Abstract
The nuclear lamina is an interconnected meshwork of intermediate filament proteins underlying the nuclear envelope. The lamina is an important regulator of nuclear structural integrity as well as nuclear processes, including transcription, DNA replication and chromatin remodeling. The major components of the lamina are A- and B-type lamins. Mutations in lamins impair lamina functions and cause a set of highly tissue-specific diseases collectively referred to as laminopathies. The phenotypic diversity amongst laminopathies is hypothesized to be caused by mutations affecting specific protein interactions, possibly in a tissue-specific manner. Current technologies to identify interaction partners of lamin A and its mutants are hampered by the insoluble nature of lamina components. To overcome the limitations of current technologies, we developed and applied a novel, unbiased approach to identify lamin A-interacting proteins. This approach involves expression of the high-affinity OneSTrEP-tag, precipitation of lamin-protein complexes after reversible protein cross-linking and subsequent protein identification by mass spectrometry. We used this approach to identify in mouse embryonic fibroblasts and cardiac myocyte NklTAg cell lines proteins that interact with lamin A and its mutant isoform progerin, which causes the premature aging disorder Hutchinson-Gilford progeria syndrome (HGPS). We identified a total of 313 lamina-interacting proteins, including several novel lamin A interactors, and we characterize a set of 35 proteins which preferentially interact with lamin A or progerin.
Key words: laminopathies, lamin A, progerin, Hutchinson-Gilford progeria syndrome, protein interactions, mass spectrometry, oneSTrEP pull-down
Introduction
The nuclear lamina lines the nuclear envelope (NE), which is composed of an outer and inner nuclear membrane (ONM, INM,) interrupted by nuclear pore complexes (NPC).1 The lamina is a proteinaceous structure consisting of the intermediate filament A-type (somatic: lamin A, AΔ10, C; germ cell specific: C2) and B-type lamins (somatic: lamin B1, B2; germ cell specific: B3), integral membrane proteins anchoring the lamina to the NE including LAP2, Emerin, Lamin B receptor, MAN1 and proteins involved in a variety of nuclear processes including DNA replication, chromatin remodeling, cell cycle progression and signal transduction.2 Mutations in lamins or other lamina proteins can impair specific functions or interfere with the structural integrity of the nucleus.3 Mutations in lamin components cause various human diseases referred to as laminopathies and include several types of muscular dystrophies, lipodystrophies, cardiomyopathies, neurological disorders and premature aging syndromes.2
One model for the tissue-specificity and phenotypic diversity among laminopathies is that mutations which cause laminopathies lead to differential interactions of mutant lamins with their associated proteins.2 In support, several mutations in the carboxy-terminus of lamin A that lead to Emery-Dreifuss muscular dystrophy (EDMD) or dilated cardiomyopathy 1A (CMD1A) impair binding of lamin A to emerin.4 In addition, progerin, the lamin A isoform that causes the premature aging disease Hutchinson-Gilford progeria syndrome (HGPS), is unable to interact with several members of the NURD chromatin remodeling complex, thereby contributing to HGPS characteristic chromatin defects.5 Although these observations suggest a role for differential protein interactions in the tissue-specificity of laminopathies, they are circumstantial. Systematic mapping of the complete spectrum of wild-type and mutant lamin A interacting proteins is required to test this disease model. Such approaches will also clarify the function of lamin A in biological processes and may contribute to the identification of targetable pathways relevant to specific laminopathies.
Experimental approaches to identify lamin interacting proteins are severely hampered by the unique biochemical properties of the nuclear lamina, which is classically defined as a non-ionic detergent, salt and nuclease insoluble peripheral nuclear structure.1 Solubilization of the lamina by ultra-sonication and high concentrations of ionic- or chaotropic detergents inevitably results in disruption of intact lamin-protein complexes, impeding analysis by precipitation methods. Various alternative strategies have been applied to circumvent these technical problems, including immunoprecipitation (IP) with partial solubilization under mild conditions, IP using harsh solubilization conditions followed by in vitro protein complex re-assembly under mild conditions, mass spectrometry (MS) analysis of differential extractions from isolated whole nuclear membrane fractions and yeast two-hybrid, GST-pull-downs and other techniques which do not need protein complex solubilization.6–12 Each technology has its specific drawbacks including low recovery, high false positive rates and non-physiological conditions.
Here we have developed and applied a novel method to identify wild-type and mutant lamin A interacting proteins. This approach is unbiased and overcomes many of the limitations of current methods to identify lamin A-interacting proteins. Our approach consists of expression of lamin A marked with a OneSTrEP biotin analogue tag followed by isolation of intact lamin-protein complexes after protein cross-linking for 5 minutes using 1% formaldehyde. Direct and indirect interaction partners are identified by sensitive MS after a highly specific streptactin matrix precipitation. Using this approach we identify 313 lamina-interacting proteins including numerous novel candidates and proteins that appear to associate preferentially with either wildtype lamin A (8 candidates, including Nup153, Nup98 and Translocated promoter region) or progerin (27 candidates, including Paired related homeobox 1, Proliferating cell nuclear antigen and Pinin).
Results
Characterization of OneSTrEP tagged A-type lamins.
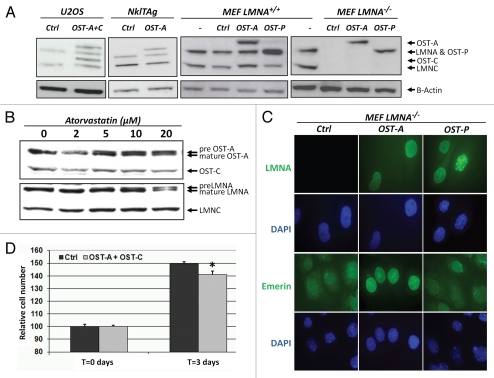
To ensure protein purification with high affinity and specificity, we made use of a modified version of the well-characterized streptavidin/biotin system.30,31 A-type lamins were tagged with the OneSTrEP (OST) tag (Supp. Fig. 1) and expressed at endogenous levels in human osteosarcoma (U2OS), mouse myocyte NklTAg or mouse embryonic fibroblasts (MEFs) from LMNA+/+ or LMNA−/− cells (Fig. 1A).14 The OST-tag is a biotin analogue, but offers the advantage of binding with high specificity and affinity (Kd = 1 µM) to the streptavidin analogue Strep-Tactin matrix.18 In addition, the OST-tag is relatively small (3 kDa) compared to other epitope tags typically used for MS purposes, thus reducing the likelihood of interference with protein function. To test for physiological functionality of OST-tagged A-type lamins, we first determined whether OST-tagged lamin A (OST-A) was farnesylated correctly in U2OS cells. Normally mature lamin A is formed after cleavage of the farnesylated C-terminal Caax motif in pre-lamin by Zmpste24.32 Atorvastatin inhibits the synthesis of farnesyl pyrophosphate and thereby impairs farnesylation and processing of pre-lamin A to mature lamin A.33 The farnesylation of both OST-A and endogenous lamin A in U2OS cells was inhibited similarly by increasing concentrations of atorvastatin (Fig. 1B). OST-tagged and wildtype lamin C do not contain a Caax motif and remained unaffected by atorvastatin (Fig. 1B).
Figure 1.
Characterization of OneSTrEP tagged A-type lamins. (A) Western blot of endogenous and tagged A-type lamin in OneSTrEP-tagged lamin A (OST-A), lamin C (OST-C), progerin (OST-P) or control virus-infected mouse cardiac NklTAg myocytes, human osteosarcoma U2OS cells and mouse embryonic fibroblasts (MEFs) from wildtype (LMNA+/+) or LMNA knock-out (LMNA−/−) embryos. The following antibodies were used: α-LaminA/C (Sc6215, Goat, Santa Cruz; detects endogenous Lamin A and C, OST-A, OST-C and OST-P) and α-Beta-Actin (Cl.AC15, A-5441, mouse, Sigma). (B) Western blot of endogenous and OST-tagged lamin A, lamin C and prelamin A in U2OS cells infected with OST-A or OST-C and treated for 48 hours with indicated concentrations of atorvastatin, an inhibitor of lamin A farnesylation. The following antibodies were used: α-LaminA/C (Sc7293, mouse, Santa Cruz; detects only endogenous Lamin A and C, and not OST-A, OST-C or OST-P) and α-Onestrep (2-1507-001, mouse, IBATagnology). (C) Immunofluorescence staining of OST-A, OST-P and control infected LMNA−/− MEFs for A-type lamins (LMNA) and emerin. DNA was stained with DAPI. The following antibodies were used: α-LMNA/C (Sc-7292, Mouse, Santa Cruz, detects OST-tagged human Lamin A and Progerin, but no endogenous murine lamins), α-emerin (Cl.4G5, mouse, Leica). (D) MTS [(3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)] assay to quantify relative cell number 62 hours after attachment of equal cell numbers of control or OST-A/C infected LMNA−/− MEFs. An asterisk indicates a significant difference (p < 0.05) between control and OST-A/C infected cells.
Next, we visualized the localization of OST-A and OST-tagged progerin (OST-P) in A-type lamin knock-out (LMNA−/−) MEFs (Fig. 1C).15 As expected, both OST-A and OST-P were detected at the nuclear rim. In contrast to OST-A, OST-P generated distortions in the nuclear lamina identical to those seen in HGPS patient cells (Fig. 1C).16 Furthermore, expression of OST-P led to a decrease of total LAP2 (all isoforms) and HP1γ protein levels in the nucleus (Supp. Fig. 2) as observed in HGPS and progerin-expressing fibroblast.17 OST-A expression further restored correct localization of endogenous emerin in LMNA−/− MEFs to the NE (Fig. 1C).34 This capacity was reduced after 3 days in culture for OST-P (Fig. 1C). Finally, combined expression of OST-A and OST-C inhibited cell proliferation, in line with previously reported endogenous lamin knock-down and HA-tagged lamin A and C overexpression studies (Fig. 1D).13 Taken together, these data demonstrate that OST-tagged A-type lamins recapitulate endogenous lamin processing, subcellular localization and functions.
OneSTrEP pull-down assay.
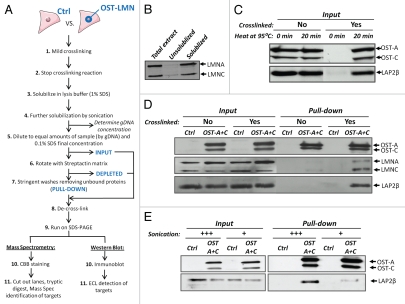
To isolate lamin A-associated proteins we used mild reversible cross-linking (5 minutes, 1% formaldehyde) to capture protein/protein and protein/DNA complexes (Fig. 2A). Cross-linking enabled us to extensively solubilize A-type lamins by using vigorous sonication in combination with high percentage ionic detergent (1% SDS; Fig. 2B). Solubilized protein complexes were diluted to comparable amounts of starting material by correcting sample volumes to reach equal genomic DNA concentration (Fig. 2A), and OST-tagged lamins were precipitated using Strep-Tactin matrix, a high-affinity streptavidin analogue.30,31 Precipitated lamin-protein complexes were eluted and de-crosslinked under conditions (20 minutes at 95°C in Laemmli Sample Buffer) that fully reversed crosslink-induced epitope masking on Western blots without detectable protein degradation (Fig. 2C). Cross-linking was crucial for co-precipitation of lamin interactors, as omitting this step resulted in loss of the known OST-A/C interaction partners wildtype lamin A/C and LAP2β in purified protein complexes (Fig. 2D).35,36 Increasing sonication strength drastically improved retrieval of the lamin A interaction partner LAP2β (Fig. 2E). These findings demonstrate that the OST-tag/Strep-Tactin pull-down approach is able to recover lamin A protein complexes after transient cross-linking and solubilization.
Figure 2.
OneSTrEP pull-down assay. (A) Schematic overview of the OST pull-down protocol for cells infected with control and OST-tagged lamin, with the input, depleted and pull-down fraction indicated in bold. Depending on the purpose of the experiment, proteins separated on SDS-Page gels were stained with Coomassie brilliant blue (CBB) (for mass spectrometry), or immunoblotted for verification of individual candidates. (B) U2OS cells were fixed in 1% formaldehyde, sonicated in SDS lysis buffer and further diluted in ChIP dilution buffer as described in Materials and Methods. Total protein extracts were spun down, separating the solubilized fraction and the insoluble fraction (i.e., pellet, twice washed with PBS). Total, unsolubilized and solubilized fractions were compared for endogenous lamin A/C protein levels by western blot. The following antibody was used: α-LaminA/C (Sc6215, goat, Santa Cruz; detects endogenous Lamin A and C). (C) Western blot of U2OS OST-A/C cell extracts (1% formaldehyde vs. no fixation) dissolved in Laemmli sample buffer (no heating vs. 20 minutes at 95°C) before loading on SDS-PAGE gels. The following antibodies were used: α-LAP2 (Y-20, Sc19783, goat, Santa Cruz; LAP2β isoform depicted) and α-Onestrep (2-1507-001, mouse, IBATagnology). (D) Western blot of endogenous lamin A/C and Lap2β after cross-linking with 1% formaldehyde in U2OS cells infected with control or OST-A/C virus. The following antibodies were used: α-LAP2 (Y-20, Sc19783, goat, Santa Cruz; LAP2β isoform depicted), α-LaminA/C (Sc7293, mouse, Santa Cruz; detects only endogenous Lamin A and C and not OST-A or OST-C) and α-Onestrep (2-1507-001, mouse, IBATagnology). (E) Western blot of OST-pull-down for lamin-A/protein complexes solubilized in SDS lysis buffer with increasing sonication strength (from 12 × 1 sec at 5 µm amplitude, +; to 6 × 5 sec at 10 µm amplitude, +++). The following antibodies were used: α-LAP2 (Y-20, Sc19783, goat, Santa Cruz; LAP2β isoform depicted) and α-Onestrep (2-1507-001, mouse, IBATagnology).
Identification of lamin A interacting proteins.
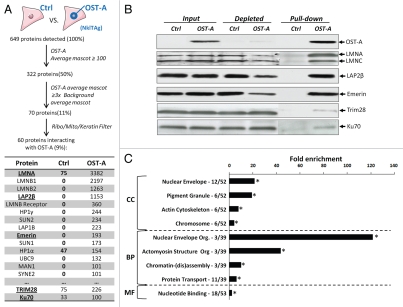
To identify and characterize lamin A-interacting proteins OST pull-downs were performed on murine cardiac myocyte NklTAg cells expressing OST-A at endogenous levels or a control virus lacking the lamin A transgene. Co-purified proteins were size-separated and retrieved from SDS-PAGE gels and analyzed by MS (Fig. 3A). In total 5,162 unique peptide fragments corresponding to 649 individual proteins were detected in two independent experiments (Fig. 3A). We reduced the list of potential interactors by stringent restrictive analysis (i.e., mascot score cut-off, minimum fold over background and filtering out non-essential peptides; see Materials and Methods; Fig. 3A) to 60 proteins. The set of identified proteins contains several known lamin A interactors including Lamin B, Lap2β, Emerin, MAN1, Sun1 and 2, as well as the muscle specific interactor Syne-2 (Fig. 3A and Supp. Table 1).37 About half of our candidate lamin A interactors were identified in two previous NE proteome studies which had identified proteins in an INM/lumen/ONM6 fraction or an INM/lamina/nucleoplasmic fraction,10 respectively, in other cell types.
Figure 3.
Identification of lamin A interacting proteins. (A) Overview of the selection criteria for identification of lamin A interacting proteins from raw mass spectrometry data. Control and OST-A pull-down average mascot scores for several lamin A interactors identified in NklTAg cells are depicted (underlined proteins are verified in B). (B) Western blot indicating protein levels of mass spectrometry identified lamin A-interactors (endogenous lamin A/C, Lap2β, Emerin, Trim28 and Ku70) precipitated in NklTAg cells infected with control or OST-A virus. The following antibodies were used: α-Onestrep (2-1507-001, mouse, IBATagnology), α-LaminA/C (Sc7293, mouse, Santa Cruz; detects only endogenous Lamin A and C, and not OST-A), α-LAP2 (Y-20, Sc19783, goat, Santa Cruz; LAP2β isoform depicted), α-Emerin (Cl.4G5, mouse, Leica), α-Trim28 (A300-767A, rabbit, Bethyl Laboratories) and α-Ku70 (Ab10878, rabbit, Abcam). (C) GO analysis of identified lamin A interactors. GO analysis was performed separately for the GO categories “cellular components” (CC), “biological process” (BP), “molecular function” (MF) and compared against a random protein set.22,23 The number of lamin A interacting proteins identified within a specific GO category versus the total amount of annotated lamin A interacting proteins is indicated next to the significantly enriched GO categories (p < 1 × 10−3 indicated by asterisk; statistical tests include using a kappa statistic measurement and is further described elsewhere).71
We identified 29 novel lamin A interactors (Supp. Table 1). These include proteins implicated in muscle differentiation (Luc1-like, Serpine1 mRNA binding protein), transcription factors (Strap, Similar to calponin 3), DNA repair (Ku70) and heterochromatin maintenance (Trim28).38–43 Several known (Emerin, Lap2β) as well as previously unidentified lamin A interactors (Trim28, Ku70) were chosen for validation of the MS data by targeted co-precipitation. Trim28 and Ku70 were specifically chosen as they represented the two proteins that were closest to the selection cut-off and therefore were most likely to represent false positives. OST-A co-precipitated considerable amounts of endogenous Lamin A/C, Lap2β, Emerin, as well as Trim28 and Ku70 (Fig. 3B). In contrast no interactions were observed in control pull-downs from cells only expressing OST (Fig. 3B). In comparison to Lap2β and Emerin, Ku70 and Trim28 appear to be weaker interactors with OST-A (Fig. 3B). In addition to Lap2β we identified interactions of lamin A with Lap2α, γ, δ and ε isoforms in U2OS cells by OST pull-down and immunoblotting (Supp. Fig. 3). Taken together these data demonstrate that the OST pull-down/MS assay and bioinformatical selection criteria can be used for identification of lamin A interacting proteins.
Functional annotation of lamin A interacting proteins.
To globally characterize the identified lamin A interactors, we performed a GO analysis to describe the subcellular localization (CC, cellular component), biological processes (BP) and molecular functions (MF) of identified interactors (Fig. 3C). As expected, compared to random proteins lamin A interactors were 22-fold enriched for ‘nuclear envelope’ components (p = 5.0 × 10−12; Fig. 3C and Supp. Table 2), and to a lesser extent for other subcellular compartments. The interactors are enriched in the functional groups of NE organization' (p = 2.5 × 10−4), actomyosin structure organization (p = 9.9 × 10−3) and protein transport (p = 5.6 × 10−6), with a preferential molecular function of ‘nucleotide binding’ (p = 5.5 × 10−4). The GO analysis had high similarity to the previously identified proteins in the NE proteome (Table S2).6,10
Identification of preferential lamin A or progerin interactors.
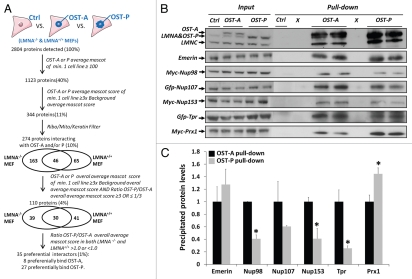
We next applied the OST pull-down method to identify proteins that bind exclusively to lamin A or progerin as well as interactors that bind to both proteins with different affinity. To this end, we expressed control virus, OST-A or OST-P in LMNA−/− or LMNA+/+ MEFs at endogenous levels (Fig. 1A). OST pull-downs were performed in duplicate in all cell lines. In total we detected 54,939 unique peptide fragments in all OST and control pull-downs, corresponding to 2,804 different proteins (Fig. 4A). Using similar selection criteria as validated for the NklTAg OST-A pull-down (mascot score cut-off, minimum fold over background and filtering out non-essential peptides; see Materials and Methods) we defined 274 lamina-interacting proteins, 209 in LMNA−/− MEFs and 111 in LMNA+/+ MEFs, of which 46 proteins were identified as interactors in both cell lines. Of these 274 interactors 150 proteins interacted with lamin A and 170 with progerin (Supp. Table 5). The relatively low number of interactors identified in both LMNA+/+ and −/− cells may be due to the very high stringency used in identifying interactors or may be due to differential effects of lamin A expression in a wild-type or null background. As many of the identified lamin A interactors were also detected by MS at levels just below inclusion criteria in progerin pull-down samples and vice versa, we used additional criteria to identify preferential interactors (minimal fold difference between OST-A and OST-P mascot scores and similarity of results in MEFs−/− and MEFs+/+; see Materials and Methods; Fig. 4A). 110 proteins interacted with both lamin A and progerin and applying these criteria resulted in a final selection of 35 proteins candidates that preferentially interact with either wild-type lamin A (8 candidates, including Nup153, Nup98 and Translocated promoter region) or progerin (27 candidates, including Paired related homeobox 1 (Prx1), Proliferating cell nuclear antigen (Pcna) and Pinin).
Figure 4.
Identification of preferential lamin A/progerin interacting proteins. (A) Overview of the selection criteria applied to identify preferential laminA/progerin interacting proteins (See also Material and Methods). (B) Verification of lamin A and progerin preferential interactors in U2OS cells transfected with GFP-Nup107, GFP-Tpr, Myc-Nup98, Myc-Nup153 or Myc-Prx1. OST pull-down was performed in U2OS cells infected with control, OST-A or OST-P virus. The following antibodies were used: α-LaminA/C (Sc6215, goat, Santa Cruz; detects endogenous Lamin A and C, OST-A and OST-P), α-Emerin (Cl.4G5, mouse, Leica), α-c-Myc (MS-139-P, mouse, Labvision) and α-GFP (Ab6673, Goat, Abcam). (C) Quantification of western blot protein levels in OST-A and OST-P pull-down fractions (average ± standard deviation) corrected for input signal intensity. Relative protein levels for OST-A pull-down fractions were set arbitrarily at 1. Protein levels identified by western blot in OST-P pull-down fractions (B) are indicated. Asterisks indicate a significant (p < 0.05; Student's t test) difference between OST-A and OST-P pull-down fraction.
To validate preferential interactors we performed OST pull-downs in U2OS cells expressing control virus, OST-A or OST-P at endogenous levels (Fig. 4B) and transiently expressed several potential differential interactors (Myc-tagged Nup98, Nup153, Prx1 and GFP-tagged Tpr). In support of the pull-down results, we found weaker interaction of OST-P with Nup98, Nup153 and Tpr (all p < 0.05, Fig. 4B and C). The NPC protein Nup107 also showed reduced, albeit statistically not significant, binding to OST-P. Prx1 was confirmed to have increased, but not unique, affinity for OST-P (p < 0.05, Fig. 4B and C). Finally, immunoblot detection confirmed the predicted interaction of endogenous emerin with OST-tagged lamin A and progerin (p > 0.05, Fig. 4B and C). These data show that lamin A and progerin have distinct protein-interactions that can be identified by OST pull-down assays.
Functional analysis of preferential lamin A or progerin interactors.
A GO-signature comparison of 150 identified lamin A-associated proteins and 170 progerin-associated proteins, shows that both protein sets were over 50-fold enriched for INM and nuclear lamina proteins, are preferentially involved in NE organization, mRNA and protein transport and enriched for proteins with GTP binding capacity (Supp. Fig. 4 and Supp. Table 3). Lamin A interactors were preferentially enriched (p < 1 × 10−4) in NPC-related nucleocytoplasmic and protein transport proteins and to a lesser extent (1 × 10−2 < p > 1 × 10−3) in proteins of the NE, the cytoskeleton and ECM (‘sarcomere’ and ‘PDGF binding’ GO-terms). Progerin interactors showed enrichment for endoplasmatic reticulum (ER) membrane proteins involved in protein folding and processing (‘cell redox homeostasis’, ‘intramolecular oxidoreductase’ and ‘signal peptidase processing’ GO-terms) as well as proteins involved in chromatin organization (mainly histone 1 and 2A variants). ER-localized A-type lamin interacting proteins were preferentially membrane-associated (43 out of 46, Supp. Table 4) and not lumenal localized (3 out of 46, Supp. Table 4). To further assess the functional context of lamin A and progerin interactors we used the Reactome protein interaction database, which contains maps of known protein complexes, as a reference.44 This analysis showed that 10 proteins of the 26S proteasome complex were identified as progerin interactors (Supp. Fig. 4 and Supp. Table 3). These GO and Reactome profiles for lamin A and progerin interactors resemble those found in the previously identified NE proteome (Supp. Table 3).
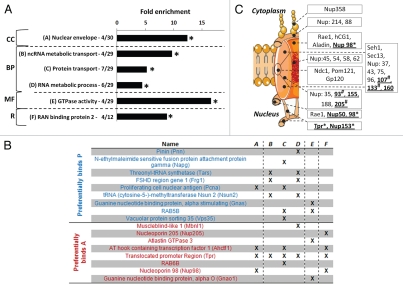
GO-analysis of the 35 preferential lamin A/progerin interactors showed enrichment of NE proteins, proteins involved in RNA metabolism (‘ncRNA metabolic transport’ and ‘RNA metabolic process’ GO terms), general protein transport components, including members of the NPC (‘Ran binding protein 2’ Reactome term) and proteins with GTPase activity (All p < 1 × 10−2; Fig. 5A and B and Supp. Table 2).44 NPC lamin-interacting proteins were preferentially located at the nucleoplasmic site of the NPC (Fig. 5C), of which a vast majority showed an overall diminished interaction with progerin in both LMNA−/− and LMNA+/+ MEFs, including 3 proteins identified in our assay as preferential lamin A interactors (Tpr, Nup98, Nup153; Fig. 5C and Table 1).
Figure 5.
Functional annotation of preferential lamin A/progerin interactors. (A) GO profile of preferential interactors specified for cellular components (CC), biological process (BP), molecular function (MF) GO categories. Representation of protein complexes was analyzed using the Reactome database (R).44 Significant categories based on comparision are indicated by asterisks (p < 1 × 10−3; random proteins used as background; reviewed in ref. 71), as well as the number of proteins within the specific category out of the total number of annotated preferential interactors. (B) Overview of GO annotated lamin A/progerin preferential interactors in significant categories. Individual categories are marked A–F and correspond to (A). (C) Schematic illustration of the nuclear pore complex (NPC) indicating the location of lamin A-interacting NPC proteins identified by mass spectrometry analysis (underlined).72 Identified interactors preferentially localize in the nuclear basket (Nup153, Tpr), nuclear ring (Nup50, Nup98) or scaffold (Nup-93, -107, -133, -155, -160 and -205). Nup98 has been described to localize in both the nuclear basket as well as the NPC central channel. Asterisks indicate proteins identified to preferentially interact with lamin A. Hashes indicate proteins with MS data indicative of diminished interaction with progerin as well (similar direction change for ratio OST-P/OST-A overall average mascot ratio's in both MEF LMNA−/− and MEF LMNA+/+).
Table 1.
Overview of preferential lamin A/progerin interactors
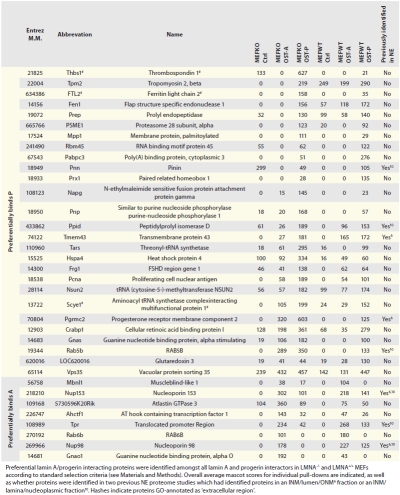 |
Discussion
The systematic mapping of lamin A interaction partners is currently hampered by the insoluble nature of lamina proteins. We have developed and applied a novel pull-down approach based on the One-STrEP tag to identify in vivo A-type lamin interactors, of which a subset interacts preferentially with lamin A compared to its mutant isoform progerin.
To isolate lamin-associated protein complexes we took advantage of the OneSTrEP-tag/Strep-Tactin matrix system, an engineered version of the strongest non-covalent biological interaction known (biotin/streptavidin).18,45 We first verified that the addition of a 3 kDa small OST-tag to lamin A recapitulated endogenous lamin A as judged by farnesylation, subnuclear localization, emerin NE localization and cell proliferative effects. Lamin interactors were identified by subsequent solubilization of the majority of A-type lamins, high affinity precipitation of OST-tagged lamins, high stringency washes of precipitated protein complexes and a highly restrictive selection based on MS results.46 We identify a large number of known direct A-type lamin interactors (including Emerin, Ubc9, LAP2α, LAP1β, LMNB1, Rbbp7), as well as known indirect interactors (including Nesprin/Syne-1, Nesprin/Syne-2; HP1γ/CBX3, HP1α/CBX5, MAN1), therefore directly validating our approach.5,12,47–52 Our method is able to identify physical associations which occur via multiple hierarchical interactions as indicated by the identification TorsinA, which is indirectly connected to the lamina via Nesprins and Sun proteins.52 The identification and verification of two previously unidentified lamin A interactors (Trim28 and Ku70) that met the inclusion criteria justifies the applied selection threshold. Comparative analysis with two published NE proteomes further validates the OST approach.6,10 Although these studies differ in their starting materials with one using isolated nuclear envelopes and the other isolated lamina preparations, they identify a significant part of the NE proteome, including laminar proteins and therefore are considered of value for a cross-comparison. About half of the OST pull-down identified lamin A interactors in NklTAg cells were identified in these previous studies and the GO signature of our hits highly resembles that of NE proteins identified in these analyses.6,10
Our OST pull-down method differs from previously published approaches. First, in comparison to approaches that biochemically fractionate nuclear membranes or the lamina, we specifically precipitate lamin A interactors, regardless of their subcellular localization. Secondly, the inclusion of a cross-linking step preserves short-lived lamin-protein interactions and thereby contributes to detection of identified transient and indirect interactors. Thirdly, we use a highly sensitive and stringent MS analysis approach. Our validation indicates a low level of false positives. However, we cannot formally rule out that a small percentage of identified hits reflect non-specific interactions. Especially various extracellular matrix (ECM) identified interactors and proteins involved in Golgi related vesicle trafficking might reflect non-specific interactions. ER proteins form a group of identified interactors that have been considered background in previous NE proteomic studies.10 Although this exclusion is correct for lumenal ER proteins, which are not directly accessible to lamins, membrane-associated ER proteins could reflect bona fide interactors of OST-tagged lamins involved in protein folding and processing during their synthesis and post-translational modification or diffuse from the ER to the continuous INM and anchor to the lamina.53 In accordance with this, our data indicate that almost all identified A-type lamin interactors residing in the ER are membrane-associated and not lumenal.
Our method combines beneficial aspects of several methods used to identify protein-content of and interactions at the nuclear lamina. IP studies have the advantage of enriching for the interactome of a protein of interest and the ability to analyze the impact of mutations on protein interactions, but are hampered by the need to solubilize intact NE protein complexes.54 In contrast, differential extraction from isolated whole nuclear membrane fractions permits disruption of protein complexes prior to MS analysis, but identifies general protein composition at a subnuclear level based on similarity of proteins in biochemical characteristics.6,8,10 Yeast two-hybrid screens allow comparison of protein interactions for wild-type and mutated protein fragments without solubilization, but have the drawback of high false positive-rates and non-physiological conditions.55–57 The OST pull-down approach combines advantages of identifying and comparing the interactome for a wild-type and mutated protein of interest with a low false positive rate under physiological conditions, with complete extraction of intact lamina protein complexes. These unique characteristics complement current technology to identify lamin A interactors.
We identified both interactors of lamin A and of progerin and compared their GO profiles. Differences in profiles indicate a loss of interactions of progerin with the cytoskeleton in comparison to lamin A, in agreement with reported disruption of nuclear-cytoskeletal coupling by progerin.58 Furthermore, GO analysis suggests a preferential interaction of progerin with protein folding and processing enzymes as well as proteasome complex members. This observation is intriguing in the light of the reported degradation in HGPS patient cells of a large set of nuclear proteins, particularly chromatin-associated proteins.5,16 To further investigate differences between lamin A- and progerin-interacting proteins we identified preferential interactors by comparing OST pull-downs in both LMNA−/− MEFs, in the absence of endogenous lamin A/C which could mask distinct interactions due to dimerization with progerin, as well as biological more relevant LMNA+/+ MEFs.36 Several identified distinct lamin A/progerin interactors preferentially possessed GTPase activity and others were involved in RNA metabolism or nucleocytoplasmic transport.
We found reduced interaction of Nup153 with progerin compared to wild-type lamin A. Nup153 anchors NPCs to the nuclear lamina and directly interacts with Tpr, which is involved in formation of NPC-associated heterochromatin exclusion zones.59–61 Interestingly, HGPS is associated with peripheral loss of heterochromatin and clustering of Nup153 and other NPC proteins.62,63 A diminished interaction between OST-P and Nup153 could explain a loss of NPC anchoring to the nuclear lamina, as well as clustering and defective NPC-mediated nuclear import which have all been implicated in HGPS.62,63
We found an increased interaction of progerin with paired related homeobox 1 (Prx1 or Prrx1), a transcriptional co-activator involved in skeletogenesis and vasculogenesis.28,64 Loss of Prx1 results in lethal vertebrae and craniofacial skeletal defects, while overexpression induces vascular smooth muscle cell (VSMC) proliferation and ECM remodeling by direct upregulation of tenascin C expression.28,64 These phenotypes highly resemble HGPS etiology as Zmpste24 knock-out mice, which serve as a model system for HGPS, suffer from osteogenesis imperfecta, manifested in vertebrae and craniofacial bone fractures.65 Furthermore progerin drastically downregulates tenascin C expression and progerin transgenic mice suffer from a progressive loss of VSMCs and changes in ECM composition.66,67 An increased interaction of progerin with Prx1 could impair transcriptional activation of tenascin C and cause consecutive HGPS characteristic ECM, VSCM and bone related disorders.
A number of differential interactors link lamin A and progerin to previously reported abnormal cell biology in the context of LMNA mutations. We find an increased interaction of Proliferating cellular nuclear antigen (Pcna) with progerin. Pcna, as a replication fork processivity factor, regulates genomic replication and has been reported to redistribute upon introduction of progerin.68 Several studies report abnormal proliferation in LMNA mutant cells.69 Pcna is thought to become trapped in intranuclear mutant lamin aggregates, thereby ultimately blocking DNA replication and causing HGPS-associated DNA damage.63,68 Of relevance in this context is also the identification of Trim28 and Ku70 as interaction partners of lamin A. Both are associated with DNA damage response and/or repair and Trim28 is a reported interaction partner of heterochromatin protein 1.70 While the contribution of these preferential interactions will need to be elucidated in targeted studies, the global identification of novel lamin A and progerin interactors using a novel pull-down method has generated several leads for studies of specific interaction partners of lamin A and its mutants with the goal of uncovering the molecular mechanisms of tissue-specific laminopathies.
Materials and Methods
Cell lines and culture.
A lentiviral expression plasmid for N-terminal OneSTrEP tagged progerin (OST-P) was created by PCR amplification of lamin A, omitting amino acids 609–658 using the previously described pCDH MCSNard OST-A plasmid as template.5 Forward and reverse primers, containing a BamHI and EcoRI restriction site, were used for subsequent ligation of the PCR product in BamHI/EcoRI digested pCDH MCSNard OST-A plasmid. For the cloning of pCDH MCSNard OST-Lamin C (OST-C), lamin C was PCR amplified from the previously described pBabe purox HA-Lamin C plasmid, adding an in frame 5′ SalI and a 3′ EcoRI site, which were used for ligation into equally digested pCDH MCSNard OST-A plasmid.13 Lentivirus was produced in 293 FT cells (Invitrogen, Carlsbad, USA) co-transfected with pCDHblast MCSNard OST-A, OST-P or OST-C in combination with pSPAX and pMD2.G vectors (Trono Lab). The mouse cardiac myocyte cell line NklTAg, human osteosarcoma cell (U2OS), as well as mouse embryonic fibroblasts (MEFs) of wildtype (LMNA+/+) and LMNA knock-out (LMNA−/−) embryos were infected with various concentrations of lentivirus and 48 hours post infection selected with blasticidin.14,15 Infected cells with OST-tagged lamin protein amounts comparable to endogenous lamin protein levels were further cultured and grown in Dulbecco's modified Eagle's/F-12 media (for NklTAg cells) or Dulbecco's modified eagle medium (for U2OS and MEFs) both supplemented with 2 mM L-glutamine, 10 mM non-essential amino acids, 110 mg/l pyruvate, 10% fetal bovine serum and antibiotics in a humidified atmosphere at 37°C and 5% CO2. NklTAg cells were plated on 12.5 µg/ml fibronectin and 0.1% gelatin coated plates.
Immunofluorescence.
For immunofluorescence microscopy LMNA−/− MEFs infected with control, OST-A or OST-P lentivirus were grown on 0.1% gelatin/PBS solution coated multi-wells glass slides (MP Biomedicals). After fixation (4% paraformaldehyde/PBS; 15 min), cells were permeabilized (0.5% Triton X-100/PBS; 5 min), blocked (1% BSA, 1% sucrose in milliQ; 1 hour) and incubated for one hour with appropriate primary antibodies (α-LMNA/C, Sc-7292, mouse, Santa Cruz, detects OST-tagged and endogenous human, but not mouse, lamin A, Lamin C and Progerin; α-emerin, Cl.4G5, mouse, Leica; α-HP1γ, mab3450, Chemicon, Temecula, USA; α-LAP2, kind gift from K. Wilson, raised against residues 1–187 of human LAP2 and suitable for detection of all LAP2 isoforms). Primary antibodies were detected with an Alexa Fluor 488 chromophore labeled secondary antibody (Invitrogen). Cells were mounted in Vectashield containing 10 g/ml DAPI and observed on a Nikon E800 microscope. Quantitative microscopy measurements were performed as previously described.16,17 Exposure times and acquisition settings were established at the beginning of the experiment and kept constant for all samples. The average nuclear intensity of total LAP2 (all isoforms included) and HP1γ minus average background signal was measured for at least 200 cells per sample in duplicate using Metamorph software (Molecular Devices). The criteria to define cells with reduced levels of chromatin proteins were previously described.16,17
OneSTrEP-lamin A farnesylation and proliferative effects.
To determine whether OST-A was correctly farnesylated U2OS cells expressing OST-A and C at endogenous levels were treated with indicated concentrations of Atorvastatin (10 mM stock dissolved in DMSO was further diluted in cell culture medium) for 48 hours. Samples were harvested in Laemmli Sample Buffer and further analyzed by western blotting. Proliferative effects of OST-tagged lamin A/C were determined with the MTS assay, as described elsewhere.13 LMNA−/− MEFs infected with empty control or OST-A and C were seeded at 1,000 cells per well in 96 wells plates.15 Sixty-two hours post attachment 20 ml 2 mg/ml 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (Promega CellTiter 96w AQueous MTS) and 1 ml 0.92 mg/ml phenazine methosulfate (Merck) was added for 3 hours. The tetrazolium is reduced by the living cells into a colored formazan product, which absorbance was measured at 490 nm and is directionaly proportional to the cell number. Statistical analysis was performed by Student's t-test.
OneSTrEP pull-down assay and mass spectrometry.
U2OS, NklTAg and MEF cell lines were grown until confluency in 150 mm dishes. Plates were left for several minutes at ambient temperature before protein complexes were mildly cross-linked by direct addition of formaldehyde to the culturing medium to a final concentration of 1% (10 minutes, ambient temperature). The cross-linking reaction was stopped by addition of glycine (125 µM final conc.) and gentle shaking for 5 minutes at ambient temperature. Fixed cells were scraped in PBS, pelleted, dissolved in ice-cold SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8.1), snap frozen in dry ice once, thawed, left for 30 minutes on ice and finally sonicated for 2 times 30 seconds at 10 µm amplitude using a tip sonicator. Endogenous biotinylated proteins were blocked by addition of avidin (10 µg/ml). As the affinity of avidin for biotin is much higher then for the OneSTrEP epitope, OST-tagged lamin pull-downs will not be impaired by the addition of avidin.18 A small sample aliquot was taken, de-cross-linked for 6 hours at 65°C in the presence of 200 mM NaCl and 30 µg/ml RNAase A, after which the genomic DNA concentration was determined with a Qiagen PCR clean-up kit (Qiagen). Next, protein samples were diluted in SDS lysis buffer to contain equal amounts of genomic DNA (typically 200 µg genomic DNA/pull-down for MS purposes) and subsequently diluted in ChIP dilution buffer (0.1% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 17 mM Tris pH 8.1, 170 mM NaCl) to reach a final concentration of 0.2% SDS, 0.1% Triton X-100, 2 mM EDTA, 20 mM Tris.HCl pH 8.1, 150 mM NaCl. Both SDS lysis buffer and ChIP dilution buffer were supplemented with 2 mM sodiumortohovanadate and protease inhibitors (Boehringer MannHeim). Undissolved fractions were discarded after a 10 minute centrifugation step (16,000 g) at 4 degrees and a small sample of the supernatant (i.e., input fraction) was stored for further analysis. ChIP dilution buffer pre-washed Strep-Tactin Matrix (IBATagnology) was added and OST-lamin/protein complexes were bound by overnight end-over-end rotation. Strep-Tactin matrix was pelleted by centrifugation and the supernatant (i.e., depleted fraction) was stored, while the pellet was washed four times with OST stringent wash buffer (2 M NaCl, 2% Trx-100, 500 mM LiCl, 0.1% SDS, 1% Sodium Deoxycholate, 20 mM Tris pH 8.1, 2 mM EDTA) and twice with ChIP dilution buffer. LiCl was included to improve the effective removal of non-specific interactors by the presence of different types of salts. Pelleted protein-chromatin complexes (pull-down fraction), input and depleted fractions were de-cross-linked by boiling for 20 minutes at 95°C in Laemmli Sample Buffer.
For pull-down verification purposes samples were analyzed by western blot (see below). For mass spectrometry (MS) pull-down fractions were separated on NuPage Novex Bis-Tris 4–20% gradient gels (Invitrogen) and stained with Coomassie Brilliant Blue (CBB). For CBB staining gels were first fixed for minimally 2 hours in solution 1 (50% v/v ethanol, 2% v/v phosphoric acid), washed 3 × 30 minutes in distilled water, incubated for 1 hour in solution 2 (34% v/v methanol, 17% w/v ammonium sulphate, 2% v/v phosphoric acid) and finally stained with CBB-G (0.66% w/v in distilled water) until desired staining intensity was reached (1–3 days). Gels were stored in distilled water and transported for subsequent MS analysis at the Erasmus MC Proteomics Center (Rotterdam, The Netherlands). Gel lanes were cut and subjected to in-gel digestion with trypsin (Promega), essentially as described previously.19 Polypeptides were identified on an LQT Orbitrap hybrid mass spectrometer (ThermoFischer), coupled to a 1,100 series capillary liquid chromatography system (Agilent Technologies) as described elsewhere.20 Data analysis was performed using a Mascot based search algorithm (version 2.2; MatrixScience).20
Mass spectrometry data analysis.
In order to compare MS results of various samples, all retrieved GI and IPI protein identifiers were converted to Mus Musculus and Homo Sapiens Entrez identifiers, using the Database for Annotation, Visualization and Integrated Discovery (DAVID), BioDBNet (http://biodbnet.abcc.ncifcrf.gov/db/db2db.php), Clone/Gene ID converter and g:profiler databases.21–24 For identifiers that could not be converted to Entrez identifier with these databases, the top hit with matching Entrez ID of a protein blast search (http://blast.ncbi.nlm.nih.gov) using a PSI-Blast algorithm was used. This created one list of all MS identified proteins in our study, which was used for alignment of individual MS sample results using excel software with a DigDB plugin (http://www.digdb.com/). To select true lamin interactors, several selection criteria were sequentially applied. First, proteins needed to be detected at high levels, i.e., average OST-A mascot score above 100, with the average calculated only in OST-A samples in which the protein was detected. Second, proteins were required to be detected at levels minimally 3-fold less in background pull-downs (based on average mascot scores, calculated as described for previous criterion). Third, keratin proteins and proteins with gene ontology (GO) codes for mitochondria and ribosome, which are all considered non-lamin A interacting proteins, were excluded. In order to compare OST-A and OST-P pull-down results in LMNA−/− and LMNA+/+ MEFs these selection criteria were first applied to identify lamina interactors, independent of cell line and OST-A or OST-P construct. Amongst this protein set of lamina interacting proteins, differential interactors were selected by several additional criteria. Proteins were only included if in either cell line the overall average mascot score (including samples in which no protein fragment was detected for calculation of the average) of OST-A or OST-P was above 100 and more than 3-fold larger than background pull-down overall average mascot scores. Next, proteins were required to have a ratio of OST-P over OST-A overall average mascot scores above 3.0 or below 0.33 in either LMNA−/− or LMNA+/+ MEFs. The final inclusion criterion was that this ratio was similar directed (both above or below 1.0) in both cell lines. Mascot scores were considered semi-quantitative only, and were used to distinguish proteins that might either have higher affinity for lamin A or progerin (or associated proteins) or were expressed more abundantly in OST-A versus OST-P cells. Whether these candidates bound lamins directly or indirectly was not revealed by this method.
GO analysis for indicated proteins subsets were performed separately for GO categories “cellular components level 5” (CC), “biological process all” (BP) and “molecular function all” (MF) using Mus Musculus Entrez Identifiers as input values and standard DAVID settings for functional annotation clustering, listing significant clusters by most significant GO term.22,23 GO analysis was performed against a random protein background consisting of all known Mus Musculus Entrez identifiers, or a set of previously identified nuclear envelope proteins.6,10 To identify enrichment of protein complexes a clustering analysis was performed with DAVID against the reactome database, using Homo Sapiens Entrez gene identifiers.
Plasmid transfection.
For verification of differential interactors U2OS Ctrl, OST-A and OST-P expressing cells were plated at 80% confluency in 10 mm-dishes and FuGENE transfected with previously described expression plasmids for Myc-Nup98, Myc-Nup153, GFP-Nup107, GFP-Tpr and Myc-Prx1 (Kindly provided by Günter Blobel, Brian Burke, Thomas Schwartz, Larry Gerace and Peter Lloyd Jones).25–29 32 hours after transfection cells were harvested for a OST pull-down assay.
Western blot.
Western blots were performed essentially as described elsewhere.5 SDS-PAGE Laemmli sample buffer was added to protein samples from various stages of the OST-pull-down (input, depleted, pull-down) and heated for 20 minutes at 95°C to de-cross-link and denature proteins. Equal amounts of protein extract were loaded and run on NuPage Novex Bis-Tris 4–20% gradient gels (Invitrogen, Carlsbad, USA), blotted on immobilon-PVDF membrane (Millipore, Billerica, USA), blocked 1 hour in 5% bovine serum albumin/TBS-T (20 mM Tris-HCl pH 7.5, 150 mM NaCl and 0.1% Tween 20). The blocked membrane was incubated at 4°C overnight in block buffer diluted primary antibodies. The following antibodies were used: α-LaminA/C (Sc6215, goat, Santa Cruz; detects endogenous Lamin A and C, OST-A, OST-C and OST-P), α-LaminA/C (Sc7293, mouse, Santa Cruz; detects only endogenous Lamin A and C, and not OST-A, OST-C or OST-P), α-LAP2 (Y-20, Sc19783, goat, Santa Cruz; detects all LAP2 isoforms), α-GFP (Ab6673, goat, Abcam), α-Onestrep (2-1507-001, mouse, IBATagnology), α-c-Myc (MS-139-P, mouse, Labvision), α-Emerin (Cl.4G5, mouse, Leica), α-Beta-Actin (Cl.AC15, A-5441, mouse, Sigma), α-Trim28 (A300-767A, rabbit, Bethyl Laboratories), α-Ku70 (Ab10878, rabbit, Abcam). After incubation with appropriate secondary HRP-labeled secondary antibodies immunoluminescence was detected with an ECL western blotting detection system from Amersham. Intensity of signals were quantified using ImageJ software and statistically analyzed with a Student's t-test.
Acknowledgements
We thank Günter Blobel, Brian Burke, Thomas Schwartz, Larry Gerace and Peter Lloyd Jones for kindly sharing expression plasmids. Thijs Konsten is thanked for his help in optimization of the OST pull-down assay. This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), NCI, Center for Cancer Research, grants from the Dutch Heart Foundation, ZonMW and the EU-KP7 grant ‘Inheritance’.
Abbreviations
- Ahctf1
AT hook containing transcription factor 1
- CBB
coomassie brilliant blue
- CMD1A
dilated cardiomyopathy 1A
- FPLD
dunnigan familial partial lipodystrophy
- EDMD
emery-dreifuss muscular dystrophy
- ER
endoplasmic reticulum
- ECM
extracellular matrix
- GO
gene ontology
- HGPS
Hutchinson-Gilford progeria syndrome
- IP
immunoprecipitation
- INM
inner nuclear membrane
- LSB
laemmli sample buffer
- LMNA+/+
lamin A/C wildtype
- LMNA−/−
lamin A/C knock-out
- MS
mass spectrometry
- MEF
mouse embryonic fibroblast
- NE
nuclear envelope
- NPC
nuclear pore complexes
- OST
oneSTrEP
- OST-A
oneSTrEP tagged lamin A
- OST-C
oneSTrEP tagged lamin C
- OST-P
oneSTrEP tagged progerin
- ONM
outer nuclear membrane
- Prx1
paired related homeobox1
- Pcna
proliferating cellular nuclear antigen
- SDS
sodium dodecyl sulfate
- Srebp1
sterol element binding protein 1
- Tpr
translocated promoter region
- VSMC
vascular smooth muscle cell
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/13512
Supplementary Material
References
- 1.Foisner R. Dynamic Connections of Nuclear Envelope Proteins to Chromatin and the Nuclear Matrix. In: Collas P, editor. Nuclear Envelope Dynamics in Embryos and Somatic Cells. 2003. [Google Scholar]
- 2.Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- 3.Chi YH, Chen ZJ, Jeang KT. The nuclear envelopathies and human diseases. J Biomed Sci. 2009;16:96. doi: 10.1186/1423-0127-16-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holt I, Ostlund C, Stewart CL, Man N, Worman HJ, Morris GE. Effect of pathogenic mis-sense mutations in lamin A on its interaction with emerin in vivo. J Cell Sci. 2003;116:3027–3035. doi: 10.1242/jcs.00599. [DOI] [PubMed] [Google Scholar]
- 5.Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, Misteli T. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreger M, Bengtsson L, Schoneberg T, Otto H, Hucho F. Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc Natl Acad Sci USA. 2001;98:11943–11948. doi: 10.1073/pnas.211201898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holaska JM, Wilson KL. An emerin “proteome”: purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing and nuclear architecture. Biochemistry. 2007;46:8897–8908. doi: 10.1021/bi602636m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korfali N, Fairley EA, Swanson SK, Florens L, Schirmer EC. Use of sequential chemical extractions to purify nuclear membrane proteins for proteomics identification. Methods Mol Biol. 2009;528:201–225. doi: 10.1007/978-1-60327-310-7_15. [DOI] [PubMed] [Google Scholar]
- 9.Lin F, Morrison JM, Wu W, Worman HJ. MAN1, an integral protein of the inner nuclear membrane, binds Smad2 and Smad3 and antagonizes transforming growth factor-beta signaling. Hum Mol Genet. 2005;14:437–445. doi: 10.1093/hmg/ddi040. [DOI] [PubMed] [Google Scholar]
- 10.Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- 11.Wilkinson FL, Holaska JM, Zhang Z, Sharma A, Manilal S, Holt I, et al. Emerin interacts in vitro with the splicing-associated factor, YT521-B. Eur J Biochem. 2003;270:2459–2466. doi: 10.1046/j.1432-1033.2003.03617.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhong N, Radu G, Ju W, Brown WT. Novel progerin-interactive partner proteins hnRNP E1, EGF, Mel 18 and UBC9 interact with lamin A/C. Biochem Biophys Res Commun. 2005;338:855–861. doi: 10.1016/j.bbrc.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Van Berlo JH, Voncken JW, Kubben N, Broers JL, Duisters R, van Leeuwen RE, et al. A-type lamins are essential for TGF-beta1 induced PP2A to dephosphorylate transcription factors. Hum Mol Genet. 2005;14:2839–2849. doi: 10.1093/hmg/ddi316. [DOI] [PubMed] [Google Scholar]
- 14.Rybkin II, Markham DW, Yan Z, Bassel-Duby R, Williams RS, Olson EN. Conditional expression of SV40 T-antigen in mouse cardiomyocytes facilitates an inducible switch from proliferation to differentiation. J Biol Chem. 2003;278:15927–15934. doi: 10.1074/jbc.M213102200. [DOI] [PubMed] [Google Scholar]
- 15.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nature medicine. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.IBA bioTAGnology, author. A comprehensive Manual. 2005. Expression and purification of proteins using Strep-tag and/or 6xHistidine-tag. [Google Scholar]
- 19.Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, et al. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 21.Alibes A, Yankilevich P, Canada A, Diaz-Uriarte R. IDconverter and IDClight: conversion and annotation of gene and protein IDs. BMC Bioinformatics. 2007;8:9. doi: 10.1186/1471-2105-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:193–200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boehmer T, Jeudy S, Berke IC, Schwartz TU. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell. 2008;30:721–731. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fontoura BM, Blobel G, Matunis MJ. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186 kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frosst P, Guan T, Subauste C, Hahn K, Gerace L. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J Cell Biol. 2002;156:617–630. doi: 10.1083/jcb.200106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones FS, Meech R, Edelman DB, Oakey RJ, Jones PL. Prx1 controls vascular smooth muscle cell proliferation and tenascin-C expression and is upregulated with Prx2 in pulmonary vascular disease. Circ Res. 2001;89:131–138. doi: 10.1161/hh1401.093582. [DOI] [PubMed] [Google Scholar]
- 29.Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Junttila MR, Saarinen S, Schmidt T, Kast J, Westermarck J. Single-step Strep-tag purification for the isolation and identification of protein complexes from mammalian cells. Proteomics. 2005;5:1199–1203. doi: 10.1002/pmic.200400991. [DOI] [PubMed] [Google Scholar]
- 31.Witte CP, Noel LD, Gielbert J, Parker JE, Romeis T. Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol Biol. 2004;55:135–147. doi: 10.1007/s11103-004-0501-y. [DOI] [PubMed] [Google Scholar]
- 32.Young SG, Fong LG, Michaelis S. Prelamin A, Zmpste24, misshapen cell nuclei and progeria—new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J Lipid Res. 2005;46:2531–2558. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Gao J, Liao J, Yang GY. CAAX-box protein, prenylation process and carcinogenesis. Am J Transl Res. 2009;1:312–325. [PMC free article] [PubMed] [Google Scholar]
- 34.Vaughan A, Alvarez-Reyes M, Bridger JM, Broers JL, Ramaekers FC, Wehnert M, et al. Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. J Cell Sci. 2001;114:2577–2590. doi: 10.1242/jcs.114.14.2577. [DOI] [PubMed] [Google Scholar]
- 35.Lang C, Krohne G. Lamina-associated polypeptide 2beta (LAP2beta) is contained in a protein complex together with A- and B-type lamins. Eur J Cell Biol. 2003;82:143–153. doi: 10.1078/0171-9335-00305. [DOI] [PubMed] [Google Scholar]
- 36.Strelkov SV, Schumacher J, Burkhard P, Aebi U, Herrmann H. Crystal structure of the human lamin A coil 2B dimer: implications for the head-to-tail association of nuclear lamins. J Mol Biol. 2004;343:1067–1080. doi: 10.1016/j.jmb.2004.08.093. [DOI] [PubMed] [Google Scholar]
- 37.Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc Natl Acad Sci USA. 2005;102:4359–4364. doi: 10.1073/pnas.0500711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haag J, Aigner T. Identification of calponin 3 as a novel Smad-binding modulator of BMP signaling expressed in cartilage. Exp Cell Res. 2007;313:3386–3394. doi: 10.1016/j.yexcr.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Kimura E, Hidaka K, Kida Y, Morisaki H, Shirai M, Araki K, et al. Serine-arginine-rich nuclear protein Luc7l regulates myogenesis in mice. Gene. 2004;341:41–47. doi: 10.1016/j.gene.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 40.Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 41.Song K, Jung Y, Jung D, Lee I. Human Ku70 interacts with heterochromatin protein 1alpha. J Biol Chem. 2001;276:8321–8327. doi: 10.1074/jbc.M008779200. [DOI] [PubMed] [Google Scholar]
- 42.Swingler TE, Kevorkian L, Culley KL, Illman SA, Young DA, Parker AE, et al. MMP28 gene expression is regulated by Sp1 transcription factor acetylation. Biochem J. 2010;427:391–400. doi: 10.1042/BJ20091798. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Kanda Y, Roberts DJ, Ecker JL, Losel R, Wehling M, et al. Expression of progesterone receptor membrane component 1 and its partner serpine 1 mRNA binding protein in uterine and placental tissues of the mouse and human. Mol Cell Endocrinol. 2008;287:81–89. doi: 10.1016/j.mce.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 44.Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, et al. Reactome: a knowledge-base of biological pathways. Nucleic Acids Res. 2005;33:428–432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holmberg A, Blomstergren A, Nord O, Lukacs M, Lundeberg J, Uhlen M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis. 2005;26:501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- 46.Yoon E, Beom S, Cheong H, Kim S, Oak M, Cho D, et al. Differential regulation of phospholipase Cgamma subtypes through FcepsilonRI, high affinity IgE receptor. Biochem Biophys Res Commun. 2004;325:117–123. doi: 10.1016/j.bbrc.2004.09.216. [DOI] [PubMed] [Google Scholar]
- 47.Dechat T, Korbei B, Vaughan OA, Vlcek S, Hutchison CJ, Foisner R. Lamina-associated polypeptide 2a binds intranuclear A-type lamins. J Cell Sci. 2000:3473–3484. doi: 10.1242/jcs.113.19.3473. [DOI] [PubMed] [Google Scholar]
- 48.Lobov IB, Tsutui K, Mitchell AR, Podgornaya OI. Specifcity of SAF-A and Lamin B binding in vitro correlates with the satellite DNA bending state. J Cell Biochem. 2001;83:218–229. doi: 10.1002/jcb.1220. [DOI] [PubMed] [Google Scholar]
- 49.Yang L, Guan T, Gerace L. Lamin-binding fragment of LAP2 inhibits increase in nuclear volume during the cell cycle and progression into S Phase. J Cell Biol. 1997;139:1077–1087. doi: 10.1083/jcb.139.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye Q, Worman HJ. Protein-protein interactions between human nuclear lamins expressed in yeast. Exp Cell Res. 1995;219:292–298. doi: 10.1006/excr.1995.1230. [DOI] [PubMed] [Google Scholar]
- 51.Mansharamani M, Wilson KL. Direct binding of nuclear membrane protein MAN1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. J Biol Chem. 2005;280:13863–13870. doi: 10.1074/jbc.M413020200. [DOI] [PubMed] [Google Scholar]
- 52.Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ostlund C, Sullivan T, Stewart CL, Worman HJ. Dependence of diffusional mobility of integral inner nuclear membrane proteins on A-type lamins. Biochemistry. 2006;45:1374–1382. doi: 10.1021/bi052156n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang SX, Garcia-Gras E, Wycuff DR, Marriot SJ, Kadeer N, Yu W, et al. Identification of direct serum-response factor gene targets during Me2SO-induced P19 cardiac cell differentiation. J Biol Chem. 2005;280:19115–19126. doi: 10.1074/jbc.M413793200. [DOI] [PubMed] [Google Scholar]
- 55.Sakaki M, Koike H, Takahashi N, Sasagawa N, Tomioka S, Arahata K, et al. Interaction between emerin and nuclear lamins. J Biochem. 2001;129:321–327. doi: 10.1093/oxfordjournals.jbchem.a002860. [DOI] [PubMed] [Google Scholar]
- 56.Goldberg M, Lu H, Stuurman N, Ashery-Padan R, Weiss AM, Yu J, et al. Interactions among Drosophila nuclear envelope proteins lamin, otefin and YA. Mol Cell Biol. 1998;18:4315–4323. doi: 10.1128/mcb.18.7.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Libotte T, Zaim H, Abraham S, Padmakumar VC, Schneider M, Lu W, et al. Lamin A/C-dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol Biol Cell. 2005;16:3411–3424. doi: 10.1091/mbc.E04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape, mechanics and mechanotransduction. Circ Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, et al. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 2010;29:1659–1673. doi: 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smythe C, Jenkins HE, Hutchison CJ. Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 2000;19:3918–3931. doi: 10.1093/emboj/19.15.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20:5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Busch A, Kiel T, Heupel WM, Wehnert M, Hubner S. Nuclear protein import is reduced in cells expressing nuclear envelopathy-causing lamin A mutants. Exp Cell Res. 2009;315:2373–2385. doi: 10.1016/j.yexcr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.ten Berge D, Brouwer A, Korving J, Martin JF, Meijlink F. Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development. 1998;125:3831–3842. doi: 10.1242/dev.125.19.3831. [DOI] [PubMed] [Google Scholar]
- 65.Bergo MO, Gavino B, Ross J, Schmidt WK, Hong C, Kendall LV, et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness and a prelamin A processing defect. Proc Natl Acad Sci USA. 2002;99:13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Csoka AB, English SB, Simkevich CP, Ginzinger DG, Butte AJ, Schatten GP, et al. Genome-scale expression profiling of Hutchinson-Gilford progeria syndrome reveals widespread transcriptional misregulation leading to mesodermal/mesenchymal defects and accelerated atherosclerosis. Aging Cell. 2004;3:235–243. doi: 10.1111/j.1474-9728.2004.00105.x. [DOI] [PubMed] [Google Scholar]
- 67.Varga R, Eriksson M, Erdos MR, Olive M, Harten I, Kolodgie F, et al. Progressive vascular smooth muscle cell defects in a mouse model of Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2006;103:3250–3255. doi: 10.1073/pnas.0600012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Musich PR, Zou Z. Genomic instability and DNA damage responses in progeria arising from defective maturation of prelamin A. Aging. 2009;1:28–37. doi: 10.18632/aging.100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nitta RT, Jameson SA, Kudlow BA, Conlan LA, Kennedy BK. Stabilization of the retinoblastoma protein by A-type nuclear lamins is required for INK4A-mediated cell cycle arrest. Mol Cell Biol. 2006;26:5360–5372. doi: 10.1128/MCB.02464-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maraldi NM, Lattanzi G, Capanni C, Columbaro M, Mattioli E, Sabatelli P, et al. Laminopathies: a chromatin affair. Adv Enzyme Regul. 2006;46:33–49. doi: 10.1016/j.advenzreg.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 71.Huang da W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.