Abstract
The nuclear envelope (NE) is a barrier that separates nuclear from cytoplasmic processes. It is composed of an inner and outer nuclear membrane (INM, ONM), separated by the perinuclear space (PNS). The ONM is contiguous with the endoplasmic reticulum (ER), and thus, the lumen of the NE and that of the ER constitute one compartment. The lamin B receptor (LBR) is a NE protein that has a central structural role as a linker of the INM, the lamina and chromatin, and a less well characterized functional role as a sterol reductase. In a recent study, we reported that the forced expression of mutant variants of LBR in some cell types induces a separation of the INM from the outer nuclear envelope concomitantly with a separation of ER membranes, whereas in other cells no separation is observed. In this extra view, we speculate about the mechanism that leads to this fundamental disruption of NE and ER structure. Our observations furthermore raise the question to what extent LBR contributes to the establishment or maintenance of the ER and PNS luminal compartment, and how a single mutant protein can so drastically interfere with its regular organization.
Key words: lamin B receptor, Pelger-Huät anomaly, greenberg skeletal dysplasia, nuclear envelope, endoplasmic reticulum
The nuclear envelope (NE) is the structure that surrounds the nucleus and physically separates nuclear from cytoplasmic functions. It consists of an inner (INM) and an outer nuclear membrane (ONM), which both contain a distinct repertoire of proteins.1 The ONM is continuous with the endoplasmic reticulum (ER), a network-like compartment in which protein folding, modification and delivery into the secretory pathway takes place. The space in between both membranes, the perinuclear space (PNS), is accordingly continuous with the ER lumen.2 Closely underneath the INM lies, at least in metazoan cells, a filamentous network termed the nuclear lamina. It is mainly composed of nucleus-specific intermediate filament proteins, hence called lamins. The lamina is tightly connected to both nuclear pore complexes and chromatin.3
The human lamin B receptor (LBR) is a fascinating multi-span integral membrane protein of 615 amino acids sitting in the inner membrane of the NE and fulfilling two distinct functions.4 First, the 208 amino acid amino-terminal part binds to B-type lamins, chromatin and chromatin-associated proteins, such as the heterochromatin protein 1 (HP1) and the methyl-CpG binding protein 2 (MeCP2).4–6 The carboxy-terminal part of 407 amino acids has been predicted to exhibit 8 transmembrane domains that anchor the protein into the membrane.4 In this way, LBR physically links this membrane to the underlying lamina and chromatin. Second, the carboxy-terminal domain has enzymatic activity, i.e., it is a sterol reductase acting at a distinct step of the cholesterol biosynthesis pathway (Fig. 1).7,8 This finding was especially surprising, since the final steps of cholesterol biosynthesis are so far believed to take place only at the ER. However, LBR is found to a significant part in the ER too. It may well be that it is not just on its way into the nucleus but that it indeed acts as sterol reductase in this compartment “in transit”.9,10 Very importantly, defective yeast and plants can be rescued for this biochemical activity by transfection of human LBR.11 It is interesting to note here too that both fungi and plants are believed to not contain lamins.12–14
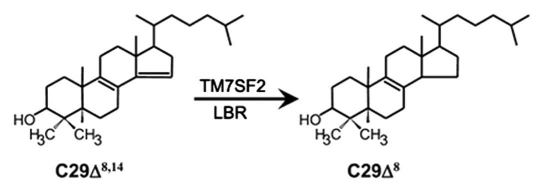
Figure 1.
The conversion of 4,4-dimethyl-5α-cholesta-8(9),14-dien-3β-ol (C29Δ8,14) to 4,4-dimethyl-5α-cholesta-8(9)-en-3β-ol (C29Δ8) is catalyzed by TM7SF2 and LBR. The image was reprinted with kind permission from Rita Roberti.34
Heterozygous mutations of LBR lead to Pelger-Huät anomaly (PHA), a mild hematological trait characterized by abnormally shaped granulocyte nuclei. Instead of adopting a lobulated shape (Fig. 2A, left), mature granulocytes of PHA individuals appear hypolobulated with typically only 2–3 lobes and coarse chromatin (Fig. 2A, middle). Besides that, no other symptoms have been described for heterozygous carriers of the gene defect. Several mutations have been characterized including frameshift, nonsense and splice site mutations.15,16 Most of them affect the carboxy-terminal domain that contains the transmembrane regions and harbors the sterol reductase enzymatic activity. PHA was so far regarded as a laminopathy, affecting basically nuclear morphology and chromatin structure, but up to date no studies on sterol metabolism of affected individuals have been reported.17
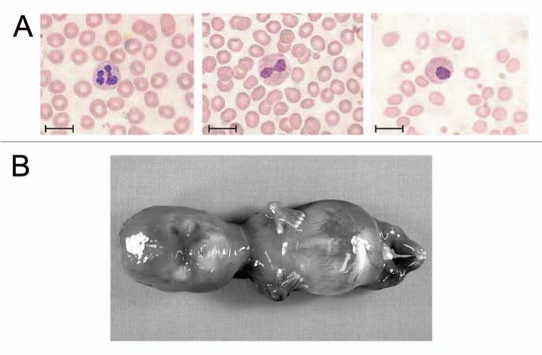
Figure 2.
Mutations in LBR can cause autosomal dominant PHA (A) or autosomal recessive Greenberg skeletal dysplasia (B). (A) Blood smears from healthy controls show normal granulocytes with multisegmented nuclei (left), heterozygous PHA carriers display granulocytes with a typical bilobed nucleus (middle). The patient homozygous for PHA displays granulocytes with round nuclei (right). Scale bar, 10 µm. (B) Photograph of fetus with Greenberg dysplasia, presenting with marked hydrops and abnormally short limbs. The images in (A) were reprinted by permission from Macmillan Publishers Ltd: Nature Genetics, Hoffmann et al. 2002,16 copyright 2002. Image in (B) is reproduced from Offiah et al. 2003,35 with permission from BMJ Publishing Group Ltd.
In contrast to the mild phenotype of a heterozygous mutation, homozygous LBR mutations are surprisingly fatal, leading to prenatal death of the fetuses. This condition is termed Greenberg skeletal dysplasia. Affected fetuses present with hydrops, short limbs and abnormal chondro-osseous calcification leading to a severe skeletal phenotype (Fig. 2B).8 In the case of Greenberg skeletal dysplasia it is still not clear whether the complete loss of functional LBR or a dominant-negative mutant protein is causing disease, nor if the INM-lamina-chromatin-“linking” function or the sterol reductase activity is responsible for lethality. In principle, the very same step in cholesterol synthesis that is catalyzed by LBR can also be catalyzed by a closely related sterol reductase, TM7SF2.18 Recent studies by Wassif and coworkers showed that the two proteins are largely redundant, and the authors therefore concluded that Greenberg skeletal dysplasia is rather a laminopathy than an inborn error of cholesterol biosynthesis.19 Hence, it may be concluded that the loss of LBR affects either the organization of chromatin or the anchorage of protein factors to the nuclear periphery, or both. On the other hand, the INM contains a large ensemble of proteins that interact in multiple ways with lamins and chromatin.20 For example, emerin interacts with A-type lamins, but in addition also binds, like the other members of the LEM family (Lap2 and MAN1), to chromatin via a small dimeric, chromatin-binding protein termed barrier-to-autointegration factor (BAF).21 On the basis of these multiple connections between the membrane, lamina and chromatin, the fatal outcome of homozygous mutations appears surprising.
It is of note that an “intermediate” phenotype exists between the heterozygous and the homozygous state of an LBR mutation. One patient has previously been described to be homozygous for a splice acceptor mutation leading to the loss of exon 13. This patient presented with completely ovoid granulocyte nuclei, mental retardation and skeletal defects (Fig. 2A, right). Western blot analysis of protein extracts derived from lymphoblastoid cells of the patient revealed the complete absence of the mutant protein in this cell-type, however, wildtype LBR was present in trace amounts indicating that a small part of the mutant mRNA is spliced correctly despite the splice site mutation.16 Further homozygous PHA patients have been described in the literature with ovoid neutrophil nuclei as well as varying degrees of developmental and skeletal abnormalities, but in none of those cases genetic analyses were performed.22
The little that is known about the molecular mechanisms causing PHA and Greenberg skeletal dysplasia prompted us to investigate the effect of the forced expression of disease causing mutations on nuclear structure and chromatin organization of various types of human cells. As a first approach, we chose to test LBR mutations in human cultured cells on the background of endogenous LBR. We tested six protein variants, namely LBR wildtype, one protein variant corresponding to a Greenberg skeletal dysplasia mutation and four protein variants corresponding to mutations that were found in individuals with PHA. Amino-terminally fused to YFP, we overexpressed each of these mutant proteins in six different human cell lines of different origin, including MCF7, HeLa and U2OS (osteosarcoma) cells. In some cell types such as MCF7, we found all protein variants to localize at the nuclear rim and also in the ER, most probably as a result of overexpression. In these cells, none of the mutants significantly altered nuclear morphology. In other cells such as U2OS, however, we made a striking observation with three of the disease causing mutants: Nuclei appeared to have compacted the entire chromatin to one “half”, with a clear and abrupt border on one side.23 The other “half” was DAPI-negative, and with differential interference contrast (DIC) microscopy, this chromatin-free region appeared as a distinct “vacuole-like” structure (Fig. 3). Using either DIC or phase contrast microscopy, we also detected smaller ovoid “vacuole-like” areas within the cytoplasm. The mutants that induced this phenotype were YFP-LBR (1–533), corresponding to a mutation detected in a fetus with Greenberg skeletal dysplasia, YFP-LBR (Δ523–563) and YFP-LBR (1–435), corresponding to mutations detected in the homozygous PHA patient and in a heterozygous PHA individual, respectively.
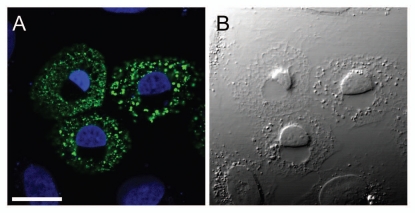
Figure 3.
Human U2OS cells transiently expressing the LBR mutant YFP-LBR (1–533). Overexpression of this mutant leads to a separation of the INM from the ONM and dilation of the ER lumen concomitantly with nuclear compaction in susceptiple cell lines. (A) Fluorescence signal of YFP-LBR (1–533) in green, merged with DAPI-stained chromatin in blue. (B) Corresponding DIC micrograph. Scale bar, 20 µm.
We next performed immunofluorescence studies on U2OS cells expressing one of those mutants. The cells were stained with antibodies against different NE proteins (lamin A/C, emerin, nuclear pore complexes and endogenous LBR), components of the linker of nucleoskeleton and cytoskeleton (LINC) complex (Sun1, Sun2 and nesprin2-giant) and the ER (calnexin). The LINC complex comprises a class of proteins that physically couple cytoskeletal structures such as actin filaments to components within the nucleus, e.g., the lamina. Among these factors, SUN proteins in the INM and nesprins in the ONM have been demonstrated to directly interact with each other in the PNS.24,25 We found both Sun1 and Sun2 as well as nesprin2-giant localized as expected at the nuclear rim but they were strikingly displaced from the NE at the interface between nucleus and the vacuolar structure. Moreover, nuclear pore complexes (NPCs) were largely absent in this boundary but distributed normally at the remaining NE. All other proteins investigated localized more or less as expected, although a significant fraction of emerin was redistributed to the cytoplasm. In essence, mainly those components disappeared from the flattened side of the nucleus that function in tethering the INM to the ONM. Using electron microscopy, we finally elucidated the nature of the chromatin-free “half” flanking the nucleus: this area represents a strongly dilated PNS, with INM and ONM having separated over large distances of up to 10 µm (Fig. 4). This dilation occurs progressively approximately 8 to 12 hours after transient transfection of the above-named LBR mutants. The separation usually starts at one distinct site of the nucleus and progressively continues along the NE. With increasing dilation of the two membranes, NPCs and a number of LINC complex components are displaced from the nuclear membranes, and the nucleus becomes pushed inward and flattened at this site of the NE. The dilation can become extremely expanded in late stages of this process. In such cells, the nucleus is usually sickle-shaped and the chromatin is extremely compacted, i.e., up to tenfold as concluded from the area occupied by DAPI stained material (Fig. 4B). Eventually, the cells die 6 to 30 hours after the first dilations are detectable.
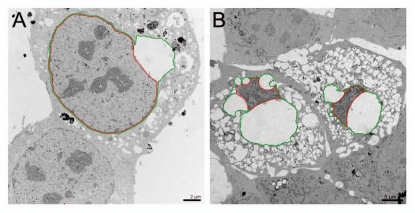
Figure 4.
Electron microscopy images of ultrathin sections showing U2OS cells transiently expressing the mutant YFP-LBR (1–533) after 10 h (A) and after 24 h (B). INM and ONM are highlighted in red and green, respectively. During the process of membrane dilation, the distance of the nuclear membranes and dilation of the ER lumen constantly increases (reviewed in ref. 23).
Concomitantly with NE separation, ER membranes started to separate at multiple sites in the cytoplasm. Apparently, the initial step of membrane dilation can occur either at the NE or the ER, since we found cells with only dilated nuclear membranes and cells exhibiting dilations within the ER before those of the NE appeared (Fig. 5). Importantly, the effects described here for U2OS are not due to unspecific cellular stress responses such as apoptosis, autophagy or ER stress induced by the unfolded protein response (UPR) pathway.
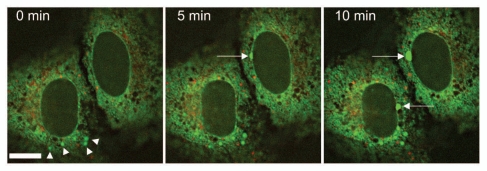
Figure 5.
Live cell imaging of U2OS cells 12 hours after cotransfection of mCherry-LBR (1–533) (red) and SP-GFP (green), a marker construct for the ER lumen and the PNS. Images were taken every 5 minutes, revealing the progressively dilating ER lumen (arrowheads) as well as the dilation of the PNS (arrows). Scale bar, 10 µm.
As mentioned above, only three of the five tested mutants induced NE and ER dilation as well as nuclear compaction. We generated various constructs of different protein length in order to elucidate the protein segments important for these effects. Most interestingly, the loss of the very last amino acid of LBR, a tyrosine, or its mutation to alanine was sufficient for induction of the effects described above. Moreover, the mere attachment of a GFP moiety to the carboxy-terminus of the full-length LBR was sufficient to elicit the full destructive effect. In contrast, mutation of tyrosine to phenylalanine did exhibit no effect at all indicating it is not the loss of a potential phosphorylation site for tyrosine kinases that causes the effect. The presence of an aromatic amino acid at this position is obviousy essential for the proper sterical conformation of the carboxy-terminus and protein function as a whole, possibly by establishing interactions with other proteins, or for the enzymatic activity of LBR. Since the carboxy-terminus of LBR was the crucial domain for NE and ER integrity or disruption, a malfunction in the proteins' sterol reductase activity, leading to the accumulation of cholesterol precursors, could be a part of the molecular mechanism accounting for the disturbances within the internal membrane systems. To test this hypothesis, we performed two experiments: first, the sterol content of untransfected or YFP-LBR wt expressing U2OS cells was compared to that of U2OS cells transfected with YFP- LBR (1–533). No significant differences in sterol composition and no accumulation of cholesterol intermediates were detected. Secondly, we tested if two related sterol reductases, TM7SF2 and DHCR7, could induce similar effects when expressed as truncated variants.26 Strikingly, not only those truncated variants, but even the full length TM7SF2 and DHCR7 proteins induced an identical phenotype, including membrane dilation and nuclear compaction, when overexpressed in U2OS cells. In summary, LBR and the related sterol reductases TM7SF2 and DHCR7 contain a carboxy-terminal domain that can critically affect membrane integrity: In the case of LBR, loss of the last amino acid or improper folding leads to massive disturbance of the integrity of NE and ER, in the case of the two related sterol reductases, overexpression is yet sufficient. Notably, TM7SF2 and DHCR7 may not even have to enter the nucleus to cause nuclear compaction, if membrane integrity as such is affected.
Since our studies are based on overexpression of LBR, TM7SF2 and DHCR7 protein variants on the basis of endogenous protein, it is important to consider the relevance and specificity of the observed effects. Indeed, overexpression of certain proteins can lead to various effects that would not occur if the protein was expressed on physiologic levels. For example, high abundance of transmembrane proteins in the ER can induce extensive membrane growth, whereas low expression does not alter ER morphology.27 However, overexpression studies with various LBR constructs have also turned out to be a valuable tool to elucidate protein function.10,28,29 In our study, we carefully tested transfected cells for indications of unspecific stress reactions that might occur as a result of exogenous protein overload, such as apoptosis, autophagy or ER stress due to UPR induction.30–32 None of these pathways was activated. Regarding the reaction of cells towards LBR mutants, namely membrane dilation and nuclear compaction, several observations underline the specificity of this cellular reaction. Firstly, only a part of the tested cell lines “reacted” to LBR mutants, whereas in MCF7 or PLC—despite high levels of expression—membrane dilation events never occurred. We conclude that the above mentioned LBR mutants are “toxic” only under distinct conditions. These may represent a certain composition of ER, NE and/or LINC complex proteins, or the presence of factors that antagonize ER lumen and PNS enlargement. Secondly, the phenotypic difference between cells expressing YFP-LBR wt with 615 amino acids and YFP-LBR (1–614), lacking only one amino acid, respectively, reveals that indeed an important protein function was disrupted due to loss of the last tyrosine. In the case of the other two sterol reductases, high abundance within the ER is sufficient to affect ER and PNS luminal integrity. The three proteins seem to have a distinct function within the membrane that is—in a well-regulated fashion—most probably crucial for NE and ER function. Yet this function—altered or unregulated—might have a strongly negative impact on these compartments at high abundance of the respective proteins.
We assume that membrane dilation and nuclear compaction result from gross increase of PNS and ER luminal volume. In the process of the membrane dilation effect, the nucleoplasm becomes highly compacted and the cytoplasm is widely displaced by large luminal arrays, as observed by electron microscopy (Fig. 4B). The proteins that generally confine the constraints of the ER and NE and shape these compartments might not be directly affected but loose their attachment to the membrane or their association to each other due to an increased pressure inside the lumen. This might explain why we detected SUN proteins, nesprin2-giant and NPCs at the NE of cells with PNS dilations, but all of these factors were lacking at the area where INM and ONM were separated over large distances. Similarly, nuclear compaction might not be a direct effect of LBR mutants appearing in the INM but rather occur due to pressure exerted from the PNS. Given the massive luminal expansion, we assume that LBR, TM7SF2 and DHCR7 might not only act as sterol reductases but may additionally be involved in regulating luminal ion concentrations and eventually osmotic potential.
LBR was previously reported to oligomerize and to form distinct microdomains within the membrane.33 It is well possible that LBR oligomers form channels through the membrane, and loss of a single amino acid might alter properties of channel permeability. In line with this notion, also point mutations within the membrane domain, i.e., glutamic acid at position 407 to alanine, cause the very same effect as the mutation of the tyrosine at position 615. TM7SF2 and DHCR7 might also form channels, yet their conduction rates might be regulated solely by protein abundance. It is not clear, however, what molecule could pass such channels, and how altered channel properties would lead to such a strong increase in PNS and ER lumen that it would exert pressure on the nucleoplasm and cytoplasm.
Our study aimed on elucidating the effect of disease causing LBR mutants on chromatin and nuclear structure of cultured cells. To our surprise, overexpression of three of the investigated LBR mutants as well as TM7SF2 and DHCR7 induced the large-scale separation of the INM from the ONM as well as of ER membranes, implicating an additional function of these proteins in maintaining the integrity of these compartments. Surely, our assumptions are still highly speculative but may indeed provide hints for further investigations.
Acknowledgements
This work was supported by European Union's FP6 Life Science, Genomics and Biotechnology for Health area (LSHM-CT-2005-018690 to H.H. and M.Z.). The authors thank Peter Lichter for continuous interest and support.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/11801
References
- 1.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 2.Lusk CP, Blobel G, King MC. Highway to the inner nuclear membrane: rules for the road. Nat Rev Mol Cell Biol. 2007;8:414–420. doi: 10.1038/nrm2165. [DOI] [PubMed] [Google Scholar]
- 3.Dwyer N, Blobel G. A modified procedure for the isolation of a pore complex-lamina fraction from rat liver nuclei. J Cell Biol. 1976;70:581–591. doi: 10.1083/jcb.70.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye Q, Worman HJ. Primary structure analysis and lamin B and DNA binding of human LBR, an integral protein of the nuclear envelope inner membrane. J Biol Chem. 1994;269:11306–11311. [PubMed] [Google Scholar]
- 5.Guarda A, Bolognese F, Bonapace IM, Badaracco G. Interaction between the Inner Nuclear Membrane Lamin B Receptor and the Heterochromatic Methyl Binding Protein, MeCP2. Exp Cell Res. 2009 doi: 10.1016/j.yexcr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 6.Pyrpasopoulou A, Meier J, Maison C, Simos G, Georgatos SD. The lamin B receptor (LBR) provides essential chromatin docking sites at the nuclear envelope. EMBO J. 1996;15:7108–7119. [PMC free article] [PubMed] [Google Scholar]
- 7.Silve S, Dupuy PH, Ferrara P, Loison G. Human lamin B receptor exhibits sterol C14-reductase activity in Saccharomyces cerevisiae. Biochim Biophys Acta. 1998;1392:233–244. doi: 10.1016/s0005-2760(98)00041-1. [DOI] [PubMed] [Google Scholar]
- 8.Waterham HR, Koster J, Mooyer P, Noort Gv G, Kelley RI, Wilcox WR, et al. Autosomal recessive HEM/Greenberg skeletal dysplasia is caused by 3beta-hydroxysterol delta 14-reductase deficiency due to mutations in the lamin B receptor gene. Am J Hum Genet. 2003;72:1013–1017. doi: 10.1086/373938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreger CK, Konig AR, Spring H, Lichter P, Herrmann H. Investigation of nuclear architecture with a domain-presenting expression system. J Struct Biol. 2002;140:100–115. doi: 10.1016/s1047-8477(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 10.Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, et al. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye Q, Callebaut I, Pezhman A, Courvalin JC, Worman HJ. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J Biol Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- 12.Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell. 2007;19:2793–2803. doi: 10.1105/tpc.107.053231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erber A, Riemer D, Hofemeister H, Bovenschulte M, Stick R, Panopoulou G, et al. Characterization of the Hydra lamin and its gene: A molecular phylogeny of metazoan lamins. J Mol Evol. 1999;49:260–271. doi: 10.1007/pl00006548. [DOI] [PubMed] [Google Scholar]
- 14.Rose A, Schraegle SJ, Stahlberg EA, Meier I. Coiled-coil protein composition of 22 proteomes—differences and common themes in subcellular infrastructure and traffic control. BMC Evol Biol. 2005;5:66. doi: 10.1186/1471-2148-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Best S, Salvati F, Kallo J, Garner C, Height S, Thein SL, et al. Lamin B-receptor mutations in Pelger-Huet anomaly. Br J Haematol. 2003;123:542–554. doi: 10.1046/j.1365-2141.2003.04621.x. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann K, Dreger CK, Olins AL, Olins DE, Shultz LD, Lucke B, et al. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huet anomaly) Nat Genet. 2002;31:410–414. doi: 10.1038/ng925. [DOI] [PubMed] [Google Scholar]
- 17.Speeckaert MM, Verhelst C, Koch A, Speeckaert R, Lacquet F. Pelger-Huet anomaly: a critical review of the literature. Acta Haematol. 2009;121:202–206. doi: 10.1159/000220333. [DOI] [PubMed] [Google Scholar]
- 18.Bennati AM, Castelli M, Della Fazia MA, Beccari T, Caruso D, Servillo G, et al. Sterol dependent regulation of human TM7SF2 gene expression: role of the encoded 3beta-hydroxysterol Delta14-reductase in human cholesterol biosynthesis. Biochim Biophys Acta. 2006;1761:677–685. doi: 10.1016/j.bbalip.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Wassif CA, Brownson KE, Sterner AL, Forlino A, Zerfas PM, Wilson WK, et al. HEM dysplasia and ichthyosis are likely laminopathies and not due to 3beta-hydroxysterol Delta14-reductase deficiency. Hum Mol Genet. 2007;16:1176–1187. doi: 10.1093/hmg/ddm065. [DOI] [PubMed] [Google Scholar]
- 20.Schirmer EC, Gerace L. The nuclear membrane proteome: extending the envelope. Trends Biochem Sci. 2005;30:551–558. doi: 10.1016/j.tibs.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Margalit A, Brachner A, Gotzmann J, Foisner R, Gruenbaum Y. Barrier-to-autointegration factor—a BAFfling little protein. Trends Cell Biol. 2007;17:202–208. doi: 10.1016/j.tcb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Oosterwijk JC, Mansour S, van Noort G, Waterham HR, Hall CM, Hennekam RC. Congenital abnormalities reported in Pelger-Huet homozygosity as compared to Greenberg/HEM dysplasia: highly variable expression of allelic phenotypes. J Med Genet. 2003;40:937–941. doi: 10.1136/jmg.40.12.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zwerger M, Kolb T, Richter K, Karakesisoglou I, Herrmann H. Induction of a massive endoplasmic reticulum and perinuclear space expansion by expression of lamin B receptor mutants and the related sterol reductases TM7SF2 and DHCR7. Mol Biol Cell. 21:354–368. doi: 10.1091/mbc.E09-08-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmer L, Pezhman A, Worman HJ. The human lamin B receptor/sterol reductase multigene family. Genomics. 1998;54:469–476. doi: 10.1006/geno.1998.5615. [DOI] [PubMed] [Google Scholar]
- 27.Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, et al. Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith S, Blobel G. The first membrane spanning region of the lamin B receptor is sufficient for sorting to the inner nuclear membrane. J Cell Biol. 1993;120:631–637. doi: 10.1083/jcb.120.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soullam B, Worman HJ. The amino-terminal domain of the lamin B receptor is a nuclear envelope targeting signal. J Cell Biol. 1993;120:1093–1100. doi: 10.1083/jcb.120.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 31.Hacker G. The morphology of apoptosis. Cell Tissue Res. 2000;301:5–17. doi: 10.1007/s004410000193. [DOI] [PubMed] [Google Scholar]
- 32.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makatsori D, Kourmouli N, Polioudaki H, Shultz LD, McLean K, Theodoropoulos PA, et al. The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J Biol Chem. 2004;279:25567–25573. doi: 10.1074/jbc.M313606200. [DOI] [PubMed] [Google Scholar]
- 34.Bennati AM, Schiavoni G, Franken S, Piobbico D, Della Fazia MA, Caruso D, et al. Disruption of the gene encoding 3beta-hydroxysterol Delta-reductase (Tm7sf2) in mice does not impair cholesterol biosynthesis. FEBS J. 2008;275:5034–5047. doi: 10.1111/j.1742-4658.2008.06637.x. [DOI] [PubMed] [Google Scholar]
- 35.Offiah AC, Mansour S, Jeffrey I, Nash R, Whittock N, Pyper R, et al. Greenberg dysplasia (HEM) and lethal X linked dominant Conradi-Hunermann chondrodysplasia punctata (CDPX2): presentation of two cases with overlapping phenotype. J Med Genet. 2003;40:129. doi: 10.1136/jmg.40.12.e129. [DOI] [PMC free article] [PubMed] [Google Scholar]