Root-specific expression of a cytokinin-degrading CKX gene induces the formation of a larger root system, whereas growth and development of the shoot remain similar. Transgenic plants exhibiting a larger root system had a higher survival rate after severe drought treatment and an increased leaf element content, suggesting the approach might be useful to optimize crop plants.
Abstract
Optimizing root system architecture can overcome yield limitations in crop plants caused by water or nutrient shortages. Classic breeding approaches are difficult because the trait is governed by many genes and is difficult to score. We generated transgenic Arabidopsis thaliana and tobacco (Nicotiana tabacum) plants with enhanced root-specific degradation of the hormone cytokinin, a negative regulator of root growth. These transgenic plants form a larger root system, whereas growth and development of the shoot are similar. Elongation of the primary root, root branching, and root biomass formation were increased by up to 60% in transgenic lines, increasing the root-to-shoot ratio. We thus demonstrated that a single dominant gene could regulate a complex trait, root growth. Moreover, we showed that cytokinin regulates root growth in a largely organ-autonomous fashion that is consistent with its dual role as a hormone with both paracrine and long-distance activities. Transgenic plants had a higher survival rate after severe drought treatment. The accumulation of several elements, including S, P, Mn, Mg, Zn, as well as Cd from a contaminated soil, was significantly increased in shoots. Under conditions of sulfur or magnesium deficiency, leaf chlorophyll content was less affected in transgenic plants, demonstrating the physiological relevance of shoot element accumulation. Our approach might contribute to improve drought tolerance, nutrient efficiency, and nutrient content of crop plants.
INTRODUCTION
Growth of plant organs is regulated by environmental inputs (e.g., nutrient availability and light) as well as by endogenous factors, such as plant hormones. Mechanisms regulating organ size in plants are just beginning to be revealed, and only a few genetic factors controlling organ size have been identified (Gonzalez et al., 2009; Krizek, 2009). In the past, changes in growth of crop plants obtained by mutations in genes controlling the hormonal status have contributed significantly to increasing yield and were the basis for the green revolution (Hedden, 2003; Salamini, 2003). Therefore, a better understanding of the regulation of organ growth and size is relevant to improve rational approaches in plant breeding.
Most studies on organ size control investigate growth of aerial organs (Mizukami and Fischer, 2000; Hu et al., 2006; Li et al., 2008), whereas roots are only rarely considered. However, the development of plant root systems is an important agronomic trait. Plant roots perform many essential functions, including water and nutrient uptake, storage of reserves, synthesis of specific compounds, anchorage to the soil, and the establishment of biotic interactions in the rhizosphere (López-Bucio et al., 2003). The size and the architecture of the root system determine the plant’s ability to access water and nutrients, factors that limit growth and, thus, yield in many agricultural ecosystems (Lynch, 1995; Price et al., 1997). As water and nutrient shortages pose increasing problems in agriculture, understanding and manipulation of root architecture becomes more important. Fertilization can supply limiting nutrients, but it is expensive, energy-consuming, and often causes environmental problems such as eutrophication. The negative impact of the aforementioned factors may be reduced by optimizing root system architecture.
A correlation between root system size and resistance to water stress has been found in several crop plants, and breeding attempts have focused on obtaining cultivars with larger root systems (Price et al., 2002; Tuberosa et al., 2002). For example, drought-resistant rice (Oryza sativa) varieties have a deeper and more highly branched root system than drought-sensitive varieties (Price et al., 1997). Plants with larger root systems have an increased ability to compete for nutrients and to survive under conditions of nutrient deficiency (Hodge et al., 1999; Liao et al., 2001; Coque and Gallais, 2006). Arabidopsis thaliana varieties with a high root-to-shoot biomass ratio show the highest phosphate acquisition (Narang et al., 2000), and increased accumulation of specific elements in the shoot has been found to correlate with higher root-to-shoot biomass ratio in comparative studies of Thlaspi spp metal hyperaccumulator and nonaccumulator species (Krämer et al., 1997; Roosens et al., 2003). Moreover, several investigations report the overlap between quantitative trait loci determining root features and those for plant productivity, suggesting a causal link between these traits (de Dorlodot et al., 2007).
The development of the root system is determined by intrinsic genetic factors and modulated by numerous environmental cues, such as the local availability of nutrients and water. Root system architecture, including root elongation and the frequency of lateral root formation, is governed by many genes. Quantitative trait loci associated with root system architecture have been described, but most of the corresponding genes remain unidentified (de Dorlodot et al., 2007). The genetic complexity and the difficulty in scoring and selecting traits that are hidden underground have made it difficult to breed for improved root systems. Here, an alternative approach for root enhancement is presented; this approach is based on the genetic engineering of the hormone status of the root.
The plant hormone cytokinin is recognized as an essential regulator of plant root systems (reviewed in Mok and Mok, 2001; Werner and Schmülling, 2009). It is known that cytokinin inhibits root elongation and branching (Skoog and Miller, 1957; Cary et al., 1995). More recently, it was shown that a reduction of the cytokinin status in planta causes the formation of a larger root system. Different approaches have been used to reduce the cytokinin status, including constitutive overexpression of cytokinin-degrading cytokinin oxidase/dehydrogenase (CKX) genes (Werner et al., 2001, 2003; Yang et al., 2003), reduction of cytokinin biosynthesis (Miyawaki et al., 2006), mutation of cytokinin receptor genes (Riefler et al., 2006), or suppression of the cytokinin signaling pathway (Mason et al., 2005; Heyl et al., 2008). It was shown that a reduced cytokinin status retards the exit of dividing cells from the root meristem, thus increasing the number of cells in the meristem (Werner et al., 2003; Dello Ioio et al., 2007). Genetic analyses have shown that cytokinin acts at the transition zone via the transcription factor IAA3/SHY2 (Dello Ioio, 2008).
Collectively, this work demonstrated that cytokinin is a negative regulator of root growth and that its physiological concentration is supraoptimal for root growth under standard conditions. Therefore, manipulation of the cytokinin status might be a feasible approach to genetically engineer plants with an enhanced root system. However, cytokinin is often not considered specific enough to be used for targeted plant growth regulation (Busov et al., 2008). For example, cytokinin is required for shoot growth, and a systemic reduction of the cytokinin status reduces sink strength in the young shoot tissues and thus inhibits their growth (Werner et al., 2008). Therefore, the detrimental consequences of cytokinin deficiency in the shoot need to be avoided. In this respect, it is also noteworthy that organ growth in plants is, at least in part, under correlative control (i.e., growth of one organ may influence the growth of a distant organ) (Bögre et al., 2008). Cytokinins have locally restricted (paracrine) functions in growth regulation but are also transported and may contribute to growth control of distant organs of the plant (Hirose et al., 2008; Argueso et al., 2009; Werner and Schmülling, 2009). Thus, it is uncertain whether the consequences of a localized change in cytokinin status can be limited to a particular organ or tissue. It is also not clear to what extent a plant would benefit from an increased root system under conditions of limiting water or soil nutrients.
Here, we describe how root-specific expression of CKX genes allows the production of transgenic plants with an enlarged root system without negative side effects in the aerial organs. This also increases our understanding of the paracrine and long-distance hormonal activities of cytokinin in root-to-shoot communication. It is further shown that plants with an increased root system have an improved drought tolerance and increased leaf element contents, the latter allowing the maintenance of higher chlorophyll concentrations under nutrient limitation. This establishes an approach for modulating root system architecture and might be useful for generating crop plants optimized to grow in difficult agricultural environments.
RESULTS
Grafting Experiments Demonstrate that Morphological Changes Caused by an Altered Cytokinin Status Are Organ Autonomous
To explore the feasibility of limiting the consequences of a reduced cytokinin status to a specific part of the plant, we first analyzed reciprocal grafts between wild-type and 35S:CKX transgenic plants showing all the aspects of the cytokinin deficiency syndrome in roots and shoots (Figure 1). The aerial morphology of wild-type scions grafted onto 35S:CKX transgenic rootstocks was similar to wild-type scions grafted onto a wild-type rootstock (Figure 1A). This indicates that the stronger sink activity caused by the enhanced growth of the root stock does not negatively influence the development of the concurrent sink, the growing shoot. Interestingly, the senescence of the basal leaves of wild-type scions grafted onto transgenic rootstocks was retarded, giving the scions a healthier appearance than the scions of the control grafts. The morphology of 35S:CKX transgenic scions grafted onto wild-type rootstocks did not change and resembled that of shoots of 35S:CKX transgenic plants (Werner et al., 2001), indicating that the shoot phenotype is autonomous and not caused by the enhanced root growth (Figure 1B). Wild-type root growth was not altered when carrying a 35S:CKX transgenic scion. Similarly, improved growth of 35S:CKX transgenic roots is autonomous and does not depend on the 35S:CKX transgenic shoot (Figure 1C). Taken together, the results of the grafting study suggested that the effects of a reduced cytokinin content can be limited to the site of enhanced CKX expression. An attempt was then made to construct transgenic tobacco (Nicotiana tabacum) and Arabidopsis plants with an enhanced expression of a CKX gene in the root. For this purpose, the CKX1 and CKX3 genes of Arabidopsis, which both cause strong symptoms of cytokinin deficiency when expressed under control of the 35S promoter (Werner et al., 2001, 2003), were chosen, and two different promoters, which were reported to mediate expression predominantly in the root, were tested.
Figure 1.
Reciprocal Grafts of 35S:CKX2 Transgenic Tobacco Plants and Wild-Type Plants.
(A) Control grafts (wild-type scion on a wild-type root stock; two plants on the left) and wild-type scions grafted on a 35S:CKX transgenic rootstock (two plants on the right).
(B) Control graft (wild-type scion on a wild-type root stock; left) and 35S:CKX scion grafted on wild-type rootstock (right).
(C) Close-up of the root system of plants from (A). Wild-type root stock carrying a wild-type scion (left) and 35S:CKX2 rootstock carrying a wild-type scion (right).
Plants were grafted 55 DAG when the height of wild-type plants was ~15 cm. Pot diameter was 18 cm. Photographs were taken 15 weeks after grafting.
Root-Specific CKX Expression in Tobacco Results in an Enlarged Root System and Normal Shoots
To generate stable transgenic lines with enhanced CKX expression in the roots, the CKX1 gene was positioned under the transcriptional control of the WRKY6 promoter (called W6 in the following) of Arabidopsis, which has been described as strongly expressed in the root (Robatzek and Somssich, 2001). The gene construct was transformed into tobacco cv SNN, and >70 primary W6:CKX1 transformants were selected. Thirty independent lines showed a moderate to strong increase in the size of the root system on soil. Homozygous T2 progeny of two selected lines, W6:CKX1-24 and W6:CKX1-29, were analyzed in more detail.
RNA gel blot analysis showed that transgene expression was strongest in the roots of W6:CKX1 transgenic plants (Figure 2A). A weak hybridization signal was also obtained with RNA extracted from old senescing leaves, but no signal was obtained with RNA from young leaves. This result is consistent with the previously described expression characteristics of the WRKY6 promoter (Robatzek and Somssich, 2001).
Figure 2.
Tobacco Plants Expressing W6:CKX1 Form an Enhanced Root System, but Their Shoot Development Is Similar to the Wild Type.
(A) W6:CKX1 is predominantly expressed in roots. Two independent W6:CKX1 transgenic lines (24 and 29), wild-type (WT), and 35S:CKX1 plants were grown in hydroponic culture for 95 d, and total RNA was isolated from the youngest top leaves (YL), from the oldest bottom leaves (OL), and from roots (R). Twenty micrograms of RNA was separated in a 1.2% agarose gel, transferred to a nylon membrane, and hybridized with a 32P-labeled full-length CKX1 cDNA probe. rRNAs were visualized by ethidium bromide as a loading control.
(B) Primary root length of in vitro–grown plants at 15 DAG. Data represents mean values ± sd (n ≥ 20).
(C) Comparison of the root systems of wild-type (left) and W6:CKX1-29 transgenic (right) plants grown in hydroponic culture for 95 d.
(D) Root biomass of plants grown in hydroponic culture for 95 d. Values are arithmetic means ± sd (n ≥ 8). FW, fresh weight.
(E) Root-to-shoot biomass ratio in W6:CKX1-expressing plants grown in hydroponic culture for 95 d compared with the wild type. Values are arithmetic means ± sd (n ≥ 8).
(F) Shoot phenotype of adult plants grown under greenhouse conditions for 95 d. From left to right: wild-type, 35S:CKX1, and transgenic plants of line W6:CKX1-29.
(G) Time course of stem elongation of the different lines grown on soil under greenhouse conditions. Data represent means ± sd (n ≥ 18).
(H) Total number of leaves formed at 55 DAG. Data represent means ± sd (n ≥ 18).
(I) Onset of flowering, given as the plant age when the first fully developed flower formed. Data represent means ± sd (n ≥ 18).
Student’s t tests were used to compare values obtained from transgenic lines to the wild type. *P < 0.05. Bars = 5 cm for (C) and 15 cm for (F).
[See online article for color version of this figure.]
Elongation of primary roots in W6:CKX1 plants increased by ~50% in comparison to the wild type (Figure 2B). This is similar to plants with constitutive 35S:CKX1 expression, which showed an ~60% increased root elongation rate. To test whether the root enhancement was maintained during subsequent plant development, plants were cultivated in a hydroponic system. Figure 2C shows that mature transgenic plants also possess a larger root system than wild-type plants. Ninety-five days after germination (DAG), the fresh biomass of the root systems of the two tested W6:CKX1 transgenic lines was 27 and 39% higher, respectively, than that of wild-type plants (Figure 2D). Consequently, the root-to-shoot biomass ratios were increased by up to ~40% in W6:CKX1 transgenic lines (Figure 2E).
In contrast with the strong changes in root development, the development of W6:CKX transgenic shoots was comparable to the wild type. In W6:CKX1 plants, shoot development was normal, despite a somewhat slower growth in the beginning, in contrast with the strong impairment of shoot growth noted in 35S:CKX transgenic plants (Figure 2F). A detailed quantitative analysis of stem elongation (Figure 2G), number of leaves formed 55 DAG (Figure 2H), and onset of flowering (Figure 2I) revealed no, or only minor, differences between the wild type and each of the two W6:CKX transgenic lines, in marked contrast with the differences seen in the 35S:CKX transgenic plants.
Enhanced Root Expression of a CKX Gene Causes Increased Root Growth in Arabidopsis
To determine whether the conclusion from experiments described above in tobacco is applicable to other plant species, an Arabidopsis CKX3-GFP (for green fluorescent protein) fusion gene was positioned under the transcriptional control of the PYK10 promoter of Arabidopsis (here called P10:CKX3). The PYK10 promoter is known to mediate high expression in the root (Nitz et al., 2001).
About half of the 40 primary transformants developed larger root systems than the wild type on soil, and most of them did not display any obvious changes in shoot development with respect to overall morphology and rosette size. RNA gel blot analysis of four independent lines showed that the PYK10 promoter conferred mostly predominant expression in root tissue, although the level of expression varied between individual lines (Figure 3A). By contrast, transgene expression in rosettes was very low.
Figure 3.
Arabidopsis Plants Expressing P10:CKX3 Show Enhanced Elongation of the Primary Root, Increased Lateral Root Formation, and Higher Total Root Biomass.
(A) Predominant expression of CKX3-GFP in roots of P10:CKX3 transgenic plants. Four independent P10:CKX3 transgenic lines and wild-type (WT) plants were grown in hydroponic culture for 26 d, and the total RNA was isolated from all rosette leaves (S) and roots (R). Fifteen micrograms of RNA was separated in a 1.2% agarose gel, transferred to a nylon membrane, and hybridized with a 32P-labeled full-length CKX3 cDNA probe. rRNAs were visualized by ethidium bromide as a loading control.
(B) Elongation of the primary root in vitro between 2 and 9 DAG.
(C) Number of emerged lateral roots per seedling in vitro at 9 DAG.
(D) Root biomass of plants grown in hydroponic culture for 34 d. FW, fresh weight.
(E) Root systems of wild type (left) and P10:CKX3-10 plants (right) grown on soil for 40 d.
Independent homozygous P10:CKX3 transgenic lines were analyzed. Values are means ± sd (n ≥ 20). Transgenic lines differed significantly from the wild type for all parameters measured (Student’s t test, P < 0.001). FW, fresh weight. Bar = 2 cm.
A more detailed morphological analysis was performed with homozygous plants of the T3 generation. First, the effects of the enhanced CKX expression on root growth and architecture were analyzed. The elongation rate of primary roots in P10:CKX3 seedlings was increased by up to 37% in comparison to the wild type (Figure 3B). Plants with 35S-driven CKX expression displayed an ~50% increase in root elongation rate. Additionally, the formation of lateral and adventitious roots was significantly enhanced in P10:CKX3 plants. The number of emerged lateral roots had increased by up to 75% in individual P10:CKX3 lines 9 DAG compared with the wild type (Figure 3C). This corresponded to an increase of lateral root density by up to 30% (4.5 ± 0.9 lateral roots cm−1 primary root in P10:CKX3-1 versus 3.4 ± 0.4 in the wild type; n ≥ 20), indicating that cytokinin regulates the distance between established and newly initiated lateral root primordia. We noted that a low level of CKX3 transgene expression is sufficient to induce the formation of a larger root system (Figure 3A). To analyze the root biomass at a later developmental time point, three independent lines (P10:CKX3-1, -10, and -16) were grown in a hydroponic system. At 34 DAG, P10:CKX3 plants had formed 40 to 60% more root biomass than the wild-type control plants (Figure 3D). An increased size of the root system of the transgenic plants was also clearly visible in soil-grown plants (Figure 3E). These results demonstrated that the targeted increase of CKX activity predominantly in roots can positively influence their growth in largely a tissue-autonomous fashion also in Arabidopsis. Interestingly, root growth was comparable in all P10:CKX3 transgenic Arabidopsis lines, despite the fact that the transgene expression levels in the root differed significantly between lines (Figure 2). This suggests that a moderate CKX expression suffices to reach a cytokinin level that is sufficiently low to significantly enhance root growth.
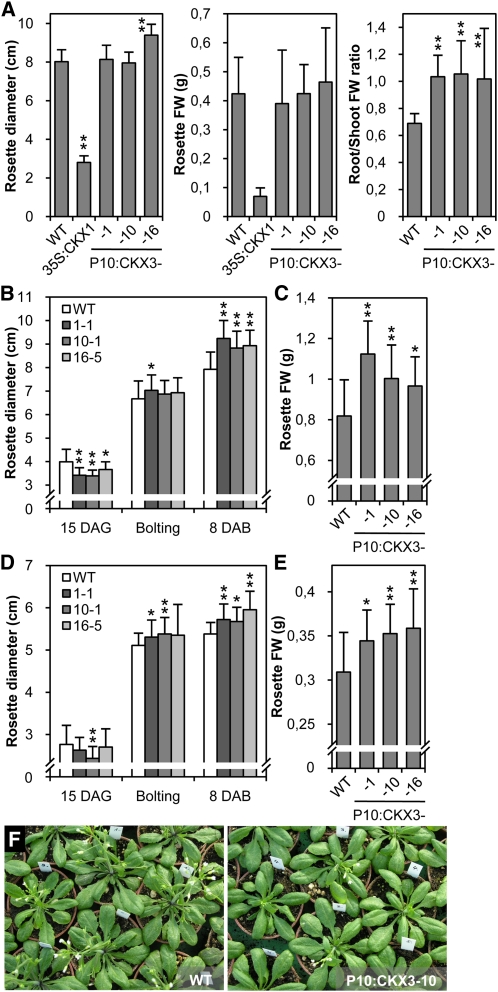
To assess the effect of enhanced root CKX activity on the growth of the shoot in Arabidopsis, several morphological parameters were scored under different growth conditions. After 34 d in hydroponic culture, control plants constitutively expressing the CKX3 gene (35S:CKX3) showed an ~85% reduction in rosette diameter and fresh biomass in comparison to the wild type (Figure 4A). By contrast, these parameters were not significantly lowered in the P10:CKX3 transgenic lines (Figure 4A). The root-to-shoot biomass ratio was strongly increased in P10:CKX3 lines owing to the increased root biomass, which was about equivalent to the shoot biomass (Figure 4A).
Figure 4.
Shoot Development of P10:CKX3 Transgenic Arabidopsis Plants.
(A) Rosette diameter, fresh weight (FW), and root-to-shoot biomass ratio in independent P10:CKX3 lines and control plants grown in hydroponic culture for 34 d. WT, wild type.
(B) and (C) Rosette size (B) and fresh weight (C) in P10:CKX3 and control plants grown on fertilized soil. Rosette size was scored 15 DAG, at the time of bolting of individual plants, and 8 DAB. Fresh weight was determined 8 DAB.
(D) and (E) Results of equivalent experiments as in (B) and (C), performed on nonfertilized soil.
(F) Shoots of wild-type (left) and P10-CKX3 transgenic plants (right) grown on conventionally fertilized soil for 33 d. Independent P10:CKX3 homozygous lines were analyzed. Pot diameter was 6 cm.
Data in (A) to (E) represent means ± sd (n ≥ 20). Student’s t test was used to compare values to the wild type. *P < 0.05; **P < 0.01. FW, fresh weight.
[See online article for color version of this figure.]
Plants were grown on a conventionally fertilized soil under standard greenhouse conditions to analyze shoot growth and development in more detail. During the initial vegetative growth phase, the rosette diameter of P10:CKX3 plants was slightly reduced in comparison to the wild type because of a lower initial growth rate (Figure 4B). However, no difference in size was observed at the time of bolting (Figures 4B and 4F). Eight days after bolting (DAB), the rosette diameter and fresh weight were up to 17 and 37% larger, respectively, in P10:CKX3 plants compared with wild-type plants (Figures 4B and 4C). Interestingly, an increase in rosette diameter and fresh weight of up to 11 and 17%, respectively, was also observed when plants were grown on nonfertilized soil (Figures 4D and 4E). P10:CKX3 plants bolted and terminated flowering 2 d later than the wild type (n ≥ 30, P < 0.001).
The Cytokinin Content of CKX Transgenic Plants
In light of the classic view that a substantial part of the plant cytokinin pool is root borne (Letham and Palni, 1983), we investigated the extent to which enhanced degradation of cytokinin in the root affects local cytokinin homeostasis and whether distant effects in the shoot could be found. Roots and shoots of 12-d-old P10:CKX3 Arabidopsis seedlings were harvested separately, and their cytokinin content was compared with that of wild-type and 35S:CKX3 transgenic seedlings. The analysis showed that in both roots and shoots of wild-type seedlings, trans-zeatin (tZ)-type cytokinins were more abundant than the isopentenyladenine (iP)-type cytokinins (see Supplemental Table 1 online). Interestingly, the content of the most biologically effective metabolites (tZ, tZ riboside, and iP) was higher in the root, whereas the content of inactive cytokinins, such as glucose conjugates and cytokinin nucleotides (tZMP and iPRMP), was higher in the shoot. Almost all analyzed cytokinin metabolites were significantly reduced in roots and shoots of both types of CKX transgenic plants, although to a different extent (see Supplemental Figure 1 and Supplemental Table 1 online). The reduction to ~35% of the total cytokinin content of wild-type roots was similar in roots of both transgenic genotypes (Figure 5), consistent with the high expression of the PYK10 promoter in the root. By contrast, shoots of 35S:CKX3 plants showed a reduction of the total cytokinin content to ~12% of the wild-type level, whereas the cytokinin content of P10:CKX3 shoots was lowered to ~30% of the wild type. In both organs, the reduction of tZ-type cytokinins was stronger than that of iP-type cytokinins, which was particularly pronounced in P10:CKX3 shoots (Figure 5).
Figure 5.
Cytokinin Concentrations in Roots and Shoots of P10:CKX3 Transgenic Plants.
Schematic representation of changes in the concentration of tZ- and iP-type cytokinins in roots and shoots of P10:CKX3 and 35S:CKX3 transgenic plants compared with the respective wild-type tissues. A detailed compilation of the data is shown in Supplemental Figure 2 and Supplemental Table 1 online.
Transgenic Plants with an Enhanced Root System Show Increased Drought Tolerance
To investigate whether an enlarged root system improves drought tolerance, the survival rates of W6:CKX1 and wild-type tobacco plants were scored after a period of complete water deprivation and subsequent water resupply. In a mixed population of plants that were not watered for 26 d, 39 to 42% of the transgenic plants started growing again after rewatering, whereas on average, only 25% of the wild-type plants survived (Figure 6). The higher survival rate of transgenic plants suggested that the transgenic tobacco plants competed more successfully than wild-type plants for limited water resources and thus possessed higher drought tolerance.
Figure 6.
Improved Resistance of W6:CKX1 Transgenic Tobacco Plants to Drought Stress.
(A) Survival rates of wild-type (WT) and W6:CKX1 transgenic tobacco plants. Fifty transgenic and 50 wild-type plants were grown together in one tray in a regular alternate pattern. At least 200 plants per genotype were analyzed. A representative result of two independent experiments is shown.
(B) Representative example of plant survival after drought stress, as shown in (A). Plants were photographed 13 d after the beginning of rewatering. Crosses indicate the surviving wild-type plants; open circles indicate the surviving W6:CKX1-24 plants. Bar = 2 cm.
Mineral Accumulation Is Altered in Plants with Enhanced Root Systems
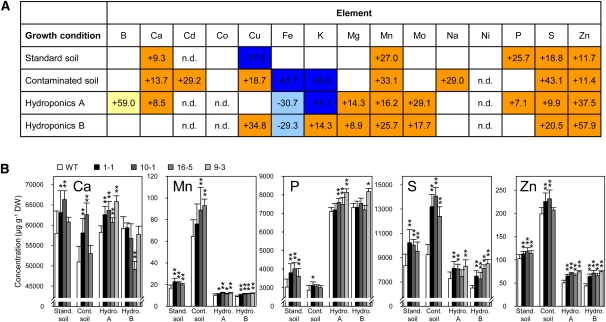
To test whether an enlarged root system influences the uptake and accumulation of nutrients, the concentrations of a range of elements in the leaves of P10:CKX3 Arabidopsis and W6:CKX1 tobacco plants were analyzed and compared with levels in the corresponding wild-type controls. Plants were grown on two different soils, standard soil and a contaminated soil. In addition, Arabidopsis plants were grown in hydroponic cultures with high or low N supply. In all transgenic Arabidopsis lines, leaf concentrations of several elements were significantly higher than in the wild type on all growth substrates (Figure 7). Large and consistent increases by up to 57, 43, and 33%, respectively, were found for Zn, S, and Mn. The content of these three elements was also strongly increased in W6:CKX1 tobacco lines under at least two different growth regimes (see Supplemental Figure 2 and Supplemental Table 3 online). Leaf phosphate concentrations were increased by up to 25% in P10:CKX3 transgenic Arabidopsis and by 45% in W6:CKX1 transgenic tobacco plants compared with the respective wild type. Similarly, leaf concentrations of Ca, Mo, and Mn were higher in the transgenic lines than in the corresponding wild types under several different conditions. Leaf concentrations of Fe and K were generally reduced in the transgenic plants compared with the wild-type plants, suggesting that a negative regulation of the accumulation of these elements was operating in the transgenic plants, either at the level of root uptake or at the level of root-to-shoot translocation.
Figure 7.
Elemental Concentrations in Rosettes of P10:CKX3 Transgenic Arabidopsis Plants and in Wild-Type Plants.
(A) Summary of changes in the concentrations of all analyzed elements in plants grown under different conditions. Analyses were performed on plants grown on a standard soil, an industrially contaminated soil, and in hydroponic cultures supplied with different nitrate concentrations. Three and four independent P10:CKX3 lines were compared with the wild type in experiments performed on soil and in hydroponics, respectively. Mean differences between the elemental concentrations in individual transgenic lines and the wild type are given as a percentage of wild-type values. Color coding for experiments performed on soil is as follows: orange, the mean concentration (n ≥ 6 per line) in a given element was significantly higher than that of the wild type at P < 0.05 in at least one transgenic line, and at least one additional line showed an increase at the significance level P < 0.1; dark blue, the mean concentration (n ≥ 6 per line) of a given element was significantly lower than that of the wild type at P < 0.05 in at least one transgenic line, and at least one additional line showed a decrease at the significance level P < 0.1. Color coding for experiments performed in hydroponics is as follows: orange, the mean content (n ≥ 6 per line) of a given element was significantly higher than in the wild type at P < 0.05 in at least two independent transgenic lines, and at least one additional line showed an increase at significance level P < 0.1; yellow, the mean content in one transgenic line was significantly higher than that of the wild type at P < 0.05, and at least one additional line showed an increase at significance level P < 0.1. Dark-blue and light-blue indicate a decrease of element concentration according to the criteria applied to orange and yellow, respectively. White indicates no significant change; n.d., not detected. The complete data set is provided in Supplemental Table 2 online.
(B) Concentrations of elements showing the most prominent changes in accumulation in rosette leaves of transgenic lines when compared with wild-type (WT) plants. Independent homozygous P10:CKX3 transgenic lines were analyzed. Data represents mean values ± sd (n ≥ 6). Student’s t test was used to compare values to the wild type. *P < 0.1; **P < 0.05. Cont. soil, contaminated soil; DW, dry weight; Hydro. A, Hydroponics A; Hydro. B, Hydroponics B; Stand. soil, standard soil.
P10:CKX3 Arabidopsis plants were also grown on an industrially contaminated soil (19.0 ± 1.3 ppm Cd), and W6:CKX1 tobacco plants on soil artificially spiked with 50 ppm of Cd to also test the ability of the plants to accumulate heavy metals from contaminated soils. The concentration of cadmium in the leaves of the transgenic lines had increased by ~30% compared with the wild type in both Arabidopsis and tobacco (Figure 7; see Supplemental Figure 2 online). The increased cadmium content in the soil did not negatively influence the accumulation of most other elements.
Improved Leaf Chlorophyll Content under Nutrient Limitation
Next, we investigated whether the ability of transgenic lines to accumulate higher element concentrations in the leaves becomes physiologically relevant under nutrient-limiting conditions. To this end, we performed growth tests on media containing a series of reduced sulfur or magnesium concentrations, respectively. Tobacco plants were chosen for these experiments because under our growth conditions they appeared more sensitive than Arabidopsis plants. Sulfur and magnesium deficiency cause a strong decrease in leaf chlorophyll content (Marschner, 1995). Therefore, we compared the leaf chlorophyll content of W6:CKX1 and wild-type tobacco plants grown in a hydroponic medium supplemented with suboptimal concentrations of sulfate or magnesium.
Cultivation of tobacco plants with a suboptimal sulfate supply led to stunted growth and uniform leaf yellowing. The leaf chlorophyll content decreased gradually with decreasing sulfate content in the medium (Figure 8A; see Supplemental Figure 3 online). By comparison, the reduction in leaf chlorophyll content in W6:CKX1 transgenic plants was significantly less pronounced than in wild-type plants. The chlorophyll content of leaves of wild-type plants grown on medium lacking added sulfate was reduced to 33% of the sulfate-replete control treatment. In comparison, the content was 25 and 60% higher in the two W6:CKX1-expressing tobacco lines (Figure 8A; see Supplemental Figure 3 online). Similarly, transgenic leaves of different developmental stages contained significantly more chlorophyll when plants were grown on sulfate-deficient medium supplemented only with 30 or 75 μM sulfate (Figure 8A; see Supplemental Figure 3 online) or when they were exposed to long-term sulfate deprivation (Figure 8B).
Figure 8.
Leaf Chlorophyll Content of W6:CKX1 Transgenic Tobacco Plants Grown under Nutrient-Limited Conditions.
Leaf chlorophyll content as indicated by SPAD readings (see Methods) in plants grown on a perlite substrate and Hoagland nutrient solution supplemented with different concentrations of sulfate ([A] and [B]) or Mg2+ ([C] and [D]). SPAD readings were taken on plants that were exposed to the indicated nutrients regimes for 4 ([A] and [C]) and 11 weeks ([B] and [D]), respectively. Leaf number 3 (counted from the apex down) ([A] and [C]) and indicated older leaves ([B] and [D]) were analyzed. Data represent means ± sd (n = 10). Student’s t test was used to compare values from transgenic lines to the corresponding wild-type (WT) values. *P < 0.05; **P < 0.01; ***P < 0.001 for (A) and (C). For (B) and (D), all changes in transgenic lines were highly significant (P < 0.01), except for leaves 11 and 13 of line W6:CKX1-29 in (B).
Similar to sulfate deprivation, Mg2+ starvation caused typical deficiency symptoms, including slowed growth, reduction of leaf chlorophyll contents, and development of interveinal chlorosis in older leaves of wild-type plants. Without supplemental Mg2+ in the medium, leaf chlorophyll content was reduced to between 33 and 48% of the control treatment in the wild type (Figure 8C; see Supplemental Figure 3 online). In comparison, transgenic plants retained 15 to 60% more chlorophyll than the corresponding wild-type leaves (Figure 8C; see Supplemental Figure 3 online). Leaves of W6:CKX1 retained higher chlorophyll contents than the wild type also on media supplemented with 15 and 30 μM Mg2+ (Figure 8C; see Supplemental Figure 3 online) as well as in plants exposed to long-term Mg2+ deficiency (Figure 8D). Similar relative differences were found in leaves of different developmental stages of W6:CKX1 plants (Figure 8D).
Expression of Genes Encoding Proteins Involved in Nutrient Transport
To better understand the mechanism of enhanced accumulation of specific elements in shoots of lines with an enhanced root system, we analyzed the expression of genes encoding proteins involved in the acquisition of these elements. Sulfur is primarily taken up from the soil as inorganic sulfate, and the uptake is mediated in root cells by proton/sulfate cotransporters (SULTR) (Buchner et al., 2004). Using quantitative RT-PCR (qRT-PCR), we compared the transcript abundance of seven SULTR genes in roots and shoots of two independent P10:CKX3 transgenic and wild-type Arabidopsis plants grown in vitro on media with the same nutrient composition as in Hydroponics A in Figure 7. In most cases, there were only subtle differences in transcript levels between the analyzed genotypes. The transcript levels of SULTR1;2 and SULTR5;1 were weakly (up to 1.7-fold) increased in P10:CKX3 roots compared with wild-type roots (Figure 9A). On the other hand, steady state transcript levels of two analyzed genes, SULTR3;5 and the vascular tissue-associated SULTR2;1 (Buchner et al., 2004), were significantly reduced in transgenic roots. Weak increases of the transcript levels of three SULTR genes were detected in shoots of the P10:CKX3 lines (Figure 8A).
Figure 9.
Steady State Transcript Levels of Genes Encoding Nutrient Transporters.
Transcript abundance of selected transporter genes was determined by qRT-PCR in roots and shoots of the wild type and two independent P10:CKX3 transgenic Arabidopsis lines grown in vitro for 12 d. Transcript levels are given as relative values compared with the corresponding wild-type tissue. Results represent mean values ± sd from two independent biological replicates with two technical replicates for each. Relative expression levels were normalized using At UBC10 (At5g53300) as an internal control. Student’s t test was used to compare values to the corresponding wild-type tissue. *P < 0.05; **P < 0.01. Only gene family members showing significant differences to the wild type are presented. Other genes are shown in Supplemental Figure 4 online.
(A) Expression of genes coding for proteins involved in sulfate acquisition and metabolism.
(B) Expression of genes coding for proteins involved in phosphate acquisition and metabolism.
(C) Expression of genes coding for proteins involved in acquisition and metabolism of other elements.
The phosphate oxoanion is the predominant form of phosphorus available in soil. The uptake of inorganic orthophosphate (Pi), which is the main form assimilated by plants, is mediated by three families of Pi transporters: Pht1, Pht2, and Pht3 (Bucher, 2007). Members of the largest family (Pht1) are most relevant for Pi uptake by roots. We analyzed the transcript levels of four representative genes from this family and detected an up to 2.7-fold increase in the level of PHT1;9 expression and a less pronounced increase of PHT1;1 expression in P10:CKX3 transgenic roots (Figure 9B). By contrast, the expression was very similar in transgenic and control shoots (Figure 9B). Also, the transcript level of the central regulator gene PHO2 (Aung et al., 2006; Bari et al., 2006) was increased in roots but remained unchanged in shoots of P10:CKX3 transgenic lines (Figure 9C).
Mg2+ ions are transported in plants by proteins of the MRS2/MGT family (Gebert et al., 2009). qRT-PCR analysis revealed that none of the eight analyzed MRS2/MGT genes showed a significantly and consistently increased expression level in roots of transgenic plants when compared with wild-type controls (see Supplemental Figure 4 online), suggesting that the increased content of Mg2+ in transgenic leaves and their higher chlorophyll content under Mg2+ limitation was not caused by an enhanced expression of the transporting machinery.
The concentration of Mn was consistently increased in lines with root-specific CKX expression grown under different conditions (Figure 7; see Supplemental Figure 2 online). A recent study showed that Arabidopsis relies on a high-affinity uptake system to acquire Mn from the soil in conditions of low Mn availability and that this activity is catalyzed by the major transporter NRAMP1 (Cailliatte et al., 2010). Consistent with the observed enhancement of Mn content, we detected a 1.9-fold increase of the level of NRAMP1 transcript in P10:CKX3 roots (Figure 9C). An increase, though less pronounced, was detected also in the transgenic shoots. This result suggests that the higher level of NRAMP1 expression in transgenic roots may be causally involved in the enhanced Mn accumulation in the shoot. Similarly, expression of ZIP4 of the zinc transporter gene family was increased in roots of P10:CKX3 plants (Figure 9C).
The high-affinity iron uptake system of Arabidopsis consists of the root surface ferric chelate reductase FRO2 and the plasma membrane ferrous iron importer IRT1 (Robinson et al., 1999; Vert et al., 2002). Transcript levels of both genes are known to be increased in roots of iron-deficient plants and repressed in response to exogenous cytokinin (Séguéla et al., 2008). In agreement with these results, FRO2 transcript levels were ~2.5-fold higher in the P10:CKX3 transgenic roots than in the wild-type control (Figure 9C). However, in contrast with Mn and Zn, shoot concentrations of Fe were lower in transgenics than in the wild type (Figure 7). Transcript levels of COPT1, which encodes a plasma membrane transport protein contributing to copper uptake (Kampfenkel et al., 1995), were not increased in transgenic lines (Figure 9C). Taken together, these data suggest that for S, P, Mn, and Zn, transcriptional regulation of the uptake machinery contributes to the increase in nutrient accumulation observed in the shoots of P10:CKX3 lines.
DISCUSSION
Here, we show that targeted root-specific expression of CKX genes confines the morphological effects of cytokinin deficiency to the root system. In this way, root biomass enhancement was achieved without detrimental effects on other plant parts. The results have implications for the understanding of regulation of carbon allocation, the organ-specific activity of cytokinin, and the relevance of the root system to cope with water and nutrient limitation.
Increase of the Root-to-Shoot Biomass Ratio
The root-to-shoot biomass ratio of transgenic Arabidopsis and tobacco plants was strongly enhanced by root-specific expression of CKX genes, indicating that the source capacity of the shoot was sufficient to supply additional sink tissue in the root with fixed carbon. This is interesting in view of different concepts of the regulation of fixed carbon allocation to distinct plant organs. The mechanisms of regulation of carbon allocation are only poorly understood, but both growth conditions as well as genotype are important (Marcelis and Heuvelink, 2007). The functional equilibrium model proposes that carbon allocation is regulated by an equilibrium between root activity and shoot activity, implying that the root may only increase its growth on the expense of the shoot and vice versa (Wilson, 1988). Our results are more compatible with models in which carbon allocation and growth is primarily or partly determined by the sink strength of the root (Marcelis, 1996; Farrar and Jones, 2000). Under our conditions, assimilate supply would not be a limiting factor, and the root is capable of realizing a higher growth rate because restraints by the otherwise inhibitory cytokinin concentrations are lowered. A small growth reduction noted during the early vegetative shoot development may be due to a residual activity of the promoters in shoot tissue and/or a relatively higher sink demand of the root compared with the shoot during early developmental stages.
Cytokinin Regulates Growth in an Organ-Autonomous Fashion
The organ-autonomous consequences of a reduced cytokinin content shown in this study are notable because cytokinin has not only local functions but also is transported in the vascular system and plays a role in root-to-shoot communication (Sakakibara, 2006; Werner and Schmülling, 2009). Root-specific enhancement of cytokinin degradation lowered the root cytokinin content to ~30% of the wild-type level. This caused the formation of a larger root system due to an increased elongation of the primary root as well as increased branching. Increased elongation is likely due to the formation of larger root meristems because cytokinin controls the exit of cells from the zone of cell division into cell differentiation (Werner et al., 2003; Dello Ioio et al., 2007). Cytokinin negatively regulates the initiation of lateral roots by blocking the first divisions of xylem-pole pericycle cells (Li et al., 2006; Laplaze et al., 2007). Hence, a reduction of the cytokinin status causes a more frequent initiation of lateral roots.
The cytokinin content was also lowered in the shoots of P10:CKX3 plants, although to a lesser extent than in 35S:CKX plants. This was surprising because of the much lower activity of the PYK10 promoter in the shoot and its normal appearance. Interestingly, in the transgenic plants, concentrations of the predominantly root-borne tZ-type cytokinins were reduced much more strongly than those of iP-type cytokinins, which are preferentially synthesized in the shoot (Hirose et al., 2008). We hypothesize that the lower cytokinin content in P10:CKX3 shoots is primarily a consequence of reduced cytokinin export from the roots. In addition, it could be that the cytokinin-depleted roots act as a sink for shoot-produced cytokinin, thus lowering the cytokinin content of the latter. The absence of strong morphological changes in the shoot indicates that growth regulation does not depend on root-borne cytokinin but that the shoot is self-sufficient. This may be particularly true for the young growing tissues where cytokinin is synthesized locally (Miyawaki et al., 2004). It has been argued previously that root-derived cytokinin may be less relevant for shoot functions than originally thought (Faiss et al., 1997). However, it was shown that the activity of the cambium in the shoot may be maintained by cytokinin produced in the root and vice versa (Matsumoto-Kitano et al., 2008). In addition, the shoot is capable of buffering alterations of its cytokinin status quite well. For example, a single out of three cytokinin receptor genes is sufficient to maintain normal shoot growth under standard conditions (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). Together, the results are consistent with the idea that roots and shoots exchange cytokinins by long-range transport but that each organ has the capacity to regulate growth largely in an autonomous fashion.
Increased Root System Size Improves Drought Resistance
Water limitation is the most significant limitation to crop productivity (Fischer and Turner, 1978; Boyer, 1982). Because of global climate change, it is predicted that drought periods will be more frequent in the future. Here, it is shown that an enhanced root system could contribute to limiting drought-induced yield losses. The survival rate of transgenic plants was higher than that of their nontransgenic counterparts in a mixed planting under severe drought stress. This indicates that a larger root system provided better access to limited water in this competitive situation. We have shown, using near-isogenic lines, that the size of the root system makes a significant contribution to drought resistance. This approach, possibly in combination with other strategies (Nelson et al., 2007; Rivero et al., 2007; Castiglioni et al., 2008), can contribute to improve crop productivity with limited water. A larger root system may be particularly advantageous to facilitate the absorption of water in deep soil layers, which is considered a drought avoidance strategy (Fukai and Cooper, 1995; Yue et al., 2006).
Cytokinin Status and Root System Size Influence Acquisition of Soil Nutrients
To improve agricultural performance, a further important goal is enhanced nutrient acquisition (Boyer, 1982). Correlative evidence suggested a causal link between the size of the root system and nutrient acquisition (Marschner, 1995). Here, it is shown that the reduced cytokinin content associated with an increased root system led to a significantly increased accumulation of soil-derived nutrients in the shoots of both Arabidopsis and tobacco. Similar patterns of changes were observed in both species and under different growth regimes, indicating that the factors determining nutrient uptake and distribution are similar and robust and are altered by a reduced cytokinin status. This suggests that there is a chance for a successful implementation in crops under field conditions. Importantly, the transgenic plants showed a less severe reduction in chlorophyll content when grown under sulfur or magnesium deficiency, confirming that the enhanced ability to accumulate nutrients in the leaves is physiologically relevant under these conditions.
Several factors may contribute to the changed element content of the leaves. Enhanced root elongation and branching increases the absorptive surface of the root and makes nutrients available in an enlarged soil volume. It has been shown that a larger size of the root system is advantageous to access nutrients with limited diffusion in the soil (e.g., phosphate) (Marschner, 1995). The increased expression of genes encoding sulfate, phosphate, Mn, and Zn transporters observed in cytokinin-deficient roots could additionally contribute to shoot accumulation of these nutrients. A direct influence of the cytokinin status on the expression of genes involved in the uptake and transport of nutrients is consistent with the well-documented role of the hormone in the response to phosphorus, sulfur, and iron deficiency (Franco-Zorrilla et al., 2002, 2005; Maruyama-Nakashita et al., 2004; Séguéla et al., 2008). Contrasting with the above nutrients, leaf Fe concentrations were generally reduced in CKX transgenic lines, indicating that additional levels of regulation have a decisive influence on leaf Fe accumulation in CKX transgenic lines.
Soil nutrient deficiencies are a widespread agricultural problem, in particular deficiencies in nitrogen, phosphorus, potassium, and sulfur. Mn deficiency is prevalent in large parts of the agricultural soils of Scandinavia, China, and tropical areas (Marschner, 1995; Hebbern et al., 2005; Yang et al., 2007), and Zn deficiency is widespread in Asian and Australian soils (Grotz and Guerinot, 2006; Broadley et al., 2007). Increased leaf nutrient accumulation may help the plant to maintain efficient photosynthesis, metabolism, and, thus, biomass production during episodes of nutrient deficiency. To avoid a decrease in yield, nutrients need to be added by soil or foliar application. An advantage of plants with a larger root system could be a reduced need to add fertilizer because it is taken up more efficiently and/or a reduced loss of fertilizer from the soil. A proper soil management strategy obviously will be required to allow for sustainable use of an enhanced root system. Moreover, the human diet is often deficient in trace elements, such as Zn (Lönnerdal, 2003). Our results indicate that root enhancement is a viable strategy in biofortification (i.e., improvement of micronutrient supply of limited elements, such as Zn, through food crops) (Palmgren et al., 2008). Evidently, it remains to be shown that the enhancement seen in the plants described here can also be obtained in the relevant organs of food crops. The fact that the uptake of Cd from contaminated soil is increased in the P10:CKX and W6:CKX transgenic plants indicates that an enhanced root system may also be applicable in phytoremediation (i.e., the cleanup of contaminated soils using plants) (Krämer and Chardonnens, 2001; Krämer, 2005), providing independent support for previous merely correlative evidence (Krämer et al., 1997; Roosens et al., 2003).
Root Engineering Could Improve the Performance of Crop Plants
An improvement of water and nutrient use efficiency is of considerable interest to enhance the productivity of crop plants. Formation of an enhanced root system to improve access to limited soil resources could contribute to a practical solution. This study shows, in two different species, the general feasibility of constructing plants with an enhanced root system. The strategy employs a single dominant gene to profoundly alter the complex trait of root growth, which is difficult to achieve by conventional breeding methods. Consequently, this approach will be valuable to explore the relevance of an enhanced root system in a variety of crop plants for the performance in nonoptimal agricultural environments.
However, it should be noted that numerous other factors regulate root architecture, including various nutrients (Casimiro et al., 2003; Malamy, 2005; Gojon et al., 2009), and it will be interesting to study these regulatory pathways in cytokinin-deficient roots. The responsiveness of roots to environmental changes appears to determine plant performance under drought or nutrient-limiting conditions (Hartung and Turner, 1997; Narang et al., 2000; Robinson, 2001). Thus, for agricultural purposes, it might be necessary to refine the strategy described here, e.g. by introducing a CKX gene under the control of a promoter that responds to a given environmental stimulus.
METHODS
Plasmid Construction
The 35S promoter of plasmid pBinSMGFP (Werner et al., 2003) was excised with EcoRI and KpnI and replaced by a synthetic double-stranded oligonucleotide (link-plus 5′-CGGAATTCCTAGGCTTCTGCCCGGGCTTCTGGGTACCCC-3′ and link-minus 5′-GGGGTACCCAGAAGCCCGGGCAGAAGCCTAGGAATTCCG-3′) containing EcoRI-AvrII-SmaI-KpnI sites. An Arabidopsis thaliana CKX3 genomic fragment was subcloned via KpnI/XhoI restriction sites from pBS-AtCKX3 (Werner et al., 2001) to generate an N-terminal fusion with GFP. Next, a 1456-bp promoter fragment of PYK10 was amplified by PCR from the DNA of Arabidopsis Columbia (Col-0) using the forward primer 5′-GACCCGGGACTGCAACGAAGTGTA-3′ and the reverse primer 5′-CAGGTACCTTTTTGTTTGTAATTCTG-3′. The fragment was first cloned into pCR2.1-TOPO (Invitrogen) and further subcloned in SmaI/KpnI sites upstream of the CKX3-GFP fusion gene, resulting in vector pBinP10:CKX3-GFP.
A 1311-bp long promoter fragment of WRKY6 was amplified from the plasmid pUC9-WRKY6-GUS (Robatzek and Somssich, 2001) using the forward primer 5′-GGAATTCCAATTAAGCTTTCACGTGGAA-3′ and the reverse primer 5′-GGGGTACCCCAGAAAAAGAAAGAGATCACG-3′. The fragment was cloned in the EcoRI/KpnI sites of pBINHygTx (Gatz et al., 1992) replacing the 35S promoter. Subsequently, At CKX1 was subcloned from pUC19-AtCKX1 (Werner et al., 2001) into a SalI site downstream of the WRKY6 promoter, yielding the vector pBinWRKY6:CKX1, which was used for plant transformation.
Transgenic Plants and Growth Conditions
Arabidopsis accession Col-0 was used for transformation by the floral dip method (Clough and Bent, 1998), and 40 independent P10:CKX3 transgenic lines were selected. Plants were grown in a greenhouse under 16-h-light/8-h-dark cycles at 21°C (light period) and 18°C (dark period). P soil, T soil (Einheitserde), and perlite (Knauf Perlite) in a volume ratio of 4:4:1 were used as standard soil. Soil used for phytoremediation experiments with Arabidopsis was obtained from an industrially contaminated site and contained 19.0 ± 1.3 ppm Cd. In vitro experiments were performed on half-strength Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with 1% sucrose. The hydroponic system used was basically as described previously (Tocquin et al., 2003). Microfuge tubes (0.7 mL) filled with quarter-strength MS medium and 0.8% agar, with their conical ends cut off, were used as seed holders. Arabidopsis plants were cultured in quarter-strength modified Hoagland nutrient solution containing 1.25 mM KNO3, 1.5 mM Ca(NO3)2, 0.75 mM MgSO4, 0.28 mM KH2PO4, 50 μM KCl, 25 μM H3BO3, 5 μM MnSO4, 1.0 μM ZnSO4, 0.5 μM CuSO4, 0.1 μM Na2MoO4, 25 μM FeNaEDTA, and 3 mM MES, pH 5.7 (Hydroponics A). Then, 0.2 mM NH4NO3 was added and the concentration of KNO3 was increased to 3.0 mM, providing a total concentration of 6.2 mM NO3− to test the influence of higher N supply (Hydroponics B).
For tobacco (Nicotiana tabacum) experiments, homozygous plants of line 35S:CKX2-38, described previously (Werner et al., 2001), were used for the grafting experiments. To generate transgenic tobacco plants, N. tabacum cv Samsun NN leaf explants were transformed and regenerated as described (Horsch et al., 1985). Characterization of transgenic tobacco was performed on homozygous T2 progeny obtained by selfing. Plants were cultured in vitro on full-strength MS medium or in a greenhouse under 15-h-light/9-h-dark cycles at 24°c (light period) and 20°C (dark period). T soil and perlite in a volume ratio of 4:1 was used as a standard soil. A perlite or sand/perlite mixture fertilized with half-strength Hoagland solution was used for hydroponic growth.
For experiments with limited nutrient supply, seeds were germinated on perlite with half-strength Hoagland solution. After 2 weeks, 10 seedlings per genotype were planted together in a regular alternate pattern in a tray filled with perlite and grown on modified nutrient solution for 4 or 11 weeks, respectively. Control plants were fed with half-strength Hoagland solution (1.5 mM MgSO4), whereas plants cultivated under Mg deficiency were grown on media supplemented with 0, 15, 30, or 75 μM MgSO4. To maintain a constant sulfur supply, the reduced MgSO4 was replaced by equivalent concentration of K2SO4. Sulfate deficiency was studied on 0, 30, and 75 μM MgSO4 and compared with control (1.5 mM MgSO4). MgSO4 was compensated with an equivalent concentration of MgCl2 to avoid Mg deficiency. Plants were irrigated with an equal volume of nutrient solution every 4 d.
Gene Expression
Total RNA from tobacco and Arabidopsis tissue was extracted according to Verwoerd et al. (1989) and Brenner et al. (2005), respectively. RNA gel blot analysis was performed as described (Brenner et al., 2005).
For qRT-PCR analysis, RNA was extracted from dissected roots and shoots of Arabidopsis plants grown in vitro for 12 d on media with the same nutrient composition as in Hydroponics A (see above). RNA was purified using the RNeasy Mini Kit (Qiagen) including DNaseI digestion. Five micrograms of total RNA were used as template for first-strand cDNA synthesis with Superscript III (Invitrogen) according to the instruction manual. cDNA (200 ng) was used for qPCR with SYBR Green reagent and Immolase DNA Polymerase (Bioline) as hot start enzyme. qPCR was performed on a 7500 Fast Real-Time PCR system (Applied Biosystems), and data were analyzed with 7500 Software V2.0.3 evaluation software. UBC10 was used as reference gene for normalization, and the relative expression was determined using the ΔΔCT method (Livak and Schmittgen, 2001). Primer sequences for all genes analyzed are listed in Supplemental Table 4 online. Two biological replicates were performed with the wild type and two independent transgenic lines.
Quantification of Endogenous Cytokinins
The concentrations of endogenous cytokinins were determined in roots and shoots of 12-d-old Arabidopsis plants grown in vitro on media with the same nutrient composition as in Hydroponics A (see above). Roots and shoots (100 to 200 mg) were pooled for each sample. Three independent biological replicates were analyzed for each genotype and tissue. Extraction, purification, and quantification of endogenous cytokinins by ultraperformance liquid chromatography–electrospray tandem mass spectrometry was performed according to the method described by Novák et al. (2003), including modifications described by Novák et al. (2008).
Morphometric Analyses
For in vitro root assays, Arabidopsis seedlings were grown on vertical plates, and the elongation of the primary root between 2 and 9 DAG was measured from digital images using Scion Image software. Root length of tobacco seedlings was determined at 15 DAG. The number of emerged lateral roots was scored under a stereomicroscope. Onset of flowering in tobacco was defined as the plant age when the first fully developed flower was formed. Bolting time in Arabidopsis was defined as the plant age at which an inflorescence of 0.5 cm was apparent. Rosette size and plant height were determined using a ruler.
Test for Drought Tolerance
The 5-d-old transgenic tobacco seedlings (lines W6:CKX1-24 and W6-CKX1-29) and wild-type seedlings were planted together in a regular alternate pattern with 2-cm spacing in 28 × 46-cm trays containing a 4:1 mixture of soil and sand to test the drought tolerance. The substrate was watered to saturation, and the plants were subsequently grown for 26 d without an additional water supply, at which time extensive signs of wilting and desiccation were observed on both genotypes. Watering was then resumed and, after a recovery phase of 11 d, the survival rates of the transgenic lines and the wild-type control were scored.
Chlorophyll Measurement
Total chlorophyll content was measured in tobacco leaves as defined in Figure 8 (10 biological replicates) after 4 or 11 weeks of growth on nutrient-deficient medium. A 0.5-cm2 disc was cut from the middle of the leaf blade, weighted, and extracted with 100% methanol overnight. Measurement and calculation were performed according to Lichtenthaler (1987). In addition, the chlorophyll content was determined with a Minolta SPAD-502 chlorophyll meter (Spectrum Technologies). Three readings were taken in the middle part of each leaf blade half, with care taken to avoid placing the meter over major leaf veins. Linear regression analysis revealed a strong correlation (R2 = 0.927) between single photon avalanche diode (SPAD) readings and extractable chlorophyll under different growth conditions.
Determination of Element Concentrations
Three (P10:CKX3-1, -10, and -16) and four (P10:CKX3-1, -9, -10, and -16) independent Arabidopsis lines were analyzed in experiments performed on soil and in hydroponics, respectively. Plants were harvested 37 and 34 DAG in soil and hydroponics experiments, respectively. For tobacco experiments, the transgenic lines W6:CKX1-24 and W6:CKX1-29 were grown for 58 d on the indicated soil (see Supplemental Figure 2 online). On soil artificially spiked with 50 ppm Cd, plants were cultivated for 37 d after they were grown on a standard soil for 3 weeks. The entire shoots of tobacco and all the rosette leaves of Arabidopsis were used respectively to determine the fresh weight after harvest. Subsequently, shoot material was washed, dried at 80°C for 2 d, weighed, and ground. Subsamples of between 18 and 60 mg dry biomass were digested in 2 mL 65% (w/w) nitric acid (Suprapur; Merck) in a Mars 5 microwave accelerated reaction system (CEM Corp.). Digests were filled up with ultrapure water to 10 mL. Elemental concentrations were determined by inductively coupled atomic emission spectrometry using an IRIS Advantage Duo ER/S (Thermo Fisher Scientific). Digests of tobacco certified reference material VTL-2, as well as a liquid calibration standard, were run once every 20 samples for quality control.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database or the Arabidopsis Genome Initiative database under the following accession numbers: CKX1 (At2g41510), CKX3 (At5g56970), PYK10 (At3g09260), WRKY6 (At1g62300), UBC10 (At5g53300), COPT1 (At5g59030), FRO2 (At1g01580), MRS2;1 (At1g16010), MRS2;2 (At5g64560), MRS2;3 (At3g19640), MRS2;4 (At3g58970), MRS2;5 (At2g03620), MRS2;7 (At5g09690), MRS2;10 (At1g80900), MRS2;11 (At5g22830), NRAMP1 (At1g80830), SULTR1;1 (At4g08620), SULTR1;2 (At1g78000), SULTR2;1 (At5g10180), SULTR3;4 (At3g15990), SULTR3;5 (At5g19600), SULTR4;1 (At5g13550), SULTR5;1 (At1g80310), PHO2 (At2g33770), PHT1;1 (At5g43350), PHT1;4 (At2g38940), PHT1;7 (At3g54700), PHT1;9 (At1g76430), and ZIP4 (At1g10970).
Supplemental Data
The following materials are available in the online version of the article.
Supplemental Figure 1. Cytokinin Concentration in Roots and Shoots of P10:CKX3 Transgenic Plants Compared with the Wild Type.
Supplemental Figure 2. Elemental Concentrations in Leaves of W6:CKX1 Transgenic Tobacco Plants and in Wild-Type Plants.
Supplemental Figure 3. Leaf Chlorophyll Content of W6:CKX1 Transgenic Tobacco Plants Grown under Nutrient-Limited Conditions.
Supplemental Figure 4. Steady State Transcript Levels of Genes Encoding Nutrient Transporters.
Supplemental Table 1. Cytokinin Concentration in Roots and Shoots of Wild-Type and CKX3 Transgenic Plants.
Supplemental Table 2. Elemental Concentrations in Rosette Leaves of P10:CKX3 Transgenic Arabidopsis Plants and in Wild-Type Plants.
Supplemental Table 3. Elemental Concentrations in Leaves of W6:CKX1 Transgenic Tobacco Plants and in Wild-Type Plants.
Supplemental Table 4. Primers Used for qRT-PCR Analysis.
Acknowledgments
We thank the Deutsche Forschungsgemeinschaft for financial support, Verena Schade and Hana Martinková for excellent technical assistance, and Imre Somssich for a plasmid harboring the WRKY6 promoter. U.K. acknowledges a Heisenberg Fellowship and funding through the FP6 EU Grant PHIME (FOOD-CT-2006-016253).
References
- Argueso C.T., Ferreira F.J., Kieber J.J. (2009). Environmental perception avenues: The interaction of cytokinin and environmental response pathways. Plant Cell Environ. 32: 1147–1160 [DOI] [PubMed] [Google Scholar]
- Aung K., Lin S.-I., Wu C.-C., Huang Y.-T., Su C.L., Chiou T.-J. (2006). pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Datt Pant B., Stitt M., Scheible W.-R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L., Magyar Z., López-Juez E. (2008). New clues to organ size control in plants. Genome Biol. 9: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J.S. (1982). Plant productivity and environment. Science 218: 443–448 [DOI] [PubMed] [Google Scholar]
- Brenner W.G., Romanov G.A., Köllmer I., Bürkle L., Schmülling T. (2005). Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Broadley M.R., White P.J., Hammond J.P., Zelko I., Lux A. (2007). Zinc in plants. New Phytol. 173: 677–702 [DOI] [PubMed] [Google Scholar]
- Bucher M. (2007). Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol. 173: 11–26 [DOI] [PubMed] [Google Scholar]
- Buchner P., Takahashi H., Hawkesford M.J. (2004). Plant sulphate transporters: Co-ordination of uptake, intracellular and long-distance transport. J. Exp. Bot. 55: 1765–1773 [DOI] [PubMed] [Google Scholar]
- Busov V.B., Brunner A.M., Strauss S.H. (2008). Genes for control of plant stature and form. New Phytol. 177: 589–607 [DOI] [PubMed] [Google Scholar]
- Cailliatte R., Schikora A., Briat J.-F., Mari S., Curie C. (2010). High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 22: 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary A.J., Liu W., Howell S.H. (1995). Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol. 107: 1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I., Beeckman T., Graham N., Bhalerao R., Zhang H., Casero P., Sandberg G., Bennett M.J. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Castiglioni P., et al. (2008). Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiol. 147: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coque M., Gallais A. (2006). Genomic regions involved in response to grain yield selection at high and low nitrogen fertilization in maize. Theor. Appl. Genet. 112: 1205–1220 [DOI] [PubMed] [Google Scholar]
- de Dorlodot S., Forster B., Pagès L., Price A., Tuberosa R., Draye X. (2007). Root system architecture: Opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 12: 474–481 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M.T., Aoyama T., Costantino P., Sabatini S. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Faiss M., Zalubìlová J., Strnad M., Schmülling T. (1997). Conditional transgenic expression of the ipt gene indicates a function for cytokinins in paracrine signaling in whole tobacco plants. Plant J. 12: 401–415 [DOI] [PubMed] [Google Scholar]
- Farrar J.F., Jones D.L. (2000). The control of carbon acquisition by roots. New Phytol. 147: 43–53 [Google Scholar]
- Fischer R.A., Turner N.C. (1978). Plant productivity in the arid and semiarid zones. Annu. Rev. Plant Physiol. 29: 277–317 [Google Scholar]
- Franco-Zorrilla J.M., Martín A.C., Leyva A., Paz-Ares J. (2005). Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 138: 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Martin A.C., Solano R., Rubio V., Leyva A., Paz-Ares J. (2002). Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation responses in Arabidopsis. Plant J. 32: 353–360 [DOI] [PubMed] [Google Scholar]
- Fukai S., Cooper M. (1995). Development of drought-resistant cultivars using physiomorphological traits in rice. Field Crops Res. 40: 67–86 [Google Scholar]
- Gatz C., Frohberg C., Wendenburg R. (1992). Stringent repression and homogeneous de-repression by tetracycline of a modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J. 2: 397–404 [DOI] [PubMed] [Google Scholar]
- Gebert M., Meschenmoser K., Svidová S., Weghuber J., Schweyen R., Eifler K., Lenz H., Weyand K., Knoop V. (2009). A root-expressed magnesium transporter of the MRS2/MGT gene family in Arabidopsis thaliana allows for growth in low-Mg2+ environments. Plant Cell 21: 4018–4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojon A., Nacry P., Davidian J.-C. (2009). Root uptake regulation: A central process for NPS homeostasis in plants. Curr. Opin. Plant Biol. 12: 328–338 [DOI] [PubMed] [Google Scholar]
- Gonzalez N., Beemster G.T.S., Inzé D. (2009). David and Goliath: What can the tiny weed Arabidopsis teach us to improve biomass production in crops? Curr. Opin. Plant Biol. 12: 157–164 [DOI] [PubMed] [Google Scholar]
- Grotz N., Guerinot M.L. (2006). Molecular aspects of Cu, Fe and Zn homeostasis in plants. Biochim. Biophys. Acta 1763: 595–608 [DOI] [PubMed] [Google Scholar]
- Hartung W., Turner N.C. (1997). Abscisic acid relations in stressed roots. Biology of Root Formation and Development, Altman A., Waisel Y., (New York: Plenum Press; ), pp. 125–132 [Google Scholar]
- Hebbern C.A., Pedas P., Schjoerring J.K., Knudsen L., Husted S. (2005). Genotypic differences in manganese efficiency: Field experiments with winter barley (Hordeum vulgare L.). Plant Soil 272: 233–244 [Google Scholar]
- Hedden P. (2003). The genes of the green revolution. Trends Genet. 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Heyl A., Ramireddy E., Brenner W.G., Riefler M., Allemeersch J., Schmülling T. (2008). The transcriptional repressor ARR1-SRDX suppresses pleiotropic cytokinin activities in Arabidopsis. Plant Physiol. 147: 1380–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N., Takei K., Kuroha T., Kamada-Nobusada T., Hayashi H., Sakakibara H. (2008). Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59: 75–83 [DOI] [PubMed] [Google Scholar]
- Hodge A., Robinson D., Griffiths B.S., Fitter A.H. (1999). Why plants bother: Root proliferation results in increased nitrogen capture from an organic patch when two grasses compete. Plant Cell Environ. 22: 811–820 [Google Scholar]
- Horsch R.B., Fry J.E., Hoffmann N.L., Eichholtz D., Rogers S.G., Fraley R.T. (1985). A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Hu Y., Poh H.M., Chua N.-H. (2006). The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J. 47: 1–9 [DOI] [PubMed] [Google Scholar]
- Kampfenkel K., Kushnir S., Babiychuk E., Inzé D., Van Montagu M. (1995). Molecular characterization of a putative Arabidopsis thaliana copper transporter and its yeast homologue. J. Biol. Chem. 270: 28479–28486 [DOI] [PubMed] [Google Scholar]
- Krämer U. (2005). Phytoremediation: Novel approaches to cleaning up polluted soils. Curr. Opin. Biotechnol. 16: 133–141 [DOI] [PubMed] [Google Scholar]
- Krämer U., Chardonnens A.N. (2001). The use of transgenic plants in the bioremediation of soils contaminated with trace elements. Appl. Microbiol. Biotechnol. 55: 661–672 [DOI] [PubMed] [Google Scholar]
- Krämer U., Smith R.D., Wenzel W.W., Raskin I., Salt D.E. (1997). The role of metal transport and tolerance in nickel hyperaccumulation by Thlaspi goesingense Halacsy. Plant Physiol. 115: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B.A. (2009). Making bigger plants: key regulators of final organ size. Curr. Opin. Plant Biol. 12: 17–22 [DOI] [PubMed] [Google Scholar]
- Laplaze L., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letham D.S., Palni L.M.S. (1983). The biosynthesis and metabolism of cytokinins. Annu. Rev. Plant Physiol. 34: 163–197 [Google Scholar]
- Li X., Mo X., Shou H., Wu P. (2006). Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol. 47: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Li Y., Zheng L., Corke F., Smith C., Bevan M.W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22: 1331–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H., Rubio G., Yan X., Cao A., Brown K.M., Lynch J.P. (2001). Effect of phosphorus availability on basal root shallowness in common bean. Plant Soil 232: 69–79 [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148: 350–382 [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lönnerdal B. (2003). Genetically modified plants for improved trace element nutrition. J. Nutr. 133(5 suppl. 1): 1490S–1493S [DOI] [PubMed] [Google Scholar]
- López-Bucio J., Cruz-Ramírez A., Herrera-Estrella L. (2003). The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6: 280–287 [DOI] [PubMed] [Google Scholar]
- Lynch J. (1995). Root architecture and plant productivity. Plant Physiol. 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J.E. (2005). Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 28: 67–77 [DOI] [PubMed] [Google Scholar]
- Marcelis L. (1996). Sink strength as a determinant of dry matter partitioning in the whole plant. J. Exp. Bot. 47: 1281–1291 [DOI] [PubMed] [Google Scholar]
- Marcelis L.F.M., Heuvelink E. (2007). Concepts of modelling carbon allocation among plant organs. Functional-Structural Plant Modelling in Crop Production, Vos J., Marcelis L.F.M., de Visser P.H.B., Struik P.C., Evers J.B., (Dordrecht, The Netherlands: Springer; ), pp. 103–111 [Google Scholar]
- Marschner H. (1995). Mineral Nutrition of Higher Plants. (London: Academic Press; ). [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Yamaya T., Takahashi H. (2004). A novel regulatory pathway of sulfate uptake in Arabidopsis roots: Implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 38: 779–789 [DOI] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Kitano M., Kusumoto T., Tarkowski P., Kinoshita-Tsujimura K., Václavíková K., Miyawaki K., Kakimoto T. (2008). Cytokinins are central regulators of cambial activity. Proc. Natl. Acad. Sci. USA 105: 20027–20031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K., Matsumoto-Kitano M., Kakimoto T. (2004). Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Tarkowski P., Matsumoto-Kitano M., Kato T., Sato S., Tarkowska D., Tabata S., Sandberg G., Kakimoto T. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 103: 16598–16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y., Fischer R.L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok D.W., Mok M.C. (2001). Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 89–118 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Narang R.A., Bruene A., Altmann T. (2000). Analysis of phosphate acquisition efficiency in different Arabidopsis accessions. Plant Physiol. 124: 1786–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.E., et al. (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 104: 16450–16455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., Ueguchi C. (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitz I., Berkefeld H., Puzio P.S., Grundler F.M.W. (2001). Pyk10, a seedling and root specific gene and promoter from Arabidopsis thaliana. Plant Sci. 161: 337–346 [DOI] [PubMed] [Google Scholar]
- Novák O., Hauserová E., Amakorová P., Doležal K., Strnad M. (2008). Cytokinin profiling in plant tissues using ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69: 2214–2224 [DOI] [PubMed] [Google Scholar]
- Novák O., Tarkowski P., Tarkowska D., Doležal K., Lenobel R., Strnad M. (2003). Quantitative analysis of cytokinins in plants by liquid chromatography-single-quadrupole mass spectrometry. Anal. Chim. Acta 480: 207–218 [Google Scholar]
- Palmgren M.G., Clemens S., Williams L.E., Krämer U., Borg S., Schjørring J.K., Sanders D. (2008). Zinc biofortification of cereals: Problems and solutions. Trends Plant Sci. 13: 464–473 [DOI] [PubMed] [Google Scholar]
- Price A.H., Cairns J.E., Horton P., Jones H.G., Griffiths H. (2002). Linking drought-resistance mechanisms to drought avoidance in upland rice using a QTL approach: Progress and new opportunities to integrate stomatal and mesophyll responses. J. Exp. Bot. 53: 989–1004 [DOI] [PubMed] [Google Scholar]
- Price A.H., Tomos A.D., Virk D.S. (1997). Genetic dissection of root growth in rice (Oryza sativa L.) I: A hydroponic screen. Theor. Appl. Genet. 95: 132–142 [Google Scholar]
- Riefler M., Novak O., Strnad M., Schmülling T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero R.M., Kojima M., Gepstein A., Sakakibara H., Mittler R., Gepstein S., Blumwald E. (2007). Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. USA 104: 19631–19636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Somssich I.E. (2001). A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 28: 123–133 [DOI] [PubMed] [Google Scholar]
- Robinson D. (2001). Root proliferation, nitrate inflow and their carbon costs during nitrogen capture by competing plants in patchy soil. Plant Soil 232: 41–50 [Google Scholar]
- Robinson N.J., Procter C.M., Connolly E.L., Guerinot M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Roosens N., Verbruggen N., Meerts P., Ximénez-Embún P., Smith J.A.C. (2003). Natural variation in cadmium tolerance and its relationship to metal hyperaccumulation for seven populations of Thlaspi caerulescens from western Europe. Plant Cell Environ. 26: 1657–1672 [Google Scholar]
- Sakakibara H. (2006). Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57: 431–449 [DOI] [PubMed] [Google Scholar]
- Salamini F. (2003). Plant Biology. Hormones and the green revolution. Science 302: 71–72 [DOI] [PubMed] [Google Scholar]
- Séguéla M., Briat J.-F., Vert G., Curie C. (2008). Cytokinins negatively regulate the root iron uptake machinery in Arabidopsis through a growth-dependent pathway. Plant J. 55: 289–300 [DOI] [PubMed] [Google Scholar]
- Skoog F., Miller C.O. (1957). Chemical regulation of growth and organ formation in plant tissue cultured in vitro. Symp. Soc. Exp. Biol. 11: 118–131 [PubMed] [Google Scholar]
- Tocquin P., Corbesier L., Havelange A., Pieltain A., Kurtem E., Bernier G., Périlleux C. (2003). A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana. BMC Plant Biol. 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuberosa R., Sanguineti M.C., Landi P., Giuliani M.M., Salvi S., Conti S. (2002). Identification of QTLs for root characteristics in maize grown in hydroponics and analysis of their overlap with QTLs for grain yield in the field at two water regimes. Plant Mol. Biol. 48: 697–712 [DOI] [PubMed] [Google Scholar]
- Vert G., Grotz N., Dédaldéchamp F., Gaymard F., Guerinot M.L., Briat J.-F., Curie C. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd T.C., Dekker B.M.M., Hoekema A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Holst K., Pörs Y., Guivarc’h A., Mustroph A., Chriqui D., Grimm B., Schmülling T. (2008). Cytokinin deficiency causes distinct changes of sink and source parameters in tobacco shoots and roots. J. Exp. Bot. 59: 2659–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Motyka V., Strnad M., Schmülling T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 98: 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Schmülling T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Wilson J.B. (1988). A review of evidence on the control of shoot: root ratio, in relation to models. Ann. Bot. (Lond.) 61: 433–449 [Google Scholar]
- Yang S., Yu H., Xu Y., Goh C.J. (2003). Investigation of cytokinin-deficient phenotypes in Arabidopsis by ectopic expression of orchid DSCKX1. FEBS Lett. 555: 291–296 [DOI] [PubMed] [Google Scholar]
- Yang X.-E., Chen W.-R., Feng Y. (2007). Improving human micronutrient nutrition through biofortification in the soil-plant system: China as a case study. Environ. Geochem. Health 29: 413–428 [DOI] [PubMed] [Google Scholar]
- Yue B., Xue W., Xiong L., Yu X., Luo L., Cui K., Jin D., Xing Y., Zhang Q. (2006). Genetic basis of drought resistance at reproductive stage in rice: Separation of drought tolerance from drought avoidance. Genetics 172: 1213–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]