto A, Nishikawa M, Fujisawa JI. Rev-dependent association of the intron-containing HIV-1 gag mRNA with the nuclear actin bundles and the inhibition of its nucleocytoplasmic transport by latrunculin-B. Genes Cells. 2000;5:289–307. doi: 10.1046/j.1365-2443.2000.00326.x. [DOI] [PubMed] [Google Scholar]
- 38.Percipalle P, Zhao J, Pope B, Weeds A, Lindberg U, Daneholt B. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J Cell Biol. 2001;153:229–236. doi: 10.1083/jcb.153.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cisterna B, Necchi D, Prosperi E, Biggiogera M. Small ribosomal subunits associate with nuclear myosin and actin in transit to the nuclear pores. Faseb J. 2006;20:1901–1913. doi: 10.1096/fj.05-5278fje. [DOI] [PubMed] [Google Scholar]
- 40.Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 41.Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, et al. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X, Yoo Y, Okuhama NN, Tucker PW, Liu G, Guan JL. Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat Cell Biol. 2006;8:756–763. doi: 10.1038/ncb1433. [DOI] [PubMed] [Google Scholar]
- 43.Yoo Y, Wu X, Guan JL. A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J Biol Chem. 2007;282:7616–7623. doi: 10.1074/jbc.M607596200. [DOI] [PubMed] [Google Scholar]
- 44.Louvet E, Percipalle P. Transcriptional control of gene expression by actin and myosin. Int Rev Cell Mol Biol. 2009;272:107–147. doi: 10.1016/S1937-6448(08)01603-1. [DOI] [PubMed] [Google Scholar]
- 45.Guilly MN, Bensussan A, Bourge JF, Bornens M, Courvalin JC. A human T lymphoblastic cell line lacks lamins A and C. EMBO J. 1987;6:3795–3799. doi: 10.1002/j.1460-2075.1987.tb02715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart C, Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 1987;51:383–392. doi: 10.1016/0092-8674(87)90634-9. [DOI] [PubMed] [Google Scholar]
- 47.Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci. 2004;117:4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- 48.Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, et al. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mislow JM, Holaska JM, Kim MS, Lee KK, Segura-Totten M, Wilson KL, et al. Nesprin-1alpha self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett. 2002;525:135–140. doi: 10.1016/s0014-5793(02)03105-8. [DOI] [PubMed] [Google Scholar]
- 51.Libotte T, Zaim H, Abraham S, Padmakumar VC, Schneider M, Lu W, et al. Lamin A/C-dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol Biol Cell. 2005;16:3411–3424. doi: 10.1091/mbc.E04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strelkov SV, Schumacher J, Burkhard P, Aebi U, Herrmann H. Crystal structure of the human lamin A coil 2B dimer: implications for the head-to-tail association of nuclear lamins. J Mol Biol. 2004;343:1067–1080. doi: 10.1016/j.jmb.2004.08.093. [DOI] [PubMed] [Google Scholar]