Abstract
The combination of targeted muscle reinnervation (TMR) and pattern classification of electromyography (EMG) has shown great promise for multifunctional myoelectric prosthesis control. In this study, we hypothesized that surface EMG recordings with high spatial resolution over reinnervated muscles could capture focal muscle activity and improve the classification accuracy of identifying intended movements. To test this hypothesis, TMR subjects with transhumeral or shoulder disarticulation amputations were recruited. Spatial filters such as single differential filters, double differential filters, and various two-dimensional, high-order spatial filters were used, and the classification accuracies for fifteen different movements were calculated. Compared with monopolar recordings, spatially localized EMG signals produced increased accuracy in identifying the TMR patients’ movement intents, especially for hand movements. When the number of EMG signals was constrained to 12, the double differential filters gave 5–15% higher classification accuracies than the filters with lower spatial resolution, but resulted in comparable accuracies to the filters with higher spatial resolution. These results suggest that double differential EMG recordings may further improve the TMR-based neural interface for robust, multifunctional control of artificial arms.
Keywords: Electromyography, Targeted muscle reinnervation, Spatial filter, Neural–machine interface
INTRODUCTION
Electromyographic (EMG) signals recorded from the residual limb of individuals with arm amputations can be effective neural control signals for powered prostheses.1 In the past 40 years, substantial research effort has been carried out in the field of myoelectric control, and significant progress has been made.18 Myoelectric control has found widespread use for individuals with amputations or congenital upper limb deficiencies. Several myoelectric control systems are now available and are capable of controlling a single device in a prosthetic limb, such as a hand, an elbow, or a wrist.20 These systems typically extract control information based on the amplitude or rate of change of the EMG signals, and are directly controlled by these signals, either in proportional or on/off mode. However, although these systems have been successful, they do not provide sufficient information to reliably control more than one degree of freedom at a time.
Pattern recognition has been proposed as a potential method to increase the information extracted from EMG signals and improve the dexterity of myoelectric control. The goal of pattern recognition is to discriminate among desired classes of limb movement based on the assumption that patterns of EMG signal at each location will be repeatable for a given motion but different between motions.4,5 Advances in EMG feature extraction and classifier design have resulted in high classification accuracies for different movement patterns.4,8
The steady progress of EMG signal processing technology suggests great promise for multifunction prosthesis control using EMG information from residual muscles. However, for patients with amputations above the elbow, the potential EMG recording sites are limited while the number of functions to control is increased.11 To address this challenge, a novel neural-machine interface based on targeted muscle reinnervation (TMR) has been developed and applied to patients with transhumeral or higher-level amputations.13,16,17,21 TMR is a surgical technique that transfers the nerves originally controlling the missing limb to remaining muscles which are no longer biomechanically functional. After successful reinnervation, the voluntary neural control signals propagate along the transferred nerves, activate these muscles, and provide additional EMG recording sites for multifunction prosthetic arm control. In previous studies, we used a high-density electrode array (up to 128 monopolar electrodes) to collect EMG signals from the reinnervated muscles of TMR patients.21 Using an EMG pattern recognition (PR) algorithm, we found an average accuracy of 96% for classifying intended movements of the elbow, wrist, thumb and fingers. A classification accuracy of 90% could still be achieved when EMG signals were recorded by only 12 bipolar electrodes.13 These results suggest that a clinically practical number of EMG signals from TMR muscles contain tremendous neural control information which can be successfully extracted by a PR algorithm. Therefore, TMR combined with EMG pattern recognition opens up possibilities for further improving control dexterity for individuals with transhumeral or higher-level amputations.
The majority of classification errors in previous studies were due to misclassification of hand (thumb and finger) movements.13,21 This may be related to the high information density (defined as the amount of information per muscle volume) in the reinnervated muscle.15 The median, radial, and ulnar nerves in the brachial plexus consist of many motoneurons that normally innervate dozens of muscles in the forearm and hand to control the wrist, thumb, and fingers. When such a nerve is transferred to one muscle compartment, all of the neural control information is mixed together and presented via surface EMG within a limited muscle space, resulting in high information density. In order to clearly separate the compound neural control sources and accurately identify the user’s movement intentions, it may be necessary to use localized EMG recordings. Spatial filtering has shown promise as one of the ways to improve the spatial resolution of EMG signals. One-dimensional (1D) or two-dimensional (2D) high-pass spatial filters have been successfully used with surface EMG signals to investigate single motor unit activities, 2,6 and to reduce cross-talk in surface EMG signals. 19 In the present study, we hypothesized that using spatial filters for more focal recording of TMR muscle activity would allow better control source separation and increased classifier performance. Various spatial filters were applied to high-density EMG signals and a reduced set of EMG signals recorded from TMR muscles. The classification performance of these spatial filters was compared, with the aim of exploring an efficient and practical spatial filtering strategy for further improvement of multifunctional myoelectric prosthesis control following TMR.
METHODS
Participants with TMR Surgeries
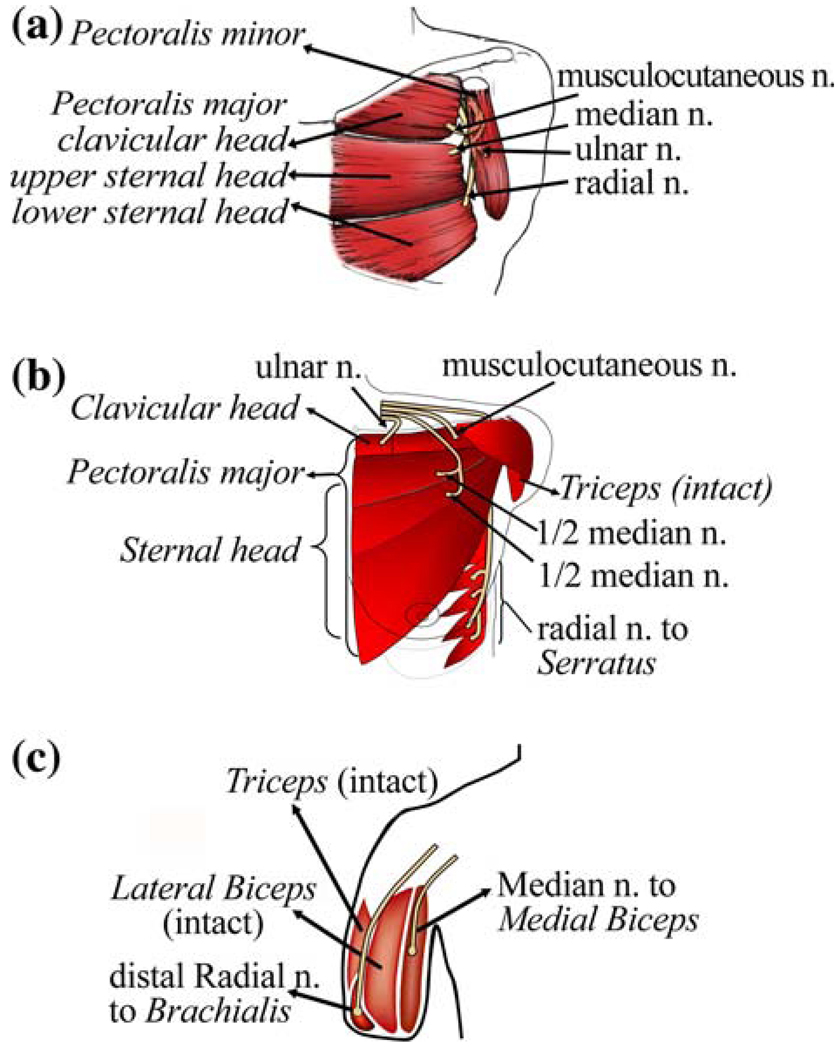
The procedures in this study were approved by the Northwestern University Institutional Review Board (IRB), and written informed consent was obtained from all subjects. Three subjects with successful TMR surgeries were recruited. The first subject was a 54-year-old man with bilateral shoulder disarticulation amputations (subject SD). Four brachial plexus nerve transfers had been performed on his left side.10,16 The musculocutaneous, median, and radial nerves had been transferred to the upper, middle, and lower regions of the pectoralis major muscle, respectively (Fig. 1a). The ulnar nerve had been transferred to the pectoralis minor muscle, which was moved out to the lateral chest wall. TMR had also been performed on a 23-year-old woman with a very short transhumeral amputation (subject STH) (Fig. 1b).17 The ulnar nerve had been transferred to the medial region of the upper pectoralis muscle, the musculocutaneous nerve had been transferred to the lateral region of the upper pectoralis muscle, the median nerve had been transferred to the middle and lower pectoralis muscles, and the distal radial nerve had been transferred to the serratus anterior muscle. The third TMR subject was a 42-year-old male with a long transhumeral amputation (subject LTH) (Fig. 1c). The median and distal radial nerves had been transferred to the medial biceps and the brachialis muscles, respectively. The residual biceps and triceps muscles were intact and could be used to control elbow motion.
FIGURE 1.
Schematic description of targeted muscle reinnervation techniques on (a) a subject (SD) with a shoulder disarticulation, (b) a subject (STH) with a short transhumeral amputation, and (c) a subject (LTH) with a long transhumeral amputation.
Muscle reinnervation was observed 3–5 months after the surgery; it provided additional EMG signals for intuitive prosthesis control. For example, when subject STH attempted to close her missing hand, the efferent neural signals traveled along the median nerve and activated the lower pectoralis muscle for contraction. When she attempted to open her hand, the neural control signals propagated along the radial nerve and caused contraction of the serratus muscle. The resulting EMG signals could be used to drive prosthetic hand closing and opening.
Experimental Protocol and Data Collection
High-density surface EMG signals were recorded over reinnervated muscles and residual biceps and triceps muscles, if present, for each subject.21 EMG signals were collected using 128 13-mm-diameter monopolar electrodes with a center-to-center distance of 15 mm, and a 128-channel Refa EMG recording system (TMS International BV, Amsterdam, Netherlands). For each channel, the surface EMG signals were sampled at 2 kHz, with a hardware low-pass filter setting the −3 dB point at 500 Hz. Before electrode placement, the skin was shaved, lightly abraded, and cleaned with alcohol pads. A ground electrode was placed on the bony area of the shoulder.
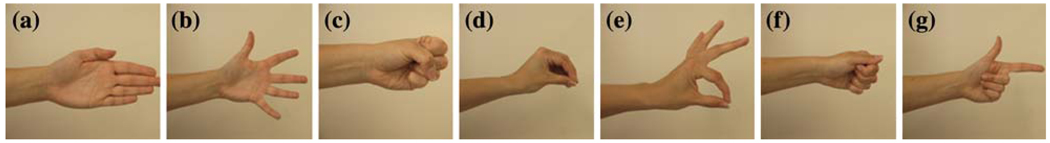
Each subject was comfortably seated and asked to follow a video demonstration and attempt 15 different movements with the amputated limb. The movements included elbow flexion/extension, wrist flexion/extension, pronation/supination, ulnar/radial deviation, two hand-opening patterns, and five functional hand-closing (grasping) patterns (Fig. 2). The two hand-opening patterns involved finger abduction and finger adduction, respectively. The five hand-closing patterns were power grip, prehensile (3-jaw chuck) grip, fine pinch grip, key grip, and trigger grip. Note that this study included hand-grasp patterns in lieu of individual finger movements because they are more functionally related to and easy-to-realize in prosthetic hand control. Each trial contained 10 repetitions of the same movement. For each repetition, the subjects were instructed to attempt the movement with a comfortable level of effort and to hold each contraction for approximately 4–5 s. There was a 5-s relaxation period between repetitions, and subjects rested between trials to avoid muscle and mental fatigue. The sequence of prompted movements was random.
FIGURE 2.
Hand movements, including two hand-open patterns: (a) hand open with finger adduction, (b) hand open with fingers abduction, and five functional hand grasping patterns: (c) power grip, (d) prehensile grip, (e) fine pinch, (f) key grip, (g) trigger grip.
EMG Analysis and Pattern Recognition
The raw EMG signals were filtered by a fifth-order Butterworth high-pass filter with a 5 Hz cutoff frequency to remove movement artifacts. Then, the data recorded during the subjects’ movement attempts were applied for EMG pattern recognition.20
EMG pattern classification was conducted within sequential 200 ms analysis windows. In each window, four time-domain features were extracted from each EMG signal. These time-domain features included (1) the mean absolute value of the EMG signal, (2) the number of zero crossings, (3) the number of slope sign changes, and (4) the waveform length4,14,21; their mathematical definitions can be found elsewhere.4,14,21 A vector, f̄, composed of the four features, was obtained for each signal, and a feature vector was formulated by F̄ = [f̄1, f̄2, f̄3, …,f̄L]1×4L, where L denotes the total number of EMG signals. In this study, a linear discriminant analysis (LDA) pattern classifier was used to classify the EMG signal features. The LDA algorithm was chosen due to its simplicity, its comparable performance to other more complex algorithms, such as the artificial neural network, hidden Markov model, and Gaussian Mixed Model,4,5,9,12 and its previously demonstrated computational efficiency in real-time prosthesis control.4 In discriminant analysis, the observed data is assigned to the movement class with the highest posteriori probability (i.e. the class with maximum probability given the observed feature vector F̄).3,13 The first half of the EMG data was used to train the LDA classifier by estimating the mean and covariance matrices of the posteriori probability for each movement class. The second half of the EMG data was used as a test set to evaluate the classifier’s performance.13
The overall classification accuracy3 (CA) (Eq. 1) was used to measure the performance of the classifier:
| (1) |
In addition, the confusion matrix (C), listing the test results between the targeted classes and the estimated task classes, was computed as
| (2) |
where the element
| (3) |
The diagonal of the confusion matrix, aii, is the classification accuracy of task class i. Other elements indicate the misclassification of EMG patterns.
Spatial Filters Applied to the High-Density EMG Signals
Classification was also performed on the surface EMG signals processed by various high-pass spatial filters (Table 1). The filters, listed in order of increasing spatial localization, included one-dimensional (TSD, LSD, BipD, BipRD, BipT, BipL, DDT, and DDL) and two-dimensional (LapD, IR, and IB2) filters. Each spatial filter corresponded to an electrode configuration (Table 1) comprised of several neighboring monopolar electrodes with different weights. These weight values can also be presented as a matrix (S) as in Table 1.
TABLE 1.
Applied spatial filters.
| Name | Description | Electrode configuration | Matrix (S) | |||
|---|---|---|---|---|---|---|
| TSD | Single differential filter in transverse direction with large inter-electrode distance (i.e. center to center inter-electrode distance of 30 mm) | 1• | •−1 |
|
||
| LSD | Single differential filter in longitudinal direction with large inter-electrode distance (i.e. center to center inter-electrode distance of 30 mm) | 1• |
|
|||
| −1• | ||||||
| BipD | Single differential filter in diagonal direction (i.e. center to center in ter-electrode distance of around 21 mm) | 1• |
|
|||
| −1• | ||||||
| BipRD | Single differential filter in reverse diagonal direction (i.e. center to center inter-electrode distance of around 21 mm) | 1• |
|
|||
| −1• | ||||||
| BipT | Single differential filter in transverse direction (i.e. center to center inter-electrode distance of 15 mm) | 1• | •−1 |
|
||
| BipL | Single differential filter in longitudinal direction (i.e. center to center inter-electrode distance of 15 mm) | 1• |
|
|||
| −1• | ||||||
| DDT | Double differential filter in transverse direction | −1• | 2• | •−1 |
|
|
| DDL | Double differential filter in longitudinal direction | −1• |
|
|||
| 2• | ||||||
| −1• | ||||||
| LapD | 5-point Laplace filter | −1• |
|
|||
| −1• | 4• | •−1 | ||||
| −1• | ||||||
| IR | Inverse rectangle filter | −1• | −1• | •−1 |
|
|
| −1• | 8• | •−1 | ||||
| −1• | −1• | •−1 | ||||
| IB2 | Inverse binomial filter of order 2 | −1• | −2• | •−1 |
|
|
| −2• | 12• | •−2 | ||||
| −1• | −2• | •−1 | ||||
The spatially filtered signal was obtained by weighted summation of neighboring monopole recordings. The filtering procedure applied to the high-density EMG signals was equivalent to the two-dimensional (2D) convolution for each sample time. The high-density EMG electrode array was first organized into a 2D electrode map based on the experimental setup. At each sample time tj, a unfiltered, monoploar EMG map (a 2D image I[n1, n2, tj] as shown in Fig. 3b) with size of N1 × N2 was obtained; the value of the pixel at location [na, nb] was the MN-EMG signal reading from the corresponding monopolar electrode in the high-density EMG array at time sample tj. Next, the spatial filter matrix (S in Table 1) of size M1 × M2 was applied to the MN-EMG map as (4)
| (4) |
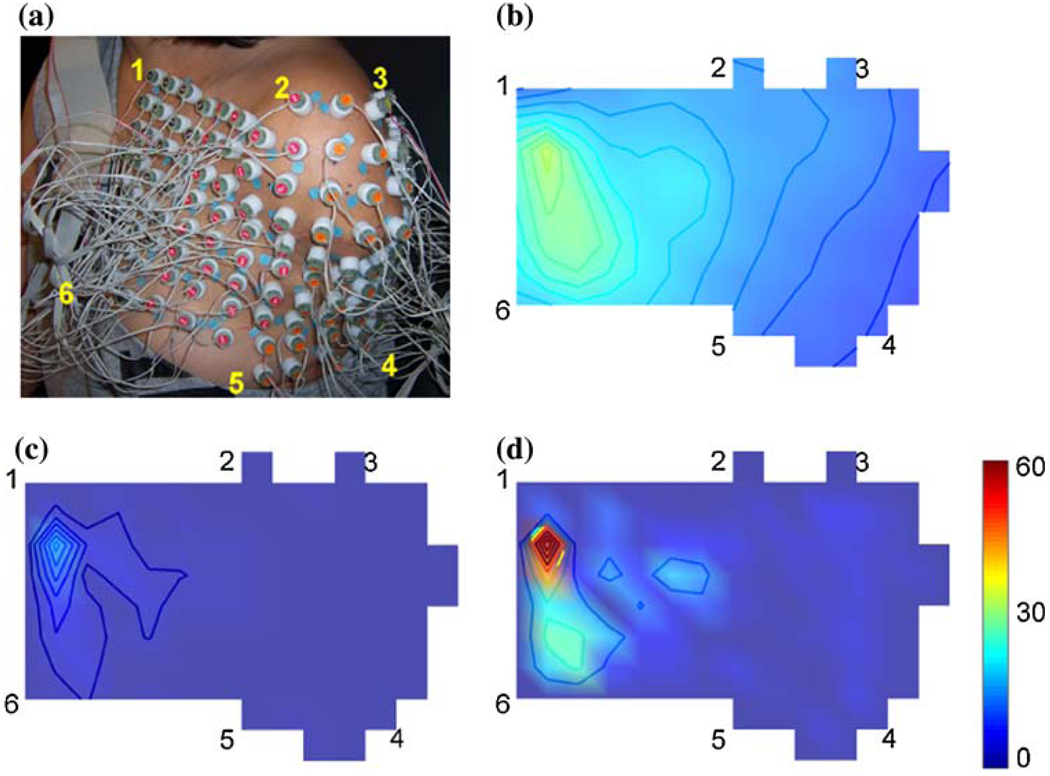
FIGURE 3.
(a) HD EMG electrode placement on the STH subject; (b) map of the EMG root mean square (RMS) using unfiltered monopolar EMG recordings when the subject performed “fine pinch”; (c) map of the RMS of BipT-filtered EMG recordings; (d) map of the RMS of IB2-filtered EMG recordings.
The double asterisks (**) denote 2D convolution. Then, the spatially filtered EMG signal at location [na, nb] of the high-density EMG array was reacquired by reading the associated pixel values from the sequence of output images {Ic[na, nb, t1], Ic[na, nb, t2], … Ic[na, nb, tj], … Ic[na, nb, tT]} along the time domain. Note that the edge electrodes (pixels) in the high-density electrode array (map) were convoluted with zero padding, and were therefore eliminated. Finally, the pattern recognition algorithm described in the previous section was applied to the spatially filtered EMG signals. The classification performances derived from different spatially filtered EMG signals were calculated.
Spatial Filters Applied to a Reduced Number of EMG Signals
The effect of spatial filtering on the classification performance was also investigated using a clinically feasible number of EMG signals. Based on the number of electrodes that could reasonably be embedded in a prosthetic socket, and the signal processing capabilities of embedded processors, no more than 12 EMG signals were considered practical in this study.13 Two methods were used to reduce the number of spatially filtered EMG signals based on our previous study13: the sequential forward searching (SFS) method and clinical selection. The SFS algorithm is an iterative searching procedure that selects the subset of EMG signals that provides the highest classification accuracy. However, the SFS algorithm may result in different EMG channels for different spatial filters and is not practical for clinical use.13 In clinical selection, the subset of electrodes is chosen based on knowledge of the TMR surgery scheme and outcome. The locations of clinically selected EMG recordings were the same for all spatial filters. After fixing the number of EMG signals, we then evaluated classification accuracy.
RESULTS
Spatial Filters Applied to the High-Density EMG Signals
The spatial filters resulted in localized recordings of muscle activity (Fig. 3). For example, clear muscle activity was observed in the middle of the pectoralis muscle reinnervated by the median nerve when subject STH performed fine pinch grip (Fig. 3). Compared to the unfiltered EMG (NM-EMG) map (Fig. 3b), the BipT-EMG map (Fig. 3c) provided greater spatial resolution in the horizontal direction. The IB2-EMG map (Fig. 3d) showed highly localized activation spots; it recorded more spatially selective muscle activity than the unfiltered EMG and BipT-EMG.
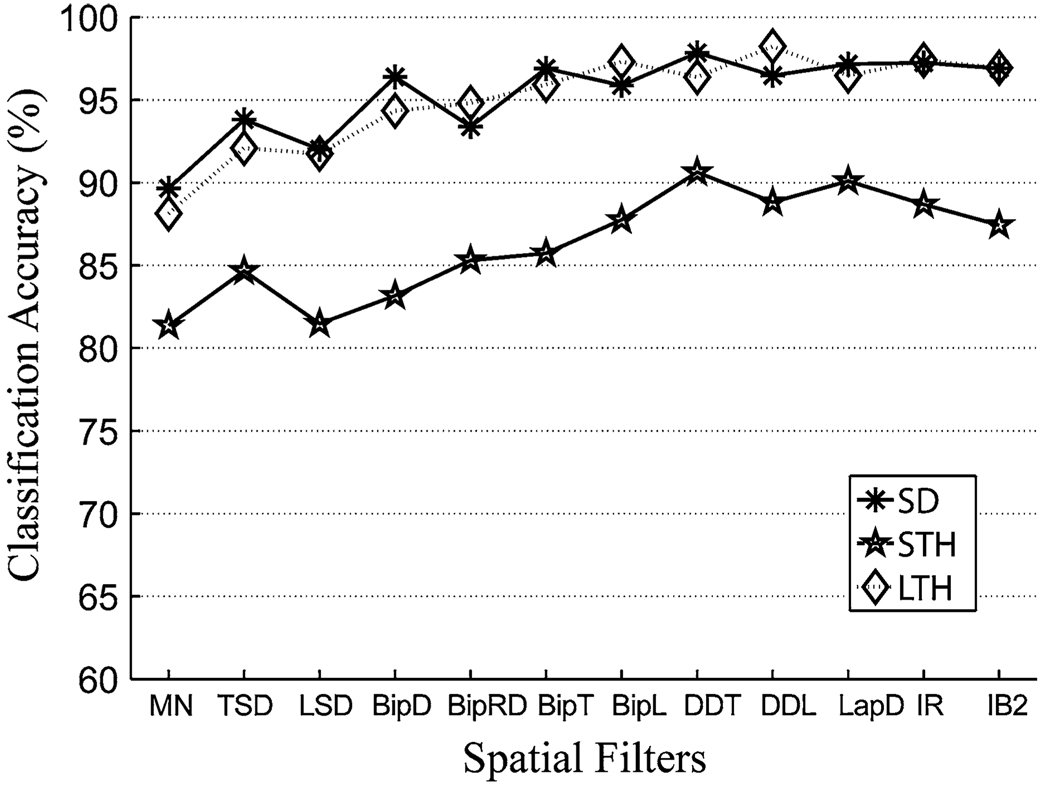
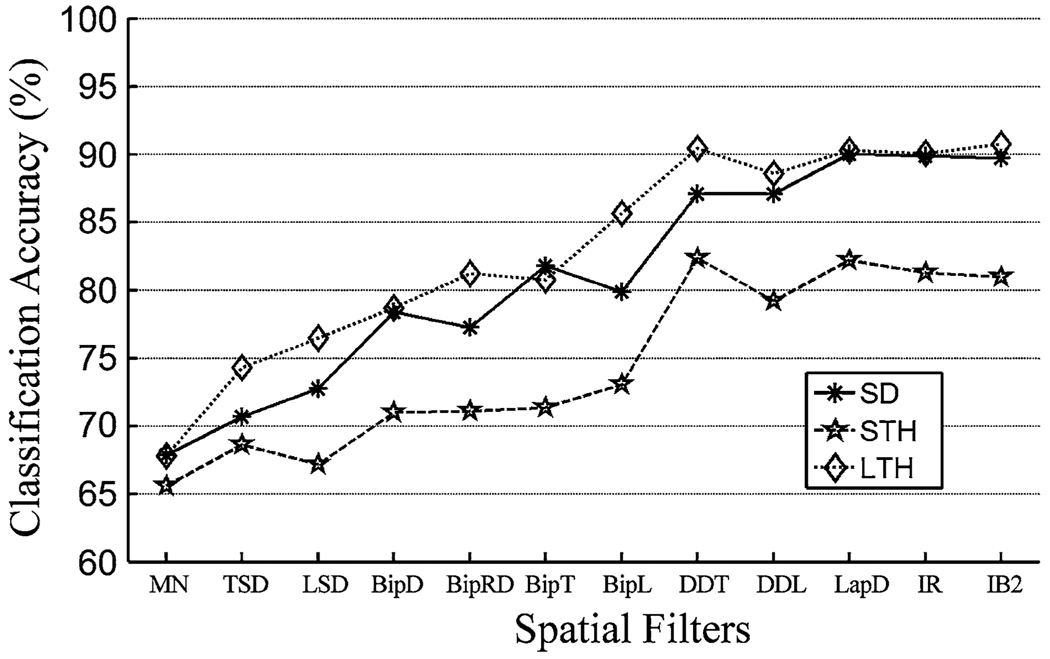
The spatially filtered EMG signals led to more accurate classification than the unfiltered EMG recordings (Fig. 4). In addition, classification accuracy as a result of using single differential filters with short inter-pole distances (15 mm for BipT, and BipL), double differential filters (DDL and DDT), and 2D filters (LapD, IR, and IB2) were comparable; their variation was within 3% for subjects SD and LTH and within 5% for subject STH.
FIGURE 4.
The influence of spatial filtering on the classification accuracy based on high-density EMG recordings.
Spatial Filters Applied to a Reduced Number of EMG Channels
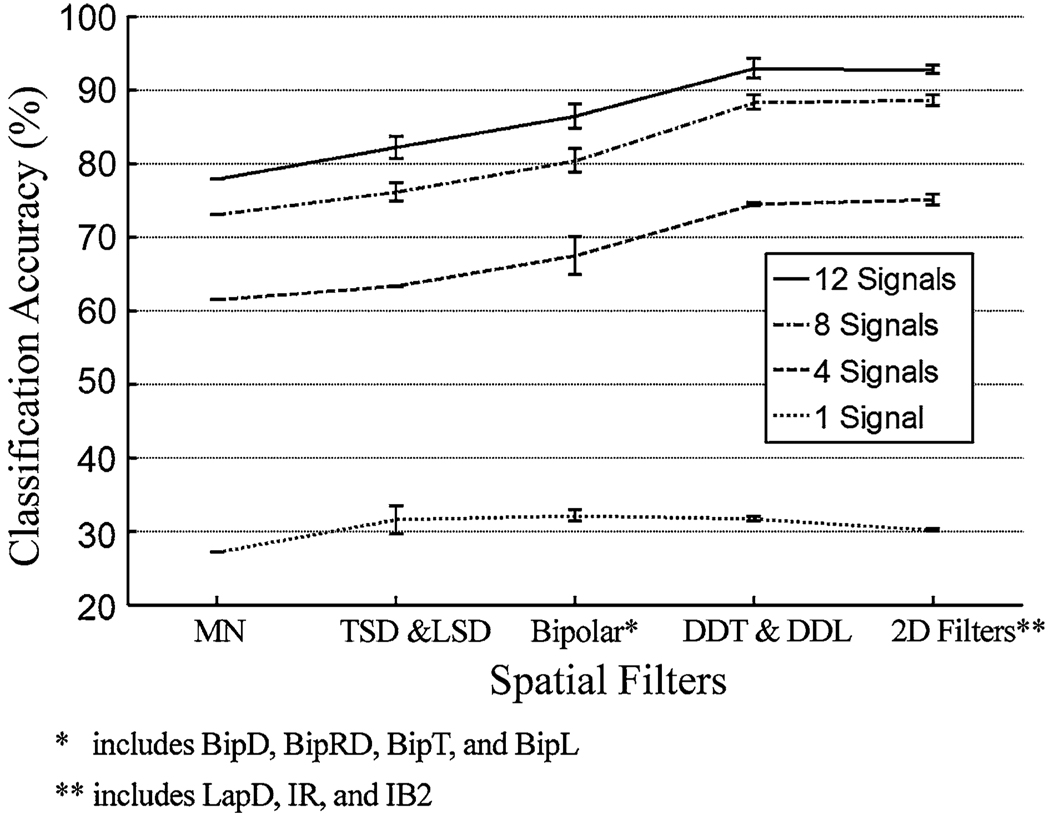
Using only one EMG signal gave low classification accuracy, regardless of the filter used. Increasing the number of applied signals to 8 or greater dramatically improved the accuracy, especially for the spatially localized EMG recordings (see Fig. 5 for an example). The accuracies derived from 12 SFS-selected double differential or 2D spatially filtered EMG signals were similar to those derived from the high-density signals. In addition, the double differential filters and the 2D spatial filters outperformed other studied filters; they improved the classification accuracy up to 9.6% over that derived from the clinically relevant single differential filters (BipD, BipRD. BipT, and BipL). The classification performances of DDT, DDL, and 2Dfilters were similar.
FIGURE 5.
The influence of spatial filtering on the classification accuracy based on differing numbers of EMG signals. Data shown are from subject SD. The spatial filters were grouped based on their spatial resolution. The bar indicates the range of classification accuracy derived from a group of spatial filters.
Fixing the number of SFS-selected EMG signals at 12, we compared the confusion matrices derived from the TSD-EMG and from the DDT-EMG in Table 2. For the TSD-EMG, most of the classification errors occurred between the hand patterns; the accuracy for classifying the key grip, trigger grip, and hand open with fingers abduction was only 60–65% (Table 2). The EMG signals with higher spatial resolutions such as DDT greatly improved the accuracy of identifying the user’s intent to perform hand patterns. With the double differential spatial filter the classification accuracy for each movement tested (see the shaded diagonal cells of Table 2) ranged from 85 to 100%.
TABLE 2.
The confusion matrix derived from using 12 SFS-selected TSD-EMG signals (the matrix elements are in the un-shaded
| Estimated Tasks | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Elbow flexion | Elbow extension | Wrist flexion | Wrist extension | Supination | Pronation | Ulnar deviation | Radial deviation | Hand open 1* | Hand open 2** | Power grip | Prehensile grip | Fine pinch | Key grip | Trigger grip | ||
| Targeted Tasks | Elbow flexion | 97.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.2 | 1.2 | 0 | 0 | 0 |
| 97.5 | 0 | 2.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Elbow extension | 0 | 0 | 89.4 | 0 | 8.0 | 0.8 | 0 | 0 | 0 | 0 | 1.8 | 0 | 0 | 0 | 0 | |
| 0 | 0 | 92.0 | 0 | 1.8 | 3.5 | 0 | 0 | 0 | 0 | 2.7 | 0 | 0 | 0 | 0 | ||
| Wrist extension | 0 | 0 | 0 | 97.6 | 0 | 0 | 2.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Supination | 0 | 0 | 1.6 | 0 | 96.8 | 1.6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Pronation | 0 | 0 | 1.2 | 0 | 1.2 | 92.7 | 0 | 0 | 3.7 | 0 | 0 | 0 | 0 | 0 | 1.2 | |
| 0 | 0 | 0 | 1.2 | 1.2 | 92.7 | 0 | 0 | 4.9 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Ulnar deviation | 0 | 6.0 | 1.5 | 0 | 1.5 | 0 | 91.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 0 | 0 | 0 | 0 | 0 | 0 | 93.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.8 | ||
| Radial deviation | 0 | 0 | 0 | 2.7 | 0 | 1.4 | 0 | 79.4 | 0 | 15.1 | 0 | 0 | 0 | 0 | 1.4 | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 87.5 | 0 | 0 | 2.1 | 10.4 | 0 | 0 | 0 | ||
| Hand open 1* | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 84.4 | 4.4 | 0 | 0 | 0 | 0 | 1.2 | |
| 0 | 0 | 0 | 0 | 0 | 5.6 | 0 | 0 | 94.4 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Hand open 2** | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.6 | 19.7 | 60.6 | 0 | 0 | 0 | 0 | 11.1 | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | ||
| Power grip | 0 | 0 | 4.8 | 0 | 0 | 2.4 | 0 | 0 | 0 | 0 | 86.8 | 1.2 | 1.2 | 3.6 | 0 | |
| 0 | 0 | 1.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 96.4 | 2.4 | 0 | 0 | 0 | ||
| Prehensile grip | 0 | 0 | 3.5 | 0 | 0 | 3.5 | 0 | 1.1 | 0 | 0 | 0 | 72.9 | 5.9 | 8.3 | 4.8 | |
| 0 | 0 | 0 | 0 | 1.2 | 0 | 0 | 0 | 0 | 0 | 0 | 95.3 | 3.5 | 0 | 0 | ||
| Fine pinch | 0 | 0 | 1.6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.6 | 84.3 | 12.5 | 0 | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.8 | 0 | 0 | 0 | 3.1 | 89.1 | 0 | 0 | ||
| Key grip | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.1 | 18.7 | 16.7 | 62.5 | 0 | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.4 | 0 | 84.9 | 13.7 | ||
| Trigger grip | 0 | 0 | 1.4 | 0 | 9.6 | 0 | 0 | 4.1 | 9.6 | 23.3 | 0 | 0 | 0 | 0 | 52.0 | |
| 0 | 0 | 0 | 0 | 0 | 0 | 6.2 | 0 | 1.2 | 0 | 0 | 0 | 0 | 2.5 | 90.1 | ||
Hand open 1 is the hand open movement with fingers adduction.
Hand open 2 is the hand open motion movement with fingers abduction.
Using 12 clinically selected EMG signals, the double differential filters and the 2D filters outperformed the other types of filters for all three subjects (Fig. 6). The EMG signal locations were the same for all tested filters. In this case, the difference between the highest and lowest accuracies was 20%, averaged over 3 subjects.
FIGURE 6.
The influence of spatial filtering on the classification accuracy. Twelve clinically selected EMG signals were used.
DISCUSSION
In the present study, we utilized various spatial filters to enhance the spatial selectivity of EMG recordings and studied the effect of spatial filtering on the performance of EMG pattern classification. When spatial filters were applied to high-density EMG recordings from TMR muscles, classification accuracy was enhanced. When BipT, BipL, or higher-order filters were applied, the classification accuracy for 15 movements was above 95% for patients SD and LTH, and above 85% for patient STH. The results demonstrate once again that tremendous neural control information was successfully transferred to TMR muscles. Although the high-density monopolar EMG array should have been able to record most of the spatial information in the surface EMG, the monopolar, TSD, LSD, BipRD, and BipD filters produced lower classification accuracies than other high-order spatial filters. This may be because the high-density EMG signals with lower spatial resolution contained somewhat larger common-mode signal components, which reduced the classification performance.
When the number of EMG signals was reduced, the type of spatial filter used considerably influenced the classification accuracy: localized EMG recordings provided higher accuracy than EMG signals with low spatial resolutions. This outcome is inconsistent with the results of previous reports.7,9 Previous studies compared localized EMG recordings (targeted, intramuscular EMG) to EMG signals with global information (untargeted, surface bipolar EMG) and reported that the two measures provided comparable classification accuracies for identifying user intent.7,9 They concluded that the EMG PR algorithm can extract information from multi-channel EMG signals, regardless of whether recordings are localized or global. This may be true for normally innervated muscles in able-bodied subjects or subjects with transradial amputations, but does not appear to be true for reinnervated muscles. Because of the high information density of EMG signals from TMR muscles, especially those reinnervated by the median and radial nerves, wrist and hand movements were heavily misclassified when EMG signals with lower spatial resolutions were used (Table 2); in contrast, spatially localized EMG signals gave much more accurate classifications of user intent, especially for hand movements. In addition, 12 EMG signals can extract a similar amount of neural control information as highdensity EMG arrays. Thus, the EMG recording system that provides greater spatial resolution has the potential to further improve the TMR-based neural control interface for robust prosthesis control.
Although 2D spatial filtering provides high spatial resolution for EMG recordings and improves classification accuracy, the design and construction of such a system are too complicated to be practical. For example, one EMG electrode with inverse rectangle (IR) filtering required 9 monopoles and at least 8 amplifiers. Twelve EMG recordings would therefore need as many as 108 monopoles to be mounted on the socket and 96 amplifiers in the EMG recording system. Hence, a trade-off is necessary to design a practical EMG recording system with high spatial selectivity for reliable classification.
A double differential EMG system may offer an acceptable trade-off of increased accuracy with a modest increase in electrode pole and amplifier number. The double differential filters (DDL and DDT) provided better classification performance than the most clinically-relevant single differential filters (LSD, TSD, BipT, BipL, BipD, and BipRD) (Fig. 5), while the double differential filters produced equivalent classification performances to the filters with higher orders and dimensions (LapD, IR, and IB2). Although one double differential EMG electrode requires two pre-amplifiers and three contacts, as opposed to the one amplifier and two contacts required for a bipolar EMG electrode, the complexity of design and realization of a double differential recording system is moderate; in fact, the double differential EMG recording system is commercially available (e.g. Delsys Inc, Boston, MA). In addition, compared to single differential signals, fewer double differential EMG signals were required to achieve a comparable classification performance. For example, in order to produce 85% classification accuracy for 15 movements for subject SD (Fig. 5), 12 bipolar EMG signals (24 monopoles) were necessary, while only 8 double differential EMG signals (24 monopoles) were required. One of the direct benefits of using fewer EMG inputs is the improved computational efficiency. For 12 inputs, the feature space dimension is 48 (four time domain features per channel), but the dimension for 8 inputs is only 32. Reducing the number of EMG inputs from 12 to 8, therefore, decreases the covariance matrix in the LDA algorithm from 48 × 48 to 32 × 32, which reduces the computational burden on real-time EMG pattern recognition. Furthermore, the locations of double differential electrodes can be selected based on clinical knowledge of TMR surgery, as described by Huang et al.13 When using 12 clinically selected double differential signals, the classification accuracies for the three subjects ranged from 80 to 90%s. Therefore, the double differential EMG recording system is both technically and clinically viable, and could be used to further improve the accuracy of multifunctional prosthesis control.
A limitation of this study was the availability of only three TMR subjects for testing. This was because TMR is a novel and unique procedure which has only been applied to select patients with transhumeral or higher-level amputations. However, the consistent results among the tested subjects provide insight into further improving the accuracy of EMG PR-based control by using double differential EMG recordings. Continuing efforts will include the design of prosthetic sockets fit with the double differential EMG recording interface, and evaluation of the reliability of this interface for real-time EMG PR-based prosthesis control on amputee patients with TMR surgery.
CONCLUSION
Spatially selective EMG recording can measure more separable neural control information from TMR muscles because of the high information density of these muscles. By investigating the influence of various spatial filters on the performance of EMG pattern classification, we found that double differential EMG recordings provided higher classification accuracy than conventional bipolar EMG signals and were more clinically practical than 2D spatial filters. Therefore, a TMR-based neural-machine interface may be further improved by using double differential EMG electrodes for reliable multifunctional prosthetic arm control.
ACKNOWLEDGMENTS
The EMG signal classification code used in this study was provided by Professor Kevin B. Englehart, PhD, PE, of the Institute of Biomedical Engineering, University of New Brunswick, Canada. We thank Aimee Schultz, M.S. for editing the manuscript. This work was supported by the National Institute on Disability and Rehabilitation Research (Grant # H133F060029 & H133F080006), the NIH National Institute of Child and Human Development (Grants # R01 HD043137-01, #R01 HD044798, and # NO1-HD-5-3402), and the Defense Advanced Research Projects.
REFERENCES
- 1.Basmajian JV, De Luca CJ. Muscles Alive : Their Functions Revealed by Electromyography. Vol. 561. Baltimore: Williams & Wilkins; 1985. p. xii. [Google Scholar]
- 2.Disselhorst-Klug C, Silny J, Rau G. Improvement of spatial resolution in surface-EMG: a theoretical and experimental comparison of different spatial filters. IEEE Trans. Biomed. Eng. 1997;44:567–574. doi: 10.1109/10.594897. [DOI] [PubMed] [Google Scholar]
- 3.Duda RO, Hart PE, Stork DG. Pattern Classification. Vol. 654. New York: Wiley; 2001. p. xx. [Google Scholar]
- 4.Englehart K, Hudgins B. A robust, real-time control scheme for multifunction myoelectric control. IEEE Trans. Biomed. Eng. 2003;50:848–854. doi: 10.1109/TBME.2003.813539. [DOI] [PubMed] [Google Scholar]
- 5.Englehart K, Hudgins B, Parker PA, Stevenson M. Classification of the myoelectric signal using time-frequency based representations. Med. Eng. Phys. 1999;21:431–438. doi: 10.1016/s1350-4533(99)00066-1. [DOI] [PubMed] [Google Scholar]
- 6.Farina D, Cescon C. Concentric-ring electrode systems for noninvasive detection of single motor unit activity. IEEE Trans. Biomed. Eng. 2001;48:1326–1334. doi: 10.1109/10.959328. [DOI] [PubMed] [Google Scholar]
- 7.Farrell TR, Weir RF. A comparison of the effects of electrode implantation and targeting on pattern classification accuracy for prosthesis control. IEEE Trans. Biomed. Eng. 2008;55:2198–2211. doi: 10.1109/TBME.2008.923917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graupe D, Salahi J, Kohn KH. Multifunctional prosthesis and orthosis control via microcomputer identification of temporal pattern differences in single-site myoelectric signals. J. Biomed. Eng. 1982;4:17–22. doi: 10.1016/0141-5425(82)90021-8. [DOI] [PubMed] [Google Scholar]
- 9.Hargrove LJ, Englehart K, Hudgins B. A comparison of surface and intramuscular myoelectric signal classification. IEEE Trans. Biomed. Eng. 2007;54:847–853. doi: 10.1109/TBME.2006.889192. [DOI] [PubMed] [Google Scholar]
- 10.Hijjawi JB, Kuiken TA, Lipschutz RD, Miller LA, Stubblefield KA, Dumanian GA. Improved myoelectric prosthesis control accomplished using multiple nerve transfers. Plast. Reconstr. Surg. 2006;118:1573–1578. doi: 10.1097/01.prs.0000242487.62487.fb. [DOI] [PubMed] [Google Scholar]
- 11.Hoffer JA, Loeb GE. Implantable electrical and mechanical interfaces with nerve and muscle. Ann. Biomed. Eng. 1980;8:351–360. doi: 10.1007/BF02363438. [DOI] [PubMed] [Google Scholar]
- 12.Huang H, Kuiken T, Lipschutz RD. A strategy for identifying locomotion modes using surface electromyography. IEEE Trans. Biomed. Eng. 2009;56:65–73. doi: 10.1109/TBME.2008.2003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H, Zhou P, Li G, Kuiken TA. An analysis of EMG electrode configuration for targeted muscle reinnervation based neural machine interface. IEEE Trans. Neural Syst. Rehabil. Eng. 2008;16:37–45. doi: 10.1109/TNSRE.2007.910282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hudgins B, Parker P, Scott RN. A new strategy for multifunction myoelectric control. IEEE Trans. Biomed. Eng. 1993;40:82–94. doi: 10.1109/10.204774. [DOI] [PubMed] [Google Scholar]
- 15.Kuiken TA, Childress DS, Rymer WZ. The hyper-reinnervation of rat skeletal muscle. Brain Res. 1995;676:113–123. doi: 10.1016/0006-8993(95)00102-v. [DOI] [PubMed] [Google Scholar]
- 16.Kuiken TA, Dumanian GA, Lipschutz RD, Miller LA, Stubblefield KA. The use of targeted muscle reinnervation for improved myoelectric prosthesis control in a bilateral shoulder disarticulation amputee. Prosthet. Orthot. Int. 2004;28:245–253. doi: 10.3109/03093640409167756. [DOI] [PubMed] [Google Scholar]
- 17.Kuiken TA, Miller LA, Lipschutz RD, Lock BA, Stubblefield K, Marasco PD, Zhou P, Dumanian GA. Targeted reinnervation for enhanced prosthetic arm function in a woman with a proximal amputation: a case study. Lancet. 2007;369:371–380. doi: 10.1016/S0140-6736(07)60193-7. [DOI] [PubMed] [Google Scholar]
- 18.Parker P, Englehart K, Hudgins B. Myoelectric signal processing for control of powered limb prostheses. J. Electromyogr. Kinesiol. 2006;16:541–548. doi: 10.1016/j.jelekin.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 19.van Vugt JP, van Dijk JG. A convenient method to reduce crosstalk in surface EMG. Cobb Award-winning article, 2001. Clin. Neurophysiol. 2001;112:583–592. doi: 10.1016/s1388-2457(01)00482-5. [DOI] [PubMed] [Google Scholar]
- 20.Williams TW., 3rd Practical methods for controlling powered upper-extremity prostheses. Assist. Technol. 1990;2:3–18. doi: 10.1080/10400435.1990.10132142. [DOI] [PubMed] [Google Scholar]
- 21.Zhou P, Lowery MM, Englehart KB, Huang H, Li G, Hargrove L, Dewald JP, Kuiken TA. Decoding a new neural machine interface for control of artificial limbs. J. Neurophysiol. 2007;98:2974–2982. doi: 10.1152/jn.00178.2007. [DOI] [PubMed] [Google Scholar]