This work shows that auxin binds to the cell cycle F-box SKP2A to regulate its proteolysis and DPPB and E2FC degradation. Auxin seems to regulate the interaction between SKP2A and DPB. Mutant SKP2A proteins that do not bind auxin are unable to interact with DPB and to promote cell division. These findings provide evidence that SKP2A directly connects auxin and cell division.
Abstract
Arabidopsis thaliana S-Phase Kinase-Associated Protein 2A (SKP2A) is an F-box protein that regulates the proteolysis of cell cycle transcription factors. The plant hormone auxin regulates multiple aspects of plant growth and development, including cell division. We found that auxin induces the ubiquitin-dependent degradation of SKP2A both in vivo and in vitro, suggesting that this hormone acts as a signal to trigger SKP2A proteolysis. In this article, we show that auxin binds directly and specifically to SKP2A. By TIR1-based superposition and docking analyzes, we identified an auxin binding site in SKP2A. Mutations in this binding site reduce the ability of SKP2A to bind to auxin and generate nondegradable SKP2A forms. In addition, these non-auxin binding proteins are unable to promote E2FC/DPB degradation in vivo or to induce cell division in the root meristem. Auxin binds to TIR1 to promote its interaction with the auxin/indole-3-acetic acid target proteins. Here, we show that auxin also enhanced the interaction between SKP2A and DPB. Finally, a mutation in SKP2A leads to auxin-resistant root growth, an effect that is additive with the tir1-1 phenotype. Thus, our data indicate that SKP2A is an auxin binding protein that connects auxin signaling with cell division.
INTRODUCTION
In multicellular organisms, cell proliferation must be coordinated with cell differentiation to achieve proper growth and development. Plants have evolved toward a postembryonic growth and development, requiring a continuous balance between cell proliferation and differentiation (Gutierrez et al., 2002). Genomic analyses of several plants species have shown that the majority of the cell cycle core genes have been conserved (Vandepoele et al., 2002). However, to assure correct integration of the cell division process with growth and development, plants have acquired specific regulatory pathways that allow them to grow efficiently. It has been well established that hormone signaling pathways play a key role in these integrations. Among the different plant hormones, auxin is one of the most important ones, since it controls nearly every aspect of plant development, including cell division (Davies, 1995; Leyser, 2002; Benjamins and Scheres, 2008; Mockaitis and Estelle, 2008). Auxin response is controlled by two large families of transcription factors, auxin/indole-3-acetic acid (Aux/IAA) and auxin response factors (ARFs). Auxin induces gene transcription by promoting the degradation of the Aux/IAA repressors and thereby allowing ARFs to activate the transcription of auxin-responsive genes (Gray et al., 2001; Zenser et al., 2001). Recent studies have clearly shown that auxin stimulates the interaction between Aux/IAA proteins and the SCFTIR1 complex (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005) or its homologous SCFAFBs (Dharmasiri et al., 2005b). Auxin binds to TIR1 to fill a hydrophobic gap stabilizing the interaction between this F-box and the Aux/IAA targets (Tan et al., 2007), indicating that TIR1 acts as an auxin receptor. Although it is clear that transcriptional and developmental responses to auxin depend on degradation of Aux/IAA proteins, the molecular events that connect auxin perception with cell division are poorly understood.
Control of cell division is ensured through a number of regulatory mechanisms, including phosphorylation and ubiquitin-dependent degradation of key regulatory proteins (Hershko, 2005; Nakayama and Nakayama, 2006; Johnson, 2009). The selective degradation of proteins by the ubiquitin-proteasome system (UPS) is an irreversible and highly precise mechanism that ensures the correct transition between cell cycle phases (Hershko, 2005; Nakayama, and Nakayama, 2006). In mammals, the SCFSkp2, an E3 ubiquitin ligase complex, plays a central role in controlling the stability of several cell cycle regulators (Frescas and Pagano, 2008). However, in the majority of cases, the molecular mechanisms that trigger the degradation of these targets in a cell type–specific manner or in a given window of time are still unknown. In Arabidopsis thaliana, SKP2A and SKP2B proteins, which contain an F-box and leucine-rich repeat (LRR) domains, were identified based on sequence similarity to the human Skp2 (del Pozo et al., 2002). SKP2A is a cell cycle–regulated protein that controls the stability of at least two cell division transcriptional factors, E2FC and DPB (del Pozo et al., 2006; Jurado et al., 2008). On the other hand, SKP2B, but not SKP2A, orchestrates the degradation of the cyclin-dependent kinase inhibitor KRP1 (Ren et al., 2008), indicating that, despite their high similarity, both SKP2A and SKP2B have distinct target specificities. Biochemical analyses showed that SKP2A is posttranslationally regulated by modification with polyubiquitin in vivo, indicating that this protein is degraded through the UPS (Jurado et al., 2008). Several lines of evidences have shown that auxin signaling controls the degradation of SKP2A (Jurado et al., 2008). For example, auxin promoted SKP2A degradation in vivo and Terfestatin A (Yamazoe et al., 2005), a compound that blocks auxin signaling, increased SKP2A levels (Jurado et al., 2008). In addition, axr2-1 and axr3-1 mutants, which have an altered auxin response, accumulated far less SKP2A protein than control plants (Jurado et al., 2008). However, we do not understand the molecular mechanisms that connect this hormone with SKP2A proteolysis.
Here, we report that auxin promotes SKP2A degradation in a cell-free system, suggesting a direct effect on SKP2A stability. Indeed, we found that auxin binds directly and specifically to SKP2A, but not to its closest homolog SKP2B. Using computational analyses, we have been able to identify a novel auxin binding site in SKP2A. Mutations in residues of this site reduce the ability of SKP2A to bind to auxin and generate nondegradable SKP2A forms. In addition, overexpression of this SKP2A mutant does not promote cell division in the root meristem, likely because these mutated versions are unable to promote the degradation of cell cycle repressors. Similarly to the TIR1 model, where auxin acts as molecular glue that favors the interaction between TIR1 and Aux/IAA protein, we found that auxin enhanced the interaction between SKP2A and DPB. Finally, we also found that a mutation in SKP2A confers auxin-resistant root growth, an effect that is additive to the tir1-1 mutant phenotype, indicating that SKP2A also participates in the auxin response.
RESULTS
Auxin-Dependent Degradation of SKP2A Is Not Mediated by TIR1
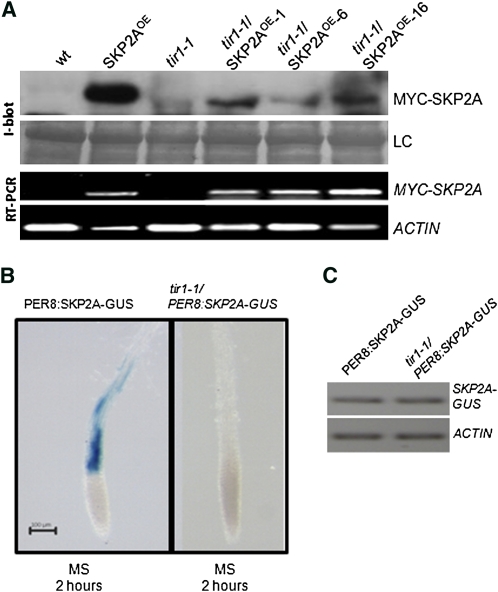
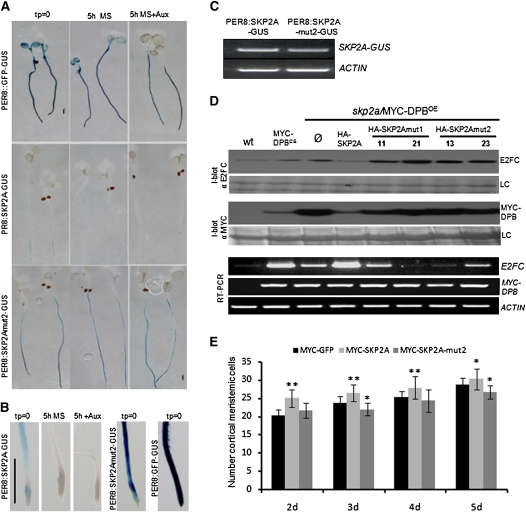
After auxin perception by TIR1, the Aux/IAA proteins are quickly recognized and labeled with ubiquitin for their degradation (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005; Maraschin et al., 2009). In a previous study, we showed that auxin triggers SKP2A proteolysis in vivo, with an estimated half-life of 45 min (Jurado et al., 2008). Reasoning that TIR1 is involved in auxin-dependent protein degradation, we wondered if it was involved in SKP2A proteolysis. To answer this, a MYC-SKP2A transgene was crossed into the tir1-1 mutant background (tir1-1/MYC-SKP2AOE). By immunoblotting, we found that the tir1-1 mutant accumulated less MYC-SKP2A protein, while expression of the MYC-SKP2A mRNA was similar (Figure 1A). These results indicate that SKP2A turnover is faster in the tir1-1 mutant than in wild-type plants and also that the SCFTIR1 complex is not the E3 involved in SKP2A degradation.
Figure 1.
SKP2A Is Degraded in Response to Auxin in a TIR1-Independent Manner.
(A) Immunoblotting analyses of the MYC-SKP2A levels in the tir1-1 mutant. Total protein from wild-type (wt), tir1-1, MYC-SKP2A–overexpressing plants (SKP2AOE), and two different lines tir1-1/SKP2AOE was extracted in the presence of MG132 and immunoblotted with anti-MYC. LC is the loading control, corresponding to the Ponceau-stained blot. The bottom panel shows the expression of the MYC-SKP2A transgene analyzed by RT-PCR. As a control, the expression of the ACTIN gene was analyzed.
(B) Five-day-old PER8:SKP2A-GUS and tir1-1/PER8:SKP2A-GUS transgenic seedlings were incubated with 10 μM estradiol in a liquid medium for 16 h to induce the expression of the chimeric protein. After induction, these seedlings were incubated in MS medium for 2 h and then stained for GUS activity. Bar = 0.2 mm.
(C) Expression of the SKP2A-GUS transgene analyzed by RT-PCR using primers from SKP2A and GUS coding regions in 5-d-old PER8:SKP2A-GUS and tir1-1/PER8:SKP2A-GUS transgenic seedlings that were incubated with 10 μM estradiol in a liquid medium for 16 h. As a control, the expression of the ACTIN gene was analyzed.
[See online article for color version of this figure.]
To analyze this degradation in planta, we fused SKP2A to the β-glucuronidase (GUS) reporter protein and expressed this chimeric protein in Arabidopsis plants under the control of an inducible promoter (PER8:SKP2A-GUS). To verify that SKP2A-GUS behaves like MYC-SKP2A protein, after the induction of SKP2A-GUS with estradiol, seedlings were treated with the proteasome inhibitor MG132 or terfestatin A (TerfA), a compound that blocks auxin signaling (Yamazoe et al., 2005) and leads to a high accumulation of MYC-SKP2A (Jurado et al., 2008). In both cases, SKP2A-GUS protein accumulated to higher levels than in the nontreated seedlings, indicating that this SKP2A-GUS is degraded through the UPS and that auxin signaling control such proteolysis (see Supplemental Figure 1 online). Then, SKP2A-GUS was introduced into the tir1-1 mutant by genetic crossing (tir1/PER8:SKP2A-GUS). Five-day-old seedlings were induced with estradiol for 16 h and then transferred for 2 h to a noninducible medium. Afterwards, these seedlings were stained for GUS activity. The SKP2A-GUS fusion protein was clearly observed in the root of wild-type plants, while it was not detected in tir1-1 roots (Figure 1B). The mRNA expression was similar in both the control and tir1-1 (Figure 1C), indicating that the tir1-1 mutation affects the stability of SKP2A in planta.
Auxin Promotes SKP2A Degradation
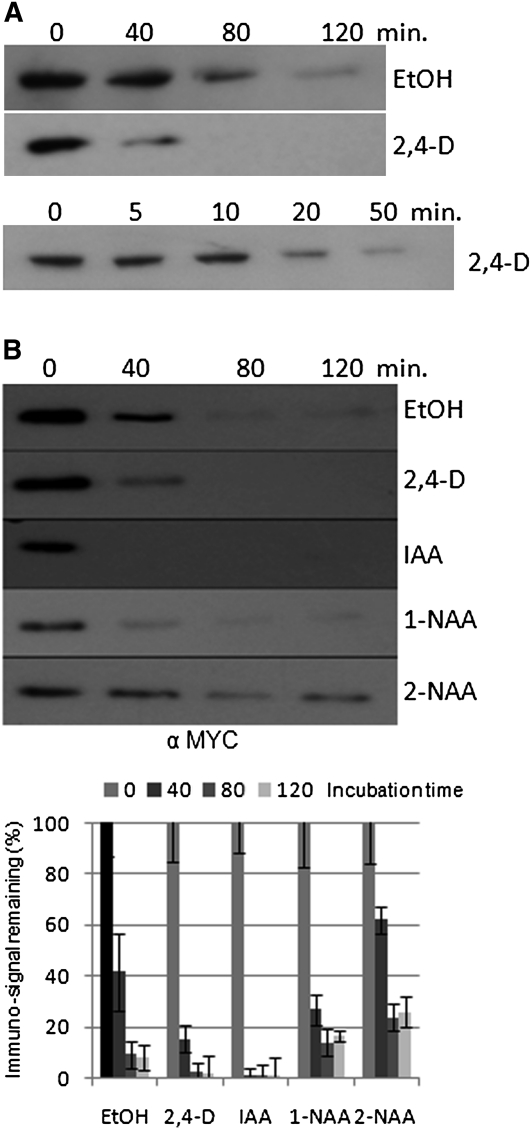
Since auxin stimulates SKP2A degradation in a TIR1-independent manner, we wanted to analyze if auxin regulates its degradation directly. To test this, we performed degradation assays in a cell-free system, in which 2,4-D (a synthetic auxin) was added. As shown in Figure 2A, without 2,4-D, the MYC-SKP2A protein is largely degraded after 80 min. However, when the reaction contained 1 μM 2,4-D, the MYC-SKP2A protein is degraded faster than in the control reaction (Figure 2A). We also tested the effect of active auxin compounds (IAA and 1-naphthaleneacetic acid [1-NAA]) or an inactive auxin-related compound (2-naphthaleneacetic acid [2-NAA]) on SKP2A degradation. We found that the natural auxin IAA promoted a faster degradation of SKP2A than 2,4-D or 1-NAA. By contrast, the inactive auxin 2-NAA, rather than stimulating SKP2A degradation, seems to delay it, suggesting that only biologically active auxin promotes the proteolysis of SKP2A (Figure 2B).
Figure 2.
Auxin Directly Promotes SKP2A Degradation.
(A) Degradation assay using crude plant protein extract from 5-d-old MYC-SKP2AOE seedlings. The protein extracts were incubated in presence of the control ethanol solvent (EtOH) or 1 μM 2,4-D. These reactions were incubated at 30°C during the indicated time and then stopped by adding loading buffer and boiling the sample for 5 min. The levels of MYC-SKP2A were analyzed by immunoblotting with anti-MYC.
(B) SKP2A is degraded in response to active auxins. Degradation assays using crude plant extract from 5-d-old MYC-SKP2AOE seedlings. The protein extracts were incubated in presence of the control ethanol solvent (EtOH) or 1 μM 2,4-D, IAA, 1-NAA, and 2-NAA. These reactions were incubated at 30°C during the indicated time points and then they were stopped by adding loading buffer and boiling the sample for 5 min. The levels of MYC-SKP2A were analyzed by immunoblotting with anti-MYC. The bands were quantified by a Bio-Rad molecular imager, relative to a 100% value at time zero. The image presented is from one representative experiment of three, and the graph is a compilation of these experiments. Each value is the mean of the three degradation assays, and the error bars correspond to the sd.
Auxin Binds Directly to SKP2A
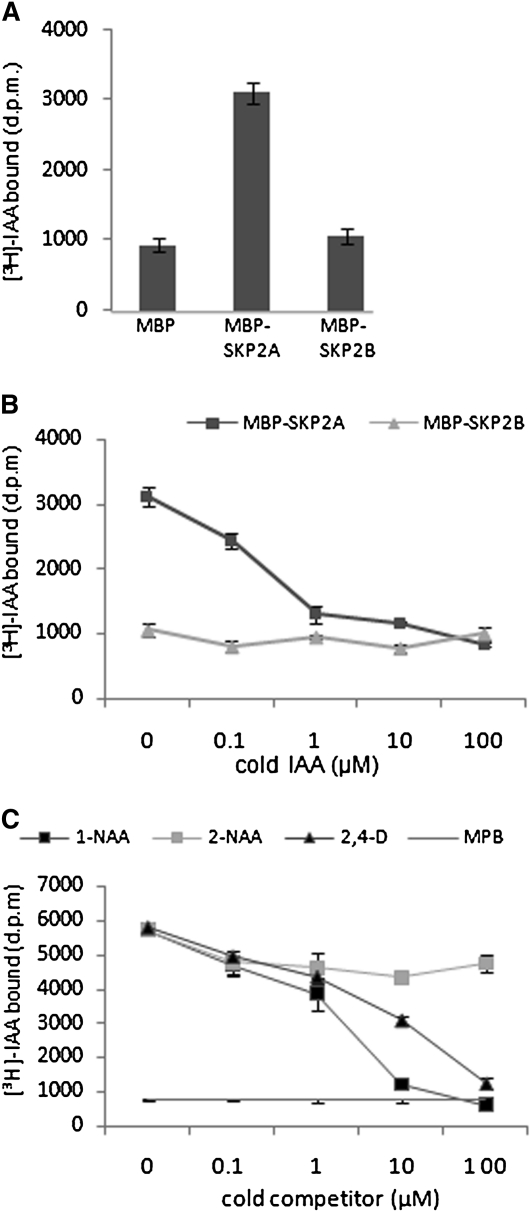
Since auxin directly stimulates SKP2A degradation in a cell-free system in a TIR1-independent manner, it could be reasonable to believe that auxin binds to SKP2A. To test this possibility, we performed pull-down assays using recombinant maltose binding protein (MBP)-SKP2A expressed in bacteria to avoid the participation of other plant proteins or endogenous auxin in binding. MBP-SKP2A protein, bound to amylose beads, was incubated with [3H]-IAA, and, after several washes, the radioactivity retained in the beads was measured by scintillation counting. As a control, we used MBP or MBP-SKP2B, which is 83% identical to SKP2A (see Supplemental Figure 2 online). As shown in Figure 3A, MBP-SKP2A retained significant amounts of [3H]-IAA compared with the amount retained by either MBP-SKP2B or MBP. To assure that this binding is specific, we performed competitive binding experiments with active auxin (IAA, 1-NAA, or 2,4-D) or with a related 2-NAA compound that is biologically inactive. We found that active auxins efficiently compete with [3H]-IAA for binding to SKP2A (Figures 3B and 3C), while the 2-NAA did not compete (Figure 3C). Scatchard analysis indicated that the estimated dissociation constant Kd of SKP2A for IAA is ~200 nM (see Supplemental Figure 3 online). The estimated median inhibitory concentration IC50 for IAA is 0.65 μM, while for 1-NAA is 2.5 μM, values that are a slightly higher than those obtained for TIR1-Aux/IAA coreceptors (Dharmasiri et al., 2005a). Taken together, our results indicate that SKP2A binds to auxin and might act as an auxin receptor, similar to the TIR1-Aux/IAA coreceptors (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005; Tan et al., 2007).
Figure 3.
IAA Binds to SKP2A, but Not to SKP2B.
(A) Pull-down reactions were performed with either MBP-SKP2A or MBP-SKP2B bound to amylase beads in the presence of 50 nM [3H]-IAA. The retained [3H]-IAA in the amylose beads after three washes was measured by scintillation counting. Each value is the mean of three independent measures, and the error bars correspond to the sd. d.p.m., disintegrations per minute.
(B) Competitive binding of [3H]-IAA to MBP-SKPA and MBP-SKP2B in the presence of increasing concentrations of unlabeled IAA. Pull-down reactions were performed in the presence of 50 nM [3H]-IAA and the indicated amount of competitor. Each value corresponds to the mean of three independent experiments. Error bars represent the sd.
(C) Competitive binding of [3H]-IAA to MBP-SKPA in the presence of increasing concentrations of unlabeled 1-NAA (black squares), 2-NAA (gray squares), or 2,4-D (black triangles). MBP alone was used as a control of nonbinding protein. Pull-down reactions were performed in the presence of 50 nM [3H]-IAA and the indicated amount of cold competitor. The inactive auxin 2-NAA does not compete with [3H]-IAA. Values are the mean of three independent experiments. Error bars represent the sd.
Identification of the Auxin Binding Site in SKP2A
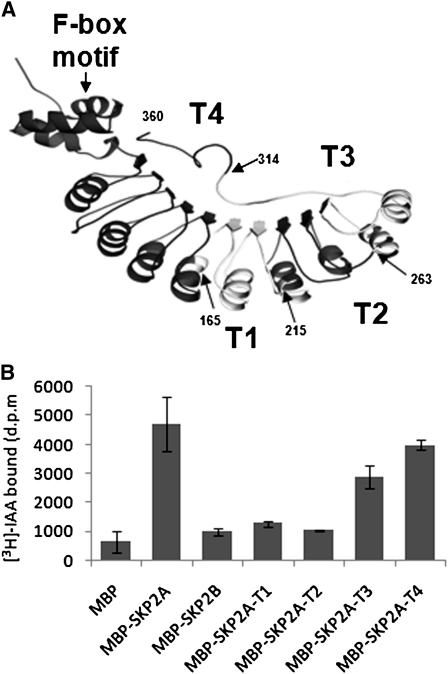
In an effort to identify which regions in SKP2A might be involved in auxin binding, we performed pull-down assays using deleted versions of MBP-SKP2A and [3H]-IAA. First, we generated four truncated proteins (T1 to T4) that have progressive deletions coming from the C terminus (Figure 4A). Deletion of the last 46 amino acids in the C terminus of SKP2A (MBP-SKP2A-T4) had a slight effect on the amount of [3H]-IAA retained, while MBP-SKP2A-T3 retained ~40% less radioactivity than the wild-type protein (Figure 4B). When we analyzed the T2 and T1 deletions, we found that these deleted proteins have severely compromised the binding ability of [3H]-IAA, retaining only control levels (Figure 4B). These results suggest that the residues necessary for auxin binding might be included between amino acids 215 and 263 or that the deletion of a large fragment of the protein, as it is the case for MBP-SKP2A-T1 or MBP-SKP2A-T2, alters the structure of SKP2A and eliminates its ability to bind to auxin.
Figure 4.
Identification of the Auxin Binding Region in the SKP2A Structure.
(A) An orthogonal view of a ribbon diagram of the modeled structure of SKP2A using the structure of human Skp2. Arrows indicate the different deletions generated for auxin binding analyses (T4, T3, T2, and T1), and the last amino acid of the truncated proteins is indicated by a number.
(B) These truncated versions (MBP-SKP2A-T1, T2, T3, and T4), MBP, MBP-SKP2A, and MBP-SKP2B were expressed in bacteria and then incubated in the presence of 50 nM [3H]-IAA. The retained [3H]-IAA in the amylose beads after three washes was measured by scintillation counting. Each value is the mean of three independent measures, and the error bars correspond to the sd. d.p.m., disintegrations per minute.
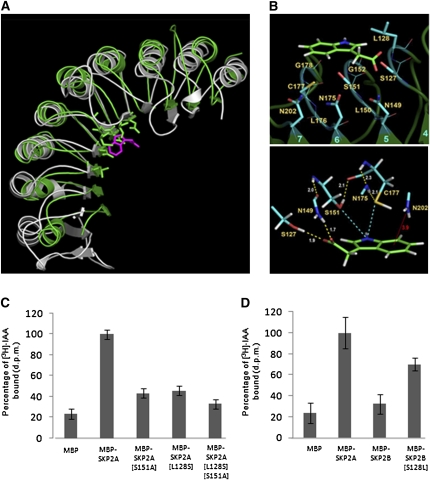
To precisely identify the auxin binding site in SKP2A, we generated its three-dimensional structure (Figure 4A), which was modeled onto the human Skp2 chain (Schulman et al., 2000). To identify a putative auxin binding site, we used as a guide the crystal structure of the auxin receptor TIR1 bound to an IAA molecule (Tan et al., 2007). With the structures of SKP2A and the C-terminal half of TIR1 (residues 300 to 594) superposed, we selected the residues in SKP2A structure that were located in a neighborhood of 4.0 Å surrounding the IAA molecule bound to TIR1 (Figure 5A). This procedure gave rise to a set of 11 residues that we used as the initial site for docking the IAA molecule (Figure 5B). Quantum calculations were then performed with an IAA molecule docked at this site, while keeping fixed the conformation of protein backbone to further refine the geometry of the site and to explore electron effects in the interaction. These calculations rendered a refined binding site composed of all the atoms of Ser-127, Leu-128, Asn-149, Ser-151, Gly-152, Asn-175, Cys-177, and Asn-202 as well as the backbone-only atoms of Leu-150, Gly-178, and Leu-176. In the presence of auxin, Ser-151 and Cys-177 residues in the docking site undergo a torsion motion situating side chain OH and SH groups at hydrogen bond distance of auxin indole N atoms, thus stabilizing the complex. In addition, the auxin phenol ring and one NH2 group of Asn-202 residue in SKP2A are positioned at a distance of 3.9 Å (Figure 5B), allowing an N-H···π interaction, a type of hydrogen bond, which is known to contribute to stabilize local structures in proteins (Steiner and Koellner, 2001; Weiss et al., 2001).
Figure 5.
Model Structure of SKP2A and Identification of the Auxin Binding Site.
(A) Superposition of the LRR domain of SKP2A (green) and the half side of TIR1 protein that binds auxin (gray) shown as ribbon diagrams. Residues of SKP2A in a neighborhood of 4.0 Å around auxin molecule (magenta) of the TIR1 structure are depicted as sticks.
(B) Auxin molecule (green frame) docked to a binding site composed of the 11 residues (cyan frames) indicated. Numbers colored cyan refer to β-strands of the LRR domain, which is shown as a ribbon diagram in dark green. Lines represent bonds between backbone atoms, and sticks correspond to bonds between side chain atoms involved in the interaction. The bottom panel shows a close-up view of the nearest groups to auxin revealed by quantum calculations. Yellow dashed lines represent hydrogen bonds with numerical labels indicating H···X distances in Å. Cyan dashed lines represent hydrogen bonds that depend on internal rotation of S151 OH group and C177 SH group. Red line represents a putative interaction of the type N-H···π between a NH2 side group of N202 and the π electron cloud of the aromatic ring of auxin. The red label gives the distance in Å between N atom and the center of this ring. Atoms are colored: C, light green and cyan; N, blue; O, red; S, yellow; H, white.
(C) Recombinant MBP, MBP-SKP2A, and two SKP2A mutants, which have replaced the Ser-151 for Ala (MBP-SKP2A[S151A]), the Leu-128 for Ser (MBP-SKP2A[L128S]), or double mutant (MBP-SKP2A[L128S; S151A]), were incubated in the presence of 50 nM [3H]-IAA. The retained [3H]-IAA in the amylose beads after three washes was measured by scintillation counting. Each value is the mean of three independent measures, and the errors bars correspond to the sd. The results were normalized relative to the amount of [3H]-IAA retained in the MBP-SKP2A beads.
(D) Recombinant MBP, MBP-SKP2A, MBP-SKP2B, and a SKP2B mutant that has replaced the Ser-128 for Leu (MBP-SKP2B[S128L]) were incubated in the presence of 50 nM [3H]-IAA. The retained [3H]-IAA in the amylose beads after three washes was measured by scintillation counting. Each value is the mean of three independent measures, and the errors bars correspond to the sd. The results were normalized relative to the amount of [3H]-IAA retained in the MBP-SKP2A beads and are presented as a percentage.
To validate these docking analyses, we generated SKP2A mutant proteins, with point mutations in residues of the auxin binding site, and then we analyzed their ability to retain [3H]-IAA. As mentioned before, Ser-151 is not only involved in direct hydrogen bonding with auxin, its surface area also forms the bottom surface of the binding site and it is adjacent to the surface of Leu-128, a residue particularly relevant in auxin binding site (see below). To confirm the importance of Ser-151, we replaced it with Ala (MBP-SKP2A-[S151A]). This mutant protein, bound to beads, was incubated with [3H]-IAA, and after several washes, the radioactivity retained was measured by scintillation counting. As shown in Figure 5C, the amount of radioactivity retained by the MBP-SKP2A-[S151A] was reduced by almost 60% compared with the wild-type protein. These data indicates that a Ser in this specific position is important to have an active auxin binding site in SKP2A. It is remarkable that among all the putative residues involved in auxin binding, only the one in the position 128 is different between SKP2A (Leu) and SKP2B (Ser) (see Supplemental Figure 2 online). To analyze the importance of this Leu-128 in the auxin binding, we generated a SKP2A point mutant in which Leu-128 was replaced with a Ser (SKP2A-[L128S]). This point mutant was used for pull-down assays in the presence of [3H]-IAA. Figure 5C shows that MBP-SKP2A-[L128S] had a significant effect on the amount of [3H]-IAA retained, since this mutant bound ~60% less radioactivity than the wild-type protein. We also generated the double SKP2A mutant MBP-SKP2A-[L128S; S151A], which retained only 30% of radiolabeled IAA compared with the wild-type protein (Figure 5C). These data indicate that Leu-128 in SKP2A might be the critical residue in terms of auxin binding. To confirm this hypothesis, we constructed a SKP2B mutant protein in which Ser-128 was replaced with Leu (MBP-SKP2B-[S128L]), generating a SKP2A-like auxin binding site. As shown in Figure 5D, now the amount of radioactivity retained by MBP-SKP2B-[S128L] was significantly increased compared with the MBP-SKP2B protein.
Auxin Binding Regulates SKP2A Stability
Our data suggest that auxin regulates SKP2A stability by binding to the protein. To test this hypothesis in vivo, we generated transgenic plants that express SKP2A or SKP2Amut2 (SKP2A-[L128S; S151A]; see Methods), fused to the GUS reporter protein under the control of an estradiol-inducible promoter. We analyzed several independent lines, and all of them rendered comparable staining patterns. After induction of the chimeric proteins, seedlings were transferred to Murashige and Skoog (MS) medium with or without auxin for 5 h and then stained for GUS activity. As shown in Figure 6A, SKP2Amut2-GUS accumulated to higher levels than SKP2A-GUS in the roots, while the mRNA expression was similar (Figure 6C). Opposite to the wild-type protein, SKP2Amut2-GUS was not degraded either in the absence or in the presence of auxin in the medium (Figure 6A). It is remarkable that SKP2Amut2-GUS accumulated in the root meristem, where SKP2A-GUS was never found (Figure 6B), except when seedlings were treated with TerfA (see Supplemental Figure 1 online). In addition, SKP2Amut2-GUS did not accumulate to higher levels following MG132 or TerfA treatment (see Supplemental Figure 1 online), suggesting that this protein did not suffer any UPS-auxin-dependent degradation or regulation by the auxin signaling. As a control, we used a green fluorescent protein (GFP)-GUS protein, which did not change its levels with any treatment (Figure 6A).
Figure 6.
Mutations in the Auxin Binding Site Stabilize SKP2A and DPB and Affect Cell Division.
(A) Five-day-old PER8:SKP2A-GUS, PER8:SKP2Amut2-GUS, and the control PER8:GFP-GUS transgenic seedlings were transferred to MS plates containing 15 μM estradiol to induce the expression of the proteins for 16 h. After induction, these seedlings were stained for GUS activity (tp = 0) or incubated during 5 h in a MS medium (5 h MS) or MS containing 5 × 10−7 M 2,4-D (5 h+Aux) and then stained for GUS activity. Bar = 500 μm.
(B) Higher magnification of PER8:SKP2A-GUS, PER8:SKP2Amut2-GUS, and the control PER8:GFP-GUS root tip shown in (A). Bar = 500 μm.
(C) Expression of the SKP2A-GUS and SKP2Amut2-GUS transgenes analyzed by RT-PCR using primers from SKP2A and GUS coding regions. As a control, the expression of the ACTIN1 (ACT) gene was analyzed.
(D) Immunoblotting analyses of the E2FC or MYC-DPB levels into the skp2a/MYC-DPB plants. Total protein was extracted from wild-type, MYC-DPBOE, skp2a/MYC-DPBOE, or skp2a/MYC-DPBOE seedlings that overexpress HA-SKP2A, HA-SKP2Amut1 (two independent lines), or HA-SKP2mut2 (two independent lines) and immunoblotted (I-blot) with anti-E2FC or anti-MYC. LC is the loading control corresponding to the Ponceau-stained blot. Ø indicates skp2a/MYC-DPBOE plants that overexpress the HA tag alone. The expression levels of both E2FC and MYC-DPB genes were analyzed by RT-PCR. As a control, the ACTIN gene levels were analyzed.
(E) Root meristem cell number of control MYC-GFP, MYC-SKP2AOE, or MYC-SKP2Amut2OE plants grown in MS medium. For monitoring root meristem growth, cortex meristematic cells were counted at the indicated days. Error bars represent se (n = 15). Asterisks indicate statistically significant difference compared with wild-type meristems as determined by Student´s t test (*P < 0.02 and **P < 0.001, respectively).
[See online article for color version of this figure.]
Mutations in the SKP2A Auxin Binding Site Reduce Cell Division
Our data clearly indicate that SKP2A binds auxin. To understand the biological meaning of this binding and its relation with cell division, we analyzed the effect of overexpressing SKP2A or SKP2A mutants that do not bind to auxin into the skp2a/MYC-DPB genotype. Previously, we showed that skp2a accumulated higher levels of E2FC and DPB, cell cycle transcription factors, than wild-type plants (del Pozo et al., 2006; Jurado et al., 2008). Overexpression of SKP2Amut1 or SKP2Amut2 did not reduce these higher E2FC or DPB levels, while the overexpression of wild-type SKP2A did (Figure 6D). These data clearly show that the auxin binding site in SKP2A is necessary to promote E2FC and DPB degradation in vivo. It is remarkable that overexpression of DPB increased the expression levels of E2FC, levels that were counterbalanced by a mutation in SKP2A (Figure 6D). In addition, we found that overexpression of SKP2A in skp2a/MYC-DPBOE restored the levels of E2FC to those found in MYC-DPBOE plants. However, overexpression of SKP2Amut1 or SKP2Amut2 did not restore these levels, indicating that these mutant versions of SKP2A are not completely functional.
We also analyzed the effect of overexpressing SKP2A or SKP2Amut2 on cell division in the root meristem. To measure the meristem size, we counted the number of meristematic cortical cell in the root tip. As shown in Figure 6E, overexpression of SKP2A led to more root meristematic cortical cells than the wild type, while overexpression of the SKP2Amut2, a protein unable to bind to auxin, did not increase the number of meristematic cortical cells, but slightly reduced it (Figure 6E).
Auxin Controls the SKP2A–DPB Interaction
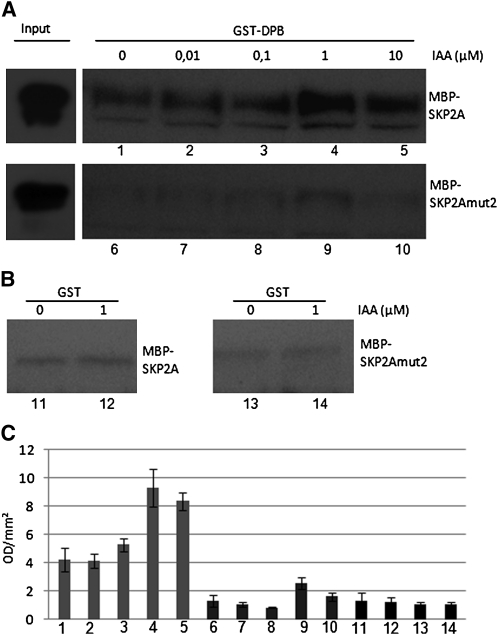
To analyze whether auxin affects the interaction between SKP2A and its targets, we performed pull-down experiments in absence or in presence of auxin. For these experiments, we used glutathione S-transferase (GST) or GST-DPB bound to beads and purified recombinant MBP-SKP2A or MBP-SKP2A mut2, which do not bind to auxin. We found that GST-DPB (Figure 7A), but not GST alone (Figure 7B), was able to interact with wild-type SKP2A. It is notable that the presence of auxin in the buffer improved the interaction, suggesting that auxin enhances recognition of DPB by SKP2A (Figures 7A and 7C). To check that this interaction was not due to the presence of the MBP tag in the SKP2A protein, we generated a 6xhistidine-tagged version of SKP2A (His-SKP2A). We found that GST-DPB interacted with His-SKP2A, and auxin enhanced this interaction (see Supplemental Figure 4 online). On the other hand, we showed that mutations in SKP2A that prevent auxin binding reduced its interaction with GST-DPB.
Figure 7.
Auxin Regulates SKP2A and DPB Interaction.
(A) Recombinant MBP-SKP2A or MBP-SKP2A[L128S; S151A] proteins were incubated with GST-DPB bound to beads. When indicated, different concentrations of IAA were added during the incubation and the washing steps. The pulled-down proteins were analyzed by immunoblotting using purified IgGs against the SKP2A protein. The image presented is from one representative experiment of three.
(B) Recombinant MBP-SKP2A or MBP-SKP2A[L128S; S151A] proteins were incubated with GST bound to beads. When indicated, 1 μM IAA was added during the incubation and the washing steps. The pulled-down proteins were analyzed by immunoblotting using purified IgGs against the SKP2A protein.
(C) The bands detected in (A) and (B) were quantified using a Bio-Rad molecular imager in optical density units per square millimeter (OD/mm2). Each value is the mean of the three different experiments, and the error bars correspond to the sd. Numbers shown in the base correlate with the different bands labeled in (A) and (B).
SKP2A Contributes to the Auxin Response
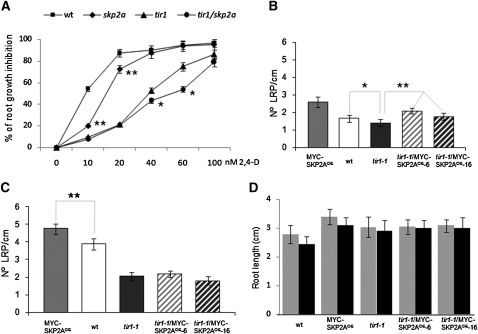
To further investigate whether SKP2A gene contributes to the auxin response, we analyzed the effect of auxin on root growth inhibition in the skp2a mutant. We found that skp2a roots were slightly more resistant to the inhibitory effect of low concentrations of 2,4-D than wild-type roots (Figure 8A). To analyze a possible genetic interaction between SKP2A and TIR1, we generated the double mutant tir1-1 skp2a and examined root growth in medium containing auxin. We found that tir1-1 skp2a seedlings displayed enhanced resistance to the inhibition of root elongation by 2,4-D compared with tir1-1 (Figure 8A), suggesting a cooperative function between these two genes in the root growth inhibition promoted by this hormone.
Figure 8.
Mutation in SKP2A Enhances tir1 Auxin Resistance.
(A) Wild-type (wt) and skp2a, tir1-1, and tir1-1 skp2a seedlings were grown in MS plates for 5 d. Afterwards, seedlings were transferred to a MS medium or MS medium containing different concentrations of 2,4-D. After 3 d, root elongation of at least 50 seedlings from two independent experiments was measured. The values are represented as the percentage of root growth inhibition. **P < 0.0001, statistically significant different for skp2a compared with the wild type as determined by Student’s t test. *P < 0.0005, statistically significant different for tir1-1 skp2a compared with tir1-1 as determined by Student’s t test.
(B) Arabidopsis wild-type, MYC-SKP2AOE, tir1-1, and tir1-1/MYC-SKP2AOE seedlings were grown for 5 d in MS medium and then 1 extra day in MS. Afterwards, seedlings were collected and incubated for 24 h in a solution of chloral hydrate:glycerol:water (80:10:10) in ice. Root lengths were measured and the number of nonemerged LRP was counted using a Leica 2000 microscope. Error bars represent se (n ≥ 30). *P < 0.01, statistically significant difference for tir1-1 compared with wild-type values as determined by Student’s t test.**P < 0.0001, statistically significant difference for tir1-1/MYC-SKP2AOE compared with tir1-1 values as determined by Student’s t test.
(C) Arabidopsis wild-type, MYC-SKP2AOE, tir1-1, and tir1-1/MYC-SKP2AOE seedlings were grown for 4 d in MS medium and then 1 extra day in MS supplemented with 50 nM 2,4-D. Afterwards, seedlings were treated as in (A), and root lengths were measured and the number of nonemerged LRP was counted. Error bars represent se (n ≥ 30). In this case, statistically significant differences for tir1-1/MYC-SKP2AOE compared with tir1-1 were not found. **P < 0.0001, statistically significant difference for MYC-SKP2AOE compared with wild-type values as determined by Student’s t test.
(D) Root length of 5-d-old seedlings grown in MS medium (gray bars) or 4 d in MS and 1 d in MS plus 50 nM 2,4-D (black bars). Error bars represent se (n ≥ 30).
It has been reported that tir1-1 has a reduced number of emerged lateral roots (Ruegger et al., 1998). Here, we show that 5-d-old tir1-1 seedlings also have fewer lateral root primordia (LRP) that have not emerged from the main root than wild-type seedlings (Figure 8B). Since overexpression of MYC-SKP2A increases the number of LRP (Jurado et al., 2008), we wondered whether overexpression of SKP2A might increase the number of LRP in tir1-1. As shown in Figure 8B, overexpression of SKP2A in tir1-1 slightly, but statistically significantly, increased the number of LRP with respect to the tir1-1 mutant, while the root length remained similar (Figure 8D). When tir1-1 seedlings were treated with a low concentration of auxin for 24 h, the number of LRP increased to the same level as untreated tir1-1/MYC-SKP2AOE seedlings (Figure 8C). These data suggest that overexpression of SKP2A seems to be sufficient to induce cell division in the founder cells to initiate the formation of lateral root primordia in the tir1-1 mutant at the levels similar to those initiated by low concentrations of auxin, but not to promote the emergence of these primordia.
DISCUSSION
In this work, we show that auxin regulates the proteolysis of the cell cycle F-box protein SKP2A through the ubiquitin proteasome system by binding to SKP2A. Using protein structure modeling, we identified a novel auxin binding site. The integrity of this site is necessary for SKP2A UPS-dependent degradation. In addition, mutations in this SKP2A auxin binding site block E2FC and DPB degradation and limit the ability of SKP2A to promote cell division in the root meristem. Finally, genetic studies show that SKP2A participates in the auxin signaling. Taken together, these data indicate that SKP2A connects the auxin response with cell division.
SKP2A Binds Auxin
The plant hormone auxin stimulates the interaction of the F-box TIR1 (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005) or its homologous AFBs (Dharmasiri et al., 2005b) with the AUX/IAA proteins. The crystal structure of the ASK1-TIR1-auxin complex showed that auxin binds to TIR1 filling a hydrophobic gap, favoring the interaction with the Aux/IAA proteins, demonstrating that the TIR1-Aux/IAA complex acts as an auxin coreceptor (Tan et al., 2007). The results presented here provide convincing evidence that SKP2A, a cell cycle F-box, also binds to auxin. Using a TIR1-based protein superposition and docking analyses, we identified a novel auxin binding site in SKP2A. Different mutations in this binding site eliminate the ability of SKP2A to retain radiolabeled IAA. Upon analyzing the electrostatic potential mapped onto the protein surface, it was noticed that this binding site, located just at the rim of a positive area, exhibits a neutral potential in the region closest to the apolar rings of the IAA, whereas its positive border is at the region nearest to auxin carboxylate. It is worth highlighting that although the topography of the site and the hydrophobic interactions are different, a number of links between SKP2A and IAA (Figure 5B) show similar features to those found in the TIR1-IAA complex (Tan et al., 2007). For example, auxin carboxylate acts as the anchor to the TIR1 site by forming two hydrogen bonds with two nearby residues, Arg-403 and Ser-438. The NH group of the auxin indole ring also makes two hydrogen bonds with backbone oxygens of Leu-404 and Leu-439 in TIR1. In addition, the nearby residue TIR1-S462 is also oriented so as to allow an O-H···π hydrogen bond with the π cloud of auxin phenyl (Tan et al., 2007).
Unexpectedly, SKP2B, which shares >80% sequence identity with SKP2A, does not bind to auxin. Among the residues identified in the SKP2A auxin binding site, only the amino acid in position 128 is different between SKP2A and SKP2B (Leu and Ser, respectively). This strongly suggests that Leu-128 is a critical residue for auxin binding. Indeed, a mutation in Leu-128 of SKP2A compromises its ability to bind to auxin, and a single mutation in SKP2B, in which the Ser in position 128 is replaced by a Leu, generates a SKP2B-like protein that now binds to auxin. The surface of Leu-128 in SKP2A generates a neutral electrostatic potential wall to shape a pocket (Figure 5B; see Supplemental Figures 5 and 6 online) that interacts with the nonpolar moiety of auxin. By contrast, in SKP2B, the residue 128 is a Ser, which offers a smaller surface than the Leu and almost removes the wall of the pocket. In addition, Ser-128 generates a strong negative electrostatic potential that hinders the interaction with the aromatic ring of auxin (see Supplemental Figure 5 online). These modeling data might explain the differences in auxin binding that we found between SKP2A and SKP2B. We are aware that to precisely define all the residues involved in the auxin binding requires a crystal structure, but taken together, our data positively indicate the existence of a novel auxin binding site in the SKP2A protein.
At present, there are some examples of proteins with LRR domains that are able to bind to hormones. The TIR1-auxin crystal has led the identification of all the residues involved in auxin binding (Tan et al., 2007). Recent reports showed that the jasmonate signal transduction pathway, which involves the F-box COI1 and the transcription factor repressor JAZ, shares similar features to the TIR1-auxin signaling (Chini et al., 2007; Thines et al., 2007). Biochemical analyses have shown that the COI1-JAZ dimer pulls down radiolabeled coronatine, indicating that the F-box COI1 likely binds to jasmonate similarly to TIR1 binding to auxin (Katsir et al., 2008). Recently, it has been reported that the BRI1 receptor binds to brassinosteroids through its LRR and adjacent sequences (Kinoshita et al., 2005). Thus, it is tempting to speculate that many other F-box or LRR-containing proteins might bind small molecules to sense and transduce endogenous and external signals.
Auxin Binding Regulates SKP2A Degradation
The auxin receptors described to date, TIR1 and AFB, mediate rapid degradation of Aux/IAA proteins to release the transcriptional potential of ARF proteins that induce the expression of auxin-regulated genes (Gray et al., 2001; Zenser et al., 2001; Dharmasiri et al., 2005b). It is now clear that auxin does not promote a conformational change in TIR1 to favor the interaction with its targets (Tan et al., 2007). Rather, auxin stimulates the interaction between SCF-TIR1 and Aux/IAA targets, acting as molecular glue. We found that SKP2A is degraded through the UPS. This feature is also shared by its human counterpart, Skp2, with stability that is controlled by a mechanism that involves phosphorylation by the Akt1 kinase and degradation by the APC-Cdh1 ubiquitin ligase complex (Gao et al., 2009). It is well known that many targets have to be posttranslationally modified to be recruited by the E3 ligases. There have been several types of modification described, such as phosphorylation, hydroxylation, or glycosylation, among others (Ravid and Hochstrasser, 2008). Our data show that auxin acts as a signaling molecule that triggers SKP2A degradation. It is likely that auxin might mimic a covalent posttranslational modification, acting as signal that triggers SKP2A degradation. Although this hypothesis has to be fully corroborated, it could provide a wide, fast, and efficient system to regulate the stability of proteins in response to different stimuli, as it does not require the participation of additional proteins. At this point, we do not know whether SKP2A is degraded by an autoubiquitination mechanism or through an E3-dependent process. It is possible that auxin generates a conformational change in SKP2A that is necessary for E3 recruitment, or auxin may act as a signal that favors the recognition of SKP2A. Nevertheless, TIR1 does not seem to be the E3 responsible for this proteolysis since SKP2A is highly unstable in the tir1-1 mutant. However, since there are other TIR1-like AFBs that also function as auxin receptors (Dharmasiri et al., 2005b), we cannot rule out the possibility that one of these AFBs could be involved in SKP2A proteolysis. Using the SKP2A-GUS reporter protein, we found that SKP2A does not accumulate either in the aerial part of the plant, in the most upper region of the root, nor in the root meristem, where the maximum concentration of auxin localized. Since auxin promotes the fast degradation of SKP2A, this might explain the lack of GUS signal in the root tip.
By competition analyses, we show that 2-NAA does not bind to SKP2A. Similarly, 2-NAA does not bind to TIR1 (Kepinski and Leyser, 2005), despite its promiscuity in binding to different auxins (Tan et al., 2007), but it binds to another auxin binding protein, ABP1 (Löbler and Klämbt, 1985). Computational modeling was not conclusive enough to disclose the reasons for the lack of binding of 2-NAA to SKP2A. Although without a crystal structure it is difficult to know with high precision why 2-NAA establishes less or weaker interactions with SKP2A than active auxin molecules, this difference is likely due to the distinct orientation of the aromatic rings with respect to the carboxyl group. It is reasonable to conjecture that this structural feature might affect the stability of the interaction of 2-NAA with any binding site for active auxin molecules. In addition, 2-NAA slightly reduces the SKP2A proteolysis in vitro. This delay might be the consequence of a partial interference of 2-NAA with active auxin present in the extract. Another possibility is that 2-NAA, which has been reported to have an antiauxin function (Dahlke et al., 2009), blocks the auxin signaling needed for SKP2A proteolysis. Thus, 2-NAA might exert a similar effect to TerfA, reducing auxin signal and leading to SKP2A accumulation. We have shown that TerfA does not compete with IAA for binding to SKP2A, but it blocks SKP2A degradation. A similar effect has been found for Aux/IAA degradation by TIR1, since TerfA blocks proteolysis without affecting the formation of the TIR1-auxin-Aux/IAA complex (Yamazoe et al., 2005). Our data suggest that TerfA blocks the auxin signaling needed for SKP2A degradation, likely by affecting the activity/induction of the E3 needed for SKP2A degradation.
SKP2A Auxin Binding Is Needed for Cell Division
SKP2A is a cell cycle–regulated protein that participates in the control of cell division by degrading, at least, two cell division transcriptional factors, E2FC and DPB (del Pozo et al., 2006; Jurado et al., 2008). Transcriptomic analyses have shown that E2FC is also important in the control of C-compounds and carbohydrates, metabolic pathways, and light signaling, suggesting a role of E2FC in other developmental processes in addition to cell division (de Jager et al., 2009). It is remarkable that SKP2B, but not SKP2A, promotes the degradation of the cyclin-dependent kinase inhibitor KRP1 (Ren et al., 2008). Previous work has shown that the skp2a mutant is unable to degrade DPB and accumulates higher levels of this protein than control plants. This DPB accumulation was reduced by overexpression of SKP2A but not by SKP2B, clearly indicating that, despite of their high sequence similarity, both proteins have distinct target specificity (Jurado et al., 2008). In this work, we have shown that overexpression of SKP2A mutant proteins that do not bind to auxin does not reduce the E2FC or DPB levels, indicating that auxin binding is needed for their degradation. In the TIR1-Aux/IAA system, auxin acts as molecular glue between the F-box and the target proteins, favoring the interaction between the F-box and the targets. Using in vitro assays, we found a slight interaction between SKP2A and DPB that was enhanced in the presence of auxin. In this regard, it is remarkable that mutations in SKP2A that reduce auxin binding also limit its interaction with DPB, suggesting that the contact between both proteins occurs through the auxin binding site (see Supplemental Figure 7 online).
There are other examples were auxin affects the stability of cell cycle proteins. E2FB is a cell cycle transcription factor that is also degraded through the UPS and auxin stabilizes it (Magyar et al., 2005). However, how auxin affects E2FB stability at the molecular level is still unknown. It is tempting to speculate that auxin might promote the degradation of the E3 responsible of E2FB proteolysis, causing its stabilization. Since auxin promotes the degradation of SKP2A, it was tempting to speculate that SKP2A might target E2FB for degradation. We conducted immunoblotting analyses to explore this possibility, and it seems that neither SKP2A nor SKP2B regulate the stability of E2FB factor (see Supplemental Figure 8 online).
Based on our data, we think that SKP2A is one of the proteins that directly connects the auxin response with cell proliferation. This is supported by the fact that SKP2A binds to auxin to regulate the stability of cell cycle transcription repressors, and overexpression of SKP2A promotes cell division in meristems and increases the auxin response (Jurado et al., 2008). However, overexpression of SKP2A mutants that do not bind to auxin reduces cell division in the root meristem and does not degrade cell cycle repressors. This could be explained by a dominant-negative effect of these mutant proteins that might affect the activity of the endogenous SCFSKP2A, blocking the degradation of its targets. In addition, the number of mitotic cells in root meristems in response to auxin is reduced in the skp2a mutant compared with the wild type (see Supplemental Figure 9 online). Taken together, these data indicate that auxin, at least partially, regulates cell division through the SKP2A pathway. In addition to this, it is tempting to speculate that in the presence of auxin, SKP2A promotes the degradation of cell cycle targets; subsequently, auxin also enhances SKP2A proteolysis to prevent its overfunction (see Supplemental Figure 7 online).
SKP2A Is a Positive Regulator of the Auxin Response
Auxin is one of the most important hormones since it plays a crucial role in almost all the development and growth processes in plants, including tropic responses, apical dominance in the shoot, lateral root formation, differentiation of the vascular system, and cell division and elongation (Davies, 1995; Leyser, 2002; Benjamins and Scheres, 2008; Mockaitis and Estelle, 2008). Previous results have shown that overexpression of SKP2A leads to an enhanced auxin response in the roots, since overexpression of SKP2A increases the expression of the synthetic auxin response marker DR5 (Jurado et al., 2008). Here, we show that a mutation in SKP2A leads to auxin-resistant root growth, suggesting that SKP2A is a positive regulator of the auxin response. In an attempt to elucidate the pathway through which SKP2A might function, we examined genetic interactions between skp2a and tir1-1. TIR1 encodes an F-box protein that promotes Aux/IAA protein degradation to release the auxin response (Gray et al., 2001). The tir1-1 mutant was identified based on its abnormal auxin responses (Hobbie et al., 1994) and leads to auxin-resistant root growth. The double mutant tir1-1 skp2a displayed a longer primary root than the single tir1-1 mutant in an auxin-containing medium, indicating that SKP2A also contributes to the auxin response. This might be explained by the positive function of SKP2A in cell proliferation. It is possible that cells in the root meristem that lack SKP2A divide less in response to auxin but elongate more than wild-type cells, contributing to a longer growth in presence of the hormone. Indeed, using the mitotic reporter CYB1:CYB1-GUS, we found significant fewer mitotic cells in the skp2a root meristems in response to auxin than in control roots (see Supplemental Figure 9 online). In addition, tir1-1 develops a lower number of lateral roots than wild-type plants (Ruegger et al., 1998). We also found that tir1-1 has a lower number of nonemerged LRPs, and overexpression of SKP2A partially rescues this defect, likely by promoting cell division in the pericycle founder cells to initiate LRPs. Genetic analyses of the double mutant tir1 ibr5 showed that IBR5, a dual phosphatase, regulates auxin responses differently than the TIR1-Aux/IAA pathway (Strader et al., 2008). Thus, our genetic analysis of skp2a tir1-1 is fully compatible with the idea that other pathways, besides the TIR1/AFB-dependent pathway, contribute to the final response to auxin, and the SKP2A pathway might be one of them.
METHODS
Plant Material and Growth Conditions
Wild-type (Columbia ecotype) MYC-SKP2AOE plants (del Pozo et al., 2002), tir1-1 (Ruegger et al., 1998), skp2a (Ren et al., 2008), or tir1-1 skp2a were grown under sterile conditions on vertically oriented MS (half MS salts, 1% sucrose, and 1% plant-agar; Duchefa) plates at 22°C with 16 h light and 8 h dark. For root growth assays and LRP measurements, seedlings were grown in MS plates for 5 d and afterwards transferred to MS or MS containing 50 nM 2,4-D plates. To analyze the skp2a auxin growth resistance, wild-type or skp2a, tir1-1, or tir1-1 skp2a seedlings were grown on MS plates for 5 d and afterwards transferred to MS alone or MS containing 20, 40, 60, or 100 nM 2,4-D for 3 d. Pictures were taken and root length was measured with Image J software (http://rsb.info.nih.gov/ij). All data are the mean value of at least 50 plants, and these experiments were repeated twice, obtaining similar values in each experiment. Data values were statistically analyzed using the Student’s t test.
Constructs and Recombinant Proteins
SKP2A and SKP2B coding regions were amplified by RT-PCR (Invitrogen) and cloned using Gateway technology into pDONR221 (Invitrogen). To generate the four truncated SKP2A versions (T1 to T4) with progressive deletions coming from the C terminus, the coding region was amplified by PCR using primers that amplified the corresponding cDNA fragment: T1 (containing from nucleotide 1 of the ATG of the cDNA to nucleotide 495), T2 (1 to 645), T3 (1 to 789), and T4 (1 to 492). These truncated versions were cloned into pDONR221. To generate point mutant proteins, we used the Quick Change Multi site-directed mutagenesis kit (Stratagene). After mutagenesis, the whole coding region was sequenced to discard undesired mutations in other parts of the proteins. All these clones were transferred to a gateway-adapted MPB vector (New England Biolabs) by LR recombination. All these constructions were expressed as recombinant in Escherichia coli Bl21 and purified using amylose beads according to standard protocols. The GST or GST-DPB proteins were expressed and purified as described by del Pozo et al. (2006). To generate SKP2A-GUS or SKP2Amut2 (Leu-128 was change to Ser; and Ser151 was changed to Ala) chimeric proteins, the coding regions of SKP2A or SKP2Amut were fused in frame to the N terminus of the GUS coding sequence in a pBluescript plasmid. Afterwards, these chimeric cDNAs were amplified by PCR using the high-fidelity Pfx polymerase (Invitrogen) and cloned into the pDONOR221 and then transferred into PER8-GW, a Gateway version of estradiol-inducible expression vector PER8 (Zuo et al., 2000). Trangenic plants were generated as described by del Pozo et al. (2002) and selected by hygromycin resistance. Several independent lines were analyzed, and we found the same expression pattern and behavior for the SKP2A-GUS and SKP2Amut-GUS proteins in all of them. The expression was induced with 10 μM estradiol during 16 h in 5-d-old seedlings, and then GUS staining was performed as described del Pozo et al. (2002).
To analyze the accumulation of MYC-DPB, we used the previously described plant skp2a/MYC-DPB (Jurado et al., 2008). This line was transformed with the pGWB14 vector in which the wild-type SKP2A or mutant version, SKP2A[L128S] (HA-SKP2Amut1) or SKP2A[L128S;S151A] (HA-SKP2Amut2), cDNAs were cloned. Several independent lines, which express similar HA-SKP2A levels, were analyzed by immunoblot to check the MYC-DPB levels. To generated transgenic plants that overexpress MYC-GFP or MYC-SKP2Amut2, the GFP and SKP2Amut2 coding regions were fused to the MYC epitope (del Pozo et al., 2002) and then mobilized to the pGWB2 vector (Nakagawa et al., 2007) by recombination. Several independent lines were selected and analyzed by immunoblots to analyze the expression of the tagged proteins.
SKP2A and DPB Interaction
For the pull-down experiments, 2 μg of GST or 1 μg GST-DPB (del Pozo et al., 2002) proteins bound to beads were incubated with 10 μg of MBP-SKP2A or MBP-SKP2A[L128S;S151A] in PBS plus 0.1% Tween 20 (PBS-T) at 4°C for 1.5 h. Afterwards, the beads were washed three times with PBS-T for 15 min each. The precipitated proteins were released by boiling the beads in SDS-Laemmli sample buffer for 10 min. The proteins were analyzed by immunoblotting with purified IgG anti-SKP2A (del Pozo et al., 2002). When indicated, different concentrations of IAA were added to the buffer either during the incubation or the washing steps. The Anti-SKP2A did not show any cross-reactivity against GST or GST-DPB recombinant proteins (see Supplemental Figure 4 online).
Immunoblotting and Auxin Binding
Detection of MYC-SKP2A or MYC-DPB proteins were performed as described by del Pozo et al. (2002, 2006). Degradation assays using crude plant extracts were performed as described by del Pozo et al. (2006). MYC-SKP2A protein was detected by immunoblotting using enhanced chemiluminescence (Millipore) and X-film. We took multiple exposures, ranging from 1 to 10 min, to assure that detection of MYC-SKP2A was in the linear range. The bands corresponding to the MYC-SKP2A protein were quantified by scanning with the Bio-Rad Molecular Imager GS-800 calibrated densitometer. For quantification, we only used those films in which the signal at time zero was not saturated according to the densitometer measures. These bands were analyzed using Quantity One software. The values were represented as percentage relative to time zero (100%) for each experiment. The values are the mean of three different degradation assays. Protein extraction and detection of E2FC was performed using an affinity-purified IgG against E2FC as described by del Pozo et al. (2002).
To analyze auxin binding, pull-down assays were done with the bacterial expressed proteins bound to amylose beads (4 μg) and 0.5 μL of [3H]-IAA (specific activity 20 mCi/mmol, with a final concentration of 50 nM [3H]-IAA) in PBS-T. All assays were incubated for 30 min at 4°C. Afterwards, the amylose beads were washed three times for 3 min each with 1 mL of ice-cold PBS-T and resuspended in 100 μL of scintillation fluid. The radioactivity of the bound [3H]-IAA was measured using a scintillation counter during 1 min in a microplate in a Perkin-Elmer MicroBeta TriLUX 1450 LSC and luminescence counter. All data points are the mean of at least three independent experiments.
Structure Modeling and Theoretical Calculations
The Template Identification Tool of the Swiss-Model Server (Arnold et al., 2006) was used to select possible templates for SKP2A and SKP2B. Model structures were obtained with the Swiss Model Project Mode using the Skp2 molecule (Schulman et al., 2000) of the Skp2-Skp1 complex (PDB entry 1FQV, chain A) as a template upon preparing project files with the program Swiss-PdbViewer (Guex and Peitsch, 1997). Structural superpositions, selection of spatial neighborhoods, and Coulomb potentials were also obtained with Swiss-PdbViewer. The geometry of the IAA molecule was optimized in B3LYP/6-311G(d,p) quantum calculations with the Gaussian03 package (Frisch et al., 2004). Dockings of IAA to model structures of SKP2A and SKP2B were accomplished using DOCK 6.1 (Moustakas et al., 2006). Refinements of dockings were first done with AutoDock 4.2 (Morris et al., 2009) working with 60 × 60 × 60 grids with grid spacing of 0.200 Å and 50 docking runs performed using the Lamarckian Genetic algorithm and then optimized by minimizing structures with Chimera 1.3 (Pettersen et al., 2004), using AMBER parameters for energy calculations (Case et al., 2005). Two-layer ONIOM(B3LYP/6-311G(d,p):HF/6-31G) quantum calculations were finally performed for the IAA/binding site complex with Gaussian03 (Frisch et al., 2004).
The electrostatic Poisson-Boltzmann (PB) potential was obtained with APBS 1.1.0 (Baker et al., 2001) assigning AMBER atomic charges and radii including hydrogen added with PDB2PQR (Dolinsky et al., 2004). Fine (0.50-Å spacing) grids around the 3691 resulting atoms were used to solve the nonlinear PB equation in sequential focusing multigrid calculations for meshes of 129 × 129 × 161 points at 298.15 K with dielectric constants of 2 for proteins and 78.54 for water. Potential values are given in units of kT per unit charge (k, Boltzmann’s constant; T, absolute temperature). PB potentials mapped onto protein surfaces and molecular graphics were rendered with PyMOL 1.1 (DeLano, 2008).
Root Length and Meristem Size Analysis
The number of meristematic cortex cells was measured as described (Casamitjana-Martínez et al., 2003). Root length was measured with Image J software.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SKP2A (At1g21410), SKP2B (At1g77000), DPB (At5g03415), and E2FC (At1g47870).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. SKP2A-GUS Protein Accumulated in Response to the Proteasome Inhibitor MG132 or Terfestatin A.
Supplemental Figure 2. Alignment of SKP2A and SKP2B Protein Sequences.
Supplemental Figure 3. Scatchard Analysis.
Supplemental Figure 4. Auxin Regulates SKP2A and DPB Interaction.
Supplemental Figure 5. Geometry of Auxin at Its Binding Site and Steric Effect of Residue 128.
Supplemental Figure 6. Topography of the Auxin Binding Site in Complete SKP2A.
Supplemental Figure 7. Model of SKP2A-Auxin Regulation.
Supplemental Figure 8. E2FB Stability Is Regulated by Neither SKP2A nor SKP2B.
Supplemental Figure 9. SKP2A Is Necessary to Control Cell Proliferation in Response to Auxin.
Supplementary Material
Acknowledgments
We thank Sara Navarro for her technical assistance. We also thank W. Gray for his helpful comments and suggestions and Olga Navarro for reading and typing of the manuscript. We thank Gabino Sanchez for helping us with sequence analyses and Hiroshi Nozaki for kindly providing the TerfA compound. This research was supported by grants from the Spanish Government (BIO2008-00639 and CDS2007-00530) to J.C.D.P. and BIO2009-07050 to L.F.P. and from the Comunidad de Madrid Research Council (S-GEN-0191-2006) to J.C.D.P. S.J. was supported by a predoctoral fellowship from the Spanish MICIIN and Z.A. by a Juan de la Cierva Spanish program.
References
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. (2001). Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Casamitjana-Martínez E., Hofhuis H.F., Xu J., Liu C.M., Heidstra R., Scheres B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Case D.A., Cheatham T.E., III, Darden T., Gohlke H., Luo R., Merz K.M., Jr, Onufriev A., Simmerling C., Wang B., Woods R.J. (2005). The Amber biomolecular simulation programs. J. Comput. Chem. 26: 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Dahlke R.I., Luthen H., Steffens B. (2009). The auxin-binding pocket of auxin-binding protein 1 comprises the highly conserved boxes a and c. Planta 230: 917–924 [DOI] [PubMed] [Google Scholar]
- Davies P.J. (1995). The plant hormones: Their nature, occurrence and functions. Plant Hormones: Physiology, Biochemistry and Molecular Biology, Davies P.J., (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 1–12 [Google Scholar]
- de Jager S.M., Scofield S., Huntley R.P., Robinson A.S., den Boer B.G., Murray J.A. (2009). Dissecting regulatory pathways of G1/S control in Arabidopsis: Common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol. Biol. 71: 345–365 [DOI] [PubMed] [Google Scholar]
- DeLano W.L. (2008). The PyMOL Molecular Graphics System. (Palo Alto, CA: DeLano Scientific; ). [Google Scholar]
- del Pozo J.C., Boniotti M.B., Gutierrez C. (2002). Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo J.C., Diaz-Trivino S., Cisneros N., Gutierrez C. (2006). The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18: 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005a). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J.S., Jürgens G., Estelle M. (2005b). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dolinsky T.J., Nielsen J.E., McCammon J.A., Baker N.A. (2004). PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32(Web Server issue): W665––W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D., Pagano M. (2008). Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: Tipping the scales of cancer. Nat. Rev. Cancer 8: 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M.J., et al. (2004). Gaussian 03. Revisions C.02 and D.01. (Wallingford, CT: Gaussian; ). [Google Scholar]
- Gao D., Inuzuka H., Tseng A., Chin R.Y., Toker A., Wei W. (2009). Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat. Cell Biol. 11: 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Gutierrez C., Ramirez-Parra E., Castellano M.M., del Pozo J.C. (2002). G(1) to S transition: More than a cell cycle engine switch. Curr. Opin. Plant Biol. 5: 480–486 [DOI] [PubMed] [Google Scholar]
- Hershko A. (2005). The ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle (Nobel lecture). Angew. Chem. Int. Ed. Engl. 44: 5932–5943 [DOI] [PubMed] [Google Scholar]
- Hobbie L., Timpte C., Estelle M. (1994). Molecular genetics of auxin and cytokinin. Plant Mol. Biol. 26: 1499–1519 [DOI] [PubMed] [Google Scholar]
- Johnson L.N. (2009). The regulation of protein phosphorylation. Biochem. Soc. Trans. 37: 627–641 [DOI] [PubMed] [Google Scholar]
- Jurado S., Díaz-Triviño S., Abraham Z., Manzano C., Gutierrez C., del Pozo C. (2008). SKP2A, an F-box protein that regulates cell division, is degraded via the ubiquitin pathway. Plant J. 53: 828–841 [DOI] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Caño-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2002). Molecular genetics of auxin signaling. Annu. Rev. Plant Biol. 53: 377–398 [DOI] [PubMed] [Google Scholar]
- Löbler M., Klämbt D. (1985). Auxin-binding protein from coleoptile membranes of corn (Zea mays L.). II. Localization of a putative auxin receptor. J. Biol. Chem. 260: 9854–9859 [PubMed] [Google Scholar]
- Magyar Z., De Veylder L., Atanassova A., Bakó L., Inzé D., Bögre L. (2005). The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17: 2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraschin Fdos.S., Memelink J., Offringa R. (2009). Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 59: 100–109 [DOI] [PubMed] [Google Scholar]
- Mockaitis K., Estelle M. (2008). Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30: 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas D.T., Lang P.T., Pegg S., Pettersen E., Kuntz I.D., Brooijmans N., Rizzo R.C. (2006). Development and validation of a modular, extensible docking program: DOCK 5. J. Comput. Aided Mol. Des. 20: 601–619 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakayama K.I., Nakayama K. (2006). Ubiquitin ligases: Cell-cycle control and cancer. Nat. Rev. Cancer 6: 369–381 [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. (2004). UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Ravid T., Hochstrasser M. (2008). Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Santner A., del Pozo J.C., Murray J.A., Estelle M. (2008). Degradation of the cyclin-dependent kinase inhibitor KRP1 is regulated by two different ubiquitin E3 ligases. Plant J. 53: 705–716 [DOI] [PubMed] [Google Scholar]
- Ruegger M., Dewey E., Gray W.M., Hobbie L., Turner J., Estelle M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman B.A., Carrano A.C., Jeffrey P.D., Bowen Z., Kinnucan E.R., Finnin M.S., Elledge S.J., Harper J.W., Pagano M., Pavletich N.P. (2000). Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408: 381–386 [DOI] [PubMed] [Google Scholar]
- Steiner T., Koellner G. (2001). Hydrogen bonds with pi-acceptors in proteins: Frequencies and role in stabilizing local 3D structures. J. Mol. Biol. 305: 535–557 [DOI] [PubMed] [Google Scholar]
- Strader L.C., Monroe-Augustus M., Bartel B. (2008). The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Raes J., De Veylder L., Rouzé P., Rombauts S., Inzé D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M.S., Brandl M., Sühnel J., Pal D., Hilgenfeld R. (2001). More hydrogen bonds for the (structural) biologist. Trends Biochem. Sci. 26: 521–523 [DOI] [PubMed] [Google Scholar]
- Yamazoe A., Hayashi K., Kepinski S., Leyser O., Nozaki H. (2005). Characterization of terfestatin A, a new specific inhibitor for auxin signaling. Plant Physiol. 139: 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser N., Ellsmore A., Leasure C., Callis J. (2001). Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 98: 11795–11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.