The TCP family of transcription factors and site II promoter elements that they bind in Arabidopsis link the regulation of gene expression for mitochondrial proteins with a variety of circadian clock components to provide specific time-of-day expression for a variety of genes.
Abstract
Diurnal regulation of transcripts encoding proteins located in mitochondria, plastids, and peroxisomes is important for adaptation of organelle biogenesis and metabolism to meet cellular requirements. We show this regulation is related to diurnal changes in promoter activities and the presence of specific cis-acting regulatory elements in the proximal promoter region [TGGGC(C/T)], previously defined as site II elements, and leads to diurnal changes in organelle protein abundances. These site II elements can act both as activators or repressors of transcription, depending on the night/day period and on the number and arrangement of site II elements in the promoter tested. These elements bind to the TCP family of transcriptions factors in Arabidopsis thaliana, which nearly all display distinct diurnal patterns of cycling transcript abundance. TCP2, TCP3, TCP11, and TCP15 were found to interact with different components of the core circadian clock in both yeast two-hybrid and direct protein–protein interaction assays, and tcp11 and tcp15 mutant plants showed altered transcript profiles for a number of core clock components, including LATE ELONGATED HYPOCOTYL1 and PSEUDO RESPONSE REGULATOR5. Thus, site II elements in the promoter regions of genes encoding mitochondrial, plastid, and peroxisomal proteins provide a direct mechanism for the coordination of expression for genes involved in a variety of organellar functions, including energy metabolism, with the time-of-day specific needs of the organism.
INTRODUCTION
All eukaryotes studied to date and even some prokaryotes have evolved an internal timekeeper in the form of a circadian clock. The central oscillator in this clock is made up of a series of interlocking, autoregulatory, negative feedback loops of gene expression thath produce self-sustained rhythms with a period length of ~24 h (Harmer, 2009). In its simplest form, the clock consists of (1) input sites that entrain the clock, including light, temperature, and nutrient availability (Harmer, 2009), (2) the core oscillator itself, and (3) a myriad of output sites to temporally coordinate biological processes (Harmer, 2009). In Arabidopsis thaliana, the core clock involves the reciprocal evening phase expression of TIMING OF CAB EXPRESSION1 (TOC1) with the morning phase expression of two MYB family transcription factors, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) (Harmer, 2009). A recent study has found that the transcription factor TCP21, termed CHE (for CCA1 Hiking Expedition), binds to TOC1, providing an important missing link to explain how TOC1 can regulate expression of CCA1, as TOC1 lacks a DNA binding domain (Pruneda-Paz et al., 2009). CHE acts as a repressor of CCA1 expression, as shown in CHE deficient plants, where CCA1 expression was increased (Pruneda-Paz et al., 2009).
TCPs are a plant-specific family of transcription factors, named after the transcription factors TEOSINTE BRANCHED1 in Zea mays, CYCLOIDEA in Anthirrinum majus, and PCF (proliferating cell nuclear antigen factor) in Oryza sativa (Navaud et al., 2007; Busch and Zachgo, 2009). As suggested by their name, the founding members of this family have been characterized to be involved in growth, cell proliferation, and organ identity in plants (Doebley et al., 1995; Luo et al., 1996, 1999; Nath et al., 2003; Takeda et al., 2003). They are basic-helix-loop-helix type transcription factors that can be divided into two main groups as a result of an ancient duplication event, TCP-P and TCP-C. TCP-P genes encode proteins that act to positively regulate gene expression, while TCP-C genes encode negative regulators (Kosugi and Ohashi, 2002; Li et al., 2005). TCP transcription factors bind cis-acting regulatory elements (CAREs), known as site II, with the core binding sequence TGGGC(C/T) in the promoter regions of various genes, the proliferating cell nuclear antigen from Arabidopsis and rice (Kosugi et al., 1995; Kosugi and Ohashi, 2002; Trémousaygue et al., 2003), as well as cyclin B and ribosomal protein genes in Arabidopsis (Li et al., 2005). Site II elements have been shown to be involved in the regulation of nuclear-located genes encoding mitochondrial proteins, specifically cytochrome oxidase 6b-2 (Cyt6b-2), Cyt5b-2, cytochrome c1, and cytochrome c2 in Arabidopsis (Welchen and Gonzalez, 2005; Gonzalez et al., 2007; Comelli and Gonzalez, 2009; Mufarrege et al., 2009; Welchen et al., 2009). Site II elements have also been shown to be involved in the regulation of Alternative oxidase1c in Arabidopsis, a non-stress-inducible alternative oxidase gene in Arabidopsis (Ho et al., 2007).
Although site II elements appear to be involved in the regulation of nuclear genes encoding mitochondrial proteins, the mechanism of their action is unclear as well as the nature of the link between circadian regulation of gene expression and the expression of nuclear genes encoding organellar proteins. Thus, we investigated the activity of a variety of gene promoters containing site II elements from genes encoding organelle proteins and found their activity was dependent on the phase of the day/light cycle. This dependency correlated to the diurnal variation in transcript abundance of both the genes regulated by these promoters and a range of TCP transcription factors. In a number of cases, we could show that the mitochondrial proteins encoded by the transcripts regulated by these promoters also varied in abundance in the day/light cycle. We determined which TCP transcription factors can interact with the site II element TGGGC(C/T) using yeast one-hybrid analysis and studied the interaction of TCP transcription factors with known components of the circadian clock in Arabidopsis, using yeast two-hybrid analysis and protein–protein interaction assays. Finally, we show how mutants of TCP transcription factors influence the expression of clock components in Arabidopsis. In this way, we established the mechanistic link of TCP transcription factors with both clock function and the expression of nuclear genes encoding organelle proteins in Arabidopsis.
RESULTS
Function of Site II Elements in Nuclear Genes Encoding Organelle Proteins Depends on the Phase of the Diurnal Cycle
To analyze the function of site II elements in the regulation of transcription for nuclear-encoded organellar proteins, the promoter regions of 15 nuclear genes encoding mitochondrial, plastid, peroxisomal, and ribosomal proteins were cloned. Initial characterization of the role of these site II elements in driving reporter activity indicated that there was no consistent significant effect of site II element deletions. The effect of site II promoter deletions varied on a day by day basis for a number of promoters tested, and an example of these data, obtained for the RIP promoter, is shown in Supplemental Figure 1A online. This technique has been used extensively to define promoter activity in our laboratory (Thirkettle-Watts et al., 2003; Ho et al., 2008; Giraud et al., 2009) and in other studies (Schmidt et al., 2004; Omidvar et al., 2008), so the variation cannot be ascribed to the method of transformation. The only variation in the above experimental design that we were aware of was the time of transformation during the light cycle. Quantitative RT-PCR (qRT-PCR) of RIP was performed to determine if and when transcript abundances for this gene varied. Supplemental Figure 1B online shows that the level of RIP transcript varied considerably and was lowest in the night period.
qRT-PCR was then performed to identify any patterns of transcript cycling abundance over the diurnal period for the other genes being studied with site II promoter elements. The transcripts of eight genes encoding mitochondrial proteins involved in a number of different functional processes from import to energy were evaluated. Specifically, these were MITOCHONDRIAL CARRIER PROTEIN3 (MCP3), MCP4, RISKE IRON SULFUR COMPLEX III SUBUNIT (RIP), ATP50 COMPLEX V SUBUNIT (ATP50), SCO1 COMPLEX IV SUBUNIT (SCO1), TRANSLOCASE OF THE OUTER MEMBRANE 20.2 (TOM20.2), TOM20.3, and TOM20.4. Two genes encoding plastid proteins were also tested, TRANSLOCASE OF THE OUTER CHLOROPLAST MEMBRANE120 (TOC120) and 60S RIBOSOMAL PROTEIN L34 (60SRPL34). Transcript abundances were also determined for two genes encoding peroxisomal proteins, PEROXISOME BIOGENESIS FACTOR11 (PEX11) and ALTERNATIVE NADH DEHYDROGENASE A1 (NDA1), which is dual targeted to the mitochondrion and the peroxisome (Carrie et al., 2008), along with four genes encoding known circadian-regulated transcripts, including the two morning associated transcripts, LHY1 and CCA1, and two evening phased transcripts, PSEUDO RESPONSE REGULATOR1 (PRR1/TOC1) and PRR5. Finally, one gene encoding a cytosolic ribosomal protein (60S ACIDIC RIBOSOMAL PROTEIN 0A [RPP0A]) was also tested (Li et al., 2005). The transcript abundances for all 24 genes encoding TCP proteins were also determined as these transcription factors are reported to be the binding partners for site II elements (Kosugi and Ohashi, 2002; Welchen and Gonzalez, 2005). However, for a number of TCPs, including TCP1, TCP6, TCP12, TCP16, TCP18, and TCP23, transcript abundances were too low to be accurately detected and were therefore omitted from further analysis.
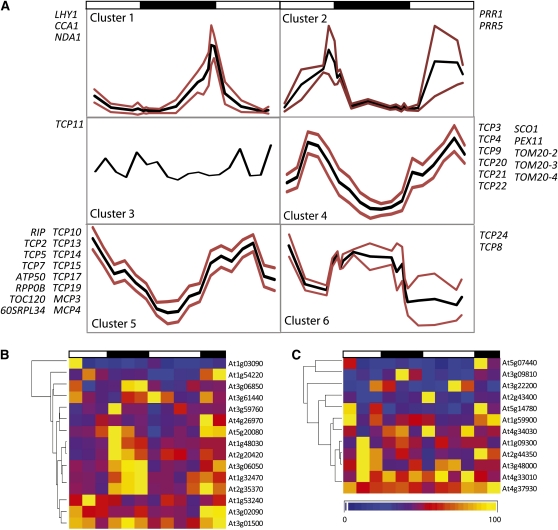
Six distinct expression patterns were observed. First, LHY1 and CCA1, which were analyzed as positive controls, along with the mitochondrial/peroxisomal NDA1, displayed classic morning phased expression that peaked as previously characterized (Elhafez et al., 2006) (Figure 1A, cluster 1; see Supplemental Figure 1C online). The second pattern represented the evening phased peak in expression levels for clock components PRR1 (TOC1) and PRR5 (Figure 1A, cluster 2). Another cluster was characterized by transcript abundances that rose during the light period and peaked at midday before decreasing to minimum levels at midnight. This pattern was evident for TOM20-2, TOM20-3, TOM20-4, SCO1, and PEX11 along with six TCP genes (Figure 1A, cluster 4; see Supplemental Figure 1C online). Transcripts in a fifth cluster group (MCP3, MCP4, RIP, 60SRPL34, RPP0B, ATP50, and TCO120, along with nine transcripts encoding TCPs) decreased throughout the day period to a minimum of roughly 3 h into the dark period and then increased throughout the night (Figure 1A, cluster 5; see Supplemental Figure 1C online). The sixth pattern (comprising TCP24 and TCP8) was opposite to that of the other clusters in that their transcript abundances were highest during the night period and there was a distinct plateau of transcript abundance during that period (Figure 1A, cluster 6). TCP11 showed relatively constitutive expression levels over the time series and no consistent diurnal pattern (Figure 1A, cluster 3).
Figure 1.
Diurnal Regulation of Transcript Abundance for Arabidopsis Genes Encoding Organelle Proteins, Core Clock Components, and TCP Transcription Factors.
(A) Transcript abundances were measured over a day/night time course for a range of transcripts encoding clock components, TCP factors, and organellar proteins with site II elements in their promoter regions. Fifteen-day-old Arabidopsis seedlings grown in a 12-h/12-h light-dark period were sampled at the times indicated. The maximum transcript abundance for each gene across the time series was set to 1, and all other values were expressed in a relative manner. Specific primer information is outlined in Supplemental Table 1B online. Transcript profiles were hierarchically clustered (shown in Supplemental Figure 1C online), and subclusters were defined as described in Methods. The trend for each subcluster (average of the cluster) is shown in black, and positive/negative limits (sd) are shown in red.
(B) and (C) Transcript abundance profiles over a 48-h day/night period for the 27 unique mitochondrial proteins that were defined as being diurnally regulated at a protein level in Lee et al. (2010). Public microarray data were taken from Smith et al. (2004) (first 24 h) and Bläsing et al. (2005) (second 24-h period). For each transcript, the maximum level across the time series was set to 1 with measurements for other time points being expressed in a relative manner. Expression levels are indicated by the scale bar below the profile in (C).
(B) Transcripts with site II elements in their promoter regions.
(C) Transcripts that lack site II elements in their promoter regions.
To further investigate the links between cycling abundances for mitochondrial located proteins and transcriptional regulation via site II CAREs in diurnal conditions, a transcriptional analysis of previously defined diurnally regulated mitochondrial proteins was undertaken. Lee et al. (2010) performed an in-depth analysis of the cycling patterns in protein abundances over day/night periods for mitochondrial located proteins. They identified 27 unique mitochondrial proteins with cycling abundances from differential two-dimensional gel electrophoresis isoelectric focusing/SDS-PAGE analysis (see Supplemental Data Set 1A online). From this list of 27 genes, we identified genes with site II elements within their 1000-bp promoter regions (Figures 1B and 1C). Over 50% of these diurnally regulated mitochondrial proteins had site II CAREs within their promoter region (15 out of 27), which is significantly more than would be expected by chance for a 6-mer in the genome. Changes in transcript abundance for these 15 genes with site II elements versus the remaining 12 genes without site II elements were visualized in diurnal microarray time series (Figures 1B and 1C, respectively). Genes with site II CAREs present in their promoter regions showed a significant cycling (one-way analysis of variance [ANOVA], P < 0.001) in transcript abundance over the day/night period (Figure 1B; see Supplemental Data Set 1B online), with generally higher levels of transcript abundance observed during the night period. By contrast, those genes without site II CAREs did not show clear cycling transcript abundance patterns (one-way ANOVA, P > 0.5) (Figure 1C; see Supplemental Data Set 1B online). A Tukey range test of the transcript patterns for the site II element-containing genes shows the variation between groups was all between dark and light periods (see Supplemental Data Set 1C online). This indicates that, while there are obviously other posttranscriptional mechanisms that also contribute to the diurnal changes observed in these mitochondrial proteins, the presence of site II elements in the promoter regions of those proteins diurnally regulated at the transcriptional level could play an important role in defining the variation observed. To analyze diurnal fluctuations in abundance of a mitochondrial protein encoded by one of the transcripts directly analyzed here, we performed immunoblots with the diurnal mitochondrial samples reported in Lee et al. (2010) using antibodies raised to TOM20-4. This also showed a significant diurnal pattern (one-way ANOVA, P < 0.001) with highest abundance early in the light period and lowest abundance early in the night (see Supplemental Data Set 1D online).
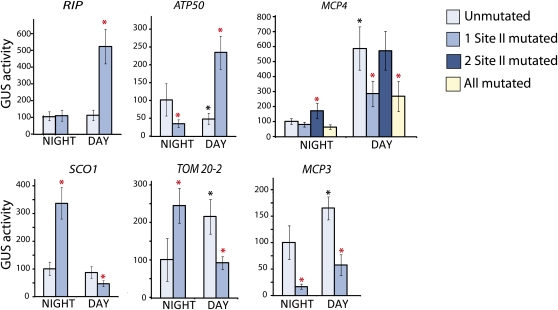
Taking into account these expression data, plants were transformed with wild-type or mutant promoter-reporter constructs lacking one or more site II elements either 2 h into the day or 2 h before the end of night conditions and harvested exactly 24 h later (in the same day/night conditions) to determine promoter activity. For the RIP promoter, it was observed that the site II element functioned as a strong repressor during the day (Figure 2, RIP), but it was not functional during the night period. Similarly for the ATP50 promoter, a 5-fold increase in promoter activity was observed under day conditions when the site II element was deleted, indicating that this element functions as a repressor during the day, and in contrast with this, during the night it acts as an activator of transcription (Figure 2, ATP50). ATP50 wild-type promoter activity was also reduced in light conditions to roughly half that of wild-type activity in the dark, consistent with these observations that site II elements present in the promoter activate expression of the gene during the night and strongly repress it during the day. In the case of the SCO1 and TOM20-2 promoters, the opposite was observed, with the site II element acting as a repressor at night and an activator during the day (Figure 2, SCO1 and TOM20-2). Altered wild-type promoter activity was also observed for TOM20-2 between day/night conditions with double the wild-type promoter activity observed during the day (also correlating with the activator activity of the site II element during the day) (Figure 2). In the MCP3 promoter, the site II element functioned as an activator both in the day and at night (Figure 2, MCP3). The MCP4 promoter was more complex due to the presence of multiple site II elements. A 5-fold increase in promoter activity was observed between night and day with the wild-type promoter, consistent with several site II elements acting as activators of transcription during the day (Figure 2, MCP4). Another site II element, which was not active during the day, acts as a repressor during the night, indicated by the slight increase in reporter gene activity when the element is mutated (Figure 2, MCP4, dark-blue bars). Therefore, in MCP4, a different set of site II promoter elements is used in dark versus light conditions rather than the same element having altered functionality at different points in the day/night cycle.
Figure 2.
Activity of the Promoter Regions of Mitochondria Gene Promoters with Site II Elements at Defined Points in the Diurnal Cycle.
GUS reporter activity of six mitochondrial promoters measured 2 h before the end of the night period and 2 h after the start of the day period are shown. The nighttime activity of the wild-type promoter was set to 100%, and other values were expressed in a relative manner for each promoter. Independent transformations using ~50 Arabidopsis seedlings were repeated at least nine times for each construct under day or night conditions, and the standard errors are indicated by error bars. Black asterisks indicate a significant difference (P < 0.05, Student’s t test) between day and night reporter activity of the wild-type promoter. Red asterisks indicate a significant difference (P < 0.05, Student’s t test) between the wild type and the promoter that contained one or more mutated site II elements.
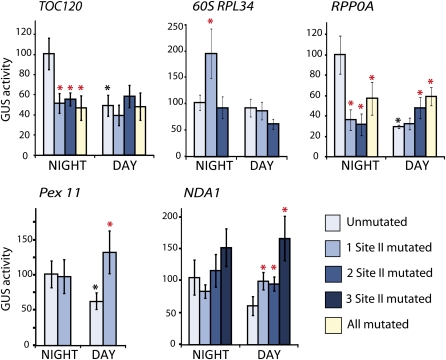
To determine whether site II elements also played a role in regulating other organellar and energy-related components in a diurnal manner, a number of other gene promoters were cloned and mutagenized for analysis in biolistic transformation assays as outlined above. Again, different site II functionality was observed under day and night conditions (Figure 3). TOC120 (chloroplastic) and RPP0A (ribosomal) promoters both showed a 2- to 4-fold decrease in wild-type promoter activity in the day compared with the night, and both promoters contain several site II elements that acted as strong activators in the dark (Figure 3, TOC120 and RPP0A). In the case of RPP0A, several of these elements also acted as repressors in the day, as indicated by increased reporter gene activity when they were mutated (Figure 3, RPP0A). The wild-type PEX11 promoter showed decreased activity during the day and contained a site II element that was not active at night but acted as a repressor during the day (Figure 3, PEX11). Similarly, in the NDA1 promoter, a number of site II elements present in the promoter were not functional during the night but acted as strong repressors during the day (Figure 3, NDA1).
Figure 3.
Activity of the Promoter Regions and Site II Elements for Genes Encoding Proteins Targeted to Peroxisomes, Plastids, or Ribosomal Proteins.
GUS reporter activity was assayed 2 h before the end of the night period and 2 h after the start of the day period. The night time activity of the wild-type promoter was set to 100%, and other values are expressed in a relative manner for each promoter. Independent transformations using ~50 Arabidopsis seedlings were repeated at least nine times for each construct under day or night conditions, and the standard errors are indicated by error bars. Black asterisks indicate a significant difference (P < 0.05, Student’s t test) between day and night reporter activity of the wild-type promoter. Red asterisks indicate a significant difference (P < 0.05, Student’s t test) between the wild-type and the promoter containing one or more mutated site II elements.
Comparison of these data sets showed that transcripts that clustered together in Figure 1A had similar functionality of the site II elements in their promoters. For example, SCO1 and TOM20-2 grouped together in cluster 3, which was characterized by decreased transcript abundances during the night (Figure 1A), and both these promoters contained site II elements that functioned as strong repressors during the night. While RIP, ATP50, MCP4, TOC132, and RPP0B4 all showed transcript abundances that generally increased throughout the night, all clustered together in cluster 4 (Figure 1A), and all have site II elements that were either not functional at night and were strong repressors during the day or acted as activators of transcription during the night (Figures 2 and 3). A number of additional promoters from genes encoding organelle proteins also showed altered functionality of the site II elements present depending on whether the elements were tested in day or night conditions (see Supplemental Figure 2 online).
TCP Proteins Are Able to Interact with Site II Elements in Yeast One-Hybrid Assays
To test the DNA binding ability and specificity of the 24 Arabidopsis TCP proteins with the characteristic site II C (TGGGCC) or site II T (TGGGCT) promoter sequences, yeast one-hybrid assays were performed (see Supplemental Figure 3 online; Table 1). Almost all of the TCPs tested were able to bind site II T or site II C sequences, with the exception of TCP8 and TCP22, which showed no interaction with the DNA sequences tested in this study. However, these factors may bind other sequences similar to site II or may bind the class II like sequences described by Kosugi and Ohashi (2002). TCP12, TCP4, TCP17, and TCP21 showed the strongest interaction with both forms of site II elements, while TCP3 and TCP11 seemed to show specificity for site II C elements, and TCP13 showed a strong interaction with site II T elements but was not able to bind site II C DNA sequences.
Table 1.
Summary of Results from Yeast One-Hybrid and Yeast Two-Hybrid Assays Showing Binding Specificity and Binding Partner Interactions for the TCP Family of Plant Transcription Factors
| Yeast One-Hybrid |
Yeast Two-Hybrid |
|||||||||||||
| Probe | AT No. | TCP Name | Type | Site II C (TGGGCC) | Site II T (TGGGCT) | PhyA | LHY | CCA1 | PIF3 | PRR1 | PRR3 | PRR5 | PRR7 | PRR9 |
| At1g67260 | AtTCP1 | CYC/TB1 | ± | – | * | * | * | * | * | * | * | * | * | |
| 254670_at | At4g18390 | AtTCP2 | CIN | + | ± | – | ο | ++ | ο | ο | ο | ο | – | – |
| 260618_at | At1g53230 | AtTCP3 | CIN | ++ | ± | – | – | – | – | ++ | – | – | – | – |
| 257267_at | At3g15030 | AtTCP4 | CIN | ++ | ++ | * | * | * | * | * | * | * | * | * |
| 247605_at | At5g60970 | AtTCP5 | CIN | – | ± | – | – | – | – | – | – | – | – | – |
| At5g41030 | AtTCP6 | PCF | + | + | – | – | – | – | – | – | – | – | – | |
| At5g23280 | AtTCP7 | PCF | + | + | – | – | – | – | – | – | – | – | – | |
| 246398_at | At1g58100 | AtTCP8 | PCF | – | – | – | – | – | – | – | – | – | – | – |
| 267515_at | At2g45680 | AtTCP9 | PCF | + | + | – | – | – | – | – | – | – | – | – |
| 266481_at | At2g31070 | AtTCP10 | CIN | ± | ± | * | * | * | * | * | * | * | * | * |
| 263888_at | At2g37000 | AtTCP11 | PCF | ++ | + | ο | ++ | ++ | – | ++ | ++ | + | – | – |
| At1g68800 | AtTCP12 | CYC/TB1 | ++ | ++ | – | – | – | – | – | – | – | – | – | |
| 259129_at | At3g02150 | AtTCP13 | CIN | – | ++ | – | – | – | – | – | – | – | – | – |
| 252425_at | At3g47620 | AtTCP14 | PCF | ± | ± | – | – | – | – | – | – | – | – | – |
| 260371_at | At1g69690 | AtTCP15 | PCF | + | – | – | – | – | – | + | – | ++ | – | – |
| At3g45150 | AtTCP16 | PCF | – | – | – | – | – | – | – | – | – | |||
| 250566_at | At5g08070 | AtTCP17 | CIN | ++ | ++ | – | – | – | – | – | – | – | – | – |
| At3g18550 | AtTCP18 | CYC/TB1 | + | + | * | * | * | * | * | * | * | * | * | |
| 248385_at | At5g51910 | AtTCP19 | PCF | + | + | – | – | – | – | – | – | – | – | – |
| 257788_at | At3g27010 | AtTCP20 | PCF | ± | + | * | * | * | * | * | * | * | * | * |
| 246011_at | At5g08330 | AtTCP21 | PCF | ++ | ++ | – | – | – | – | – | – | – | – | – |
| At1g72010 | AtTCP22 | PCF | – | – | – | – | – | – | – | – | – | – | – | |
| 262028_at | At1g35560 | AtTCP23 | PCF | + | + | – | – | – | – | – | – | – | – | – |
| 245774_at | At1g30210 | AtTCP24 | CIN | + | ± | * | * | * | * | * | * | * | * | * |
The 24 Arabidopsis TCPs were tested, and Affymetrix probe IDs are shown where available. Type refers to the class of TCP protein and is based on the founding members of the TCP family. Yeast one-hybrid assays were carried out with DNA sequences containing three copies of the classic site II sequence in tandem with either a C or a T in the last position (TGGGCC or TGGGCT). Yeast two-hybrid assays were performed to test for interactions with known components of the plant circadian clock as prey. All interactions were repeated a minimum of four times, and the strength of the interactions was graded as very strong (++), strong (+), very weak (±), or no interaction (–). For some TCP factors, consistently strong autoactivation was observed; thus, true interactions could not be determined (indicated by an asterisk). For TCP2, some autoactivation was observed when cloned into the bait vector for yeast two-hybrid assays; however, this was variable, and in some matings, interactions could be observed with LHY, PIF3, PRR1, PRR3, and PRR5 (indicated by ο).
TCP Proteins Interact with a Number of Central Clock Components in Yeast Two-Hybrid Assays
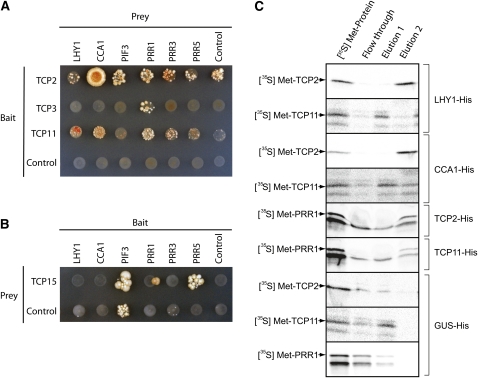
All 24 Arabidopsis TCP transcription factors were cloned for analysis in yeast two-hybrid assays to test their ability to interact with known components of the plant circadian clock. Interactions were tested with PhyA, two MYB factors that are central regulators in the circadian clock (CCA1 and LHY), PIF3, and all five PRR proteins with proposed functions in the circadian clock (PRR1, 3, 5, 7, and 9). The ability of TCP proteins to autoactivate in yeast hybrid screens has been described previously (Kosugi and Ohashi, 2002; Koyama et al., 2007); thus, the ability of all these factors to autoactivate was tested in every mating (Figures 4A and 4B; see Supplemental Figure 4 online). TCP1, 4, 10, 12, 18, 20, and 24 consistently autoactivated when cloned into both prey and bait vectors; thus, interactions with these components could not be determined (Table 1; see Supplemental Figure 4 online). TCP2 was found to interact very strongly with CCA1 (Figure 4A, Table 1). Interactions may also take place between TCP2 and LHY, PIF3, PRR1, PRR3, and PRR5; however, this cannot be fully determined as TCP2 was able to autoactivate when it was cloned into the bait vector for yeast two-hybrid assays. When cloned into the prey vector for a reversed assay, some interactions with these components were observed (Table 1). TCP3 was found to interact very strongly with PRR1 (also known as TOC1) (Figure 4A, Table 1). TCP15 was found to interact with PRR5 and to a lesser extent with PRR1/TOC1 (Figure 4B). Finally, TCP11 interacted very strongly with PRR1 (TOC1), LHY, CCA1, and PRR3 and to a lesser extent with PRR5 (Figure 4A, Table 1). Additionally, TCP11 was observed to interact with PHYA; however, this interaction could not be confirmed in matings when the yeast two-ybrid prey and bait vectors were reversed due to autoactivation (Table 1; see Supplemental Figure 4 online). No interactions were observed for TCP21, even though this factor was previously described by Pruneda-Paz et al. (2009) to interact with PRR1 (TOC1) in the central circadian clock (see Supplemental Figure 4 online).
Figure 4.
Protein Interactions between TCP Factors and Circadian Regulators.
(A) and (B) Yeast two-hybrid assays. Positive interactions were determined through auxotrophic selection media, SD-Trp/-Leu/-Ade/-His. TCP2, TCP3, and TCP11 were cloned into the bait vector and were able to activate the transcription of reporter genes.
(A) TCP proteins were cloned into the bait vector and circadian regulators were cloned into the prey vector. TCP2 interacts with CCA1. TCP3 interacts with PRR1. TCP11 interacts with LHY1, CCA1, PRR1, PRR3, and PRR5.
(B) TCP15 was cloned into the prey vector, while circadian regulators were cloned into the bait vector. TCP15 interacts with PRR1 and PRR5. Controls are bait empty vector or prey empty vector.
(C) Protein–protein interaction determined using affinity purification and His-tagged proteins. Tagged versions of LHY1, CCA1, TCP2, and TCP 11 were expressed and bound to a nickel affinity resin under native conditions. The ability to bind other proteins was determined by applying translation lysates programmed with TCP2, TCP11, and PRR1. The first lane represents 2.5% of the radiolabeled protein applied to the column, flow-through represents unbound protein, Elution 1 represents washing with 10 mM imidazole, which will not disrupt binding of His-tag protein to resin, and Elution 2 represents washing with 250 mM imidazole, which will elute the tagged protein and any protein bound. The presence of radiolabeled protein in Elution 2 represents strong binding, seen with TCP2 to LYH1, CCA1, and PRR1. TCP11 was able to bind to PRR1 and weakly to CCA1. Note that radiolabeled TCP2, TCP11, and PRR1 present in the flow-through fraction indicate no binding to the GUS-His (control). Arrows indicate the correct size, full-length translation product. Note that for PRR1, the second product in the translation mixture marked with an asterisk was also present in the flow-through fraction, but the full-length product was only evident in Elution 2 (see text).
To confirm the interactions observed in the yeast two-hybrid analyses, direct protein–protein interactions were also undertaken. This was achieved by expressing His-tagged versions of the target proteins, binding them to a nickel affinity column under native conditions, and testing their ability to bind other proteins. His-tagged versions of LYH1, CCA1, TCP2, and TCP11 were expressed in a wheat germ translation system and bound to a nickel affinity column as target genes. As a control, a similar procedure was performed for β-glucuronidase (GUS). To test for protein–protein interactions, radiolabeled proteins were synthesized and applied to columns containing the putative binding partners. The ability of the protein to interact with circadian regulators was determined by the retention of the protein on the column, which could then only be eluted under conditions that eluted the His-tagged proteins (Figure 4C, Elution 2). When TCP2, TCP11, and PRR1 were applied to the columns containing the bound GUS-HIS control protein, no radiolabeled protein was retained on the column, but instead it appeared in the flow-through and in small amounts after elution with 10 mM imidazole (Figure 4C, bottom three blot panels). By contrast, when TCP2, TCP11, CCA1, or LHY1 were bound on the column, the radiolabeled proteins were retained on the column and were absent in the flow-through, indicating protein–protein interactions. Different binding affinities were suggested by elution under different imidazole concentrations. For [35S]Met-TCP2, strong binding was evident with both LHY1-His and CCA1-His, as protein was only eluted under conditions that eluted the tag from the column (Figure 4C). In yeast two-hybrid assays, interactions between TCP2 and LHY1 could not be determined due to autoactivation (Figure 4A, Table 1). The binding of TCP2 to LHY1 was evident in the affinity purification assays as the radiolabeled TCP2 protein was only eluted from the column under conditions that eluted the HIS-tagged LHY1 protein (Figure 4C).
A strong protein–protein interaction was evident between PRR1 and both TCP2-His and TCP11-His, as the correct full-length PRR1 protein product (top band shown by arrow in gel panels 5 and 6, Figure 4C) was only eluted under conditions that elute TCP2-HIS and TCP11-HIS. Interestingly, two translation products were evident for PRR1, a full-length product (indicated with an arrow) and a second shorter translation product initiated from an internal Met codon that was 75 amino acids into the protein (indicated by an asterisk) (Figure 4C). The shorter PRR1 translation product was evident in the flow-through and the first elution wash with 10 mM imidazole; therefore, efficient binding of TCP factors requires the first 75 amino acids of PRR1. Binding of TCP11 to CCA1 and LHY1 was suggested by retention on the column and low abundance in the flow-through fraction. Some protein from these interaction assays was eluted at 10 mM imidazole and all at 250 mM imidazole, suggesting under these assay conditions that binding was weaker than the interactions outlined above. The interaction of PRR1 with TCP2 and TCP11 provides a molecular mechanism for these regulators to bind and act as transcriptional activators and further supports the hypothesis that TCP transcription factors function as DNA binding partners of PRR proteins in the circadian clock.
Transcript Abundance Profiles for Clock Components Are Altered in TCP Mutants
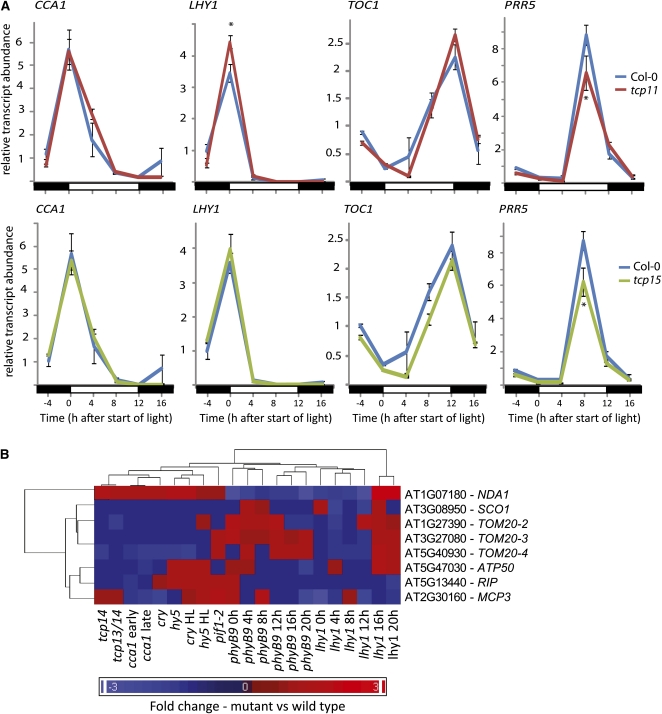
To address whether the TCP factors are necessary to correctly regulate clock function in vivo, transcript abundance profiles for several marker genes for clock function were monitored in TCP mutants grown in normal diurnal growth conditions. T-DNA insertional lines were obtained for TCP11 and TCP15 to determine if the central oscillator loop was affected in the absence of these factors (tcp11 and tcp15 plants) (see Supplemental Figure 5 online). In tcp11 (the knockout of a TCP that showed direct protein interactions with LHY1, CCA1, PRR1, PRR3, and PRR5 in Figure 4), the expected peak in LHY1 expression during the early morning was increased by 30%, indicating that TCP11 plays some role in repressing LHY1 expression levels (Figure 5A). There were no differences in phase or period of the LHY1 expression profile in tcp11 plants compared with the wild type. Transcript profiles for CCA1 and TOC1 were not significantly altered in tcp11 lines (Figure 5A). The expression profile of PRR5 was also measured, as TCP11 interacts with this PRR factor in yeast two-hybrid analyses (Figure 4). The expected peak in PRR5 expression in the late afternoon was significantly decreased by over 30% in the tcp11 lines, indicating that TCP11 also appears to have a role in the transcriptional activation of this PRR factor in a time-of-day specific manner (Figure 5A). The same four marker genes were measured in tcp15 plants, with no significant differences seen between the mutant and wild type in amplitude, phase, or period of the transcript profiles for CCA1, LHY1, or TOC1. However, a >30% decrease in the amplitude of peak expression for PRR5 was observed in tcp15 plant profiles (Figure 5A). This is in agreement with protein interaction studies, as TCP15 was only observed to interact with PRR5 (Figure 4B); thus, PRR5 regulation also appears to require the presence of TCP15.
Figure 5.
Analysis of Transcript Abundance Profiles for Core Clock Marker Genes in TCP Transgenic Plant Backgrounds.
(A) Transcript abundance profiles for CCA1, LHY1, PRR1, and PRR5 were measured via qRT-PCR over a 24-h period in 16-d-old, wild-type Arabidopsis plants (blue), plants lacking a functional TCP11 protein (tcp11- red), or plants lacking a functional TCP15 factor (tcp15, green). Plants were grown in 12-h/12-h day-night conditions, and samples were taken every 4 h over a diurnal time course (−4, 0, 4, 8, 12, and 16 h after the start of light conditions). Transcripts were measured in technical duplicate for each of the three biological replicates, consisting of pooled tissue collected from approximately five seedlings at each time point. Average relative transcript abundances are shown with standard errors for each time point. Asterisks indicate a significant difference between the abundance observed between mutant and wild-type plants (Student’s t test, P < 0.001). Black bar, night; white bar, day.
(B) Hierarchical cluster of expression profiles for the genes shown in Figure 2 in a range of mutant backgrounds at different time points after exposure to light. Fold changes are presented as values compared with the corresponding transcript abundance observed for wild-type plants. Blue indicates genes that are downregulated compared with the wild type in particular mutants, and red represents an upregulation compared with the wild type. For detailed methods on experimental data sets used and cluster generation see Supplemental Figure 6 online.
Additionally, a list of 365 nuclear genes encoding mitochondrial proteins with site II elements in their proximal promoter regions was generated. Transcript abundance profiles for these genes were investigated for 10 mutant backgrounds, namely, phyB9, lhy1, cca1, tcp14, tcp13, pif1-2, cry, hy5, phyA, and a tcp13 tcp14 double mutant (McCormac and Terry, 2002; Kleine et al., 2007; Nozue et al., 2007; Michael et al., 2008; Moon et al., 2008) (see Methods). Transcript profiles were hierarchically clustered, revealing both up- and downregulation of transcripts across the mutants (see Supplemental Figure 6 online). From this list, 293 genes were significantly upregulated or downregulated >1.5-fold in at least one of these mutant backgrounds. This represents just over 80% of the total set of 365 genes with site II elements encoding mitochondrial-located proteins. Many of the remaining 72 transcripts were very low in abundance and could not be accurately detected on the microarray chips. Additionally, 117 of the genes on this list had significantly altered expression levels in more than five clock-related mutant microarray experiments. As further confirmation that the nuclear-encoded mitochondrial transcription networks regulated by site II elements are directly affected in mutants of clock function, differences in transcript profiles for genes of the promoters that were characterized in Figure 2 were further examined in these 10 mutant backgrounds (Figure 5B). For each of these transcripts, transcript abundances different to those observed in wild-type plants were evident in at least two and up to nine of the mutant backgrounds (Figure 5B).
DISCUSSION
This study aimed to define the role of site II cis-acting regulatory elements in the promoters of nuclear genes encoding organellar proteins and to investigate whether these genes display diurnal cycling patterns of expression due to the presence of site II elements. It was important to determine if they were explicitly linked to the clock input and core feedback loops of gene expression or whether they just regulated the cycling expression of organellar proteins temporally as one of the multitude of clock-regulated output sites in plants. TCP2, TCP3, TCP11, and TCP15 were all found to interact with different components of the core circadian clock in both yeast two-hybrid assays and protein–protein interaction assays, indicating that the TCP family of transcription factors is intricately linked with circadian regulation of gene expression in Arabidopsis. TCP3 only interacts with PRR1 in the core feedback loop, and TCP2 and TCP11 interact with a variety of components, including PRR1. Notably, a second translation product for PRR1 (minus the N-terminal 75 amino acids) was observed for the column affinity purification interaction assays, and only the full-length translational product was able to interact with the TCP11 and TCP 2 factors. The CCT domain at the C-terminal of the PRR proteins is believed to mediate protein–protein interactions; however, our data indicate the N-terminal 75 amino acids of PPR1 are necessary to mediate binding to the TCP factors either directly or by providing conformational or structural support for the interaction.
TCP11 is the only Arabidopsis TCP that is not diurnally regulated at a transcript abundance level in the qRT-PCR assays performed in this study, and it was the only Arabidopsis TCP that was not defined as diurnally regulated (r > 0.8) in any public microarray data set using the DIURNAL database (Mockler et al., 2007) (http://diurnal.cgrb.oregonstate.edu/). TCP11 interacts with the greatest number of clock components in this study, including three different PRR factors. A recent study illustrated that PRR factors (TOC1 and PRR5) interact with each other in vitro and in vivo to enhance nuclear accumulation of TOC1 over 2-fold, in a time-of-day specific manner (Wang et al., 2010). PRR5 also recruits TOC1 to large subnuclear foci and promotes phosphorylation of the N-terminal region of TOC1 (the region that was found to be necessary for the TCP interactions shown in Figure 4C). Notably, these are the two PRR factors that showed the greatest ability to interact with TCP factors in this study, at least in terms of the number of TCP partners and the strength of the interactions. It is possible that TCP11 is recruited by TOC1 or PRR5 in a similar fashion and, thus, its activity would be regulated in a location-dependent manner rather than at the transcriptional level. Another possible explanation for the lack of cycling transcript abundance observed for TCP11 is that it is constitutively expressed as a transcriptional repressor of LHY1, mediated via interactions with the various PRR factors during the day time period. Traditionally, the circadian clock has been assumed to consist of a single core negative loop (CCA1, LHY1, and TOC1) (Alabadí et al., 2001) and a number of additional feedback loops and other factors associated with many oscillators integrated outside the central loop (PRR3, PRR5, PRR7, PRR9, and GI) that generally have less well known molecular functions within the clock, as reviewed by Harmer (2009). However, studies have now shown that the other PRR factors are able to act as transcriptional repressors of CCA1 and LHY1 promoter activity along with TOC1/PRR1 (Nakamichi et al., 2005b, 2010). PRR9, 7, 5, and 1 all display sequential peaks in expression throughout the day period and have been characterized as direct repressors of CCA1 and LHY1 in vitro and in vivo (Matsushika et al., 2000; Nakamichi et al., 2005a, 2005b, 2010). Binding of the constitutively expressed TCP11 to the LHY1 promoter provides a mechanism through which each of these PRR factors could associate with TCP11 to repress LHY1 throughout the day period (consistent with the timing of decreased LHY1 expression; Figure 5). This is also in agreement with the observed increase in LHY1 expression levels in plants lacking TCP11 in this study (Figure 5). Pruneda-Paz et al. (2009) similarly identified CHE as a transcriptional repressor of CCA1 in association with TOC1, but CHE does not appear to regulate LHY1 transcriptionally. Thus, the results presented in this work would represent a similar mechanism by which LHY1 is also transcriptionally repressed during the day through TCP factors associated with TOC1. Interestingly, the proposed role of TCP11 within the central clock in Arabidopsis as outlined in this study represents the involvement of a factor that does not cycle in transcript abundance itself.
TCP15 was shown to interact with PRR5, whose role in the circadian clock is not fully elucidated, but is likely to involve subsequent feedback loops to repress the expression of CCA1 and LHY (Nakamichi et al., 2005a, 2005b; Locke et al., 2006). As shown by the transcript profiles of marker clock genes analyzed in tcp15 plants, PRR5 was the only transcript that was significantly altered compared with wild-type expression peaks, again confirming interactions between PRR5 and TCP15 and the related regulation of these factors in vivo. Interestingly, the factors that exhibited the strongest interactions in the yeast two-hybrid analysis also displayed the strongest interactions in the direct protein–protein interactions tested in the affinity purification column assays (e.g., CCA1 with TCP2), reinforcing the combined use of these assays to confirm TCP binding partners for the clock components and PRR factors that lack DNA binding domains.
A number of recent metabolomic and transcriptomic studies have revealed a central role for PRR5, 7, and 9 in regulating metabolic homeostasis in the tricarboxylic acid (TCA) cycle within plant mitochondria (Fukushima et al., 2009; Nakamichi et al., 2009). In these previous studies, prr5 prr7 prr9 triple knockout mutants displayed dramatic increases in intermediates of the TCA cycle in the metabolomic analysis. In the study by Lee et al. (2010), which characterized a small but significant set of mitochondrial proteins that are diurnally regulated at the protein level, a significant number of TCA cycle components and components of energy and amino acid metabolism were also identified. An examination of these protein changes at the transcriptional level in this study indicates that site II elements may play a significant role in the diurnal transcriptional regulation of these genes. This can be observed in the diurnal cycling patterns in the transcripts with site II in contrast with a relative lack of cycling pattern in genes lacking site II promoter elements from this set of diurnal mitochondrial proteins identified by Lee et al. (2010). This also suggests the presence of additional mechanisms, separate from transcriptional control, by which mitochondrial protein abundances are altered diurnally. However, site II elements and the TCP factors that bind to them appear to link the diurnal changes in mitochondrial function, particularly TCA cycle function and core energy metabolism/amino acid metabolism, with transcriptional changes that are regulated and integrated with the central clock and PRR family function (Fukushima et al., 2009; Nakamichi et al., 2009). Many studies have identified a link between sucrose or organellar metabolism and diurnal regulation of gene expression (Smith et al., 2004; Bläsing et al., 2005; Usadel et al., 2008). From the results here, we propose that the missing molecular link between cellular and organelle metabolic activity and the circadian clock in plants is provided by the TCP family of transcription factors and the site II CAREs to which they bind.
Overall, our data show that Arabidopsis TCP transcription factors appear to be central regulators in circadian clock input sites, the clock oscillator itself, and a myriad of clock output sites, directing and regulating the expression of genes encoding cellular energy metabolism components, particularly in the plant energy organelles. The involvement of TCP transcription factors in various growth and developmental processes and the regulation of genes encoding ribosomal proteins provides a molecular link to finely coordinate metabolism, growth, and development.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana plants, ecotype Columbia (Col-0) were grown at 22°C under medium day conditions, 12 h at 100 μE m−2 s−1 light conditions and 12 h of dark, for 16 d. Plants were grown on Murashige and Skoog (MS) agar plates for transient transformation assays and all transcript analysis. Homozygous transgenic T-DNA insertion lines were obtained and confirmed for TCP 11 (SALK_003042 and SALK_020124.38.95.x) and TCP15 (SALK_011491) (see Supplemental Figure 5 online). The two independent T-DNA insertional lines for TCP11 were combined for qRT-PCR analysis so that an average response of the lines was determined compared with wild-type plants. Screening primers are shown in see Supplemental Tables 1A and 1B online.
Transient Transformation Assays
Transformation was performed using the PDS-1000 system with the Hepta adaptor according to the manufacturer’s instructions (Bio-Rad), as previously described (Thirkettle-Watts et al., 2003; Ho et al., 2008). For each of the 15 promoters analyzed, transient transformations were repeated independently over several different days a minimum of nine times. For each transformation, ~50 16-d-old Arabidopsis seedlings grown on MS plates (as described above) were transformed. Thus, for each promoter construct to be tested, ~500 plants were transformed independently. Dark transformations were performed in the dark 2 h prior to the start of light conditions, and light transformations were performed 2 h after the initiation of light conditions. Transformed seedling leaf tissue was harvested from the plates 24 h later under the same conditions as used for the transformation, for analysis of GUS and LUC reporter gene activity. Reporter gene activities for transformations performed 12 h apart could not be directly compared. Determination of GUS and Luc activities and all data processing steps were performed as described previously (Ho et al., 2008). The inclusion of a second, constitutively driven, reporter gene, LUC, in the transient transformation vector enabled the transformation efficiency for each transformation to be calculated and corrected for, thus providing a far more accurate analysis of variations in promoter activity (outlined in Ho et al., 2008).
Transcript Abundance Analysis
qRT-PCR analysis was performed on green tissue excised from approximately five 2-week-old Arabidopsis seedlings, per sample, grown on MS plates, and sampled at various time points throughout the 12-h/12-h light/dark day conditions in which the seedlings had been grown. For profiles shown in Figure 1, samples were taken at 3, 6, 10, 11, and 12 h after illumination, 0.5, 1, 3, 6, 10, 11, and 12 h after the start of dark conditions, and finally 0.5, 1, 3, 6, 10, and 11 h after illumination, resulting in an 18-point time course over 36 h. For transcript profiles of core clock components in the TCP mutant backgrounds (Figure 5), samples were taken every 4 h over a 20 h, period resulting in a six-point time series (−4, 0, 4, 8, 12, and 16 h ZT time) for wild-type and each of the mutant backgrounds. Samples collected for each time point for the two confirmed tcp15 insertional lines were combined for qRT-PCR analysis. Samples for each time point were taken in biological triplicate (from seedlings grown on separate MS plates) and snap frozen under liquid nitrogen. Total RNA isolation and cDNA synthesis was performed as described previously (Lister et al., 2004). Transcript levels were assayed using the LightCycler 480 and the LightCycler 480 SYBR Green I Master (Roche). From each independent cDNA preparation, each transcript was analyzed twice. Absolute transcript abundance measurements, determined by standard curves for each gene, were normalized to that of UBC, which was analyzed as a constitutive control. To normalize for differences in the basal level of expression between the different genes measured in qRT-PCR assays, the values of absolute transcript abundance for each gene were expressed relative to the maximum value recorded for the gene (i.e., transcript abundance values ranged from 0 to 1 for each gene measured). Hierarchically, clusters were generated using Euclidian distance and average linkage measures. The trend (average) of each subcluster was graphed (in black) along with the positive and negative standard deviation limits (in red). qRT-PCR primer sequences used for all transcripts measured are listed in Supplemental Table 1B online. Summary statistics and quality measure for each of the assays performed are shown in Supplemental Data Set 2A online. Normalized absolute transcript abundance data used to generate the cluster images in Figure 1 and Supplemental Figure 1 online are also supplied in Supplemental Data Set 2B online.
Analysis of Public Microarray Data Sets
Raw CEL file microarray data sets were downloaded from the NASCarrays repository (http://affy.Arabidopsis.info/narrays/experimentbrowse.pl), ArrayExpress (Smith et al., 2004), and from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) (Bläsing et al., 2005). The two data sets, each with a 24-h diurnal time course of plants grown under 12-h/12-h light-dark conditions, were combined to form a 48-h time series with 0-, 4-, 8-, 12-, 16-, and 20-h time points taken from Smith et al. (2004), followed by 0-, 4-, 8-, 12-, 16-, and 20-h time points taken from Bläsing et al. (2005). These two experimental data sets were taken as they most closely resemble the sampling, age of plants, and growth conditions used in Lee et al. (2010). CEL files were subjected to GC-robust multi-array average normalization in Partek Genomics Suite software version 6.4. The linear transcript abundance profiles for the 27 mitochondrial proteins defined as diurnally regulated by Lee et al. (2010) were isolated. To normalize for differences in the basal level of expression between the different genes, the fluorescence intensity profile for each gene was expressed relative to the maximum value recorded for the gene (i.e., transcript abundance values ranged from 0 to 100 for each gene measured), with 100 being the maximum expression level and all other time points expressed relative to this. The presence of site II elements or site II–like elements was searched in these 27 gene promoters using The Arabidopsis Information Resource PATMATCH function (http://www.Arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl), resulting in 15 genes out of 27. Transcript profiles over the diurnal time series for both gene sets, with or without site II elements, were hierarchically clustered using Euclidean distance and average linkage measures in Partek Genomics Suite software version 6.4.
Yeast One-Hybrid Screen
For construction of the pHIS2 bait vectors, forward and reverse oligonucleotides (see Supplemental Table 1A online) were annealed and ligated into EcoRI-SacI–linearized pHIS2 vector. Three tandem repeats of the site II C (TGGGCC) or site II T (TGGGCT) were cloned into the pHIS2 reporter vector upstream of the minimal HIS3 promoter region and HIS3 nutritional reporter gene. For transformation, competent yeast cells were prepared according to the Clontech Yeast Protocols Handbook using the Y187 yeast strain (Clontech). Yeast one-hybrid transformation screens were performed using the Clontech Matchmaker one hybrid library construction and screening kit. For each yeast one-hybrid transformation screen, 100 μL of competent yeast cells were incubated with 100 ng of pHIS2 bait vector and 100 ng of pGADT7-Rec2 prey vector, 100 μg herring testes carrier DNA (Clontech), and 0.6 mL PEG/LiAc solution. Cells were transformed according to the manufacturer’s instructions. Transformations were plated onto SD media -Leu-Trp to select for cotransformed cells and incubated at 28°C for 4 d. The pGADT7-rec2-p53 prey vector in combination with p53HIS2 was used as a positive control and pGADT7-rec2-p53 in combination with pHIS2 as negative control. Transformed yeast cells were subsequently grown overnight in YPD liquid media to an OD600 of 0.1 and diluted in a 10× dilution series. From each dilution, 5 μL was spotted on SD-Trp-Leu (DDO) and on SD-Trp-Leu-His (TDO) media plates supplemented with 90 mM 3-amino-1,2,4-triazole (Sigma-Aldrich). The plates were then incubated for 3 d at 28°C.
Yeast Two-Hybrid Assay
Protein–protein interactions were analyzed using a GAL-4–based yeast hybrid system (Matchmaker two-hybrid system; Clontech). Full-length cDNAs of all Arabidopsis TCP proteins were cloned into pGBKT7 (bait vector; Clontech) and pGADT7-Rec (prey vector; Clontech) and transformed into the Y187 yeast strain. Transformants were selected on synthetic dropout (SD)-Trp media. PHYA, LHY1, CCA1, PIF3, PRR1, PRR3, PRR5, PRR7, and PRR9 full-length cDNAs were cloned into the prey vector pGADT7-Rec (Clontech) and transformed into the AH109 yeast strain. Transformants were selected on SD-Leu medium. All cloning primers are listed in Supplemental Table 1A online. TCP16 and PRR5 cDNAs were obtained from the ABRC (clone PYAt3g45150) and Takeshi Mizuno (Laboratory of Molecular Microbiology, Nagoya University), respectively. Bait and prey strains were inoculated in YPDA and combined in a 96-well plate (NUNC) for mating, and yeast culture was then incubated at 28°C with shaking for 16 to 19 h. Successful yeast matings were confirmed on SD-Trp/-Leu, and positive interactions were selected on SD-Trp/-Leu/-His/-Ade. pGBKT7-53 and pGBKT7-Lam (Clonetech) were used as a positive and a negative bait control, while pGADT7-RecT (Clontech) was used as a prey control. Nonmated yeast cultures were also spotted on SD media as negative controls. pGBKT7 and pGADT7-Rec were used as negative controls for autoactivation. To discover interactions that might be masked from autoactivation when expressing TCP proteins as baits, TCP proteins were also cloned as preys, while potential interactors were cloned as baits for yeast matings. Each mating was performed a minimum of four times.
Protein Pull-Down Assays
For protein pull-down assays, LHY1, CCA1, TCP2, TCP11, and GUS were expressed in a cell-free wheat germ lysate with 6X HIS tags (Roche). After expression, the lysates were made up to 5 mL with native immobilized metal-ion affinity chromatography (IMAC) wash buffer 1 (300 mM KCl, 50 mM KH2PO4, and 5 mM imidazole, pH 8.0). These proteins were then bound to an IMAC purification column (Bio-Rad) using the Profinia purification system (Bio-Rad). The proteins were loaded on the column and washed once with native IMAC wash buffer 1 and once with native IMAC wash buffer 2 (300 mM KCl, 50 mM KH2PO4, and 10 mM imidazole, pH 8.0). To perform the pull-down assay, [35S]-Met radiolabeled proteins (TCP2, TCP11, and PRR1) expressed via the Promega TnT transcription/translation kit (Promega) were made up to 5 mL in native IMAC wash buffer 1 supplemented with 0.5% BSA and 2 mM MgCl2. Radiolabeled proteins were washed over the columns containing the already bound 6X HIS–containing proteins. Columns were washed once in native IMAC wash buffer 1 (termed the flow through) and then eluted first with elution buffer 1 (300 mM KCl, 50 mM KH2PO4, and 10 mM imidazole, pH 8.0) and subsequently with elution buffer 2 (300 mM KCl, 50 mM KH2PO4, and 250 mM imidazole, pH 8.0) to determine the strength of the interaction. The GUS protein was used as a control for nonspecific binding of radiolabeled proteins to the column. After each elution and wash step was performed, fractions were analyzed by SDS-PAGE and then subjected to radioautography to detect the presence of radiolabeled proteins in the various fractions.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Results for Initial Promoter Transformations and qRT-PCR Analysis for Arabidopsis Genes Encoding Organelle Proteins, Core Clock Components, and TCP Transcription Factors over a 38-h Period under Diurnal Conditions.
Supplemental Figure 2. Additional Promoter Elements Tested.
Supplemental Figure 3. Yeast One-Hybrid Assays.
Supplemental Figure 4. Yeast Two-Hybrid Analysis for Interactions between Arabidopsis TCP Family Proteins and Components of the Circadian Clock.
Supplemental Figure 5. Characterization of the T-DNA Insertional Lines Inactivating TCP11 and TCP15.
Supplemental Figure 6. Transcript Abundances for Genes Encoding Mitochondrial Proteins with Site II in a Range of Mutant Backgrounds.
Supplemental Table 1A. Primer Sequences for Cloning of All TCP Factors and Clock Components for Yeast Hybrid and Protein Interaction Studies.
Supplemental Table 1B. Primer Sequences for All Quantitative RT-PCR Assays along with Primers Used for Screening T-DNA Insertional Lines.
Supplemental Data Set 1A. Summary of the Mitochondrial Proteins That Were Found to Cycle in Protein Abundance in a Diurnal Manner, as Characterized by Lee et al. (2010).
Supplemental Data Set 1B. One-Way ANOVA Statistical Analysis for Transcript Abundance Profiles for Genes with and without Site II Elements Shown in Figures 1B and 1C, Respectively.
Supplemental Data Set 1C. Tukey Range Analysis of Diurnal Transcript Abundance Profiles for Genes with and without Site II Elements Shown in Cluster Images in Figures 1B and 1C, Respectively.
Supplemental Data Set 1D. One-Way ANOVA Statistical Analysis for Protein Abundance Profiles Shown in Figure 4 for TOM20-4 and TOM40-1.
Supplemental Data Set 2A. qRT-PCR Assay Summaries for Cluster Generation.
Supplemental Data Set 2B. Normalized Absolute Transcript Abundance Values as Determined by qRT-PCR Analysis Used for the Generation of Cluster Images in Figure 1 and Supplemental Figure 1.
Acknowledgments
This work was supported by the Australian Research Council Centre of Excellence Program CEO561495.
References
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Bläsing O.E., Gibon Y., Günther M., Höhne M., Morcuende R., Osuna D., Thimm O., Usadel B., Scheible W.R., Stitt M. (2005). Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A., Zachgo S. (2009). Flower symmetry evolution: Towards understanding the abominable mystery of angiosperm radiation. Bioessays 31: 1181–1190 [DOI] [PubMed] [Google Scholar]
- Carrie C., Murcha M.W., Kuehn K., Duncan O., Barthet M., Smith P.M., Eubel H., Meyer E., Day D.A., Millar A.H., Whelan J. (2008). Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett. 582: 3073–3079 [DOI] [PubMed] [Google Scholar]
- Comelli R.N., Gonzalez D.H. (2009). Identification of regulatory elements involved in expression and induction by sucrose and UV-B light of the Arabidopsis thaliana COX5b-2 gene, encoding an isoform of cytochrome c oxidase subunit 5b. Physiol. Plant. 137: 213–224 [DOI] [PubMed] [Google Scholar]
- Doebley J., Stec A., Gustus C. (1995). teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141: 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhafez D., Murcha M.W., Clifton R., Soole K.L., Day D.A., Whelan J. (2006). Characterization of mitochondrial alternative NAD(P)H dehydrogenases in Arabidopsis: Intraorganelle location and expression. Plant Cell Physiol. 47: 43–54 [DOI] [PubMed] [Google Scholar]
- Fukushima A., Kusano M., Nakamichi N., Kobayashi M., Hayashi N., Sakakibara H., Mizuno T., Saito K. (2009). Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc. Natl. Acad. Sci. USA 106: 7251–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Van Aken O., Ho L.H., Whelan J. (2009). The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 150: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D.H., Welchen E., Attallah C.V., Comelli R.N., Mufarrege E.F. (2007). Transcriptional coordination of the biogenesis of the oxidative phosphorylation machinery in plants. Plant J. 51: 105–116 [DOI] [PubMed] [Google Scholar]
- Harmer S.L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Ho L.H., Giraud E., Lister R., Thirkettle-Watts D., Low J., Clifton R., Howell K.A., Carrie C., Donald T., Whelan J. (2007). Characterization of the regulatory and expression context of an alternative oxidase gene provides insights into cyanide-insensitive respiration during growth and development. Plant Physiol. 143: 1519–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L.H., Giraud E., Uggalla V., Lister R., Clifton R., Glen A., Thirkettle-Watts D., Van Aken O., Whelan J. (2008). Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol. 147: 1858–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T., Kindgren P., Benedict C., Hendrickson L., Strand A. (2007). Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Kosugi S., Suzuka I., Ohashi Y. (1995). Two of three promoter elements identified in a rice gene for proliferating cell nuclear antigen are essential for meristematic tissue-specific expression. Plant J. 7: 877–886 [DOI] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.P., Eubel H., Millar A.H. (2010) Diurnal changes in mitochondrial function reveal daily optimization of light and dark respiratory metabolism in Arabidopsis. Mol. Cell Proteomics 9: 2125–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colón-Carmona A., Gutiérrez R.A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Chew O., Lee M.N., Heazlewood J.L., Clifton R., Parker K.L., Millar A.H., Whelan J. (2004). A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol. 134: 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J.C., Kozma-Bognár L., Gould P.D., Fehér B., Kevei E., Nagy F., Turner M.S., Hall A., Millar A.J. (2006). Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Copsey L., Vincent C., Clark J., Coen E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99: 367–376 [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L., Coen E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Matsushika A., Makino S., Kojima M., Mizuno T. (2000). Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 41: 1002–1012 [DOI] [PubMed] [Google Scholar]
- McCormac A.C., Terry M.J. (2002). Loss of nuclear gene expression during the phytochrome A-mediated far-red block of greening response. Plant Physiol. 130: 402–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., et al. (2008). Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler T.C., Michael T.P., Priest H.D., Shen R., Sullivan C.M., Givan S.A., McEntee C., Kay S.A., Chory J. (2007). The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 72: 353–363 [DOI] [PubMed] [Google Scholar]
- Moon J., Zhu L., Shen H., Huq E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 9433–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufarrege E.F., Curi G.C., Gonzalez D.H. (2009). Common sets of promoter elements determine the expression characteristics of three Arabidopsis genes encoding isoforms of mitochondrial cytochrome c oxidase subunit 6b. Plant Cell Physiol. 50: 1393–1399 [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Fukushima A., Kusano M., Sakakibara H., Mizuno T., Saito K. (2009). Linkage between circadian clock and tricarboxylic acid cycle in Arabidopsis. Plant Signal. Behav. 4: 660–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.-H., Sakakibara H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kita M., Ito S., Sato E., Yamashino T., Mizuno T. (2005a). The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 46: 609–619 [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kita M., Ito S., Yamashino T., Mizuno T. (2005b). PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46: 686–698 [DOI] [PubMed] [Google Scholar]
- Nath U., Crawford B.C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Hervé C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65: 23–33 [DOI] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Omidvar V., Siti Nor Akmar A., Marziah M., Maheran A.A. (2008). A transient assay to evaluate the expression of polyhydroxybutyrate genes regulated by oil palm mesocarp-specific promoter. Plant Cell Rep. 27: 1451–1459 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K., Heberle B., Kurrasch J., Nehls R., Stahl D.J. (2004). Suppression of phenylalanine ammonia lyase expression in sugar beet by the fungal pathogen Cercospora beticola is mediated at the core promoter of the gene. Plant Mol. Biol. 55: 835–852 [DOI] [PubMed] [Google Scholar]
- Smith S.M., Fulton D.C., Chia T., Thorneycroft D., Chapple A., Dunstan H., Hylton C., Zeeman S.C., Smith A.M. (2004). Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 136: 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Suwa Y., Suzuki M., Kitano H., Ueguchi-Tanaka M., Ashikari M., Matsuoka M., Ueguchi C. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33: 513–520 [DOI] [PubMed] [Google Scholar]
- Thirkettle-Watts D., McCabe T.C., Clifton R., Moore C., Finnegan P.M., Day D.A., Whelan J. (2003). Analysis of the alternative oxidase promoters from soybean. Plant Physiol. 133: 1158–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trémousaygue D., Garnier L., Bardet C., Dabos P., Hervé C., Lescure B. (2003). Internal telomeric repeats and ‘TCP domain’ protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J. 33: 957–966 [DOI] [PubMed] [Google Scholar]
- Usadel B., Bläsing O.E., Gibon Y., Retzlaff K., Höhne M., Günther M., Stitt M. (2008). Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Fujiwara S., Somers D.E. (2010). PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 29: 1903–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E., Gonzalez D.H. (2005). Differential expression of the Arabidopsis cytochrome c genes Cytc-1 and Cytc-2. Evidence for the involvement of TCP-domain protein-binding elements in anther- and meristem-specific expression of the Cytc-1 gene. Plant Physiol. 139: 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E., Viola I.L., Kim H.J., Prendes L.P., Comelli R.N., Hong J.C., Gonzalez D.H. (2009). A segment containing a G-box and an ACGT motif confers differential expression characteristics and responses to the Arabidopsis Cytc-2 gene, encoding an isoform of cytochrome c. J. Exp. Bot. 60: 829–845 [DOI] [PubMed] [Google Scholar]