This article characterizes two anther-expressed type III polyketide synthases that are related to chalcone synthase. The results support the hypothesis that the enzymes are involved in an ancient sporopollenin biosynthetic pathway that catalyzes sequential modification of fatty acid starter molecules to generate alkyl α-pyrone polyketide sporopollenin components of the pollen exine.
Abstract
Plant type III polyketide synthases (PKSs) catalyze the condensation of malonyl-CoA units with various CoA ester starter molecules to generate a diverse array of natural products. The fatty acyl-CoA esters synthesized by Arabidopsis thaliana ACYL-COA SYNTHETASE5 (ACOS5) are key intermediates in the biosynthesis of sporopollenin, the major constituent of exine in the outer pollen wall. By coexpression analysis, we identified two Arabidopsis PKS genes, POLYKETIDE SYNTHASE A (PKSA) and PKSB (also known as LAP6 and LAP5, respectively) that are tightly coexpressed with ACOS5. Recombinant PKSA and PKSB proteins generated tri-and tetraketide α-pyrone compounds in vitro from a broad range of potential ACOS5-generated fatty acyl-CoA starter substrates by condensation with malonyl-CoA. Furthermore, substrate preference profile and kinetic analyses strongly suggested that in planta substrates for both enzymes are midchain- and ω-hydroxylated fatty acyl-CoAs (e.g., 12-hydroxyoctadecanoyl-CoA and 16-hydroxyhexadecanoyl-CoA), which are the products of sequential actions of anther-specific fatty acid hydroxylases and acyl-CoA synthetase. PKSA and PKSB are specifically and transiently expressed in tapetal cells during microspore development in Arabidopsis anthers. Mutants compromised in expression of the PKS genes displayed pollen exine layer defects, and a double pksa pksb mutant was completely male sterile, with no apparent exine. These results show that hydroxylated α-pyrone polyketide compounds generated by the sequential action of ACOS5 and PKSA/B are potential and previously unknown sporopollenin precursors.
INTRODUCTION
Microsporogenesis and pollen development are complex processes that take place during flower development, starting with the initiation of stamen primordia and leading to the formation of two microgametes per mature pollen grain. These developmental events involve precisely controlled cellular processes, including cell division, cell differentiation, and cell death (Sanders et al., 1999; Scott et al., 2004; Ma, 2005). In Arabidopsis thaliana, anther development has been divided into stages based on anatomical, morphological, cellular, and molecular events (Sanders et al., 1999; Scott et al., 2004; Ma, 2005). At the beginning of anther cell differentiation, four clearly defined locules are established and visible pollen mother cells appear. Subsequent to this, the pollen mother cells undergo meiosis and tetrads are formed, connected by a callose wall. Once the callose wall degenerates to release free microspores, they become vacuolated, and the pollen wall becomes visible. Microspores continue to enlarge and develop, and the tapetum layer, a maternal cell layer that surrounds the inner side of the anther locules, starts to degenerate. At the end of anther development, the tapetum cell layer completely degenerates, and mature pollen grains fill the locules and are released by anther dehiscence (Sanders et al., 1999).
A critical event during pollen maturation is the formation of the pollen surface structure. The pollen wall represents one of the most complex plant cell walls, with contributions from both the sporophyte and gametophyte generations. The major pollen wall components are a microspore-derived cellulosic primexine synthesized by the developing haploid microspores themselves at the tetrad stage (Blackmore et al., 2007), a thick exine deposited on the outer surface of the primexine largely after the release of free microspores, and a pectocellulosic intine produced by developing microspores and male gametophytes (Blackmore et al., 2007). In contrast with the primexine and intine, the exine is maternally derived, and exine constituents are produced in the sporophytic tapetum cell layer (Piffanelli et al., 1998). These exine constituents are secreted into the locules and incorporated into the exine by polymerization on the surface of the primexine (Piffanelli et al., 1998; Scott et al., 2004; Ma, 2005), where it often assumes a characteristic reticulate pattern, consisting of baculae and tecta. The final component of the pollen wall is the lipid-rich pollen coat, or tryphine, which is deposited onto the exine surface. Pollen coat components accumulate in tapetum cells and are released into locules during the course of tapetum cell degeneration (Ma, 2005).
The main constituent of exine is sporopollenin, an extremely robust and durable biopolymer found in the spores of bryophytes and ferns and in pollen exine of seed plants (Bohne et al., 2003). The chemical composition of sporopollenin remains poorly characterized because it is extremely resistant to chemical and biological degradation procedures (Bubert et al., 2002; Vizcay-Barrena and Wilson, 2006). However, available data are consistent with a sporopollenin polymer consisting of phenolic and fatty acid–derived constituents that are covalently coupled by ether and ester linkages (Rozema et al., 2001; Bubert et al., 2002; Ahlers et al., 2003). Progress has been made in identifying genes and enzymes that are involved in sporopollenin biosynthesis and exine formation during pollen development and has revealed that enzymes involved in fatty acid metabolism play key roles in these events. For example, the Arabidopsis MALE STERILITY2 (MS2) gene, defined by the ms2 male-sterile mutation, is required for exine formation and encodes a predicted fatty acyl-CoA reductase (Aarts et al., 1997). Expression of MS2 in Escherichia coli results in the production of alcohols from endogenous E. coli fatty acids (Doan et al., 2009), supporting a function in fatty acid metabolism. Loss of function of the FACELESS POLLEN1/WAX2/YRE/CER3 gene results in defects in both exine and epicuticular wax deposition, and the gene encodes a putative enzyme of unknown function presumably involved in both wax biosynthesis and pollen wall formation (Ariizumi et al., 2003; Chen et al., 2003; Rowland et al., 2007).
Recently, Arabidopsis genes encoding the cytochrome P450 enzymes of CYP703A2 and CYP704B1 have been shown to be involved in the biosynthesis of sporopollenin, since mutants show severe to moderate effects in exine deposition and pollen grain development (Morant et al., 2007; Dobritsa et al., 2009). In vitro assays indicate that the enzymes catalyze hydroxylation of medium- to long-chain fatty acid substrates. CYP703A2 is an in-chain hydroxylase with substrate preference for lauric acid (C12:0), whereas CYP704B1 catalyzes the ω-hydroxylation of long-chain fatty acids (Morant et al., 2007; Dobritsa et al., 2009). In addition, we recently showed that ACYL-COA SYNTHETASE5 (ACOS5) encodes a fatty acyl-CoA synthetase that plays an essential role in exine formation and sporopollenin biosynthesis in Arabidopsis, since the acos5 mutant is completely male sterile, with a defective pollen wall lacking recognizable exine (de Azevedo Souza et al., 2009). In vitro assays indicate that ACOS5 acts on hydroxyl medium- to long-chain fatty acids to generate the corresponding fatty acyl-CoA esters likely to be central intermediates required for sporopollenin biosynthesis in tapetum cells (de Azevedo Souza et al., 2009).
These results are consistent with a sporopollenin structure that incorporates hydroxylated fatty acids coupled via ester and ether linkages (Guilford et al., 1988; Ahlers et al., 2000, 2003; Morant et al., 2007; de Azevedo Souza et al., 2009), but the exact chemical nature of the sporopollenin polymer and sporopollenin precursor components still remains to be elucidated. Information on such components may come from enzymes encoded by genes that are coregulated with ACOS5 and other genes involved in sporopollenin biosynthesis. Among such coregulated genes is Arabidopsis DIHYDROFLAVONOL 4-REDUCTASE LIKE1 (DRL1)/TETRAKETIDE α-PYRONE REDUCTASE (TKPR1) (Grienenberger et al., 2010), which is required for male fertility and exine formation (Tang et al., 2009). DRL1/TKPR1 encodes a reductase similar to DIHYDROFLAVONOL 4-REDUCTASE, involved in flavonoid metabolism, and plays a pivotal role in sporopollenin precursor biosynthesis as discussed in the companion article (Grienenberger et al., 2010). Recently, it was reported that the enzymes closely related to chalcone synthase (CHS) encoded by At1g02050 (LESS ADHESIVE POLLEN [LAP6]/POLYKETIDE SYNTHASE A [PKSA]) and At4g34850 (LAP5/PKSB) catalyze the sequential condensation of a starter acyl-CoA substrate with malonyl-CoA molecules to produce alkylpyrones in vitro (Mizuuchi et al., 2008; Dobritsa et al., 2010) and that the corresponding enzymes are involved in exine formation (Dobritsa et al., 2010).
Polyketide synthases (PKSs) are ubiquitous enzymes that generate a vast array of natural products and are classified as type I, II, and III enzymes based on their architectures (Austin and Noel, 2003). PKSs catalyze the decarboxylative condensation of a variety of acyl-CoA starter molecules with varying numbers of malonyl-CoA units (Austin and Noel, 2003; Saxena et al., 2003; Funa et al., 2006; Funabashi et al., 2008; Mizuuchi et al., 2008; Rubin-Pitel et al., 2008). Type III PKSs are ubiquitous in vascular plants as well as nonvascular plants such as Physcomitrella patens (Austin and Noel, 2003; Koduri et al., 2010), and plant-specific type III PKSs synthesize diverse natural products that play roles in UV photoprotection, antimicrobial defense, flower pigmentation, and pollen fertility. The best-studied type III PKS is CHS, which carries out the first committed step in flavonoid biosynthesis by catalyzing the sequential decarboxylative addition of three acetate units from malonyl-CoA to a p-coumaryl-CoA starter molecule derived from the general phenylpropanoid pathway. Moreover, other members of the type III PKS superfamily in plants use linear acyl-CoAs of varying length (C2 to C20) as starter substrates and give rise to a large variety of metabolites (Austin and Noel, 2003; Abe et al., 2004, 2005; Mizuuchi et al., 2008; Flores-Sanchez and Verpoorte, 2009).
We now significantly extend previous studies (Dobritsa et al., 2010) to show that the plant-specific CHS-like type III PKSs LAP6/PKSA and LAP5/PKSB are coexpressed with ACOS5, are specifically and transiently expressed in tapetum cells, are required for exine formation and development, and function at specific stages of microspore development. Furthermore, we show that LAP6/PKSA and LAP5/PKSB encode enzymes that catalyze the condensation of hydroxy fatty acyl-CoA esters synthesized by ACOS5 with malonyl-CoAs to yield triketide and tetraketide α-pyrones and that PKSA has a strong in vitro preference for medium-chain hydroxy fatty acyl-CoAs that, based on in vitro data, may be preferentially synthesized by ACOS5. Our genetic analyses also showed that LAP6/PKSA and LAP5/PKSB have overlapping but partially distinct functions in exine development. Combined with the finding that PKSA and PKSB preferentially localize to the endoplasmic reticulum (ER), where they may form part of a metabolon with upstream and downstream enzymes, these data illuminate the important role played by these enzymes in an ancient and evolutionarily conserved biochemical pathway or pathways required for the biosynthesis of polyketide sporopollenin precursors.
RESULTS
Analysis of ACOS5 Coexpressed Genes
Previously we reported that ACOS5 encodes a fatty acyl-CoA synthetase required for sporopollenin biosynthesis in Arabidopsis (de Azevedo Souza et al., 2009). To define other potential enzymes in the sporopollenin biosynthetic pathway, we used data mining tools to identify coexpressed genes in public global gene expression data sets. Using the Correlated Gene Search tool (http://prime.psc.riken.jp), we queried 237 microarray experiments in the Tissue and Development data set, using a cutoff Pearson coexpression coefficient (r2) of 0.80. This analysis identified 35 coexpressed genes, most of unknown function. Among these coexpressed genes, several have been shown to encode enzymes involved in sporopollenin biosynthesis, such as MS2 (At3g11980), CYP703A2 (At1g01280), and DRL1 (At4g35420) (Aarts et al., 1997; Morant et al., 2007; Tang et al., 2009). Among the coexpressed genes, we focused on those that encode enzymes that could use the potential fatty acyl-CoA product(s) generated by the ACOS5 as substrate(s) and therefore might be important in sporopollenin biosynthesis. Two of the most promising potential candidates were genes annotated as encoding plant-specific type III PKSs, LAP6/PKSA (At1g02050) and LAP5/PKSB (At4g34850), which were previously reported to generate triketide and tetraketide α-pyrone compounds using fatty acyl-CoAs (up to 20 carbon chain length) as a starter substrates (Mizuuchi et al., 2008) and were strongly coexpressed with ACOS5 (r2 for LAP6/PKSA of 0.94; r2 for LAP5/PKSB of 0.99). To facilitate their description and represent their enzymatic function for sporopollenin biosynthesis in the following paragraphs, genes and proteins corresponding to At1g02050 and At4g34850 are referred to as PKSA and PKSB, respectively, consistent with the established nomenclature of Mizuuchi et al. (2008).
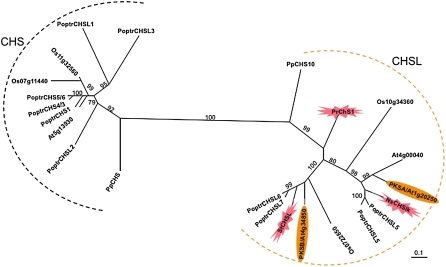
PKSA and PKSB-Like Genes Are Conserved in Land Plant Lineages
To investigate a potential conserved function for PKSA and PKSB genes in pollen wall development, we performed in silico searches of the full genome sequences of Arabidopsis, poplar (Populus spp), rice (Oryza sativa), and Physcomitrella patens using PKSA and PKSB as queries to retrieve potential PKSA- and PKSB-related PKS genes in these plants (see Supplemental Table 1 online). We also retrieved the PKS-related Nicotiana sylvestris CHSLK, Silene latifolia Chs, and Pinus radiata CHS1 genes, previously shown to have high expression in male flowers or anthers (Atanassov et al., 1998; Walden et al., 1999; Ageez et al., 2005) and generated an unrooted maximum likelihood tree of aligned CHS and PKS-related protein sequences. This analysis, shown in Figure 1, indicated that PKSA and PKSB are located in two distinct PKS subclades that are clearly distinct from the more distantly related clade containing the bona fide Arabidopsis and Physcomitrella CHS genes and other putative CHS genes from poplar and rice. Each subclade including either PKSA or PKSB contains homologs from the fully sequenced poplar and rice genomes, as well as the rice or Silene homologs known to be expressed in male organs. According to this analysis, the Pinus CHS1 gene is a PKSA/B homolog basal to the angiosperm PKSA and PKSB clades, and the Physcomitrella PKS (CHS10) and CHS genes are basal to the tracheophyte PKSA/B and CHS clades, respectively (Figure 1). These data indicate that PKSA/B clade PKS genes arose early in land plant evolution and may have common roles in male organ or spore development in land plant lineages.
Figure 1.
Phylogenetic Analysis of CHS and CHSL Protein Sequences from Land Plants.
Sequences retrieved from the complete genome sequences of Arabidopsis (At), poplar (Poptr), rice (Os), and Physcomitrella (Pp), as well as selected sequences from Silene (St), pine (Pr), and N. sylvestris (Ns) were included in the maximum-likelihood tree built using 1000 bootstrap replicates in PhyML 2.4.4. Bootstrap values are indicated on branches (out of 100). The Arabidopsis PKSA and PKSB proteins are highlighted (ovals). The proteins encoded by genes known to be expressed in tapetum cells during anther development are indicated by a flash. Protein sequences used in this analysis are given in Supplemental Data Set 1 online, and the alignment is available in Supplemental Data Set 2 online. Bar = 0.1 amino acid substitutions.
[See online article for color version of this figure.]
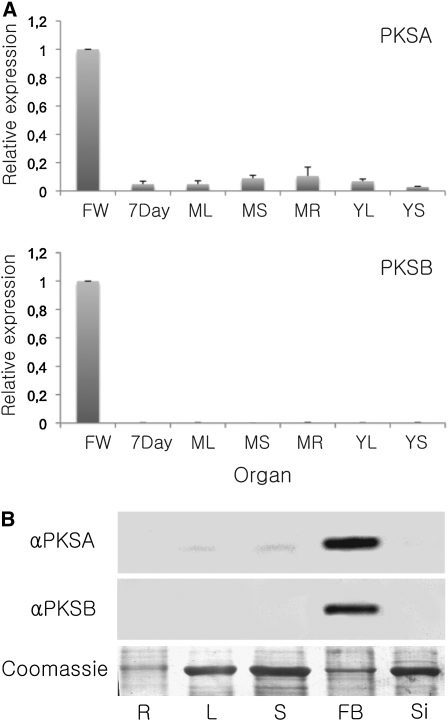
Tapetum-Specific Expression of PKSA and PKSB Proteins
To test possible functions of the Arabidopsis PKSA and PKSB proteins in male organ development, first we used quantitative RT-PCR to analyze the expression of PKSA and PKSB in different Arabidopsis organs. Expression profiles are shown in Figure 2A. Both genes were preferentially expressed in flowers, and PKSB transcripts were detected exclusively in this organ. We also assayed PKSA and PKSB protein levels in various organs using immunoblots of fractionated protein extracts reacted with specific polyclonal antibodies raised against the recombinant PKSA and PKSB proteins. Both PKSA and PKSB proteins were preferentially detected in flowers, and little or no signal could be detected in extracts from other organs (Figure 2B). Analysis of PKSA and PKSB protein accumulation over the course of flower bud development showed that the two proteins had similar but distinct accumulation patterns: both PKSA and PKSB were most abundant in the youngest flower buds tested, and levels declined as flowers matured (see Supplemental Figure 1 online). However, the PKSB protein was detected only in the earliest stage assayed, whereas PKSA protein levels decreased more slowly in more mature flowers (see Supplemental Figure 1 online). The specificity of the PKSA and PKSB antibodies was demonstrated in immunoblotting experiments with the purified recombinant proteins. Each antiserum was shown to recognize the cognate protein, and cross-reactivity was not detected with the other PKS recombinant protein (see Supplemental Figure 2 online).
Figure 2.
Developmental Expression Profiles of PKSA and PKSB.
(A) Quantitative RT-PCR analysis of relative PKSA and PKSB expression levels in various Arabidopsis organs. Expression was calculated using the ΔΔCT method and is represented relative to the organ with the highest level of expression (flowers), set at 1.0. Actin2 was used as a reference gene. Bars represent standard deviations from the means of triplicate determinations. 7day, 7-d-old seedlings; FW, flower; ML, mature leaf; MR, mature root; MS, mature stem; YL, young leaf; YS, young stem.
(B) Immunoblots of protein extracts from different Arabidopsis tissues from mature plants with polyclonal antibodies raised against each recombinant protein. Six micrograms of total protein was loaded in each lane and reacted with antibodies specific to PKSA or PKSB (α PKSA and α PKSB). The Coomassie blue–stained loading control is shown below. R, roots; L, leaves; S, stems; FB, flower buds; Si, siliques.
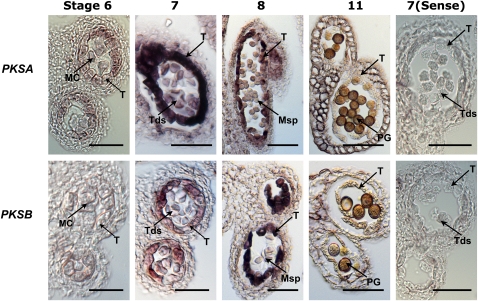
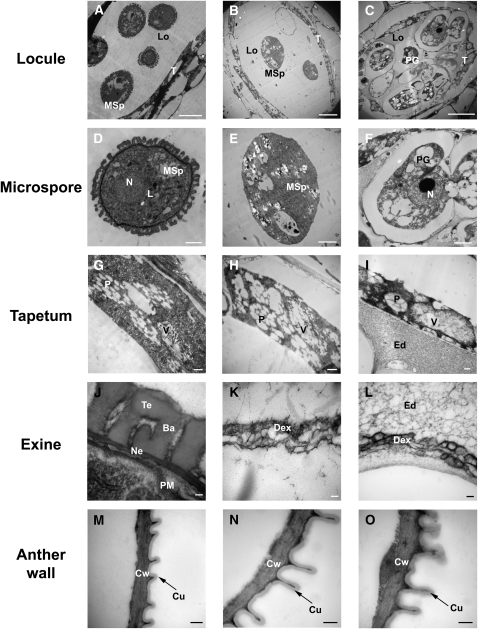
To investigate the spatio-temporal patterns of PKSA and PKSB expression and PKSA and PKSB protein accumulation in anthers, we performed both in situ hybridization and immunolocalization experiments. Because PKSA and PKSB share 62.9% nucleotide identity and could potentially cross-hybridize, the specificity of the PKSA- and PKSB-derived riboprobes used for in situ hybridization analysis was demonstrated by DNA gel blotting, showing that both probes hybridized specifically with the target templates, with no detectable cross-hybridization (see Supplemental Figure 3 online). To elucidate the specific expression patterns of PKSA and PKSB, these RNA probes were hybridized to sections of developing wild-type flowers (Figure 3) focusing on anther stages 6 to 11 as defined by Sanders et al. (1999), during which free microspores are generated and sporopollenin-containing exine is deposited. Both PKSA and PKSB were strongly and transiently expressed in the tapetum cell layer of developing anthers. Whereas the hybridization signal was largely specific to tapetal cells, low levels of signal appeared to be present in stage 7 tetrads, so that a function in microspores cannot be excluded. Interestingly, the temporal patterns of expression in the developing tapetum were slightly different. PKSA expression was first detected at stage 6, and by stage 7, strong hybridization was detected in the tapetum. By stage 8, PKSA hybridization to tapetum cells had weakened, and at stage 11, it had disappeared. By contrast, PKSB expression was initiated at stage 7, and maximal hybridization was seen in the tapetum at stage 8.
Figure 3.
Transient Tapetum-Specific Expression of PKSA and PKSB.
PKSA and PKSB mRNAs were localized by in situ hybridization to sections taken from developing anthers of wild-type (Col-0) flowers using gene-specific antisense probes for PKSA and PKSB and control sense probes. Stages of anther development are according to Sanders et al. (1999). Dark precipitates indicate hybridization of the probe. MC, meiotic cell; T, tapetum; Tds, tetrads; Msp, microspores; PG, pollen grain. Bars = 70 μm.
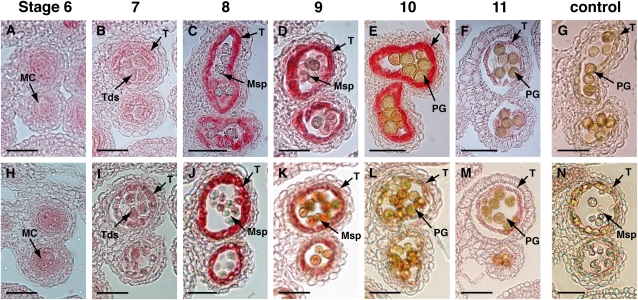
The PKSA- and PKSB-specific antibodies were used for immunolocalization experiments in which similar stages of anther development were examined. Considerable accumulation of PKSA and PKSB proteins in tapetum cells was detected by stage 8 (Figure 4). PKSA remained abundant in the tapetum through stage 10, whereas PKSB showed more transient abundance, with maximum accumulation at stage 8 and a rapid decrease in abundance by stage 9.
Figure 4.
Immunolocalization of PKS Proteins during Anther Development.
Thin sections of flower buds at different stages of development were incubated with specific antibodies raised against each purified recombinant PKS protein. Stages of anther development are indicated above sections according to Sanders et al. (1999). MC, microspore mother cells; Tds, tetrads; T, tapetum; Msp, microspores; PG, pollen grain. Bars = 70 μm.
(A) to (G) Immunolocalization of PKSA. The protein was detected at stage 7 (B) in tapetum cells surrounding tetrads. Maximum protein accumulation was observed in the tapetum at stages 9 and 10 ([D] and [E]). A section of pksa-1 mutant bud at stage 9 is shown as negative control (G).
(H) to (N) Immunolocalization of PKSB. The protein was detected at stage 7 in tapetum cells surrounding tetrads (I). Maximum protein accumulation was observed in the tapetum at stage 8 (J) and then decreased rapidly. A section of pksb-2 mutant bud at stage 8 was used as negative control (N).
Identification and Phenotypic Analysis of PKSA and PKSB Loss-of-Function Alleles
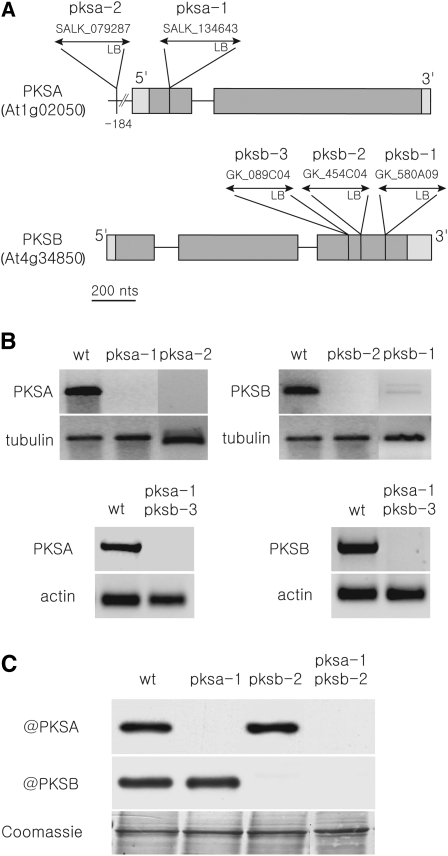
To test the roles of PKSA and PKSB in pollen development and male fertility, we obtained two independent T-DNA insertion lines (pksa-1 and pksa-2) for PKSA and three independent T-DNA insertion lines (pksb-1, pksb-2, and pksb-3) for PKSB from public collections (Alonso et al., 2003). The locations of each T-DNA insertion in the PKSA and PKSB genes were verified by sequence analysis (Figure 5A). PKSA and PKSB expression in the insertion lines was assayed by RT-PCR, using template cDNAs derived from both wild-type and mutant flowers (Figure 5B), and little or no PKSA or PKSB expression was detected. Furthermore, in pksa-1 and pksb-2 insertion lines, no protein accumulation could be detected by immunoblotting protein extracts using PKSA- and PKSB-specific antibodies (Figure 5C). This analysis suggested that each of the five alleles is a strong loss-of-function allele of PKSA or PKSB.
Figure 5.
Molecular Characterization of pksa, pksb, and pksa pksb Insertion Alleles and Effects on Gene Expression.
(A) Position of T-DNA insertions in the different mutant lines is shown. Gray boxes denote exons, with lighter gray indicating 5′ and 3′ untranslated regions. Thin horizontal lines denote introns.
(B) RT-PCR analysis of gene expression in flower buds of wild-type (Col-0) and single and double mutants. No amplicon was detectable in mutant samples. TUBULIN and ACTIN are shown as positive controls.
(C) Protein accumulation in wild-type (Col-0) and mutant bud extracts was evaluated by immunoblotting with specific antibodies as indicated. The Coomassie blue–stained loading control is shown below. No protein accumulation was detected in mutant lines, thus confirming that gene expression was knocked out in pksa-1, pksb-2, and pksa-1 pksb-2 lines. Ten micrograms of protein was loaded in each lane.
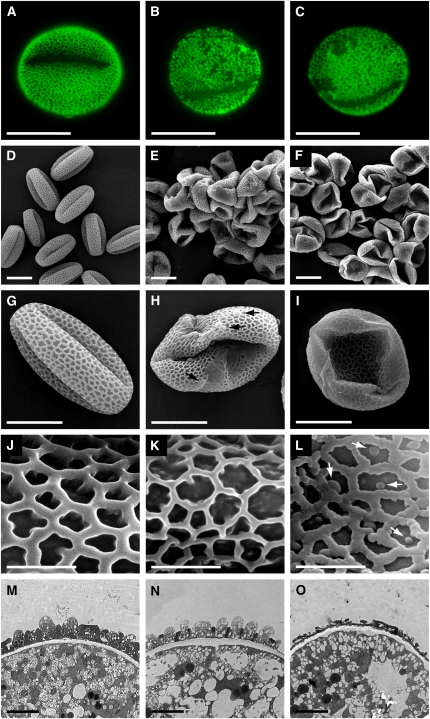
While this work was in progress, another group reported that plants homozygous for pksa or pksb loss-of-function alleles have morphological defects in exine structure but were fertile (Dobritsa et al., 2010). To examine pollen morphology of pksa and pksb mutants in more detail and at greater resolution, pollen grains from pksa-1, pksa-2, pksb-1, and pksb-2 homozygotes were obtained and compared with those from wild-type plants. Figure 6 and Supplemental Figure 4 online show the results of these analyses. When mature pollen grains were stained with auramine O, profound alterations in exine structure in pksa-1 and pksb-2 were revealed by fluorescence microscopy, with the regular reticulate pattern of wild-type exine replaced by irregular and mottled exine patterns in the pksa-1 and pksb-2 mutants (Figures 6A to 6C).
Figure 6.
Exine Defects in Pollen Grains of Single pksa-1 or pksb-2 Mutants.
Pollen grains from mature wild-type (Col-0) plants ([A], [D], [G], [J], and [M]) and plants homozygous for the pksa-1 ([B], [E], [H], [K], and [N]) and pksb-2 ([C], [F], [I], [L], and [O]) alleles were analyzed by auramine O staining, scanning electron microscopy, and TEM. Bars = 10 μm in (A) to (I), 2 μm in (J) to (L), and 1 μm in (M) to (O). Arrows in (H) indicate defective exine patterning and in (L) indicate globular protrusions observed in the pksb-2 mutant.
(A) to (C) Auramine O staining of Col-0, pksa-1, and pksb-2 pollen, respectively.
(D) to (L) Scanning electron microscopy images of pollen samples at three magnification values.
(M) to (O) TEM images of Col-0, pksa-1, and pksb-2 pollen grains, respectively.
[See online article for color version of this figure.]
Wild-type pollen grain samples prepared for scanning electron microscopy observation, including exposure to a partial vacuum, displayed a typical round shape with reticulate exine, whereas pollen grains derived from pksa-1 and pksb-2 mutant pollen, prepared identically, exhibited collapsed morphology by scanning electron microscopy, indicating that they had weakened pollen walls (Figures 6D to 6I). Higher magnification of scanning electron microscopy images from wild-type and mutant pollen showed that, whereas a conserved exine pattern was present in the mutants, the exine baculae appeared thinner or more diffuse in the mutants relative to the wild type (Figures 6J to 6L). Furthermore, amorphous extrabacular protrusions were observed in the pksb-2 mutant exine network (Figure 6L). Similar exine morphology was observed in scanning electron microscopy images of pollen from individuals homozygous for additional pska and pksb alleles (pska-2 and pksb-1; see Supplemental Figure 4 online). Finally, transmission electron microscopy (TEM) observations confirmed that the exine of mature pollen grains from pksa-1 and pksb-2 mutants was thinner, with shorter baculae and tecta compared with the wild type (Figures 6M to 6O). These results show that PKSA and PKSB both play specific roles in exine formation during pollen grain development and that each are required for exine and pollen wall integrity and exine patterning.
PKSA and PKSB Have Partially Redundant Functions in Male Fertility
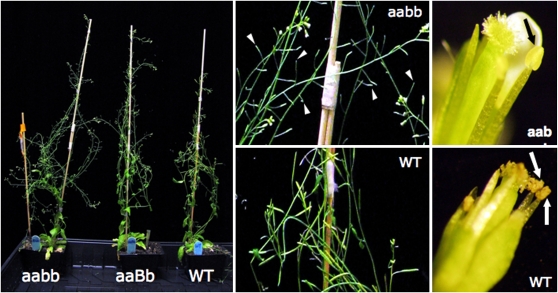
Since the Arabidopsis PKSA and PKSB genes encode PKSs with similar in vitro activities (Mizuuchi et al., 2008; Dobritsa et al., 2010), biochemical redundancy between PKSA and PKSB was a strong possibility. To test this, we generated two independent homozygous double mutants, pksa-1 pksb-2 and pksa-1 pksb-3, identified within F2 populations derived from crossing the corresponding homozygous pksa and pksb lines. Initial phenotypic examination of pksa-1 pksb-3 mutant plants (Figure 7) revealed anthers devoid of visible pollen, male sterility, and siliques devoid of seeds. Whereas no visible pollen was ever observed in plants homozygous for the pksa-1 pksb-3 alleles, pksa-1 pksb-3 flowers were female fertile when pollinated with wild-type pollen. There were no other obvious morphological differences between the pksa-1 pksb-3 mutant and wild-type plants grown to maturity (Figure 7).
Figure 7.
Phenotypic Characterization of Plants Segregating for pksa and pksb Alleles.
Plants homozygous for pksa-1 and pksb-3 alleles were crossed, and an F1 individual self-pollinated to generate a population segregating for pksa-1 and pksb-3 alleles. Plants were subjected to PCR-aided genotyping and grown to maturity. PKSA alleles are symbolized by A (WT) and a (pksa-1), and PKSB alleles are symbolized by B (WT) and b (pksb-3). Phenotypes of mature aabb, aaBb, and AABB (WT) plants are shown. Double mutant plants (aabb) had no pollen in anthers (black arrow) and undeveloped siliques (arrowheads), resulting in a complete absence of seeds, whereas Col-0 anthers had abundant pollen at this stage (white arrows). However, there are no other obvious morphological differences except that they flowered for a longer time.
[See online article for color version of this figure.]
Within the pksa-1 pksb-3 F2 population, we identified plants homozygous for pksa-1 and heterozygous for pksb-3. Interestingly, these plants displayed a partially sterile phenotype, with smaller siliques containing fewer seeds or empty siliques. Therefore, we quantified seed set in siliques taken at random from plants with different PKSA and PKSB allelic combinations (Table 1; n = 15 siliques for each genotype). pksa-1 and pksb-3 single mutants and plants heterozygous for pksa-1 and homozygous for pksb-3 show slightly reduced numbers of seeds relative to wild-type control plants, whereas plants homozygous for pksa-1 and heterozygous for pksb-3 showed clear reduction in fertility, with most siliques having 10 or fewer seeds. When placed on the wild-type (Columbia-0 [Col-0]) stigmas, both pksa-1 and pksb-2 mutant pollen grains germinated and pollen tubes grew normally (see Supplemental Figure 5 online), suggesting that the fertility differences were not due to defects in pollen germination or pollen tube growth.
Table 1.
Quantification of Seed Set in Plants with Different PKSA and PKSB Allelic Combinations
| No. of Siliques Containing the Given Numbers of Seeds |
|||||||||
| Genotypea | >71b | 70–61 | 60–51 | 50–41 | 40–31 | 30–21 | 20–11 | 10–1 | 0 |
| AABB | 6.0 | 9.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AAbb | 4.3 | 8.3 | 2.3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Aabb | 3.7 | 10.3 | 1.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AaBb | 5.3 | 9.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| aaBB | 1.0 | 10.0 | 4.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| aaBb | 0 | 0 | 0 | 0 | 0.7 | 0.7 | 2.7 | 10.0 | 1.0 |
| aabb | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15.0 |
At1g02050 (PKSA) alleles are symbolized by A (WT) and a (pksa-1), and At4g34850 (PKSB) alleles are symbolized by B (WT) and b (pksb-3).
Values represent the average number of siliques on one branch from the indicated genotype that contain seeds numbering within the given range. Genotype AABB (wild type) contained at least 61 seeds in each silique. On the other hand, genotype aabb (double mutant pksa-1 pksb-3) had no seeds in any siliques. Genotype aaBb showed clearly reduced fertility with most siliques having 10 or fewer seeds.
The pksa-1 pksb-2 double mutant also showed a strong male-sterile phenotype, similar to that of the pksa-1 pksb-3 double mutant. A few rare pollen grains were obtained from anthers of pksa-1 pksb-2 double mutant plants. Examination of these pollen grains by scanning electron microscopy revealed that they were severely distorted in shape, with no apparent reticulate exine pattern (see Supplemental Figures 6A to 6C online). After staining with auramine O, fluorescence in the pksa-1 pksb-2 mutant was attenuated and devoid of a reticulate pattern (see Supplemental Figure 6D online).
Anther and Microspore Development in the pksa-1 pksb-3 Double Mutant
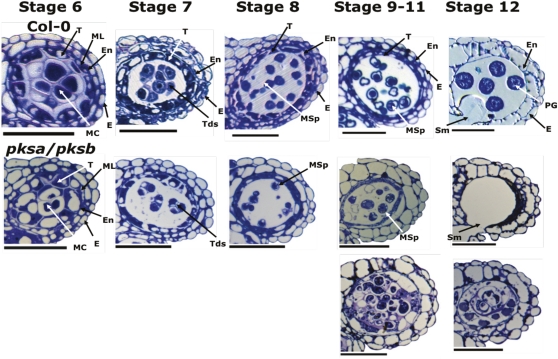
To determine the point at which pollen development was impaired in the completely male-sterile pksa-1 pksb-3 double mutant, we examined developing anthers in the double mutant background relative to the wild type by light microscopy (Figure 8), using the stages of anther development defined by Sanders et al. (1999). Microspore and anther development in pksa-1 pksb-3 plants appeared normal through stage 8, when individual microspores could be seen, indicating that the callose wall had degenerated, releasing microspores from tetrads in a normal manner. However, by stages 9 to 11, aberrant microspore development in pksa-1 pksb-3 anthers relative to the wild type was observed. Free microspores appeared to have thin walls and aberrant structures, and locules had accumulated debris of defective pollen grains (Figure 8). By stage 12, mature pollen grains were observed in locules of wild-type plants, while most pksa-1 pksb-3 anthers were devoid of pollen. In a smaller number of pksa-1 pksb-3 mutant anthers in stages 9 to 12 (bottom pksa-1 pksb-3 panels in Figure 8), some pollen grains appeared to be still in the tetrad stage or were fused with each other, and the tapetum layers were highly enlarged and vacuolated and failed to undergo programmed cell death.
Figure 8.
Phenotypic Characterization of Anther and Microspore Development in Wild-Type (Col-0) and pksa-1 pksb-3 Flowers.
Anther cross sections (1 μm) were taken from developing flowers of wild-type plants and pksa pksb mutant plants and stained with toluidine blue. Numbers indicate anther developmental stages according to Sanders et al. (1999). The two panels at the bottom illustrate anther phenotypes at stages 9 to 12 occasionally found in pksa pksb mutants. De, debris of defective pollen grains; E, epidermis; En, endothecium; MC, meiotic cell; ML, middle layer; MSp, microspores; PG, pollen grains; Sm, septum; T, tapetum; Td; tetrad. Bars =40 μm.
We used TEM to examine pksa-1 pksb-3 microspore development at greater resolution. In agreement with light microscopy observations, at stages 5 to 7, microspore and tapetum development were normal in the mutant, and free microspores were observed at stage 8 (see Supplemental Figure 7 online). However, at stage 9, pksa-1 pksb-3 microspores had thin cell walls apparently devoid of an exine, and by stage 12, locules were mostly devoid of pollen grains, although occasional defective pollen grains were observed (see Supplemental Figure 7 online).
We next used TEM to examine pksa-1 pksb-3 mutant anthers and microspores at stages 9 and 11 at greater detail, relative to the wild type. Figure 9 shows that at stage 9, wild-type anthers contained uninucleate microspores with a thick, reticulate exine, intact tapetum, and an electron-dense cuticle on the outer anther epidermis, which exhibited hair-like protrusions (Figures 9A, 9D, 9G, 9J, and 9M). By contrast, aberrant microspore development was observed at the same stage in pksa-1 pksb-3 anthers (Figures 9B, 9E, and 9K). While uninucleate microspores, an intact tapetum, and a normal anther epidermis with an intact cuticle were present (Figures 9E, 9H, and 9N), microspores were devoid of a recognizable exine (Figure 9E). At higher magnification, a defective, thin fibrillar matrix, presumably defective exine, was apparent on mutant microspores (Figure 9K). At stage 11, most pksa-1 pksb-3 mutant anthers contained only a few degenerating pollen grains (Figure 8; see Supplemental Figure 7 online). However, in a subset of pksa-1 pksb-3 mutant individuals, enlarged pollen grains were present at this stage (Figure 9C) that had an abnormal exine structure without a thick and reticulated wall (Figures 9F and 9L). Under higher magnification, it was also apparent that the locules of these anthers were filled with misshapen structures and an electron-dense network, possibly unpolymerized sporopollenin precursors (Figures 9I and 9L). In these individuals, the tapetum and anther outer wall epidermis and cuticle appeared to be normal (Figures 9I and 9O).
Figure 9.
Transmission Electron Micrographs of Wild-Type (Col-0) and pksa-1 pksb-3 Anthers and Pollen.
(A), (D), (G), (J), and (M) Microspore structure, tapetum structure, exine formation, and outer wall of anther epidermis at anther stage 9 in Col-0 wild-type plants.
(B), (E), (H), (K), and (N) Microspore structure, tapetum structure, exine formation, and outer wall of anther epidermis at anther stage 9 in pksa-1 pksb-3 plants.
(C), (F), (I), (L), and (O) Pollen grain structure, tapetum structure, exine formation, and outer wall of anther epidermis at anther stage 11 in pksa-1 pksb-3 plants.
Ba, bacula; Cu, cuticle; Cw, cell wall; Dex, defective exine structure; Ed, electron-dense material; Ex, exine; L, lipid body; Lo, locule; MSp, microspore; N, nucleus; Ne, nexine; P, plastid filled with plastoglobuli; PG, pollen grain; PM, plasma membrane; T, tapetal cell; Te, tectum; V, vacuole containing electron-dense material. Bars = 10 μm in (A) to (C), 2 μm in (D) to (F), 500 nm in (G) to (I) and (M) to (O), and 100 nm in (J) to (L).
Enzymatic Activities of Recombinant PKSA and PKSB Proteins
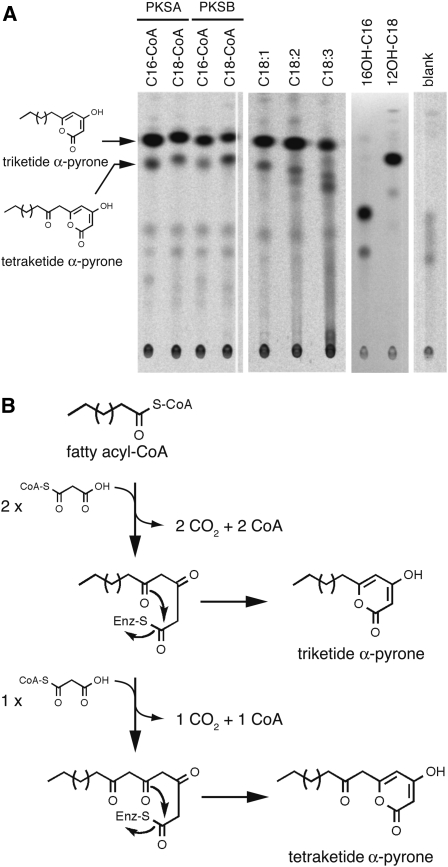
PKS activity of recombinant PKSA and PKSB proteins was assayed in the presence of various fatty acyl-CoA esters as the starter substrates and malonyl-CoA as the extender molecule. We employed 14C malonyl-CoA as a tracer to visualize radioactive condensation reaction products. Products were fractionated by thin layer chromatography (TLC) after extraction and revealed by exposure to a phosphor imager plate (Figure 10A). When synthetic C16:0-CoA or C18:0-CoA was used as a substrate (Figure 10A, left panel), two major reaction products were detected in incubation mixtures that were identified as the cognate triketide and tetraketide α-pyrone compounds by liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis (Figure 11), as previously described by Mizuuchi et al. (2008) but in contrast with Dobritsa et al. (2010), who did not observe PKSA/B conversion of fatty acyl-CoAs with chain lengths greater than C12. The same reaction products were identified upon incubation with PKSA or PKSB.
Figure 10.
TLC Analysis of Radiolabeled Reaction Products of PKSs.
Recombinant proteins produced by bacteria were incubated with different CoA esters as starters and 14C-malonyl-CoA as extender. Radioactive reaction products were separated by TLC and detected using a phosphor imager.
(A) Left panel: Chemically synthesized CoA esters used as starters are indicated above each lane. PKSA or PKSB was incubated as indicated. The structures of the major reaction products as deduced from LC-MS/MS analysis are shown on the left. Middle panel: The CoA esters synthesized after incubation of ACOS5 with unsaturated fatty acids as substrates were incubated with PKSB and malonyl-CoA. Tri- and tetraketide reaction products were identified by LC-MS/MS. Right panel: After incubation of 16-OH palmitic or 12-OH stearic acids in the presence of ACOS5, the resulting esters were condensed with malonyl-CoA by PKSB, yielding corresponding tri- and tetraketide α-pyrones. Blank was run without addition of fatty acyl-CoA. No products were observed without addition of PKSA or PKSAB.
(B) Putative mechanism of fatty acyl-CoA condensation with malonyl-CoA catalyzed by PKSs. After two rounds of extension with malonyl-CoA, the intermediate compounds may form a cyclic lactone, yielding triketide α-pyrones, or undergo an additional round of extension yielding the tetraketide α-pyrone compounds.
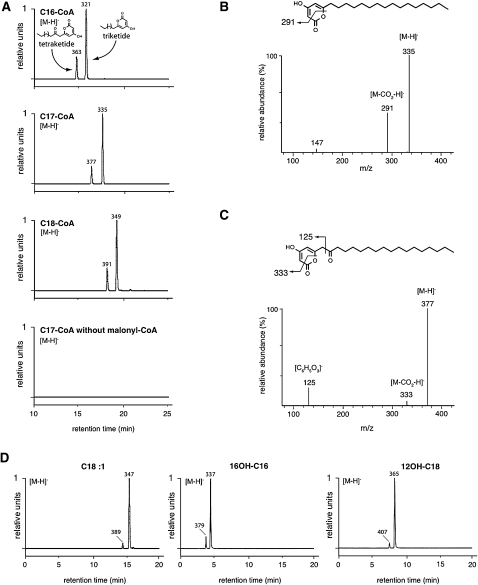
Figure 11.
Identification of PKS Reaction Products by LC-MS/MS.
Reaction products were resolved by reverse-phase UPLC and analyzed by MS/MS. Negative electrospray ionization was used, and [M-H]− ions were detected by m/z values as indicated.
(A) C16-CoA, C17-CoA, or C18-CoA was used as starter substrate as indicated, and cognate tri- and tetraketide reaction products were detected. The m/z value of [M-H]− ion of each product is indicated. A negative control incubated without malonyl-CoA is shown at the bottom.
(B) Fragmentation pattern of the [M-H]− ion of the triketide α-pyrone obtained with C17-CoA as substrate. Structure of the compound and putative fragmentation scheme are shown above.
(C) Fragmentation pattern of the [M-H]− ion of the tetraketide α-pyrone reaction product from C17-CoA incubation. Structure of the molecule and its putative fragmentation are shown.
(D) Tri- and tetraketide α-pyrone reaction products were characterized by their m/z values after incubation of the unsaturated or hydroxylated fatty acid indicated above the profile with ACOS5 and PKS enzymes (see Methods). The major peaks correspond to triketide products and the minor ones to tetraketide products.
To explore the potential sequential actions PKSA/B and ACOS5 enzyme activity in sporopollenin biosynthesis, various additional CoA esters were synthesized enzymatically by incubating recombinant ACOS5 with hydroxylated or unsaturated fatty acids in the presence of ATP and CoA and tested as potential substrates of PKSA and PKSB (Figure 10A, middle and right panels). After a 15-min incubation of ACOS5 in the presence of a given fatty acid, ATP, and CoA as substrates, PKSA or PKSB enzymes and radioactive 14C malonyl-CoA were added to the reaction medium for a further 1-h incubation. As shown in Figure 10A, radioactive reaction products were detected in all cases, thus demonstrating the promiscuous specificity of PKSA and PKSB. LC-MS/MS analysis revealed that the triketide compound was the predominant product in all incubation mixtures; however, the ratio between tri- and tetraketide α-pyrone reaction products was found to be variable, depending on the nature of the fatty acyl-CoA ester (Figures 10 and 11).
The triketide α-pyrone products resulted from the addition of 4C atoms originating from two rounds of decarboxylative condensation with malonyl-CoA, whereas the tetraketide α-pyrones are built up from the addition of 6C atoms originating from three malonyl-CoA molecules (Figure 10B). Figure 11 illustrates selected examples of the analysis of PKS products by LC-MS/MS. For instance, mass-to-charge (m/z) values of 321 and 363 were found for tri- and tetraketide products, respectively, from C16:0-CoA incubation, and 349 and 391 m/z values were measured for C18:0-CoA extension products (Figure 11A). Fragmentation patterns of tri- and tetraketide α-pyrones synthesized with C17-CoA as a substrate were obtained by collision-induced dissociation of [M-H]− ions (Figures 11B and 11C) and were characteristic of α-pyrone derivatives (Saxena et al., 2003; Funa et al., 2006). It is noteworthy that, for a given substrate, the relative amounts of reaction products varied from one enzyme preparation to another, with the tri- to tetraketide α-pyrone ratio being inversely correlated with the level of activity of the enzyme preparation. These observations suggest that the triketide α-pyrone likely represents a derailment reaction product, with poorly active enzymes becoming unable to perform the third round of condensation to malonyl-CoA that is required to build up the tetraketide α-pyrones.
When unsaturated or hydroxylated fatty acids were first incubated with ACOS5 to synthesize the cognate fatty acyl-CoA esters, all the resulting CoA esters proved to be substrates for PKS since, in all cases, the corresponding tri- and tetraketide compounds were found in the incubation medium (Figure 11D). These results suggest that tapetum-localized PKSA and PKSB enzymes use ACOS5-derived fatty acyl-CoA esters as starter substrates for the biosynthesis of triketide and tetraketide α-pyrones and that such polyketides play an essential role in generating a robust, structurally sound sporopollenin polymer during exine deposition in developing microspores.
Substrate Preference and Steady State Kinetics of PKSA
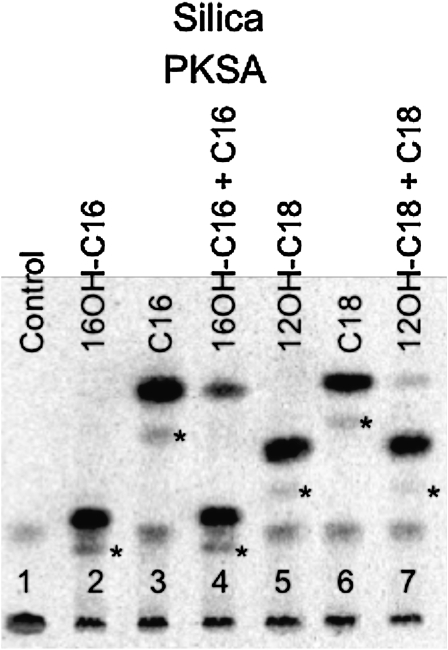
In light of the proposed involvement of hydroxy fatty acyl-CoA esters as precursors for sporopollenin biosynthesis (Morant et al., 2007; de Azevedo Souza et al., 2009; Dobritsa et al., 2009) and the ability of PKSA and PKSB to use these as substrates for generation of tri- and tetraketide α-pyrones (Figures 10 and 11), we used competition assays and enzyme kinetics to test PKSA preference for such substrates relative to unsubstituted fatty acyl-CoAs. Figure 12 shows the results of substrate competition assays using selected synthetic hydroxy fatty acyl-CoA esters (16-OH-C16-CoA and 12-OH-C18-CoA), which clearly demonstrated PKSA in vitro preference for hydroxy fatty acyl-CoA esters. When 16-OH-C16-CoA at 30 μM and C16-CoA at 100 μM were coincubated with PKSA, the production of 15-OH-C15-α-pyrone was reduced to 76% of the control level without competitor (Figure 12, lanes 2 and 4), while the production of C15-α-pyrone was reduced to 12% of the control level (Figure 12, lanes 3 and 4). Substrate preference was more drastic in the pair of 12-OH-C18-CoA and C18-CoA. Whereas C18-CoA at 100 μM reduced the production of 17-OH-C17-α-pyrone to 67% of the control level (Figure 12, lanes 5 and 7), 12-OH-C18-CoA at 30 μM almost completely inhibited the production of C17-α-pyrone (Figure 12, lanes 6 and 7), suggesting very strong preference of PKSA for 12-OH-C18-CoA relative to C18-CoA.
Figure 12.
Radio Silica-TLC of Substrate Competition Assay for PKSA.
A PKSA reaction mixture containing 50 μM [2-14C] malonyl-CoA was supplemented with 16-OH-C16-CoA (30 μM, lane 2), C16-CoA (100 μM, lane 3), or both 16-OH-C16-CoA (30 μM) and C16-CoA (100 μM) (lane 4). Similarly, the PKSA reaction was performed in the presence of 12-OH-C18-CoA at 30 μM (lane 5), C18-CoA at 100 μM (lane 6), or both 12-OH-C18-CoA and C18-CoA at 30 and 100 μM, respectively (lane 7). Each reaction produced a triketide α-pyrone as the major product and a tetraketide pyrone (denoted with an asterisk) as the minor product. To assay competition, α-pyrone product levels in lane 4 were compared with those in lanes 2 and 3 (no competitors added) and those in lanes 7 to those in lanes 5 and 6 (no competitor added). In the control lane (1) [2-14C] malonyl-CoA was incubated with PKSA in the absence of fatty acyl-CoA.
To investigate further PKSA in vitro substrate preference for hydroxy fatty acyl-CoA esters, we used purified recombinant PKSA for steady state enzyme kinetic analysis and determined the parameters shown in Table 2. The apparent Km and kcat values of PKSA for 16-OH-C16-CoA and 12-OH-C18-CoA were 25 μM and 2.8 × 10−3 s−1, and 23 μM and 5.0 × 10−3 s−1, respectively, whereas the corresponding values for C16-CoA were 48 μM and 0.13 × 10−3 s−1. Thus, the calculated values for the catalytic efficiency (kcat/Km) for 16-OH-C16-CoA and 12-OH-C18-CoA (110 and 220 M−1 s−1, respectively) are 40- to 80-fold higher than the corresponding value for C16-CoA (2.7 M−1 s−1). These results show that hydroxy fatty acyl-CoA esters such as those preferentially generated by ACOS5 are the preferred in vitro substrates of PKSA and suggest that such substrates are likely the preferred in planta substrates.
Table 2.
The Steady State Kinetic Parameters of Recombinant PKSA
| Substrate | Km (μM)a | kcat (10−3 s−1)a | kcat/Km (M−1 s−1)a |
| 16-OH-C16-CoA | 25 ± 5.0 | 2.8 ± 0.59 | 112 |
| 12-OH-C18-CoA | 23 ± 3.8 | 5.0 ± 1.1 | 217 |
| C16-CoA | 48 ± 3.2 | 0.13 ± 0.43 | 2.7 |
Values given are means ± sd; n = 3.
PKSA and PKSB Proteins Localize to the ER
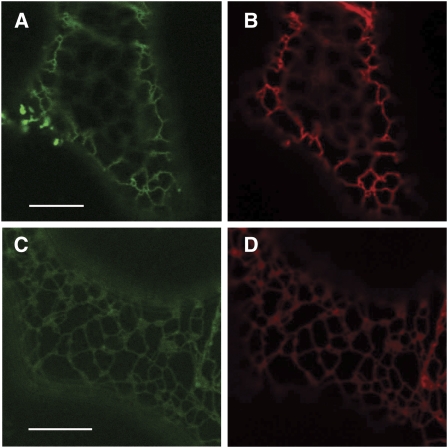
To investigate the cellular localization of PKSA and PKSB proteins, 5′ translational fusions to enhanced green fluorescent protein (eGFP) under the control of the cauliflower mosaic virus 35S promoter (p35S:PKSA/B-eGFP) were generated. The constructs were transiently expressed in Nicotiana benthamiana leaves, together with a reporter construct of an ER marker (monomeric red fluorescent protein [mRFP]-HDEL). As shown in Figures 13A and 13C, PKSA/B-eGFP displayed a reticulate expression pattern in epidermal cells. Figures 13B and 13D show that this pattern coincides with that of mRFP-HDEL, which is typical of ER localization. These data indicate that PKSA and PKSB enzymes are associated to the ER.
Figure 13.
Subcellular Localization of PKSA and PKSB.
PKSA- and PKSB-eGFP and mRFP-HDEL (ER marker protein) constructs were transiently expressed in N. benthamiana leaves by coagrotransformation. Bars = 10 μm.
(A) PKSA-eGFP expression.
(B) mRFP-HDEL coexpression coincides with PKSA-eGFP expression.
(C) PKSB-eGFP expression.
(D) mRFP-HDEL coexpression coincides with PKSB-eGFP expression.
DISCUSSION
Lipophilic polyesters such as cutin, suberin, and sporopollenin often are present at plant surfaces, provide protection from biotic and abiotic environmental factors, and are thought to have evolved early during colonization of land by plants (Pichersky and Gang, 2000; Bowman et al., 2007; Franke and Schreiber, 2007). Whereas the composition of cutin and suberin and their biosynthetic origins are relatively well characterized (Franke and Schreiber, 2007; Pollard et al., 2008), sporopollenin is poorly understood due to its heavily cross-linked nature, resistance to chemical degradation, and small amounts of biological material (Scott et al., 2004). In this article, we show that two Arabidopsis type III PKSs, PKSA and PKSB, that are related to CHS play essential roles in exine formation and sporopollenin biosynthesis during the free microspore phase of pollen development. Our data show that PKSA and PKSB are promiscuous in the substrates they accept but that these substrates include hydroxy fatty acyl-CoA products generated by anther-specific cytochrome P450s and ACOS5, key enzymes required for exine formation proposed to play roles early in a hypothetical sporopollenin precursor biosynthetic pathway (Morant et al., 2007; de Azevedo Souza et al., 2009; Dobritsa et al., 2009). The strong preference of PKSA for hydroxy fatty acyl-CoAs generated by anther-specific cytochrome P450s and ACOS5 suggests that these are used by the anther-specific PKSs PKSA and PKSB to generate polyketides that are required for sporopollenin biosynthesis. This adds a new function for this class of natural products that exhibit an amazing functional diversity. The tight and transient coexpression of ACOS5 (de Azevedo Souza et al., 2009), PKSA, and PKSB (Figure 3) in tapetum cells at the uninucleate free microspore stages of anther development, coupled with the localization of PKSA and PKSB enzymes to the tapetum at these stages (Figure 4), suggest tapetum localization of a polyketide biosynthetic pathway involving the sequential actions of ACOS5 and PKSA/PKSB, consistent with the hypothesized secretion of sporopollenin precursors from the tapetum into locules prior to their polymerization into the exine of developing free microspores (Bedinger, 1992).
PKSA and PKSB Are Specifically Required for Sporopollenin Biosynthesis
Recently, Dobritsa et al. (2010) showed that LAP5/PKSB and LAP6/PKSA are required for exine formation and male fertility, since lap5/pksa and lap6/pksb mutants are defective in exine patterning and a lap5/pksa lap6/pksb double mutant is male sterile, lacks pollen at anther maturity, and exhibits defective exine formation in developing microspores. In our work, we observed similar defects in exine formation in single and double mutants and complete loss of fertility in the double mutant. However, we also observed decreases in fertility of single mutants (Table 1), suggesting that relatively subtle defective exine phenotypes (Dobritsa et al., 2010; Figure 6) impact pollen function.
Our detailed phenotypic analysis of the double mutant revealed new features of its loss-of-function phenotype, providing insights into functions of the wild-type enzymes. In the majority of anthers observed, defective microspore development was first observed at stage 9 (Figure 8), consistent with the timing of transient PKSA and PKSB expression (Figure 3) and protein accumulation (Figure 4) in the tapetum and the timing of exine formation (Blackmore et al., 2007). High-resolution TEM images of stage 9 wild-type and double mutant anthers (Figure 9) showed that mutant microspores completely lacked exine, which was replaced by an amorphous material similar to that we observed in other mutants defective in sporopollenin biosynthesis (acos5, abcg26, and drl1/tkpr1; de Azevedo Souza et al., 2009; Grienenberger et al., 2010; Quilichini et al., 2010). No abnormalities in tapetum cells were observed, and anther epidermal wall cutin deposition was similar to that in wild-type plants. Thus, the pksa pksb defect appears to be highly specific to exine formation and sporopollenin biosynthesis, consistent with PKSA and PKSB tapetum-specific expression patterns. Thus, unlike the fatty acid hydroxylase CYP704B1, which is involved in generating both sporopollenin and cutin precursors in developing rice anthers (Li et al., 2010), PKSA and PKSB appear to be specific in generating polyketide sporopollenin constituents.
In some of the pksa pksb mutant anthers, additional more complex microspore and locule phenotypes were observed at stage 9 and later stages. The presence of aberrant microspores showing signs of developmental arrest and cell fusion suggests that lack of PKSA/PKSB-derived polyketide sporopollenin constituents affects cell surface properties of developing microspores at the tetrad and subsequent stages. The densely staining material found in the locules of such pksa pksb mutant anthers (Figure 8, stages 9 to 11, and Figures 9I and 9L, electron-dense material) was not observed in wild-type anthers and could represent high levels of unpolymerized fatty acid alkylpyrone polyketide precursors and/or material that normally copolymerizes with such polyketide sporopollenin constituents to form the exine. Abnormal accumulation of such potentially lipophilic material could also result in the abnormal microspore cellular structure and apparent cell fusions observed in these anthers, and the complex phenotype of the double mutant could explain the extensive changes in the anther metabolome observed in lap5/pksa lap6/pksb mutants (Dobritsa et al., 2010). Taken together, our results indicate an essential function for PKSA- and PKSB-derived polyketide products in sporopollenin biosynthesis and microspore development.
Different in Vivo Functions of PKSA and PKSB
Both PKSA and PKSB recombinant enzymes catalyzed the decarboxylative condensations of fatty acyl-CoAs with malonyl-CoA in vitro to generate tri- and tetraketide α-pyrones (Figures 10 and 11). Single mutants affected in either PKS gene displayed subtle changes in exine patterning and deposition (Figure 6) but were fertile. By contrast, the pksa pksb double mutant produced very small amounts of pollen and was completely male sterile (Figures 7 and 8), consistent with the observations of Dobritsa et al. (2010). Whereas these data suggest that they have partially redundant functions in exine formation, we present several lines of evidence suggesting that PKSA and PKSB could fulfill different in vivo functions. First, exine patterning defects in the pksa and pksb mutants were slightly different, with pksb mutants showing more pronounced defects, including ectopic globular exine protrusions (Figure 6). Second, results from quantification of seed set in siliques of plants with different PKSA and PKSB allelic combinations (Table 1) showed that partial addition of PKSA activity to the pksb pksb background in PKSA pksa pksb pksb plants partially restored fertility relative to fully sterile pksa pksa pksb pksb plants, whereas addition of PKSB activity to pksa pksa pksb pksb plants (pksa pksa PKSB pksb plants) did not have this effect (Table 1). One interpretation of these data is that PKSA plays a more critical role in exine formation than does PKSB.
PKSA and PKSB expression and protein accumulation patterns support differentiated in vivo roles of the two enzymes. In this respect, microarray analyses comparing gene expression in Arabidopsis wild-type anthers relative to that in sporocyteless/nozzle or excess microsporocytes1/extrasporogenous cells mutants suggest that SPL and/or EMS1 positively regulate transcription of ACOS5, PKSA, and CHSL2 (At4g00040), all of which showing highly decreased expression levels in the spl/nzz and ems1/exs mutants (Wijeratne et al., 2007). By contrast, expression of PKSB was unaffected in the mutant backgrounds (Wijeratne et al., 2007). Our results show that the timing of maximal tapetum-localized PKSA and PKSB mRNA accumulation differs over the course of anther development (Figure 3), further supporting distinct regulatory control and potential specialized functions of the two genes.
Results from immunolocalization of PKSA and PKSB proteins in the tapetum over the course of anther development indicate a pattern of PKSB accumulation that is distinct from that of PKSA. PKSB shows an earlier and more transient tapetum-localized accumulation pattern during microsporogenesis (Figure 4; see Supplemental Figure 1 online). The transient pattern of PKSB protein accumulation (Figure 4) closely mirrored the timing of transient PKSB mRNA accumulation in tapetum cells at stage 8 (Figure 3) and is indicative of rapid protein turnover in tapetum cells after this stage. This was not the case for PKSA, which showed an early, transient pattern of mRNA accumulation followed by a broad period of protein accumulation. This suggests that PKSA protein turns over at a slower rate than does PKSB in later stages of anther development, consistent with a differential role later in microspore development.
At stage 8 of anther development, following release of individual microspores from tetrads, the first exine constituents produced in the sporophytic tapetum cell layer are deposited on the primexine microspore surface (Piffanelli et al., 1998; Suzuki et al., 2008). The rapid, transient burst of PKSB expression and protein accumulation at this stage suggests that the polyketide generated by PKSB may be essential at an early step of exine formation, possibly for initiating reticulated exine patterning, in cooperation with polyketides generated by PKSA. The ectopic accumulation of globular exine protrusions outside of recognizable baculae, observed in pksb but not pksa mutants (Figure 6), is consistent with a specific function for PKSB-generated polyketides in the early stages of sporopollenin deposition, when exine patterning is initiated.
It is noteworthy that another Arabidopsis PKS gene, CHSL2 (At4g00040), is 79% similar to PKSA. However, when expressed in bacteria, the cognate recombinant protein displayed no activity (formation of polyketide products) with the same fatty acyl-CoA and malonyl-CoA substrates used by PKSA and PKB (data not shown). These results, together with expression data from public databases showing that CHSL2 is expressed at later stages of flower bud development, indicate that CHSL2 performs an unknown enzymatic function distinct from that of PKSA.
Biochemical Functions of PKSA and PKSB
Type III PKSs produce secondary metabolites that play a variety of roles in plants. Our study, together with the recent report of Mizuuchi et al. (2008), shows that both PKSA and PKSB produce triketide and tetraketide α-pyrones by condensation with long-chain fatty acyl-CoAs (up to 20-carbon chain length) and malonyl-CoA. The two PKSs have unusually broad substrate specificities and a different cyclization strategy compared with typical plant type III PKSs (Mizuuchi et al., 2008). In contrast with our results and those of Mizuuchi et al. (2008), Dobritsa et al. (2010) failed to observe activity of recombinant LAP5/PKSA or LAP6/PKSB against fatty acyl substrates greater than C12 in length. The reason for this discrepancy is not clear, but activity against C16 to C18 fatty acyl-CoA substrates was consistently observed in multiple assays using our recombinant enzyme preparations (Figures 10 to 12, Table 2).
Our work also shows that, in vitro, PKSA preferentially catalyzes condensation of hydroxy fatty acyl-CoAs, which may be sequentially generated by CYP703A2 (Morant et al., 2007) and/or CYP704B1 (Dobritsa et al., 2009) with ACOS5 (de Azevedo Souza et al., 2009) to produce corresponding hydroxy tri- and tetraketide α-pyrones (Figures 10 and 11). Since ACOS5 also accepts a broad range of fatty acid substrates (de Azevedo Souza et al., 2009), the exact nature of its in vivo products that could be used as potential in vivo PKSA and PKSB substrates remains unclear. However, ACOS5 shows highest activity against medium-chain hydroxylated fatty acids (de Azevedo Souza et al., 2009) and could thus generate medium-chain hydroxy fatty acyl-CoAs, for which PKSA has a strong substrate preference. Thus, an attractive hypothesis is that the hydroxylated acyl chains generated by ACOS5 and a suite of cytochrome P450 hydroxylases (Morant et al., 2007; de Azevedo Souza et al., 2009; Dobritsa et al., 2009; Li et al., 2010) are incorporated into multihydroxy tri- and tetraketide α-pyrones products in vivo. The preferential subcellular localization of PKSA and PKSB to the ER (Figure 13) supports this model. Since the P450 hydroxylases are likely ER localized, they may form metabolons with the ER-associated PKSs to form alkyl-α-pyrones. Consistent with this, the TKPR1 enzyme that appears to act downstream of PKSA and PKSB is also ER localized (Grienenberger et al., 2010), suggesting the presence of an ER-localized pathway for alkyl-α-pyrone sporopollenin precursor biosynthesis.
Recently, Dobritsa et al. (2010) suggested that LAP5/PKSB and LAP6/PKSA could play roles both in the synthesis of alkylpyrones and in synthesis of phenolic constituents of sporopollenin in exine (Dobritsa et al., 2010). Metabolic profiling of developing anthers indicated that several flavonoids, including chalcone, naringenin, dihydrokaempferol, and isorhamnetin 3-sophoroside, were significantly reduced in single mutants and undetectable in the double mutant. However, such flavonoids are not likely to be direct products of LAP5/PKSB or/and LAP6/PKSA. Whereas LAP5/PKSB and LAP6/PKSA are closely related to CHS, the recombinant enzymes do not exhibit CHS activity (Dobritsa et al., 2010). Furthermore, whereas CHS activity and flavonoid biosynthesis appear to be crucial to pollen development in some plants (Mo et al., 1992; van der Meer et al., 1992; Fischer et al., 1997; Höfig et al., 2006; Schijlen et al., 2007), complete disruption of CHS expression in Arabidopsis, leading to the absence of foliar anthocyanins and flower flavonoids, has no impact on male or female fertility (Burbulis et al., 1996), suggesting that CHS-generated flavonoids are not required for normal Arabidopsis pollen development.
One possible reason for reduced flavonoid levels in pksa, pksb, and pksa pksb mutants is that the defective exine walls of pollen grains in these mutants affect subsequent deposition of pollen coat tryphine that contains phenolics, flavonoids, fatty acid derivatives, and proteins (Piffanelli et al., 1998; Scott et al., 2004; Grienenberger et al., 2009). Thus, reduction or absence of flavonoids could be the indirect consequence of reduced deposition of flavonoid-containing tryphine. Consistent with this, pollen coat deposition is affected in the pksa and pksb mutants and is more severely deficient in the pksb mutant (Figures 6M to 6O). The more severe tryphine defect in lap5/pksb observed in TEM images is consistent with the much greater reduction of flavonoid accumulation lap5/pksb anthers relative to lap6/pksa anthers (Dobritsa et al., 2010).
LAP6/PKSA and LAP5/PKSB accept multiple substrates and generate multiple products in vitro (Figures 10 to 12; Mizuuchi et al., 2008; Dobritsa et al., 2010), some of which may be derailment products due to incomplete catalysis. Thus, pool size of potential substrates in vivo, together with activities of native enzymes that may function as part of a complex or complexes in vivo, may dictate the nature of in vivo products formed by these enzymes. PKSA and PKSB are strongly coexpressed in tapetum cells with ACOS5 (de Azevedo Souza et al., 2009; Figure 3), ACOS5 preferentially generates hydroxy fatty acyl-CoAs but has no activity toward hydroxycinnamic acids (de Azevedo Souza et al., 2009), and PKSA preferentially accepts hydroxy fatty acyl-CoAs to generate tri- and tetraketide α-pyrones. These observations suggest that the most plausible in vivo substrates for PKSA and PKSB are hydroxy fatty acyl-CoAs rather than phenolics, such as hydroxycinnamoyl-CoAs, and that alkyl pyrones are natural products generated by PKSA and PKSB that are required for sporopollenin biosynthesis. The biosynthesis of the tri- and tetraketide α-pyrones in plants has not been well studied, and their presence in Arabidopsis has not been reported. Thus, while our results indicate that the in vivo functions of PKSA and PKSB are to catalyze condensation of malonyl-CoA with hydroxy fatty acyl-CoAs generated by ACOS5 to generate alkyl pyrones, this hypothesis requires further testing. For example, reduction of keto functions after elongation of the chain by PKSA/PKSB would prevent the formation of the α-pyrone ring, and alkyl phloroglucinols could be formed as sporopollenin natural products. However, the strong substrate preferences for tapetum-expressed reductases DRL1/TKPR1 and TKPR2, involved in sporopollenin biosynthesis, for the in vitro tetraketide α-pyrone product (Grienenberger et al., 2010) support the hypothesis that these are true PKSA/PKSB-derived natural products.
The tri- and tetraketide α-pyrones generated in vitro by PKSA and PKSB also contain a ketone group on the α-pyrone rings and an additional ketone on the alkyl carbon chain, in the case of the tetraketide (Figures 10 and 11). These carbonyl groups could be the targets for further reduction to generate even more highly hydroxylated polyketides. Candidate enzymes for such activity are TKPR1 (DRL1) and the closely related TKPR2. Indeed, in the companion article (Grienenberger et al., 2010), we show that DRL1/TKPR1 and a close homolog encode tetraketide α-pyrone reductases that specifically catalyze the reduction of the acyl ketone group of the tetraketide α-pyrone generated by PKSA and PKSB. Polyhydroxylation of polyketide α-pyrone(s) by these mechanisms would provide multiple moieties for cross-linking by ether and ester bond formation between polyketide α-pyrone sporopollenin precursors and other potential sporopollenin constituents, such as fatty acids and/or fatty alcohols, contributing to the strong chemical and physical resistance of the sporopollenin polymer.
Conservation of PKSA and PKSB Functions in a Sporopollenin Precursor Biosynthetic Pathway
Phylogenetic analysis (Figure 1) showed that the plant PKS clade containing PKSA and PKSB is clearly distinct from the clade containing the bona fide Arabidopsis and Physcomitrella CHS genes and other putative CHS genes from poplar and rice. This relationship between the true CHS genes and the PKS CHS-like (CHSL) genes, also observed by others (Mizuuchi et al., 2008; Wu et al., 2008; Dobritsa et al., 2010), is similar to the relationship between ACOS5 and true 4CL enzymes (de Azevedo Souza et al., 2008, 2009). The CHSL clade containing PKSA and PKSB has two distinct subclades of angiosperm PKS genes that are related to either Arabidopsis PKSA or PKSB, with at least one PKSA and one PKSB homolog found in each of the fully sequenced poplar and rice genomes. In addition, the N. sylvestris PKSA homolog CHSlk (Atanassov et al., 1998) and S. latifolia PKSB homolog CHSL (Ageez et al., 2005) are known to be expressed in male reproductive organs, and the wheat and triticale homologs of the Arabidopsis PKSs also have anther and tapetum-preferred expression patterns (Wu et al., 2008).
The CHSL subclade containing Arabidopsis PKSA and PKSB contains both pine and Physcomitrella members, both of which occupy positions at or near the base of the clade (Figure 1). The P. radiata ChS1 gene is specifically expressed in male cones and is likely tapetum expressed (Walden et al., 1999). We surveyed the expression pattern of the Physcomitrella PKS (CHS10) gene (Jiang et al., 2006; Koduri et al., 2010), an apparent PKSA and PKSB homolog (Figure 1), by assessing transcript abundance in a P. patens cDNA database (http://www.ncbi.nlm.nih.gov/UniGene/library.cgi?LID=23755andPAGE=1). This analysis showed that Pp-CHS10 cDNAs are found exclusively in a library derived from RNA extracted from the sporophyte. Together, these data suggest a conserved function in sporopollenin monomer biosynthesis for PKSA and PKSB and their apparent orthologs in land plants and indicate that this function arose early in land plant evolution prior to the divergence of bryophytes and tracheophytes. The exine in pollen walls may have evolved a more elaborate structure in seed plants, based on specialized functions of the duplicated PKSA and PKSB genes found in common within the angiosperm lineage.
The remarkable conservation of Arabidopsis PKSA and PKSB, ACOS5 (de Azevedo Souza et al., 2009), and CYP703A2 (Morant et al., 2007) genes in land plants suggests that the encoded enzymes are part of an ancient sporopollenin biosynthetic pathway. The in vitro biochemical functions of PKSA and PKSB that we explored in detail, together with similar data on ACOS5 and CYP703A2, the coordinated coexpression of these genes in the tapetum during free microspore stages of anther development, and the ER localization of PKSA and PKSB support the hypothesis that this pathway catalyzes sequential modification of fatty acid starter molecules to generate integral sporopollenin components of the pollen exine. Based on these data, we propose a reaction sequence localized to the ER leading from hydroxylation of medium- to long-chain fatty acids (catalyzed by CYP703A2), to fatty acyl-CoA formation (catalyzed by ACOS5), and condensation of fatty acyl-CoAs with malonyl-CoA (catalyzed by PSKA and PSKB) to generate triketide and tetraketide α-pyrone sporopollenin precursors. The accompanying article (Grienenberger et al., 2010) extends this work and shows that reductases encoded by tapetum-expressed DRL/TKPR genes are also part of this pathway and accept the tetraketide α-pyrone product generated by PKSA and PKSB in vitro to form reduced derivatives that appear to be sporopollenin precursors.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana Columbia (Col-0) seeds were sterilized and after a cold treatment (2 d at 4°C in the dark) and germinated at 20°C under 70 μmol m−2 s−1 fluorescent lighting. Twelve days later, the plants were transferred to a growth chamber with a light/dark cycle of 16 h/8 h. T-DNA insertion mutants were obtained from SALK, SAIL (Alonso et al., 2003), and Gabi-Kat (Rosso et al., 2003) collections via The Arabidopsis Information Resource and the Nottingham Arabidopsis Stock Centre. In progeny, homozygous insertion lines SALK_079287, SALK_134643 (PKSA), GK_089C04, GK_454C04, and GK_580A09 (PKSB) were identified by PCR using gene-specific and T-DNA–specific primers (see Supplemental Table 2 online). Nicotiana benthamiana plants were grown in a growth chamber under 3000 lux lighting and a light/dark cycle of 16 h/8 h at 21 ± 2°C.
Phylogenetic and Bioinformatic Analyses
All sequences obtained are given in Supplemental Table 1 and Supplemental Data Set 1 online. Protein sequences were aligned using MUSCLE 3.6 using the default parameters (sequencing clustering; UPGMA, objective score; classic sum-of-pairs score) (Edgar, 2004), and the multiple protein sequence alignments were manually optimized. Aligned sequences are available in Supplemental Data Set 2 online. To reconstruct phylogenetic trees, maximum likelihood analyses with 1000 bootstrap replicates were performed using PhyML v2.4.4 and default settings (Guindon and Gascuel, 2003) with the JTT model of amino acid substitution.
RT-PCR
RNA quality was assessed by visual inspection of rRNA on a 1.2% formaldehyde-agarose gel and quantified spectrophotometrically, and 2.5 μg RNA/20 μL reaction was used to generate first-strand cDNA using Superscript II reverse transcriptase (Invitrogen) following the manufacturer’s protocol. For RT-PCR, gene-specific and intron-spanning primers (see Supplemental Table 2 online) were used in PCR reactions to amplify corresponding cDNA sequences under the following PCR conditions: 95°C for 3 min, followed by 35 cycles of (94°C for 30 s, 56°C for 30 s, and 72°C for 1 min) followed by 72°C for 10 min, using Taq polymerase in a 50-μL total reaction. Actin2 was used as control.
For quantitative RT-PCR analysis of PKSA and PKSB expression, 10 ng of cDNA was incubated with 10 μL iQ SYBR Green Supermix (Bio-Rad) and 5 pmol of each forward and reverse primer (see Supplemental Table 2 online) in a total volume of 20 μL. After an initial denaturation step at 95°C for 3 min, 40 cycles at 95°C for 15 s, 60°C for 15 s, and 72°C for 30 s were followed by a fluorescence reading. A melting curve was generated ranging from 95 to 60°C. Threshold cycles (CT) were adjusted manually, and the CT values for a housekeeping control Actin2 amplified in parallel on each plate were subtracted from CT values obtained for each gene of interest, thus generating normalized CT values (ΔCT). The relative starting quantities of each gene were determined by setting as a base value the gene with the highest CT value within a tissue panel or treatment series, and relative quantities were calculated using the ΔΔCT method as described by Hietala et al. (2003). ΔΔCT was calculated using immature flower buds as the highest expressing tissue.
Phenotypic Analyses
For fluorescence microscopy, pollen was stained with 0.01% auramine O in water for 5 min, washed twice with water, and examined with a Nikon E800 microscope using fluorescein isothiocyanate settings.
For scanning electron microscopy, pollen grains were coated with gold particles (S150A sputter coater; Edwards High Vacuum) for 3 min at 50 mV and viewed with a Philips XL30 ESEM (FEI) under high vacuum conditions and at 20 to 30 kV. The microscope was equipped with a Thornley-Everhart secondary electron detector and a PGT Spirit EDX microanalysis system (Princeton Gamma Tech).
To obtain cross sections of developing anthers, wild-type and homozygous double mutant inflorescences were fixed in FAA (4% paraformaldehyde, 15% acetic acid, and 50% ethanol) overnight and directly dehydrated without postfixation. Samples were then transferred to a propylene oxide solution and slowly infiltrated with Spurr’s epoxy resin (Canemco). For bright-field microscopy, 1-μm sections were cut with glass knives (Leica) on a microtome, mounted on glass slides, heat fixed to the slides, and stained with toluidine blue. Sections were photographed using a light microscope. For in vivo pollen germination assays, wild-type pistils were manually pollinated with wild-type or mutant pollen, and 24 h after pollination, the pistils were harvested and stained as described by Preuss et al. (1993).
For TEM, Arabidopsis wild-type and pksa-1 pksb-3 double mutant inflorescences were fixed with paraformaldehyde and postfixed in osmium tetraoxide. Fixed samples were dehydrated in an ethanol gradient up to 100% and then transferred to a propylene oxide solution and slowly embedded in Spurr’s resin and allowed to polymerize for at least 48 h. Thin sections (70 nm) were taken using a diamond knife microtome (Reichert Ultracut E). Sections were placed on 100-mesh copper grids and stained for 30 min with uranyl acetate, rinsed thoroughly with water, and stained for 15 min with lead citrate (Sato’s Lead). Sections were visualized using a Hitachi H7600 transmission electron microscope. For TEM analysis of pksa-1 and pksb-2 single mutants relative to the wild type, pollen was harvested by shaking open inflorescences in PBS, pH 7.0, and filtering through 50-μm nylon mesh. Pollen was centrifuged at 5000g and fixed in 3% glutaraldehyde. Fixed samples were centrifuged and pellets were taken up in 1% low melting point agarose. Samples were cut in 3-mm3 blocks with a razor blade and postfixed in osmium tetraoxide. After dehydration in an ethanol series, pollen was progressively embedded in LR white resin and allowed to polymerize for 48 h. Thin sections (70 nm) were obtained using a diamond knife microtome (Reichert-Jung Ultracut E). Sections were placed on microscope grids and stained for 30 min with uranyl acetate, rinsed with water, and observed using a Hitachi H-600 transmission electron microscope equipped with an Orca HR Hamamatsu camera.
In Situ Hybridization and Validation of RNA Probes
Arabidopsis Col-0 inflorescences were embedded in Paraplast (Sigma-Aldrich), sectioned at 8 μm thickness, and mounted on precharged slides. For sense and antisense PKSA and PKSB probe synthesis, 1209- and 1200-bp DNA template corresponding to the PKSA and PKSB coding region, respectively, were PCR amplified from flower cDNA using gene-specific forward and reverse primers (see Supplemental Table 2 online). A T7 polymerase binding site was incorporated into the forward primer for sense probe amplification and in the reverse primer for antisense probe amplification. Digoxigenin-labeled probes were transcribed off the template using T7 polymerase (Roche). Probes were shortened to 200-bp fragments by limited carbonate hydrolysis, quantified, and hybridized to slides. Tissue fixation, embedding, hybridization, and signal detection are described by Quilichini et al. (2010).
To validate specificity of RNA antisense probes, we performed DNA gel blotting. PCR-amplified cDNA clones of PKSA and PKSB (see Supplemental Table 1 online for primers under “DNA gel blotting”) were blotted to positively charged nylon membranes, Hybond-XL (GE Healthcare Life Sciences), using 10× SSC as the transfer buffer. Transfer of cDNA to the nylon membrane was monitored via ethidium bromide. The digoxigenin-labeled probes were prehybridized and hybridized using 1× Denhardt's solution, 50% deionized formamide, 10% dextran sulfate, and 0.2 mg/mL of salmon sperm DNA. The blots were washed twice using 2× SSC and 0.1% SDS at 55°C for 10 min each, three times using 2× SSC at 55°C for 30 min each, and twice using 0.2× SSC at 55°C for 10 min each. The hybridized probes were detected by the addition of BCIP/NBT color reagent (Roche).
Production of Recombinant Proteins and Specific Antibodies
Full coding sequences of genes were cloned in pGEX-KG (Guan and Dixon, 1991) after PCR amplification using forward and reverse primers and DNA restriction sites as indicated in Supplemental Table 2 online. Error-free constructs were transformed into Escherichia coli BL21-G612 strain. Conditions of expression of the recombinant proteins and purification on glutathione-agarose column were as previously described (Hoffmann et al., 2003). Analysis of PKS preparations at the different steps of purification is illustrated in Supplemental Figure 8 online.
Polyclonal antibodies were raised in 2-month-old rabbits by two injections of the purified proteins. For the first injection, 200 μg protein was emulsified with Freund’s complete adjuvant. Starting 1 month after first injection, 30-mL blood samples were taken weekly for 4 weeks and then a second injection was performed using 130 μg of protein emulsified with incomplete adjuvant. Finally, 12 d after boost injection, the animal was killed, and 130 to 150 mL of blood (70 to 80 mL of serum) was obtained.
Immunological Techniques
Before use, antibodies were preincubated overnight with an acetonic powder of Arabidopsis leaves to eliminate any nonspecific signal. For immunoblotting experiments, the procedures were as described previously (Geoffroy et al., 1990) using antibodies at 1/10,000 dilution and a chemiluminescent substrate (CDP-Star; Bio-Rad) for phosphatase activity detection. Antibody specificity for each PKS protein was enhanced by preincubation with the other recombinant protein. The specificity of the resulting antibodies was demonstrated in immunoblotting experiments with the purified recombinant proteins: each antiserum was shown to recognize the cognate protein, and no cross-reactivity was evidenced with the other PKS recombinant protein (see Supplemental Figure 2 online). For immunohistochemical analysis, flower buds were collected, fixed with an aqueous solution containing 3.7% formaldehyde, 50% ethanol, and 5% acetic acid, dehydrated in an ethanol series, and imbedded in paraplast. Ten-micrometer-thick sections were made with a RM2155 Leica microtome and dried on microscope slides, and paraffin was eliminated with Histoclear (VWR). Before immunolabeling, sections were rehydrated in PBS and then blocked for 1 h in a PBS solution containing 1% BSA, 0.05% Triton, and 5% goat normal serum. The rabbit antiserum was diluted 200-fold in the blocking solution, and sections were incubated in the serum solution overnight at 4°C. Then, after three successive washes with PBS containing 1% BSA and 0.05% Triton, sections were incubated for 1 h at room temperature with alkaline phosphatase–labeled goat anti-rabbit immunoglobulin G (Euromedex) diluted 1/1000 in PBS. Samples were then washed four times in PBS, and, after a final wash in 100 mM Tris-HCl, pH 8.1, phosphatase activity was revealed with 1 mg/mL FastRed aqueous solution (Sigma-Aldrich). Incubation was stopped by washing sections in water and sections, covered with PBS/glycerol (50/50, v/v), were observed under an E800 microscope.
Chemical Synthesis of Fatty Acyl-CoA Esters
Palmitoyl-CoA and stearoyl-CoA esters were chemically prepared from palmitic and stearic acids according to published procedures (Funa et al., 2006). CoA ester preparations were purified by reverse-phase HPLC and characterized by LC-MS/MS analysis. 16-Hydroxyhexadecanoyl-CoA (16-OH-C16-CoA) and 12-hydroxyoctadecanoyl-CoA (12-OH-C18-CoA) were synthesized using a method involving ethyl chloroformate activation of fatty acids as follows. The fatty acid (5.5 molar equivalents) was dissolved in dry THF under argon. To this solution, ethyl chloroformate (5.5 molar equivalents) and triethylamine (5.5 molar equivalents) were added at room temperature, and the resulting mixture was stirred for 30 min.
The mixture was then passed through a syringe filter into a solution of CoA trilithium salt (1 molar equivalent) and KHCO3 (5.5 molar equivalents) in water. To assure solubility of all reaction components, the final THF/water solvent mixture should be (1:1.5). This solution was then stirred for 12 h at room temperature.
The reaction mixture was filtered and purified by HPLC under basic conditions, using buffer A (5 mmol/L ammonium acetate in water) and buffer B (5 mmol/L ammonium acetate in acetonitrile) as eluents and a VP125/21 Nucleodur C18 Gravity (5 μm) column (Macherey and Nagel) as stationary phase at a flow of 25 mL/min.
Subsequent LC-MS-electrospray ionization analyses were performed on an HPLC system from Agilent (1200 series) with an Eclipse XDB-C18, 5-μm column (column dimensions: 150 × 4.60 mm) from Agilent and a Thermo Finnigan LCQ Advantage Max ESI spectrometer. For detection, absorption at 280 nm was followed. As eluents, solvent A (5 mmol/L ammonium acetate in water) and solvent B (5 mmol/L ammonium acetate in acetonitrile) were used at 1 mL/min flow rate. Gradient was as follows: 0 min/90% A/10% B → 1 min/90% A/10% B → 10 min/0% A/100% B → 12 min/0% A/100% B → 15 min/90% A/10% B.
For synthesis of 12-hydroxystearoyl-CoA, 2-hydroxystearic acid (50 mg, 0.17 mmol) was converted into 12-hydroxystearoyl-CoA using the general procedure described above. The crude product was purified by preparative HPLC (0 min/95% A/5% B → 3 min/95% A/5% B → 45 min/80% A/20% B → 50 min/75% A/25% B → 60 min/70% A/30% B → 75 min/0% A/100% B) to yield 23.4 mg 12-hydroxystearoyl-CoA as a white solid (72% yield). LC-MS was as follows: tR = 7.25, m/z calculated for C39H69N7O18P3S [M – H]− 1048.37; found 1048.33.
For synthesis of 16-hydroxyhexadecanoyl-CoA, 16-hydroxyhexadecanoic acid (10 mg, 36.8 μmol) was converted into 16-hydroxyhexadecanoyl-CoA using the general procedure described above. The crude product was purified by preparative HPLC (0 min/95% A/5% B → 3 min/95% A/5% B → 45 min/75% A/25% B → 50 min/75% A/25% B → 65 min/70% A/30% B → 75 min/55% A/45% B) to yield 14.3 mg 16-hydroxyhexadecanoyl-CoA as a white solid (40% yield). LC-MS was as follows: tR = 6.97, m/z calculated for C37H65N7O18P3S [M – H]− 1020.34; found 1020.25.
Enzyme Activity Assays and Steady State Kinetic Analysis
For ACOS5 activity, in a volume of 100 μL, 5 μg of recombinant protein was incubated in 60 mM phosphate buffer containing 10 mM MgCl2, 5 mM ATP, 2.5 mM DTT, 1 mM CoASH, and 0.1 mM fatty acid (from a 1 mM stock solution in 0.1% Triton X-100). The mixture was incubated at 30°C. For ACOS5 activity measurements, the reaction was stopped by addition of 10 μL of 1 n HCl and, after extraction of residual fatty acid with ethyl acetate, CoA esters were analyzed by LC-MS/MS. ACOS5 activity was used for the biosynthesis of various fatty acyl-CoA esters that were tested as PKS substrates.
For PKS activity, when chemically synthesized CoA esters were used, 5 μg of enzyme was incubated in 100 μL volume containing 100 μM potassium phosphate, pH 7.0, 0.1 mM fatty acyl-CoA ester, and 0.1 mM malonyl-CoA either unlabeled or [2-14C]-labeled (25 nCi). PKS activity was evaluated either after TLC and visualization of radioactive reaction products by bioimaging or by LC-MS/MS. PKS activity was also assayed with various fatty acyl-CoA esters synthesized by ACOS5. To that aim, after 15 min ACOS5 incubation, 5 μg of PKSA or PKSB protein were added to the incubation medium together with malonyl-CoA and incubated for an additional hour. Activity was measured as for chemically synthesized fatty acyl-CoA substrates.
Steady state kinetic parameters were determined using six concentrations of substrate, covering a range of 0.2 to ~4 Km, in enzyme assay buffer (100 mM potassium phosphate, pH 7.2, 0.1% Triton X-100, and 10% glycerol pH 7.2) containing 50 μM [2-14C] malonyl-CoA. Briefly, 5 μg of enzyme was incubated with appropriate substrates in a reaction volume of 50 μL at 37°C for 10 min and was terminated by acidification with 1 n HCl (3.75 μL). The triketide α-pyrone product was then extracted with ethyl acetate, and the extracts were analyzed by silica TLC (toluene/acetone/acetic acid, 85/15/1 [v/v/v]). Km and Vmax were calculated by fitting the data to the Michaelis-Menten equation using GraphPad Prism v. 5.03.
Characterization of Reaction Products by LC-MS/MS
Reaction products dissolved in methanol were injected on Acquity ultraperformance liquid chromatography (UPLC) BEH C18 column (2.1 × 100 mm, 1.7 μm) and precolumn (2.1 × 5 mm, 1.7 μm). Three-microliter samples were injected and resolved by an increasing gradient of acetonitrile in water containing 0.1% formic acid. Gradient conditions at a flow rate of 0.45 mL/min were as follows: 50 to 100% acetonitrile for 25 min, 100% for 2 min, 100 to 50% for 1 min, and column equilibration at 50% acetonitrile for 4 min.
For analysis by LC-MS/MS, an Acquity UPLC system (Waters) coupled to a Quattro Premier XE triple quadrupole MS system (Waters Micromass) was used. The mass spectrometer was run using Mass-Lynx software. The electrospray ionization source conditions in positive and negative modes were optimized by direct infusion. Nitrogen was used as the nebulization and desolvation gas at flow rates of 50 and 900 L/h, respectively. The source capillary voltage was set to 3 kV and temperature to 135°C. Cone tension was optimized at 25 V and desolvation performed at 400°C. In collision-induced dissociation experiments, argon was used as collision gas at a pressure of 3 × 10−3 mbar. Full-scan, selected ion recording, daughter scan, and multiple reaction monitoring modes were used for analysis.
Subcellular Localization
For subcellular localization of PKSA and PKSB, N-terminal fusions of the corresponding cDNAs to eGFP were PCR amplified using the primer pairs given in Supplemental Table 2 online and inserted into the expression vector pB7FWG2 (VIB) using the Gateway cloning system (Invitrogen). The mRFP fluorescent protein with the C-terminal extension-HDEL served as an ER marker protein (Robinson et al., 2007). Constructs were transiently expressed in N. benthamiana leaves by agroinfiltration and imaged as described (Grienenberger et al., 2010).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative, GenBank/EMBL, or other databases under the following accession numbers: Arabidopsis PKSA, At1g02050; Arabidopsis PKSB, At4g34850; Arabidopsis Actin2, At3g18780; Arabidopsis CHSL2, At4g00040; Oryza sativa CHSL1, LOC_Os10g34360 (http://rice.plantbiology.msu.edu/index.shtml); O. sativa CHSL2, LOC_Os07g22850 (http://rice.plantbiology.msu.edu/index.shtml); Physcomitrella patens CHS10, protein ID 149790 (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html); Pinus radiata CHS1, AAB80804; Nicotiana sylvestris CHSLK, CAA74847; Silene latifolia Chs, AB182106; Populus trichocarpa CHSL4, protein ID 551991 (http://genome.jgi-psf.org/poplar/poplar.home.html); P. trichocarpa CHSL5, protein ID 591704 (http://genome.jgi-psf.org/poplar/poplar.home.html); P. trichocarpa CHSL6, protein ID 556583 (http://genome.jgi-psf.org/poplar/poplar.home.html); and P. trichocarpa CHSL, protein ID 200918 (http://genome.jgi-psf.org/poplar/poplar.home.html).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Accumulation of PKSA and PKSB Proteins during Flower Development.
Supplemental Figure 2. Characterization of Antibodies Raised against PKS Proteins.
Supplemental Figure 3. DNA Gel Blotting with RNA Antisense Probe of Either PKSA or PKSB against PCR-Amplified PKSA and PKSB Coding Sequence.
Supplemental Figure 4. Phenotypic Effects of pksa-2 and pksb-1 Alleles on Exine Formation.
Supplemental Figure 5. Pollen Tube Growth Assay of pksa-1 and pksb-2 Mutant Pollen.
Supplemental Figure 6. Exine Defects in Pollen Grains of the Double Mutant pksa-1 pksb-2.
Supplemental Figure 7. Anther Development of Double Mutant pksa-1 pksb-3.
Supplemental Figure 8. Analysis of Recombinant Protein Preparations at Different Steps of Purification.
Supplemental Table 1. Putative PKSA and PKSB Orthologs and Expression in Other Species.
Supplemental Table 2. Primer Sequences Used in This Study.
Supplemental Data Set 1. Amino Acid Sequences Used in Construction of the Phylogenetic Tree Shown in Figure 1.
Supplemental Data Set 2. Alignment of Amino Acid Sequences Used to Generate the Phylogenetic Tree Shown in Figure 1.
Acknowledgments
The assistance of D. Meyer (Institut de Biologie Moléculaire des Plantes) in histochemical analysis, M. Alioua (Institut de Biologie Moléculaire des Plantes) in DNA sequencing, and M. Erhardt (Institut de Biologie Moléculaire des Plantes) for TEM is gratefully acknowledged. We thank J.-H. Lignot (Institut Pluridisciplinaire Hubert Curien, Strasbourg) for kind help with scanning electron microscopy, C. Ritzenthaler (Institut de Biologie Moléculaire des Plantes) for help with confocal microscopy, and D. Debayle (Institut de Biologie Moléculaire des Plantes) for assistance in LC-MS/MS analysis at the initial stages of the work. We are grateful to the Salk Genomic Analysis Laboratory (La Jolla, CA), the Institut National de la Recherche Agronomique Versailles (France), and Bielefeld University (Germany) for providing the T-DNA mutants and to the ABRC and Nottingham Arabidopsis Stock Center for distributing the seeds. The UPLC-MS/MS system was cofinanced by the Centre National de la Recherche Scientifique, the Université de Strasbourg, the Région Alsace, the Institut National de la Recherche Agronomique, and Tepral Company. We also thank Michael Friedmann and Teagen Quilichini (University of British Columbia) for helpful comments and advice and the University of British Columbia BioImaging Facility for technical assistance and advice. This work was supported by doctoral fellowships of the Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche to E.G. and B.L., and by Natural Sciences and Engineering Research Council discovery grants to C.J.D. and D.-Y.S.
References
- Aarts M.G., Hodge R., Kalantidis K., Florack D., Wilson Z.A., Mulligan B.J., Stiekema W.J., Scott R., Pereira A. (1997). The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Abe I., Oguro S., Utsumi Y., Sano Y., Noguchi H. (2005). Engineered biosynthesis of plant polyketides: Chain length control in an octaketide-producing plant type III polyketide synthase. J. Am. Chem. Soc. 127: 12709–12716 [DOI] [PubMed] [Google Scholar]
- Abe I., Watanabe T., Noguchi H. (2004). Enzymatic formation of long-chain polyketide pyrones by plant type III polyketide synthases. Phytochemistry 65: 2447–2453 [DOI] [PubMed] [Google Scholar]
- Ageez A., Kazama Y., Sugiyama R., Kawano S. (2005). Male-fertility genes expressed in male flower buds of Silene latifolia include homologs of anther-specific genes. Genes Genet. Syst. 80: 403–413 [DOI] [PubMed] [Google Scholar]
- Ahlers F., Bubert H., Steuernagel S., Wiermann R. (2000). The nature of oxygen in sporopollenin from the pollen of Typha angustifolia L. Z. Naturforsch., C, J. Biosci. 55: 129–136 [DOI] [PubMed] [Google Scholar]
- Ahlers F., Lambert J., Wiermann R. (2003). Acetylation and silylation of piperidine solubilized sporopollenin from pollen of Typha angustifolia L. Z. Naturforsch., C, J. Biosci. 58: 807–811 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Hatakeyama K., Hinata K., Sato S., Kato T., Tabata S., Toriyama K. (2003). A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol. Biol. 53: 107–116 [DOI] [PubMed] [Google Scholar]
- Atanassov I., Russinova E., Antonov L., Atanassov A. (1998). Expression of an anther-specific chalcone synthase-like gene is correlated with uninucleate microspore development in Nicotiana sylvestris. Plant Mol. Biol. 38: 1169–1178 [DOI] [PubMed] [Google Scholar]
- Austin M.B., Noel J.P. (2003). The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20: 79–110 [DOI] [PubMed] [Google Scholar]
- Bedinger P. (1992). The remarkable biology of pollen. Plant Cell 4: 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore S., Wortley A.H., Skvarla J.J., Rowley J.R. (2007). Pollen wall development in flowering plants. New Phytol. 174: 483–498 [DOI] [PubMed] [Google Scholar]
- Bohne G., Richter E., Woehlecke H., Ehwald R. (2003). Diffusion barriers of tripartite sporopollenin microcapsules prepared from pine pollen. Ann. Bot. (Lond.) 92: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Floyd S.K., Sakakibara K. (2007). Green genes-comparative genomics of the green branch of life. Cell 129: 229–234 [DOI] [PubMed] [Google Scholar]
- Bubert H., Lambert J., Steuernagel S., Ahlers F., Wiermann R. (2002). Continuous decomposition of sporopollenin from pollen of Typha angustifolia L. by acidic methanolysis. Z. Naturforsch., C, J. Biosci. 57: 1035–1041 [DOI] [PubMed] [Google Scholar]
- Burbulis I.E., Iacobucci M., Shirley B.W. (1996). A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell 8: 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Goodwin S.M., Boroff V.L., Liu X., Jenks M.A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo Souza C., Barbazuk B., Ralph S.G., Bohlmann J., Hamberger B., Douglas C.J. (2008). Genome-wide analysis of a land plant-specific acyl:coenzymeA synthetase (ACS) gene family in Arabidopsis, poplar, rice, and Physcomitrella. New Phytol. 179: 987–1003 [DOI] [PubMed] [Google Scholar]
- de Azevedo Souza C., Kim S.S., Koch S., Kienow L., Schneider K., McKim S.M., Haughn G.W., Kombrink E., Douglas C.J. (2009). A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T.T., Carlsson A.S., Hamberg M., Bülow L., Stymne S., Olsson P. (2009). Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J. Plant Physiol. 166: 787–796 [DOI] [PubMed] [Google Scholar]
- Dobritsa A.A., Lei Z., Nishikawa S., Urbanczyk-Wochniak E., Huhman D.V., Preuss D., Sumner L.W. (2010). LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol. 153: 937–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Shrestha J., Morant M., Pinot F., Matsuno M., Swanson R., Møller B.L., Preuss D. (2009). CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 151: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R., Budde I., Hain R. (1997). Stilbene synthase gene expression causes changes in flower colour and male sterility in tobacco. Plant J. 11: 489–498 [Google Scholar]
- Flores-Sanchez I.J., Verpoorte R. (2009). Plant polyketide synthases: A fascinating group of enzymes. Plant Physiol. Biochem. 47: 167–174 [DOI] [PubMed] [Google Scholar]
- Franke R., Schreiber L. (2007). Suberin—A biopolyester forming apoplastic plant interfaces. Curr. Opin. Plant Biol. 10: 252–259 [DOI] [PubMed] [Google Scholar]
- Funa N., Ozawa H., Hirata A., Horinouchi S. (2006). Phenolic lipid synthesis by type III polyketide synthases is essential for cyst formation in Azotobacter vinelandii. Proc. Natl. Acad. Sci. USA 103: 6356–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi M., Funa N., Horinouchi S. (2008). Phenolic lipids synthesized by type III polyketide synthase confer penicillin resistance on Streptomyces griseus. J. Biol. Chem. 283: 13983–13991 [DOI] [PubMed] [Google Scholar]
- Geoffroy P., Legrand M., Fritig B. (1990). Isolation and characterization of a proteinaceous inhibitor of microbial proteinases induced during the hypersensitive reaction of tobacco to tobacco mosaic virus. Mol. Plant Microbe Interact. 3: 327–333 [DOI] [PubMed] [Google Scholar]
- Grienenberger E., Besseau S., Geoffroy P., Debayle D., Heintz D., Lapierre C., Pollet B., Heitz T., Legrand M. (2009). A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J. 58: 246–259 [DOI] [PubMed] [Google Scholar]
- Grienenberger E., Kim S.S., Lallemand B., Geoffroy P., Heintz D., de Azevedo Souza C., Heitz T., Douglas C.J., Legrand M. (2010). Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22: 4067–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K.L., Dixon J.E. (1991). Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192: 262–267 [DOI] [PubMed] [Google Scholar]
- Guilford W.J., Schneider D.M., Labovitz J., Opella S.J. (1988). High resolution solid state C NMR spectroscopy of sporopollenins from different plant taxa. Plant Physiol. 86: 134–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Hietala A.M., Eikenes M., Kvaalen H., Solheim H., Fossdal C.G. (2003). Multiplex real-time PCR for monitoring Heterobasidion annosum colonization in Norway spruce clones that differ in disease resistance. Appl. Environ. Microbiol. 69: 4413–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L., Maury S., Martz F., Geoffroy P., Legrand M. (2003). Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J. Biol. Chem. 278: 95–103 [DOI] [PubMed] [Google Scholar]
- Höfig K.P., Möller R., Donaldson L., Putterill J., Walter C. (2006). Towards male sterility in Pinus radiata—A stilbene synthase approach to genetically engineer nuclear male sterility. Plant Biotechnol. J. 4: 333–343 [DOI] [PubMed] [Google Scholar]
- Jiang C., Schommer C.K., Kim S.Y., Suh D.Y. (2006). Cloning and characterization of chalcone synthase from the moss, Physcomitrella patens. Phytochemistry 67: 2531–2540 [DOI] [PubMed] [Google Scholar]
- Koduri P.K., Gordon G.S., Barker E.I., Colpitts C.C., Ashton N.W., Suh D.Y. (2010). Genome-wide analysis of the chalcone synthase superfamily genes of Physcomitrella patens. Plant Mol. Biol. 72: 247–263 [DOI] [PubMed] [Google Scholar]
- Li H., Pinot F., Sauveplane V., Werck-Reichhart D., Diehl P., Schreiber L., Franke R., Zhang P., Chen L., Gao Y., Liang W., Zhang D. (2010). Cytochrome P450 family member CYP704B2 catalyzes the omega-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22: 173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Mizuuchi Y., Shimokawa Y., Wanibuchi K., Noguchi H., Abe I. (2008). Structure function analysis of novel type III polyketide synthases from Arabidopsis thaliana. Biol. Pharm. Bull. 31: 2205–2210 [DOI] [PubMed] [Google Scholar]
- Mo Y., Nagel C., Taylor L.P. (1992). Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. USA 89: 7213–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M., Jørgensen K., Schaller H., Pinot F., Møller B.L., Werck-Reichhart D., Bak S. (2007). CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E., Gang D.R. (2000). Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 5: 439–445 [DOI] [PubMed] [Google Scholar]
- Piffanelli P., Ross J.H.E., Murphy D.J. (1998). Biogenesis and function of the lipidic structures of pollen grains. Sex. Plant Reprod. 11: 65–80 [Google Scholar]
- Pollard M., Beisson F., Li Y., Ohlrogge J.B. (2008). Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Preuss D., Lemieux B., Yen G., Davis R.W. (1993). A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes Dev. 7: 974–985 [DOI] [PubMed] [Google Scholar]
- Quilichini T.D., Friedmann M.C., Samuels A.L., Douglas C.J. (2010). ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol. 154: 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Herranz M.-C., Bubeck J., Pepperkok R., Ritzenthaler C. (2007). Membrane dynamics in the early secretory pathway. Crit. Rev. Plant Sci. 26: 199–225 [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Rowland O., Lee R., Franke R., Schreiber L., Kunst L. (2007). The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett. 581: 3538–3544 [DOI] [PubMed] [Google Scholar]
- Rozema J., Broekman R.A., Blokker P., Meijkamp B.B., de Bakker N., van de Staaij J., van Beem A., Ariese F., Kars S.M. (2001). UV-B absorbance and UV-B absorbing compounds (para-coumaric acid) in pollen and sporopollenin: the perspective to track historic UV-B levels. J. Photochem. Photobiol. B 62: 108–117 [DOI] [PubMed] [Google Scholar]
- Rubin-Pitel S.B., Zhang H., Vu T., Brunzelle J.S., Zhao H., Nair S.K. (2008). Distinct structural elements dictate the specificity of the type III pentaketide synthase from Neurospora crassa. Chem. Biol. 15: 1079–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y.C., Lee P.Y., Truong M.T., Beals T.P., Goldberg R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322 [Google Scholar]
- Saxena P., Yadav G., Mohanty D., Gokhale R.S. (2003). A new family of type III polyketide synthases in Mycobacterium tuberculosis. J. Biol. Chem. 278: 44780–44790 [DOI] [PubMed] [Google Scholar]
- Schijlen E.G., de Vos C.H., Martens S., Jonker H.H., Rosin F.M., Molthoff J.W., Tikunov Y.M., Angenent G.C., van Tunen A.J., Bovy A.G. (2007). RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol. 144: 1520–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.J., Spielman M., Dickinson H.G. (2004). Stamen structure and function. Plant Cell 16(suppl.): S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Masaoka K., Nishi M., Nakamura K., Ishiguro S. (2008). Identification of kaonashi mutants showing abnormal pollen exine structure in Arabidopsis thaliana. Plant Cell Physiol. 49: 1465–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L.K., Chu H., Yip W.K., Yeung E.C., Lo C. (2009). An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New Phytol. 181: 576–587 [DOI] [PubMed] [Google Scholar]
- van der Meer I.M., Stam M.E., van Tunen A.J., Mol J.N., Stuitje A.R. (1992). Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcay-Barrena G., Wilson Z.A. (2006). Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J. Exp. Bot. 57: 2709–2717 [DOI] [PubMed] [Google Scholar]
- Walden A.R., Walter C., Gardner R.C. (1999). Genes expressed in Pinus radiata male cones include homologs to anther-specific and pathogenesis response genes. Plant Physiol. 121: 1103–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne A.J., Zhang W., Sun Y., Liu W., Albert R., Zheng Z., Oppenheimer D.G., Zhao D., Ma H. (2007). Differential gene expression in Arabidopsis wild-type and mutant anthers: Insights into anther cell differentiation and regulatory networks. Plant J. 52: 14–29 [DOI] [PubMed] [Google Scholar]
- Wu S., O’Leary S.J., Gleddie S., Eudes F., Laroche A., Robert L.S. (2008). A chalcone synthase-like gene is highly expressed in the tapetum of both wheat (Triticum aestivum L.) and triticale (xTriticosecale Wittmack). Plant Cell Rep. 27: 1441–1449 [DOI] [PubMed] [Google Scholar]