Serine protease inhibitors (SPIs) are antidigestive proteins that presumably defend plants against herbivore attack. This work identifies and characterizes SPIs in the wild plant Solanum nigrum and evaluates the consequences of SPI silencing on plant defense, growth, and development.
Abstract
Solanaceaeous taxa produce diverse peptide serine proteinase inhibitors (SPIs), known antidigestive defenses that might also control endogenous plant proteases. If and how a plant coordinates and combines its different SPIs for the defense against herbivores and if these SPIs simultaneously serve developmental functions is unknown. We examine Solanum nigrum’s SPI profile, comprising four different active inhibitors, of which the most abundant proved to be novel, to understand their functional specialization in an ecological context. Transcript and activity characterization revealed tissue-specific and insect-elicited accumulation patterns. Stable and transient gene silencing of all four SPIs revealed different specificities for target proteinases: the novel SPI2c displayed high specificity for trypsin and chymotrypsin, while two other SPI2 homologs were highly active against subtilisin. In field and lab experiments, we found all four SPIs to display herbivore- and gene-specific defensive properties, with dissimilar effects on closely related species. However, we did not observe any clear developmental phenotype in SPI-silenced plants, suggesting that SPIs do not play a major role in regulating endogenous proteases under the conditions studied. In summary, specific single SPIs or their combinations defend S. nigrum against generalist herbivores, while the defense against herbivores specialized on SPI-rich diets requires other unknown defense mechanisms.
INTRODUCTION
Green and Ryan’s (1972) groundbreaking discovery of the wound-inducible production of protease inhibitors (PIs) that inhibit digestive herbivore gut proteases inspired the field of plant–insect interactions and became an iconic example of induced plant defenses. Since then, numerous PIs have been isolated, characterized, and tested for their potential to control herbivorous insects as well as pathogenic microorganisms, research largely motivated by the hope of engineering transgenic crop plants with increased herbivore resistance (reviewed in Ryan, 1990; Jongsma and Bolter, 1997; Mosolov and Valueva, 2005). After four decades of research, there is no doubt that plant PIs are able to negatively affect insect herbivore growth and survival and act as plant defenses (Jongsma and Bolter, 1997; Zavala et al., 2004b; Steppuhn and Baldwin, 2007). However, PI expression does not always function as a defense, increasing plant resistance. There are examples of herbivore-inducible PIs with no defensive function or even the opposite effect due to the counteradaptations of insects to the ingestion of PIs (Zhu-Salzman et al., 2008). Several studies demonstrated that some insects respond with constitutive or induced production of PI-insensitive proteases (Jongsma et al., 1995; Bown et al., 1997; Bayés et al., 2005, 2006) or by proteolytically inactivating the ingested PIs to prevent binding to sensitive proteases (Girard et al., 1998; Giri et al., 1998; Zhu-Salzman et al., 2003). For many insects, the ingestion of PI-containing tissues elicits behavioral and physiological counterresponses that increase the amount of damage they inflict on plants: sublethal PI levels stimulate feeding and induce a general overproduction of proteolytic enzymes. Such compensatory feeding responses can negate the effects of PIs and sometimes result in an even greater loss of plant biomass compared with plants not expressing PIs (De Leo et al., 1998; Winterer and Bergelson, 2001; Abdeen et al., 2005; Steppuhn and Baldwin, 2007).
As a consequence, the development of transgenic plants expressing PIs that increase crop plant resistance to herbivores has proved difficult (Gatehouse, 2008). Recent approaches using novel inhibitors, combinations of several inhibitors, or coexpression with other synergistic defense compounds might be more successful (Christou et al., 2006; Mosolov and Valueva, 2008). Dunse et al. (2010a, 2010b) recently demonstrated that the transgenic coexpression of two SPIs in cotton (Gossypium hirsutum) significantly reduced herbivory by Helicoverpa armigera. This first success of transgenic crops expressing PIs has underscored the need to understand the underlying mechanisms and interactions of multiple PIs with other defenses and with the adaptive responses of herbivores in more detail. Due to the focus on agricultural importance, most studies use insect herbivore feeding assays with transgenic crop plants ectopically overexpressing PIs or with artificial diet supplemented with isolated PIs. There are only a few publications that altered the expression of endogenous PIs in undomesticated native plants to study their relevance for plant defense in an ecological context (Zavala et al., 2004a, 2004b; Sin et al., 2006; Steppuhn and Baldwin, 2007; Steppuhn et al., 2008). In one study (Steppuhn et al., 2008), field experiments on the diverse natural herbivore community revealed that one PI is indeed an effective defense but only under specific circumstances and against particular insect species. It is also very likely that the defensive function of PIs against other native herbivore species requires coordinated expression with indirect defenses, such as the emission of volatile alarm calls that attract the predators of the herbivores (Kessler and Baldwin, 2001). PI expression is frequently not lethal to the herbivores but slows their growth and thereby extends the time that herbivores might be susceptible to predators.
Moreover, several studies highlighted another aspect that had been neglected: the in planta physiological functions of defense-related PIs. Directed and tightly controlled proteolysis is a key cellular process in all domains of life. More than 800 proteases from 60 families in Arabidopsis thaliana and more than 600 in rice (Oryza sativa) highlight the importance of proteolytic processes in plants (van der Hoorn, 2008). Such proteolytic machinery requires tight control to prevent the unwanted degradation of proteins. There is evidence that plant PIs protect specific tissues, act as storage proteins, regulate the activity of proteases, and direct their release (Ryan, 1973; Mosolov and Valueva, 2005). Plants altered in the expression of endogenous PIs display changes in growth rate (Zavala et al., 2004a; Xie et al., 2007), flower morphology and seed development (Sin et al., 2006), phloem structure (Xie et al., 2007), and nectar protein composition (Bezzi et al., 2010). These alterations in fitness-relevant traits showed that members of the potato inhibitor type II family (PI-II), formerly associated with defense, can have other physiological functions. Multiple functions have also been suggested for other PI classes: for example, cystein PIs in barley (Hordeum vulgare) interact with endogenous Cys proteases to regulate protein turnover during germination but are also supposed to defend the plant against herbivores and pathogens (Martinez et al., 2009). Philippe et al. (2009) found 31 different Kunitz PIs with tissue-specific and induction-dependent expression patterns in a genomic analysis of poplar (Populus spp), which also suggests multiple functions of this PI family.
The PI-I and the PI-II family of serine protease inhibitors (SPIs), classified as I13 and I20 in the MEROPS database (Rawlings et al., 2008), represent two of the best-described PI classes in plants and display a stunning genetic and structural diversity, particularly in the Solanaceae. Nevertheless, data on the defensive properties and other possible functions of a complete group of PI genes in a single plant species is required for a comprehensive understanding of this important group of defense proteins. Clearly, the evolutionary context of the genetic diversity makes it imperative to study undomesticated plants species that have not been shaped by breeding for particular traits (Kant and Baldwin, 2007). In our group, we established black nightshade (Solanum nigrum) as an ecological model system that allows us to use molecular tools to study gene function in the context of the natural habitat (Schmidt et al., 2004). It has been shown that S. nigrum responds typically to wounding and herbivory with the production of SPIs (Schmidt et al., 2004; Schmidt and Baldwin, 2006a; Schmidt and Baldwin, 2006b). They identified one member of the PI-II class called PIN2b, a homolog of a gene first isolated from Solanum americanum (Xu et al., 2001). Most likely additional homologs of PI-II exist in S. nigrum, as they do in many other species of the genus Solanum. Additionally, S. nigrum is hexaploid (Edmonds and Chweya, 1997; Schmidt et al., 2004), and we can assume that this has multiplied the number of PI-II genes.
Here, we identified SPIs in S. nigrum, profiled their tissue-specific expression, and investigated their substrate specificity. Furthermore, we manipulated SPI gene expression using RNA interference (RNAi)–mediated gene silencing and evaluated the consequences of PI silencing on plant defense, growth, and development. The analysis provides an understanding of the importance of SPIs for defense against herbivores, especially in natural systems, and it indicates where and which substrates they might bind to in mediating their putative in planta effects on physiological processes.
RESULTS
SPIs Are Expressed in All Aboveground Tissues but Differ Strongly in Abundance and Inducibility
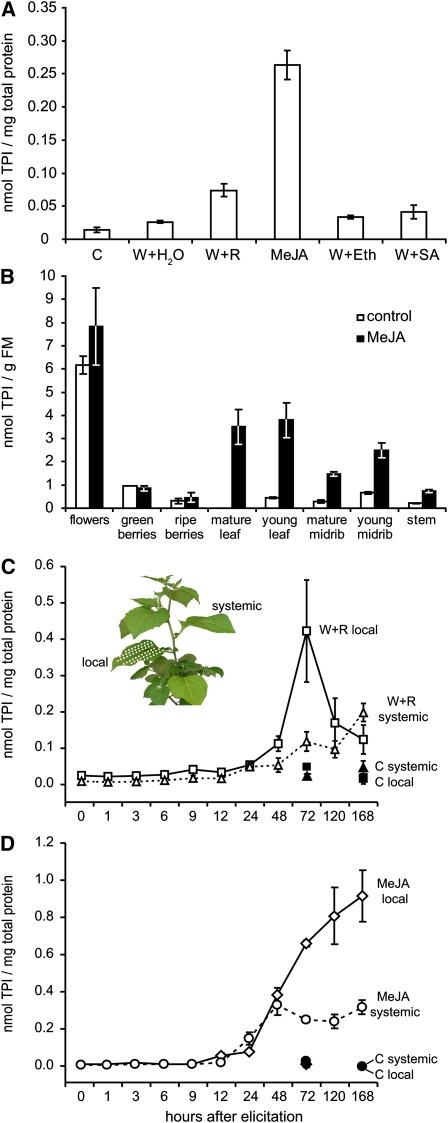
Based on preliminary experiments and previous studies (Schmidt et al., 2004; Schmidt and Baldwin, 2006a), we assumed that PI-I and PI-II-class inhibitors of S. nigrum respond to leaf-chewing herbivores. To get a better understanding of this response and the tissue-specific localization of SPIs, we measured trypsin inhibitory activity in extracts from various tissues, after different treatments, and at several time points after elicitation (Figure 1). Trypsin protease inhibitor (TPI) accumulation is slightly enhanced after mechanical wounding of leaves, but this response is strongly amplified when caterpillar feeding is mimicked through the application of regurgitates from Manduca sexta (Sphingidae) larvae to wounds (W+R). A treatment with methyl jasmonate (MeJA), which directly triggers the jasmonate-mediated responses, further increased this accumulation. By contrast, salicylic acid and ethylene, two other phytohormones related to plant defense, did not increase TPI accumulation when applied to wounds (Figure 1A).
Figure 1.
Inducibility, Tissue Specificity, and Dynamics of TPI Accumulation in S. nigrum.
(A) Mean ± se TPI concentration in leaves 48 h after different elicitor treatments (n = 4 biological replicates). C, control; W, mechanical wounds treated with water, regurgitate (R) of M. sexta larvae, ethephone (Eth), or salicylic acid (SA); MeJA, MeJA in lanolin paste.
(B) Mean ± se TPI concentration in different aboveground tissues of control and MeJA-treated plants 48 h after induction (n = 3, pooled from nine plants). FM, fresh mass.
(C) and (D) Mean ± se TPI accumulation in local and systemic leaves after a single induction with either W+R (C) or MeJA (D) (n = 5). Untreated control samples were taken after 0 h, 3 d, and 7 d. The inset shows the position of the locally elicited leaf and the harvested systemic leaf, one node above the local.
[See online article for color version of this figure.]
Constitutive and MeJA-inducible TPI activity strongly varied across tissues. It was found in all aboveground parts of the plant but was not detectable in roots. Vegetative organs contained relatively small amounts of constitutive TPIs but responded to MeJA treatment with up to 10-fold increases in TPI activity (Figure 1B). Generative organs were not or were only weakly responsive to MeJA, but flowers displayed the highest TPI activity per gram fresh mass of all tissues examined. After fertilization, these high constitutive levels decreased and by the time the berries had ripened, the levels were as low as those of constitutive vegetative organs.
The accumulation of TPIs after herbivore damage is a late defense response. Figures 1C and 1D show the temporal dynamics of TPI accumulation in local and systemic leaves after a single elicitation by wounding plus M. sexta regurgitates (W+R) or by MeJA. In both treatments, TPI levels increase in the local leaf after 12 to 24 h, reaching a maximum after 3 d in the case of W+R or rising continuously for 7 d in the case of MeJA-elicited leaves. Systemic unelicited leaves on elicited plants respond approximately in the same time frame but show a markedly lower maximum. The TPI levels in uninduced control plants were additionally recorded at days 3 and 7 but remained at the initial level throughout the experiment.
Identifying SPI Genes in S. nigrum
In a cDNA library screen, Schmidt et al. (2004) found the PI-II homolog Sn PIN2b (GenBank AY422686) using probes based on genes from S. americanum cloned by Xu et al. (2001). We suspected that another homolog, similar to Sa PIN2a from S. americanum, could also be present in S. nigrum and succeeded in identifying one clone by PCR with cDNA and genomic DNA (gDNA) as templates. To avoid confusion of these PI genes with the well-established PIN auxin transporters, we will from here on refer to the new gene as S. nigrum Serine-Protease-Inhibitor-2a (Sn SPI2a) and to Sn PIN2b as Sn SPI2b. The new clone Sn SPI2a shares 96% nucleotide sequence identity with Sa PIN2a and 78% with Sn SPI2b (which translates into amino acid sequence similarities of 98 and 73%, respectively).
We also obtained two fragments of PI-I genes through PCR-based cloning using genomic DNA as template. They share 98% identity in nucleotide sequence, which translates into two amino acid changes. Both gene sequences are 84% identical to a PI-I inhibitor from potato (Solanum tuberosum), and we refer to these two genes as Sn SPI1a and Sn SPI1b. In a DNA gel blot, we assessed the number of genes similar to SPI1, SPI2a, or SPI2b in S. nigrum using gene-specific probes and three different enzymes for digestion (see Supplemental Figure 1 online). Although the high similarity between SPI2a and SPI2b produced cross-signals that complicated the analysis, we estimate that all three SPI genes are present with three to four copies in the haploid (n = 3×) S. nigrum genome.
Silencing SPI2a and SPI2b Has No Clear Effect on Growth and Development
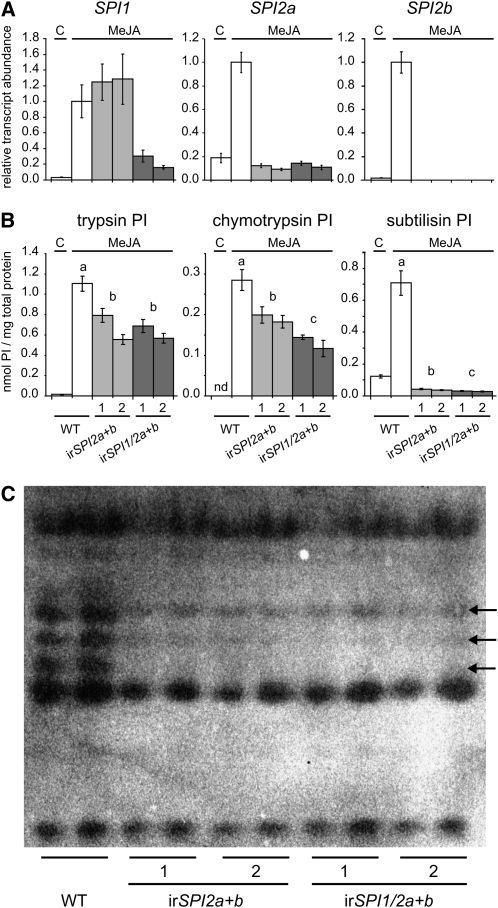
To study the function of these genes, we stably transformed S. nigrum plants with inverted-repeat (ir) RNAi constructs to silence the expression of either both SPI2 genes alone (irSPI2a+b) or additionally the two SPI1 genes (irSPI1/2a+b). For both constructs, we selected two independently transformed lines, each one containing a single T-DNA insertion (see Supplemental Figure 2 online). Figure 2A shows the mRNA levels of all three gene groups in wild-type plants and in the silenced lines 24 h after elicitation with MeJA. Both ir-constructs contained a fragment of SPI2b that reduced transcript levels to ~0.1% of wild-type levels. The high similarity of SPI2b to SPI2a resulted in a simultaneous reduction of SPI2a transcripts to 9 to 14% in all genotypes. Lines transformed with the double RNAi construct irSnSPI1/2a+b were additionally silenced in SPI1 transcripts at levels of 16 and 31%. Sin et al. (2006) reported an increase in flower size and an 80% seed abortion after silencing homologs of SPI2a and SPI2b with similar efficiency in S. americanum. However, in S. nigrum, we did not observe any effect on flower size and found that only 0.7 to 2.8% of the seeds were aborted or defective (see Supplemental Figure 3 online). Zavala et al. (2004a) and Xie et al. (2007) reported that silencing or ectopic overexpression of SPI genes can affect plant growth. In comparative growth experiments, we found no differences in plant height between wild-type and SPI-silenced plants (see Supplemental Figure 3 online). The lack of clear phenotypes suggested that the investigated SPIs have either only a minor effect on plant growth and development or that their loss might be compensated by other SPIs in S. nigrum.
Figure 2.
Silencing Efficiency and Remaining SPI Activity in Plants Silenced in the Expression of SPI1, SPI2a, and SPI2b.
(A) Mean ± se relative transcript abundance of SPI1, SPI2a, and SPI2b in leaves of wild-type plants and of transgenic lines expressing an ir-construct specific for SPI2a and SPI2b (irSPI2a+b) or a construct specific for SPI1, SPI2a, and SPI2b (irSPI1/2a+b) (n = 7). C, control; MeJA, MeJA treatment for 24 h.
(B) Mean ± se inhibitory activity against trypsin, chymotrypsin, and subtilisin in leaves of wild-type (WT) plants and of SPI-silenced lines (C, control; MeJA, MeJA treated for 3 d). Different letters indicate a significant difference (nested ANOVA, lines nested within genotypes, TPI: F2,30 = 21.95, P < 0.001, ChyPI: F2,29 = 21.42, P < 0.001, SubPI: F2,30 = 349.82 P < 0.001, followed by a Scheffé post-hoc test, P < 0.05; wild-type control plants were excluded from the analysis).
(C) Profiles of active TPI proteins present in MeJA-induced leaves of wild-type and SPI-silenced plants, visualized with GXCP after 12% native PAGE. Each genotype is represented by two pools of two biological replicates. Arrows indicate bands missing in SPI-silenced lines.
SPI2a and SPI2b Are Strong Inhibitors of Subtilisin but Account for Only One-Third of Total Trypsin Inhibition
We determined the inhibitory activity of leaf extracts from all genotypes against trypsin, chymoptrypsin, and subtilisin to find out how well the gene silencing translated into reduced PI activity and to assess the substrate specificity of the different SPIs (Figure 2B). Although the silencing reduced mRNA levels of the SPI genes efficiently, we found that extracts from leaves containing both constructs (irSPI2a+b and irSPI1/2a+b) retained two-thirds of their activity against trypsin and chymotrypsin, suggesting that another, yet unknown, SPI was likely present in the extracts. irSPI1/2a+b lines showed slightly but significantly lower inhibitory activity against chymotrypsin, indicating specificity of the SPI1 genes. The most striking effect was observed when measuring activity against subtilisin. All lines showed a drastic reduction of subtilisin-PI activity, mainly caused by the silencing of SPI2a and SPI2b (~96% reduction in irSPI2a+b) with a small contribution by SPI1 genes (~97% reduction in irSPI1/2a+b).
The high remaining activity against trypsin and chymotrypsin in the silenced lines demanded a qualitative analysis of SPIs. We used gel x-ray film contact prints (GXCPs) after native PAGE to obtain activity profiles of the inhibitors. Three proteins were indeed missing or strongly reduced in all four SPI-silenced lines (Figure 2C). A characteristic feature of PI-II proteins is that they contain one to several sequence repeats, which code for one precursor. Proteolytic cleavage of these precursors at specific linker sites results in several single or multidomain PIs with different properties (Atkinson et al., 1993; Beekwilder et al., 2000; Horn et al., 2005; Tamhane et al., 2007), which could explain the absence of more than two bands when silencing genes encoding the two-domain proteins SPI2a and SPI2b. It is possible that other peptides with low activity or concentration were not detected or resolved with the method that we used and that there are in fact more than three SPI peptides missing in the silenced lines. While the missing bands clearly represent products of SPI2a and SPI2b, no band specific for SPI1 could be identified. Three bands with high activity were not affected by the RNAi constructs. These proteins explained the majority of remaining activity of the extracts against trypsin; however, their identity remained unknown.
SPI2c, an Isoform of PI-II Inhibitors, Is the Main Inhibitor of Trypsin and Chymotrypsin
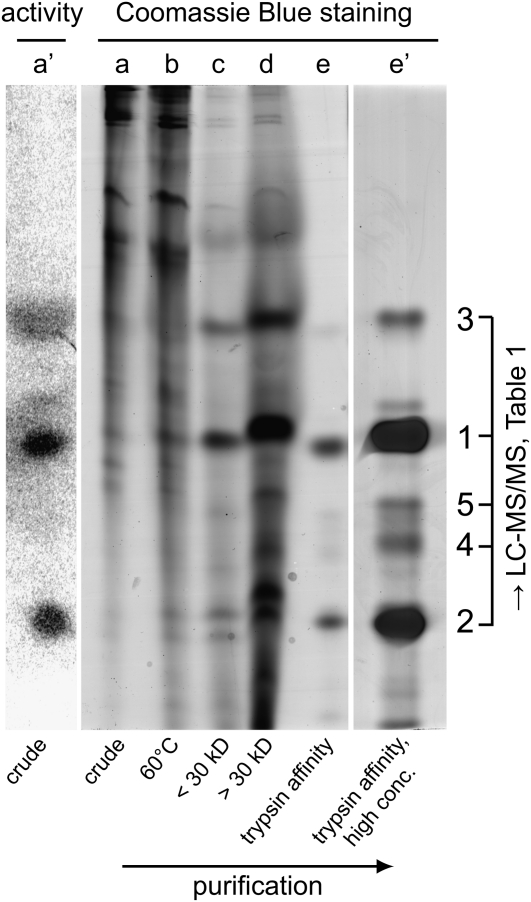
Although the x-ray film analysis revealed PIs with very high activity, we could not detect the corresponding PI proteins after staining the gels with Coomassie blue. This suggested that these active PIs occur at low abundance and require further purification. We started with 50 g of wild-type flower material because the PIs displayed high constitutive expression in these tissues. The active peptides, like many other PIs, are heat stable, which allowed them to be concentrated by incubating the extract for 30 min at 60°C. After dialysis and freeze-drying, we used ultrafiltration on the protein solution with a 30-kD molecular mass cutoff and checked for remaining activity in both fractions by native PAGE followed by GXCP. The active inhibitors were present in both, but since the filtrate contained less contamination from other proteins, we used this fraction for further purification steps. After trypsin-affinity chromatography, we separated the eluate on native PAGE and visualized the bands by Coomassie staining and GXCP (Figure 3). The active PI bands were excised and subjected to tandem mass spectrometry (MS/MS) de novo sequencing. We obtained sequences of six different peptides (Table 1) that were all similar to a protein from the PI-II family in tomato (Solanum lycopersicum; Uniprot accession: Q43710) containing three repeats.
Figure 3.
Native PAGE (12%) Representing the Purification of SPI2c Visualized with GXCP for PI Activity or Coomassie Blue Stain.
GXCP: lane a’, crude extract. Coomassie stain, lanes: a, crude extract, same as in a’; b, heat-treated crude extract; c, ultrafiltrate (< 30 kD molecular mass cutoff); d, ultrafiltration concentrate (> 30 kD molecular mass cutoff); e, eluate of c, after trypsin affinity chromatography; e’ same as e but larger amount loaded for subsequent de novo sequencing (numbered bands were excised and sequenced on liquid chromatography–MS/MS; see Table I). Protein amounts loaded: a’ and a to d, 40 μg; e and f, 1 μg; and e’, 20 μg.
Table 1.
Peptide Sequences of Protein Bands in Figure 3 (Lane e’) as Determined by Liquid Chromatography–MS/MS de Novo Sequencing
| Band No. |
|||||||
| Peptide | Peptide Massa | Peptide (Partial) Sequenceb | 1 | 2 | 3 | 4 | 5 |
| A | 2375.02 | (X)XXXYFSNDGTFLcCEGESEY | x | x | |||
| B | 1321.58 | (SD?)CTNCCAGKK | x | x | x | x | x |
| C | 1683.72 | ECDTRdLcDYGLcCPVS | x | ||||
| D | 1022.50 | LcDYGLcCPVS | x | x | |||
| E | 992.52 | LcAYGLcCPLcS | x | x | x | x | |
| F | 978.52 | LcAYGLcCPVS | x | ||||
Molecular mass of the uncharged peptide.
Given from N terminus to C terminus.
Leu and Ile are not distinguishable.
Missed cleavage site (R).
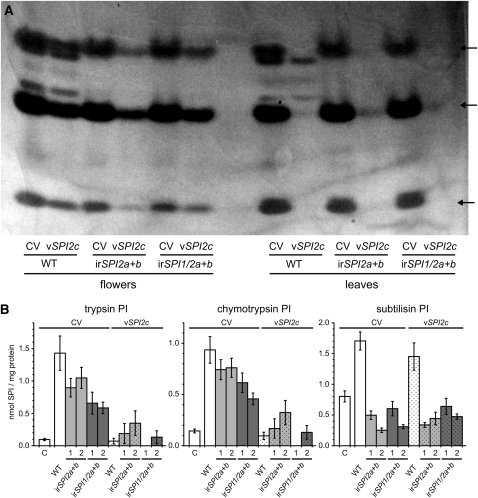
Using primers designed from the tomato sequence, we cloned several fragments of a PI-II inhibitor gene from S. nigrum by PCR with cDNA and gDNA as templates. Two different genes with high similarity to each other were found, one coding for two and one for three domains, sharing 97% amino acid sequence similarity with each other and 60 or 64% similarity with SPI2a and SPI2b, respectively (for a sequence alignment, see Supplemental Figure 4 online). We refer to these new genes as Sn SPI2c-R2 and Sn SPI2c-R3. The high similarity between the two SPI2c sequences facilitated the design of a construct for virus-induced gene silencing (VIGS), which transiently reduced the transcripts of both genes simultaneously. Figure 4A shows the active peptides present in flowers and MeJA-induced leaves from wild-type plants and stably PI-silenced lines (irSPI2a+b and irSPI1/2a+b) after silencing both SPI2c genes with the VIGS vector vSPI2c or after treatment with a control VIGS vector (CV). While the silencing in leaves was much more efficient than that observed in flowers, the results clearly demonstrated that the VIGS vector vSPI2c reduced the three most active peptides in both tissues, confirming their identity as SPI2c. In leaves, the efficient silencing also demonstrated that the three peptides remaining after silencing SPI2c in wild-type plants were SPI2a and SPI2b. VIGS silencing of SPI2c in the background of the two stably SPI-silenced lines resulted in a complete absence of active peptides. When quantifying SPI activity in leaf extracts, it became evident that SPI2c does not or only weakly binds to subtilisin but that it is a strong inhibitor of trypsin and chymotrypsin (Figure 4B), which is exactly the opposite specificity of SPI2a and SPI2b.
Figure 4.
VIGS of SPI2c in Wild-Type Plants and in Four Independently Stably Transformed Lines, Silenced Either for SPI2a and SPI2b (irSPI2a+b) or Additionally for SPI1 (irSPI1/2a+b).
Each genotype was either inoculated with tobacco rattle virus containing a CV or a vector harboring a fragment of SPI2c for gene silencing (vSPI2c).
(A) Profiles of active TPI proteins present in flowers and leaves of wild-type (WT) plants and of two SPI-silenced lines (irSPI2a+b line 1 and irSPI1/2a+b line 1) after VIGS and treatment with MeJA. The active TPIs were visualized with GXCP after 12% native PAGE. Each lane represents a pool of three biological replicates. Arrows indicate bands representing SPI2c.
(B) Mean ± se inhibitory activity against trypsin, chymotrypsin, and subtilisin in leaves of wild-type and SPI-silenced plants (n = 9). C, control; MeJA, 3 d after MeJA treatment.
SPIs Respond Differently after Simulated Herbivory
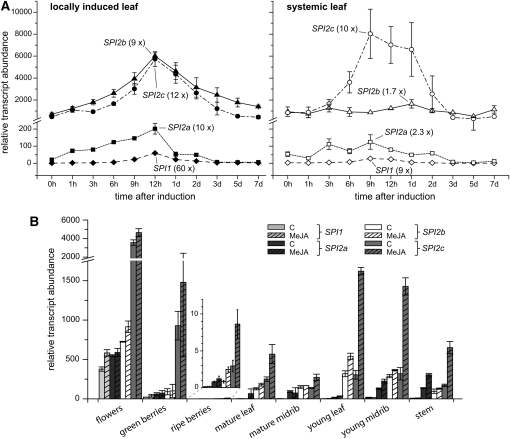
Using quantitative real-time PCR, we assessed the transcriptional contribution of the different groups of SPI genes to the W+R-induced SPI activity measured in Figure 1. In local leaves, there was a strong upregulation of all four gene types, reaching a maximum around 12 h after induction (Figure 5A). In terms of absolute transcript amounts, two groups could be distinguished: at the peak of highest accumulation, SPI2b- and SPI2c-like transcripts were ~30 or 100 times more abundant than SPI2a or SPI1, respectively. In systemic leaves, the induction of SPI1-, SPI2a-, and SPI2b-like genes was attenuated, with the latter two barely responding to the treatment. By contrast, SPI2c genes attained transcript levels in systemic untreated leaves comparable to the high levels observed in locally treated leaves. Transcript levels in untreated leaves did not vary significantly throughout the experiment and thus were not included in the graph.
Figure 5.
Transcript Levels of SPI Genes as Determined by Quantitative RT-PCR.
(A) Time series of the mean ± se relative transcript abundance of SPI1, SPI2a, SPI2b, and SPI2c in locally elicited leaves (left) and systemic untreated (right) leaves of wild-type plants after mechanical wounding and application of M. sexta regurgitant. Numbers in parentheses show the maximum fold change in transcript levels compared with untreated leaves at time point 0 h (n = 4 biological replicates per time point).
(B) Mean ± se relative transcript abundance of SPI1, SPI2a, SPI2b, and SPI2c in different aboveground tissues of control and MeJA-treated plants 48 h after induction (n = 3 biological replicates). Inset: magnified plot of the data from ripe berries.
Similarly, we analyzed the tissue-specific expression of the SPI genes. In general, the highest transcript levels of all four gene groups were found in flowers, with SPI2c being by far the most abundant (Figure 5B). MeJA treatment of flowers affected SPI expression only slightly, as we had observed for TPI activity. After fertilization, the high levels in flowers drop significantly with just SPI2c being strongly expressed in unripe fruits and almost no expression observed in ripe fruits. In vegetative tissues, SPI2c and SPI2b are the main SPI transcripts. Particularly in the leaf lamina, these two forms are predominant and strongly MeJA inducible, with higher levels in younger than in mature leaves. On the other hand, transcripts of SPI2a, although also present in the leaf lamina, were particularly abundant in leaf midribs and stems. Except for flowers, SPI1 transcripts were found in much lower amounts in all other tissues but displayed a preferential expression in midribs and stems (~4- to 8-fold higher when compared with leaf lamina), similar to SPI2a.
Silencing SPI2c Has No Effect on Development or Growth
To study the function of SPI2c in more detail and to assess if this gene could mask a potential developmental phenotype in irSPI2a+b lines, we created a double ir-construct harboring fragments of SPI2b and SPI2c to generate stable transformants silenced for all three SPI2 genes (irSPI2a+b+c). For further experiments, we selected two independently transformed lines, which exhibited transcript levels reduced to 0.3 to 2% for all three genes and TPI activity of 0.3 to 1.4% when compared with MeJA-induced wild type (see Supplemental Figure 5 online). However, irSPI2a+b+c plants were not different from irSPI2a+b lines in seed or plant development.
SPI2 Genes Defend the Plant against Natural Generalist Herbivorous Insects
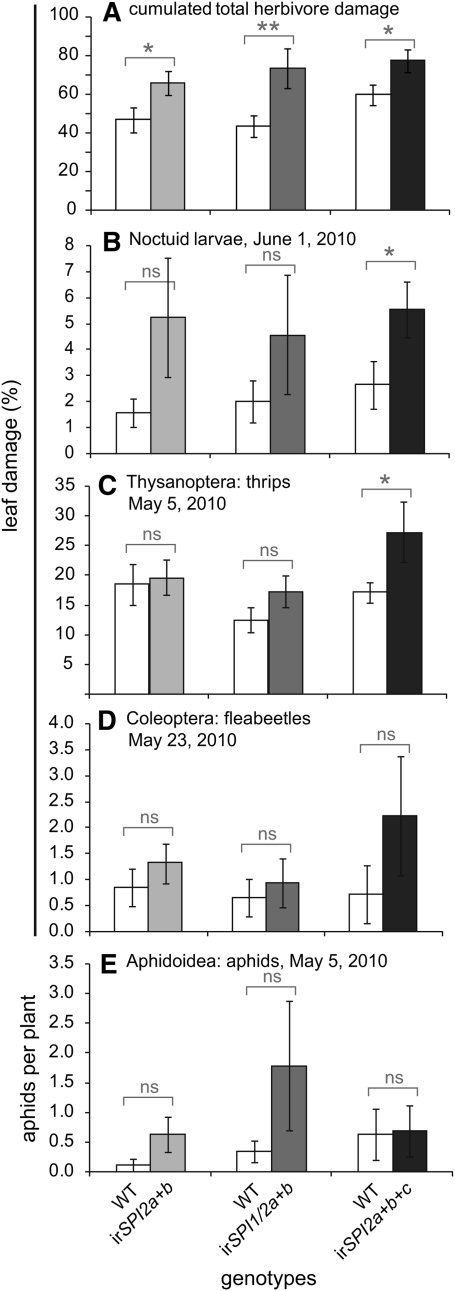
To investigate the importance of SPIs for defense against herbivores in a natural environment, we obtained permission to conduct field experiments with transgenic plants at our field station at the Lytle Ranch Preserve, Utah, in the Great Basin Desert of the southwestern US. The stability of RNAi-mediated gene silencing under field conditions is well documented in a large number of publications from our group (for example, Kessler et al., 2004; Steppuhn et al., 2008). In 2010, we planted wild-type plants and the SPI-silenced lines in pairs and monitored the percentage of loss of total leaf area, separately for different feeding guilds, over a period of 4 weeks. Figure 6 shows that all three lines suffered significantly more overall damage than the wild type. However, there were gene-specific differences for different feeding guilds. Thrips caused significantly more damage on irSPI2a+b+c plants, suggesting a defensive role of SPI2c against these herbivores. Similarly, only irSPI2a+b+c exhibited a significantly higher mean damage caused by noctuid larvae; the other two lines (irSPI2a+b and irSPI1/2a+b) also displayed a higher mean damage, but the difference was not statistically significant. Additionally, when we removed the plants from the plot while terminating the experiment, we observed a significantly higher number of an unidentified noctuid larvae feeding on roots of irSPI2a+b+c plants (see Supplemental Figure 6 online). Unfortunately, it was not possible to determine whether the higher herbivory rate or occurrence of noctuid larvae was a result of preferential oviposition by the mother or host choice by the larvae. A comparison of emitted plant volatiles as analyzed by gas chromatography–mass spectrometry did not reveal any differences among the lines (data not shown). The apparently higher damage caused by flea beetles on irSPI2a+b+c plants and the higher mean aphid number on irSPI1/2a+b plants were not statistically significant.
Figure 6.
Natural Herbivore Damage on a Planted Field Population of Wild-Type and Stably Transformed Lines Silenced for Different Combinations of SPI1, SPI2a, SPI2b, and SPI2c.
In 2010, wild-type and SPI-silenced plants were grown in pairs (n = 20) under field conditions on a plot at the Lytle Preserve field station in Utah. Asterisks indicate a statistically significant difference between groups in a paired t test (*P < 0.05; **P < 0.01). ns, not significant. In (A), irSPI2a+b, t18 = 2.48, P = 0.023; irSPI1/2a+b, t17 = 3.18, P = 0.005; irSPI2a+b+c, t18 = 2.54, P = 0.02. In (B), irSPI2a+b+c, t10 = 2.73, P = 0.021. In (C), irSPI2a+b+c, t18 = 2.15, P = 0.045.
(A) Mean ± se of the percentage of cumulated leaf damage over the entire recorded period (May 5 to June 1).
(B) to (D) Mean ± se of the percentage of leaf damage caused by different insect herbivore taxa at the time of their respective highest abundance.
(E) Mean ± se number of aphids per plant. WT, wild type.
The genotype Sn30 of S. nigrum, which we used in all experiments, originates from a collection from a native population growing near Jena, Germany. Although S. nigrum has been recently introduced to the United States (Edmonds and Chweya, 1997), which justifies the experimental setup in Utah, we were also interested in how the SPI-silenced lines perform in the original European habitat. The German release permit allowed experiments with irSPI2a+b transgenic plants at a field site near Dornburg, Germany in the years 2004 to 2006. Plants were not allowed to flower; thus, experiments were restricted to approximately 3 weeks after planting during which flower buds had to be removed regularly. Consistent with a previous study (Schmidt et al., 2004), we identified the flea beetle Epitrix pubescens (Chrysomelidae) to be the main leaf-chewing herbivore. Corresponding to its life cycle, it occurred only for a few weeks in June and July when it is able to cause heavy damage. In the 2005 season, we observed the largest population of E. pubescens, which heavily infested the planted S. nigrum population. However, the apparently slightly higher damage levels on irSPI2a+b plants were not statistically significant (see Supplemental Figure 6 online). Earlier in the season, we also encountered large numbers of the black bean aphid Aphis fabae (Aphididae) feeding on S. nigrum, but their numbers did not differ statistically between wild-type and SPI-silenced plants (data not shown).
SPIs Do Not Affect M. sexta Performance
To investigate the responses of the noctuid herbivores and the defense mechanisms of SPIs in more detail, we used the stably transformed lines to study the performance of M. sexta, the beet armyworm Spodoptera exigua (Noctuidae), and Spodoptera littoralis in laboratory experiments. Both M. sexta and a Spodoptera sp were observed to occur and feed on S. nigrum in Utah. M. sexta is a solanaceous specialist and to some extent adapted to PI-II inhibitors (Zavala et al., 2008). S. exigua is considered a generalist with a wide host range that includes solanaceous crop plants (Brown and Dewhurst, 1975). S. littoralis occurs in Africa, Europe, and Asia with an even larger recorded plant host range, which also includes S. nigrum (Brown and Dewhurst, 1975; Martins et al., 2005). There are reports that Spodoptera spp are able to cope with an SPI-rich diet by differential regulation or de novo synthesis of proteases (Jongsma et al., 1995; Brioschi et al., 2007). Unfortunately, a closer examination of the interaction with the flea beetle E. pubescens was not possible because its detailed life cycle is unknown, and we have been unsuccessful in establishing stable laboratory cultures.
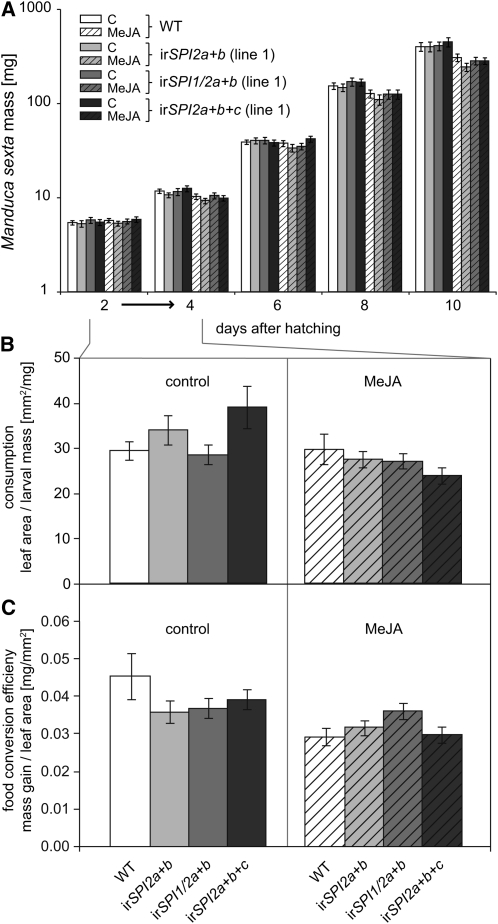
M. sexta larvae were reared on excised leaves from wild-type and SPI-silenced plants (irSPI2a+b, irSPI1/2a+b, and irSPI2a+b+c), which had been preinduced with MeJA or were left untreated. Silencing of SPIs had no effect on larval mass (Figure 7A). Induction with MeJA, however, caused a significant decrease in caterpillar mass, irrespective of the plant genotype (for statistics, see Supplemental Table 1 online). To understand this response in more detail, we measured the consumed leaf area between 2 and 4 d after hatching and related it to larval mass on day two, as a measure of consumption (Figure 7B), or to the mass gain within 48 h of feeding, as a measure of the food conversion efficiency (Figure 7C). The consumption was significantly reduced by MeJA treatment, but the plant genotype and thus the absence of the silenced PIs did not have any effect. Similarly, the MeJA treatment significantly affected the food conversion efficiency of M. sexta, but plant genotype had no effect. To summarize, MeJA induces defenses that decrease the consumption rate of M. sexta but that are independent of all four SPIs (the experiments were repeated once with two lines each of irSPI2a+b and irSPI1/2a+b and once using VIGS to silence SPI2c additionally in the background of the same lines with similar results; see Supplemental Figure 7A online).
Figure 7.
Performance of M. sexta Feeding on Excised Leaf Discs from Wild-Type Plants and from Three Independently Stably Transformed Lines Silenced for Different Combinations of SPI1, SPI2a, SPI2b, and SPI2c.
The plants were either control treated (open bars) or induced with MeJA (hatched bars).
(A) Mean ± se larval mass over time (n = 30). A repeated measures ANOVA of larval mass over all days with genotype and treatment as factors revealed day, day × treatment (within subjects), and treatment (between subjects) as significant effects (see Supplemental Table 1 online). WT, wild type.
(B) Mean ± se leaf consumption by M. sexta, measured as leaf area relative to larval mass before the feeding period of 48 h from days 2 to 4. An ANOVA model with genotype and treatment as factors indicated a clear treatment effect but no genotype or interaction effect (genotype: F3,186 = 0.169, P = 0.917; treatment: F1, 186 = 9.621, P = 0.002; genotype × treatment: F3,186 = 2.538, P = 0.058).
(C) Mean ± se food conversion efficiency of M. sexta, measured as mass gain relative to consumed leaf area during 48 h from days 2 to 4. An ANOVA, similar to the one in (B), indicates an effect of treatment but no genotype or interaction effect (genotype: F3,182 = 0.554, P = 0.646; treatment: F1,182 = 7.591, P = 0.006; genotype × treatment: F3,186 = 2.294, P = 0.079).
S. littoralis Does Not Respond to SPI2s and Is Temporarily Affected by SPI1
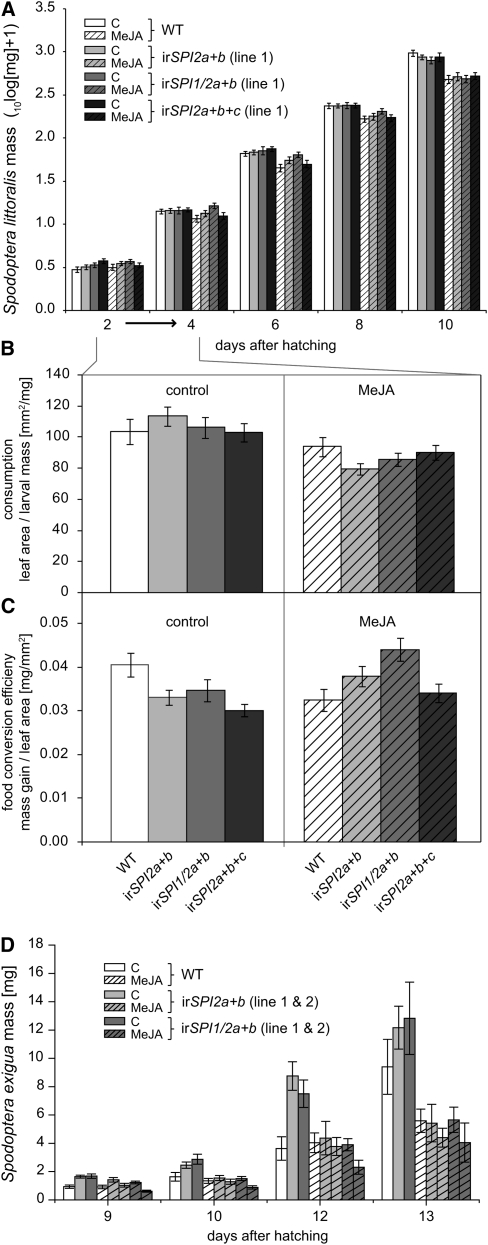
S. littoralis displayed a response very similar to M. sexta. We conducted the experiment in the same way as with M. sexta, feeding caterpillars for 3 d on uninduced and later on MeJA-induced leaf tissue. While the MeJA treatment had a clear effect on caterpillar mass, plant genotype played only a minor role (Figure 8A). A repeated measures analysis of variance (ANOVA) indicated a significant interaction of larval mass over time with genotype, which was most likely due to a transient increase in mass of larvae feeding on MeJA-induced irSPI1/2a+b lines, suggesting a mild effect of SPI1 (for statistics, see Supplemental Table 2 online). In contrast with M. sexta, only consumption was significantly reduced on MeJA-induced plants, but conversion efficiency remained largely at the same level (no effect of MeJA treatment; Figures 8B and 8C). However, in the case of larvae feeding on irSPI1/2a+b, we observed a significant change in consumption efficiency after MeJA treatment, which also correlated with the transient increase in larval mass on this line.
Figure 8.
Performance of S. littoralis and S. exigua Feeding on Excised Leaf Discs from Wild-Type Plants and from Independently Stably Transformed Lines Silenced for Different Combinations of SPI1, SPI2a, SPI2b, and SPI2c.
The plants were either control treated (open bars) or induced with MeJA (hatched bars).
(A) Mean ± se S. littoralis larval mass over time (ncontrol = 28, nMeJA = 32). A repeated measures ANOVA of larval mass over all days with genotype and treatment as factors revealed day, day × treatment, day × genotype (within subjects), and treatment (between subjects) as significant effects (see Supplemental Table 2 online). WT, wild type.
(B) Mean ± se leaf consumption by S. littoralis, measured as leaf area relative to larval mass before the feeding period of 48 h from days 2 to 4. An ANOVA model with genotype and treatment as factors indicated a clear treatment effect but no genotype or interaction effect (genotype: F3, 217 = 0.133, P = 0.940; treatment: F1, 217 = 22.989, P < 0.001; genotype × treatment: F3, 217 = 1.793, P = 0.1.49).
(C) Mean ± se food conversion efficiency of S. littoralis, measured as mass gain relative to consumed leaf area during 48 h between days 2 and 4. ANOVA indicates an effect of genotype and a significant genotype × treatment interaction but no treatment effect (genotype: F3, 217 = 3.206, P = 0.024, irSPI1/2a+b differs significantly from irSPI2a+b+c in a Tukey’s HSD post-hoc test P < 0.05; treatment: F1, 217 = 2.388, P = 0.124; genotype × treatment: F3, 217 = 5.227, P = 0.002).
(D) Mean ± se S. exigua larval mass over time (n = 21). A repeated measures ANOVA of larval mass (over all days) with genotype, line (nested within genotype), and treatment as factors revealed day, day × treatment (within subjects), and treatment (between subjects) as significant effects. Genotype had a significant effect in a reduced model on uninduced plants (see Supplemental Table 3 online).
Interestingly, another method of silencing SPI2c yielded a completely different result. The silencing of SPI2c using VIGS significantly reduced larval mass due to reduced consumption rates (see Supplemental Figures 7B to 7E online). This experiment suggested that the presence of SPI2c stimulates feeding but does not influence the digestibility of the material. Obviously the conditions necessary for VIGS (infection with Agrobacterium tumefaciens and tobacco rattle virus and different growth conditions) fundamentally influence the plant–herbivore interaction, suggesting that the defensive properties of SPIs might also depend on other, yet unconsidered, factors.
SPIs Reduce the Growth of S. exigua
In spite of its relatedness to S. littoralis, S. exigua larvae performed differently. S. exigua larvae showed a significant increase in mass when fed on uninduced tissue of SPI-silenced lines (Figure 8D; for statistical analysis, see Supplemental Table 3 online). This difference in performance was also reflected in the mortality rate: larvae on wild-type plants showed a higher mortality (38%) than larvae feeding on SPI-silenced lines (irSPI2a+b line 1, 14%; irSPI1/2a+b line 1, 10%). On MeJA-induced tissue, the difference between genotypes was lost with the larvae performing generally poorly and displaying high mortality (~50% on all genotypes). An analysis of consumption and food conversion efficiency between days 10 and 12 was complicated by the higher mortality of caterpillars feeding on wild-type and MeJA-induced plants. The resulting high variation did not allow meaningful conclusions to explain the differences between plant genotypes more precisely. Two repetitions of the experiments, which included the irSPI2a+b+c lines, with larvae from different egg batches had to be terminated prematurely because of even higher mortality. Obviously caterpillar populations vary strongly among batches concerning their performance on S. nigrum. Nevertheless, the differential performance on different SPI-silenced lines and the response to MeJA treatment demonstrates that S. exigua is affected by S. nigrum defenses, including SPIs. However, whether SPI2c has an effect on S. exigua could not be determined.
In summary, the insect performance assays revealed how diverse the responses of herbivorous insects are to plant PIs. The field experiments confirmed the defensive properties of PI-II type inhibitors against specific generalist herbivores. In particular, SPI2c protects the plant against thrips and noctuid larvae, although synergistic effects with the other SPI2s cannot be excluded in the case of the noctuid larvae. However, upon closer inspection, it became clear that herbivores specialized on solanaceous PI-rich diets, like M. sexta and E. pubescens, were not influenced in growth or feeding behavior. S. exigua, a generalist occasionally feeding on Solanaceae, performs significantly better on plants silenced for PI-II genes. By contrast, S. littoralis, a generalist with an extremely broad host range, appears to efficiently detoxify SPIs and was only transiently affected by SPI1. However, the compensatory feeding response, observed in the VIGS experiment, demonstrated that cross-reactions and influences of other factors might drastically change the effect of SPIs on herbivores.
DISCUSSION
The combination of activity-based PI profiling and specific gene-silencing enabled us to identify the SPIs of S. nigrum, which exhibit activity against the known substrates trypsin, chymotrypsin, and subtilisin. This approach led to the discovery and purification of the third and most abundant PI-II, Sn SPI2c. In total, we isolated five new expressed sequences from S. nigrum, two of them belonging to the PI-I family and the remaining three to the PI-II family. Together with the already known SPI2b, S. nigrum expresses three different forms of PI-II and one of PI-I. Each of these SPIs is represented by at least two copies in the genome, resulting in a total repertoire of likely more than 10 different SPI genes. The single or combined silencing of each SPI type in this study represents a combinatorial and functional characterization of several SPIs in a single plant species. This approach allowed us to dissect the function of native SPIs in plant defense against insect herbivores and in growth and development without having to extrapolate results from experiments with artificial diets or the ectopic overexpression of transgenes.
The analysis of Kong and Ranganathan (2008) revealed that the active sites and the linker regions of PI-II inhibitors carry the signatures of having been under positive selection pressure. We therefore expected the different PIs in S. nigrum to have different properties or even functions. Indeed, we found indications for functional diversification, and we discuss the evidence with respect to the two major functional domains of SPIs: plant development and defense.
SPIs Differ in Their Tissue Accumulation and Induction Patterns
The spatial and temporal expression patterns were consistent with a possible role in plant defense but also indicated differences between the SPIs. In the leaf lamina, the most commonly herbivore-attacked tissue, SPI2b and especially SPI2c, are most abundant and are highly amplified by W+R or MeJA application. Although SPI2a and SPI1 also respond to simulated herbivory with a strong fold increase, their transcript levels remain low compared with the other two SPIs. Why only SPI2c responds strongly in systemic tissues remains unclear, but it suggests a primary role of these genes for preparing the unattacked tissue for impending attack and correlates with its important defense function in the field. The increased accumulation of SPI2a and SPI1 in stem and midribs suggests a function in vascular tissues. Sa PIN2a, a homolog of Sn SPI2a from S. americanum, is localized in the phloem (Xu et al., 2001), and ectopic overexpression of this gene results in chloroplast formation in enucleate sieve elements (Xie et al., 2007). However, it cannot be ruled out that PIs accumulate in these tissues also as a defense response against phloem sap–feeding insects. PI-II inhibitors were shown to be elicited by aphid feeding in Capsicum annuum (Tamhane et al., 2009). We did not observe significant differences in aphid numbers on SPI-silenced lines in the field, although they were apparently higher on irSPI1/2a+b plants in Utah, but small effects on fitness or on other aphid species cannot be excluded. It is clear that mapping the expression of each gene on a finer spatial scale will yield valuable information on their possible functions. Sa PIN2b, a homolog of Sn SPI2b, was found to accumulate in trichomes, especially after MeJA treatment (Liu et al., 2006). This might be interpreted as a defense response against herbivores, but ectopic overexpression of Sa PIN2a and Sa PIN2b in tobacco (Nicotiana tabacum) also led to higher trichome densities and trichome branching (Luo et al., 2009).
SPIs Act as a Defense against Generalist Insect Herbivores
We found that the performance of solanaceous specialist herbivores like E. pubescens or M. sexta were not influenced by SPIs, but we cannot exclude more subtle fitness effects that are realized after pupation (De Leo and Gallerani, 2002). However, when trypsin PIs were silenced in N. attenuata, M. sexta mass almost doubled, demonstrating that the insect is not in general SPI insensitive (Zavala et al., 2004b). N. attenuata leaves contain around 6 to 10 times higher levels of trypsin PIs per milligram of total protein than S. nigrum (Zavala et al., 2004b; Steppuhn and Baldwin, 2007), and it is possible that M. sexta counterdefense mechanisms are sufficient to cope with levels of SPIs in S. nigrum. The decreased performance and consumption on plant material preinduced with MeJA demonstrates that S. nigrum is able to activate other yet unknown defenses, and our results suggest a lack of synergistic mechanism between these unknown defenses and the SPIs. By contrast, generalist herbivores in the field clearly preferred plants silenced in SPIs over wild-type plants. This suggests that either SPIs alone or in combination with other defenses are effective resistance mechanisms that protect S. nigrum against generalist herbivores. As the kinetics of this defense mechanism are relatively slow, it will only work when the herbivore pressure is sufficiently sustained to allow the full response to develop. Rapidly moving and feeding herbivores, such as flea beetles, could avoid these defenses by switching to other host plants before the defenses accumulate, representing a behavioral adaptation to induced defenses. Lepidopteran larvae, although sometimes mobile at later developmental stages, remain after hatching for a longer period on their host plant and are more exposed to induced systemic defenses. However, the increased occurrence of noctuid root-feeding larvae on irSPI2a+b+c plants suggests that the choice for appropriate food plants is made by the mother. If and how PIs could influence oviposition preference remains a fascinating question to be investigated in the future.
Our laboratory experiments with Spodoptera spp underscore the variable defensive value of PIs: S. exigua was particularly susceptible to the induced defenses of S. nigrum. Even constitutive levels of SPI2a and SPI2b were sufficient to affect larval growth. In combination with other induced defenses, including SPI2c, they barely managed to grow. This corresponds well to a field observation: we repeatedly found Spodoptera spp to oviposit, hatch, and feed on wild-type S. nigrum in Utah. However, after an initial feeding period, the number of larvae quickly decreases, an effect that we attribute to upregulated direct defenses, rather than top-down effects involving the attraction of predators. The problems of S. exigua to cope with SPIs of S. nigrum are remarkable because Jongsma et al. (1995) reported that the same species did not show a difference in performance when feeding on tobacco ectopically overexpressing potato inhibitor II, an effect that the authors attributed to S. exigua larvae responding to the PI-rich diet with the production of insensitive proteases. We speculate that these diet-induced proteases are sensitive to the SPIs of S. nigrum, causing a growth difference already at constitutive levels and giving the plant more time to fully activate its direct and indirect defenses. Nevertheless, the variable survivorship rate of S. exigua in several experiments demonstrated that different populations of the same species can respond in different ways. Thus, intraspecific variation should be taken into account when assessing an herbivore’s response to defenses of a particular host species.
Also, the different results in the experiments with S. littoralis caution against drawing conclusions about the effects of PIs against generalists from the findings with S. exigua. In VIGS experiments, SPI2c induced a transient compensatory feeding response in S. littoralis, which led to more rapid larval growth on wild-type plants, a phenomenon also observed in S. exigua when feeding on N. attenuata (Steppuhn and Baldwin, 2007). By contrast, the stable silencing of SPI2c had no effect on S. littoralis mass gain. The silencing of SPI1 caused a transient increase in performance, but in general, we conclude that there were no strong effects of SPIs on S. littoralis. It is known that VIGS leads to drastic changes in phytohormone levels (e.g., high levels of salicylic acid), which will likely influence a number of metabolic and defense parameters, including JA-related responses (Koornneef and Pieterse, 2008). Although we regard VIGS experiments as artifactual, the observed different response of S. littoralis suggests that the defensive potential and consequently the evolution of PIs might depend on the co-occurrence of pathogens and herbivores.
The complexity of defensive functions underscores why a clear assignment of defensive functions may be difficult. Although we see a tendency of SPI2c, and to some extent SPI2b, to behave like a typical herbivory-related gene, a single PI might not be sufficient to keep the proteolytic machinery of all naturally occurring herbivores in check. Each herbivore species may require a specific PI cocktail to be kept at bay, with evolution likely selecting for the lowest common denominator, depending on the nature and density of the herbivore populations. This need for combinatorial flexibility might have favored further domain duplication events and driven the evolution of different enzyme specificities in the single domains and of alternative posttranslational processing (Christeller, 2005). Additionally, our data demonstrate that synergistic effects of SPIs with other induced metabolites, which might include other PI classes or other toxins, are crucial for effective defense, and clearly it will be necessary to characterize these unknown defenses if we are to fully understand the defensive function of PIs.
SPIs Differ in Substrate Specificity but Play No Major Role in Growth and Development
We have shown that the SPIs in S. nigrum differ in their substrate specificity. SPI2c is a strong inhibitor of trypsin and chymotrypsin but barely interacts with subtilisin, whereas SPI2a and SPI2b show the opposite patterns of inhibition. While a large number of trypsin- and chymotrypsin-like proteases (Ser protease family S1), among others, were found in insects, only a handful of subtilase-like proteases (family S8) are known to date (Rawlings et al., 2008). By contrast, subtilases are one of the major Ser protease families in plants, whereas trypsin- and chymotrypsin-like proteases are rather rare (van der Hoorn, 2008). This suggests that while SPIs with a defensive role against insects are more likely to have an affinity toward trypsin- or chymotrypsin, SPIs that interact with plant proteases are more likely to bind to subtilases or other predominant Ser protease classes, such as S9, S10, and S33. Subtilisin PIs could also play a role in plant defense against pathogens by inhibiting bacterial or fungal proteases. For example, PI-II inhibitors in tomato were shown to be upregulated after infection with Pseudomonas syringae (Pautot et al., 1991). However, in our field experiments, we did not observe any evidence of higher susceptibility to pathogen attack in SPI-silenced plants.
As a consequence of the differences in substrate specificity, we expected to see developmental phenotypes when silencing the subtilisin-specific SPI2a and SPI2b. Sin et al. (2006) demonstrated that S. americanum silenced in the expression of PIN2a and PIN2b aborted around 80% of its seeds. Surprisingly, we found this effect to be very small in S. nigrum. A reason for this difference might be that the authors used a fragment of PIN2a for their silencing construct, while we used SPI2b to silence both SPI2a and SPI2b. However, when we compared the RNA gel blots presented in their publication to the transcript levels in our plants, it was obvious that we achieved similar levels of silencing for both genes. Alternatively, SPI2c may have compensated for the silencing effect, assuming that it is not present in S. americanum. However, the triple-silenced line irSPI2a+b+c did not show an increased abortion of seeds. To investigate the presence of SPI2c-like PIs in S. americanum, we compared its trypsin PI profile to those of S. nigrum and four other species of black nightshades (see Supplemental Figure 8 online). SPI2c-like bands were found in all investigated species, which rules out the possibility of deficiencies in the potential for compensation in S. americanum. Moreover, we could not observe any other PI-dependent developmental phenotype that had been reported for other species. There was no significant difference in flower diameter (Sin et al., 2006), number of reproductive units (Zavala et al., 2004a), plant growth (Zavala et al., 2004a; Xie et al., 2007), or trichome morphology and density (Liu et al., 2006; Luo et al., 2009). As a consequence, we infer that the developmental importance of SPI2a and SPI2b differs between species. Without knowing the exact mechanism by which PIs affect seed development in S. americanum, it will be difficult to explain these interspecific differences. Moreover, the observed subtle reduction in seed viability in S. nigrum would require further intensive experimentation to be validated as a true effect and not an artifact of transformation or other experimental procedures.
The silencing of SPI1 did not result in any phenotype except for the transient effect on S. littoralis performance, and its function in S. nigrum remains unclear. However, although often described to be wound induced, the evidence for PI-I to be an effective defense against herbivores is rather limited (Jongsma and Bolter, 1997; Mosolov and Valueva, 2005; Dunse et al., 2010b). Perhaps its higher specificity for chymotrypsin and subtilisin makes it effective against particular insect taxa or pathogens, which were not yet investigated and which did also not occur in our field studies. We found that SPI1 is preferentially expressed in flowers but also in vascular tissues, which suggests a defensive function against phloem-feeding insects or another unknown function in these tissues similar to that of SPI2a. Beuning et al. (1994) mentioned that PI-I is an ancient gene family, present in monocots and bacteria, fungi, and leech, which displays a lack of hypervariability at the active site (Functional divergence ratios at the active site did not differ significantly from 1 or were <1.) This unexpected lack of positive Darwinian selection questions the interpretation of PI-I inhibitors as being purely defense related, which would leave their putative primary function still to be discovered.
To summarize, we have shown that the SPIs of S. nigrum exhibit a certain degree of differentiation but also considerable functional overlap. The highly abundant SPI2c displays typical characteristics of a defense-related gene. The other two, SPI2a and SPI2b, show clear defensive properties but also some features that are difficult to associate with defense against herbivores. Despite their sequence similarity, they differ in their elicitation patterns and perhaps they represent an early stage of functional differentiation. Their specificity for subtilisin suggests an interaction with unknown proteases from either insects, pathogens, or the plant itself, and the identification of these target proteases will be a challenging task for future research. The different responses of the herbivores exemplified how dynamic the interaction of PIs with insect proteases can be. Given that a coevolutionary process involving several organisms creates a variety of different outcomes, this is to be expected. Such complexity underscores the challenges that the engineering of transgenic crops expressing PIs will face. Single SPIs might be effective against particular herbivore species but might be quickly overcome. Expressing more than one PI increases the chances of inhibiting insect target proteases but also increases the risk of eliciting compensatory feeding responses or of other unintended effects on plant growth and development. The likely synergisms with other defense chemicals are important to counter adaptive responses sustainably, but they also greatly complicate the engineering and application in agricultural crops. Although plant PIs have been studied for such a long time, this study shows that we have just scratched the surface of our understanding of their function and regulation. We still know little about how and to what purposes plants fine-tune PI expression and activity.
METHODS
Plant Material and Growth
All experiments were conducted with the Solanum nigrum inbred line Sn30 (Schmidt et al., 2004). Additionally, we used Solanum spp from the seed bank at IPK Gatersleben, Germany, for the comparison of PI profiles presented in Supplemental Figure 8 online. Seeds were sterilized and incubated overnight in 1 M KNO3 at 4°C before they were germinated on Gamborg B5 plant agar in Petri dishes, as described by Schmidt et al. (2004). After 10 d, seedlings were transferred to Teku trays containing a peat-based substrate (Tonsubstrat; Klasmann) and kept in growth chambers for another 10 d (16 h light/8 h dark, 26°C, 155 μmol m−2 s−1 PAR at shelf height). The plants were then transferred to 9 × 9 × 9.5-cm or 1-liter pots containing the same substrate and put in the glasshouse of the institute (16 h light, supplemental lighting by Philips Master Sun-T PIA Agro 400 W and 600 W sodium lights, 23 to 25°C, and 45 to 55% relative humidity; 8 h dark, 19 to 23°C and 45 to 55% relative humidity). The plants were automatically watered daily with 0.5 g/L of a combination fertilizer containing phosphate, potassium, and magnesium (Euflor) and 0.5 g/L Ca(NO3)2. The seedlings and plants used for VIGS were grown in climate chambers according to Hartl et al. (2008). For field experiments in Germany, plants were germinated and kept in Teku trays as described above, acclimatized for 3 to 5 d outdoors at the institute grounds and then planted at the field site 21 to 24 d after sowing. For the experiments at the field station in Utah, seeds were incubated in 1 M KNO3 as described above, germinated in Jiffy pots (Alwaysgrows Greenhouse Supplies), and kept in a shade house. After 3 weeks. the plants were transferred to 2-liter pots and hardened by reducing the shade until they were exposed to full sunlight.
The release of transformed plants at the field site near Dornburg, Germany was conducted in compliance with European Union and German regulations as administered by the Thüringer Landesverwaltungsamt and the Thüringer Landesamt für Lebensmittelsicherheit und Verbraucherschutz (release application nos. 6786-01-0156 and 6786-01-0187 [irSPI2a+b] and 6786–01-0189 [irSPI1/2ab]). The release of transgenic plants at the Lytle Ranch Preserve, Santa Clara, UT, was conducted under APHIS notification numbers 06-242-01r-a1 and 06-242-01r-a2.
In both locations, all plants were monitored daily during the whole experiment, and all flower buds of each genotype were removed before opening. After the experiments, the plants were harvested, including roots, and autoclaved or incinerated.
Plant Treatments
To mimic herbivore feeding, we punctured the youngest fully developed leaf with a fabric pattern wheel, creating four rows on each side of the midvein. Immediately after wounding, we applied 30 μL of Manduca sexta regurgitate (diluted 1:5 with deionized water) with a pipette. The regurgitate was collected from third to fourth instar larvae feeding on S. nigrum wild-type plants and centrifuged after collection to remove cell debris.
MeJA in lanolin paste was applied with a spatula either to the stem at the first internode (150 μg in 20 μL; Figures 1B, 7, and 8) or adaxially across the base of the leaf lamina (75 μg in 10 μL; Figures 1A, 1D, 2, and 4). Pure lanolin served as a control. To assess a possible response of SPIs to ethylene and salicylic acid, we applied 50 μL of the ethylene releasing compound ethephone (50 μM in 0.05% Tween 20) or 1 mM salicylic acid to leaves after mechanical wounding, as described above. All the chemicals used for elicitations were purchased from Sigma-Aldrich.
Treatments were performed between 10 and 11 am. The leaves were harvested without the midrib and, as all other tissues, frozen immediately in liquid N2. For all experiments, 4- to 5-week-old plants were used, except for the comparison of tissue specificity (Figures 1B and 5B). In this experiment, we harvested young leaves (top leaves of the primary shoot axis and of the two oldest side branches) and mature leaves (leaves at nodes 5 to 7 counted from the cotyledonary node) 5 weeks after sowing and 2 d after treatment with MeJA in lanolin paste or pure lanolin at the stem. After 11 weeks, another set of plants was treated with MeJA or lanolin and 2 d later mature buds, open flowers, and fruits were harvested. For stems, we selected a medium-aged portion spanning two internodes at the main branch. Tissues from nine plants per treatment and time point were collected, and the material from three plants was pooled to obtain three biological replicates. All other replicate numbers, as mentioned throughout the text, represent individual plants, separate for each time point or treatment.
Cloning of SPI Genes and Generation of Transgenic Lines
We used primers designed on sequences from other solanaceous species to clone fragments of Sn SPI1a, Sn SPI1b, Sn SPI2a, and Sn SPI2c by standard PCR and 3′ rapid amplification of cDNA ends (Scotto-Lavino et al., 2007) from cDNA and gDNA (all primer sequences are listed in Supplemental Table 4 online). A fragment of SPI2b was cloned into the vector pSOL3, described by Bubner et al. (2006), to generate the inverted repeat construct pSOL3PIS for silencing SPI2a and SPI2b (irSPI2a+b). Similarly, we combined fragments of SPI1a and SPI2b to form the plasmid pSOL3PIN12, silencing SPI1, SPI2a, and SPI2b simultaneously (irSPI1/2a+b), and fragments of SPI2a and SPI2c to form the plasmid pSOL8PIN2abc silencing all three SPI2 genes (irSPI2a+b+c). Agrobacterium tumefaciens–mediated plant transformation was performed as described (Krügel et al., 2002; Schmidt et al., 2004). T1 plants were screened on hygromycin-containing media, and homozygous lines were selected in T2, based on segregation analysis. For each construct, we characterized two independently transformed, homozygous lines, which harbored a single transgenic insert (as determined by DNA gel blot) and displayed reduced transcript levels (irSPI2a+b: line 1, S03-111-3; line 2, S03-114-1; irSPI1/2ab: line 1, S06-336-8; line 2, S06-356-10; irSPI2a+b+c: line 1, S09-29-6; line 2, S09-31-2).
DNA Gel Blots
We isolated genomic DNA from S. nigrum seedlings and leaves using a modified CTAB procedure (Bubner et al., 2004). The gDNA was digested with BamHI, EcoRI, and EcoRV to assess the native SPI copy numbers and with EcoRV to determine the number of transgenic inserts. The plasmid pSOL3PIS was digested with XhoI and used as a positive control on the blot containing the transgenic lines. We separated 30 μg DNA by 0.8% agarose gel electrophoresis and blotted it onto a nylon membrane (Genescreen Plus; Perkin-Elmer) according to Brown (1999). The blots were hybridized with 32P-labeled probes (Rediprime II kit; GE Healthcare) coding for the hygromycin resistance gene hptII or specifically for SPI1, SPI2a, and SPI2b (Brown, 1993; for primers used to generate probes, see Supplemental Table 4 online). The specificity of the three SPI probes was determined by hybridization to slot blots of three plasmids containing corresponding regions of the three genes (Brown, 1999; Halitschke and Baldwin, 2003; see Supplemental Figure 1 online).
VIGS
We amplified a fragment of Sn SPI2c (for primers, see Supplemental Table 4 online) and ligated it into the vector pYL156 (Liu et al., 2002) to obtain the vector pTRVPIN2c (= vSPI2c). Agrobacterium cultivation, vacuum infiltration, and plant cultivation were done as described previously (Hartl et al., 2008).
Quantitative Real-Time PCR
Total RNA was extracted from flash-frozen and ground plant material following the TRIzol (Invitrogen) protocol. Reverse transcription of 500 ng of total RNA was performed using SuperScript II reverse transcriptase (Invitrogen) and a poly-T primer to obtain cDNA. Twenty nanograms of cDNA were applied to 20 μL SYBR Green reactions (qPCR core kit for SYBR Green I; Eurogentec), which were run on an ABI PRISM 7700 sequence detection system (Applied Biosystems; cycler conditions: 10 min at 95°C, 40 cycles of 30 s at 95°C, and 30 s at 60°C). Each plate was run with a standard curve, no template control, and pools of RNA samples to check for DNA contamination. Primers were designed using Primer 3 v.0.4.0 (Rozen and Skaletsky, 2000), checked for specificity using a melting curve analysis (ABI PRISM 7700 dissociation curve software), and are summarized in Supplemental Table 4 online. For data analysis, we calculated relative transcript abundance by efficiency correcting for each primer pair and normalizing to the S. nigrum elongation factor EF1α according to Pfaffl et al. (2002).
PI Activity Assays
The samples were ground in liquid N2 and extracted with 2 mL cold extraction buffer (Jongsma et al., 1993) per gram of plant tissue by vortexing for 5 min. After repeated centrifugation and transfer of the supernatant, we determined the total protein concentration with the Bradford method (Protein Assay; Bio-Rad) using BSA (Sigma-Aldrich) as a standard. SPI activity was determined by a radial immunodiffusion assay (Jongsma et al., 1993; van Dam et al., 2001) using bovine trypsin, bovine α-chymotrypsin type II, and Subtilisin Carlsberg type VIII (all from Sigma-Aldrich). If samples from the same type of tissue were compared, the concentration of active PIs was expressed relative to milligram of total protein, if different tissues were compared relative to grams of fresh mass.
Native PAGE and GXCPs
The same extraction procedure as for the PI activity assays was used for native PAGE. We loaded equal amounts (150 μg) of total protein on 12% native vertical slab gels (Hoefer) in a discontinuous buffer system, run at 30 mA constant current. After electrophoresis, the gels were processed for activity visualization using the GXCP method (Pichare and Kachole, 1994). We washed the gels 2 × 15 min in 0.1 M Tris-HCl buffer, pH 7.8, followed by incubation in 0.1 mg/mL bovine trypsin in the same buffer for 15 min at room temperature on a rotary shaker. The gels were subsequently washed 3 × 10 min in the same buffer and then placed on x-ray film, coated with gelatin. We repeated the exposures with new film and increasing exposure times from 2 to 20 min. After exposure, the films were immediately rinsed in warm tap water until the hydrolyzed gelatin was washed off and bands appeared as unhydrolyzed gelatin against the background. We photographed the dried films under a strong uniform light source, with bands reflecting light differently than the remaining area. The pictures were converted to grayscale and inverted, and contrast and brightness were adjusted equally for the whole picture using Adobe Photoshop CS.
SPI2c Purification and MS/MS de Novo Sequencing
For the isolation of SPI2c, we collected 50 g of flowers from wild-type plants, froze and ground them in liquid N2, and extracted them with 250 mL PI extraction buffer by stirring for 1 h on ice. After repeated centrifugation (10,000g, 30 min) to clear the extract from cell debris, we incubated the supernatant for 30 min at 60°C in a water bath and centrifuged again to precipitate the denatured proteins. The supernatant was dialyzed against deionized water at 4°C overnight (Spectra/Por 7, 1 kD MWCO; Spectrum Europe) and then freeze-dried. We took up the remaining protein in 10 mL of deionized water and ultrafiltrated the solution through centrifugation columns with a 30 kD MWCO (Vivaspin 15; Sartorius). The filtrate and the concentrate were tested for TPI activity and analyzed using native PAGE followed by GXCP. Since the filtrate contained the unknown bands and showed high activity and less contamination by other proteins, we continued with this fraction. After freeze-drying, we dissolved the sample in 150 mM KCl and 10 mM Tris-HCl buffer, pH 8, incubated it with trypsin affinity agarose (Sigma-Aldrich) on a rotary shaker for 30 min at room temperature, and then loaded the slurry on a gravity column. After washing with 10 volumes of incubation buffer, we eluted with 5 volumes of 7 M urea, pH 3, and neutralized the eluate immediately with 1 M Tris-HCl, pH 8. The eluate was again dialyzed, freeze dried, dissolved in deionized water, and concentrated in a vacuum concentrator (Eppendorf) before electrophoresis. The sample was loaded together with aliquots of the purification steps on a 12% discontinuous native PAGE. The gel was stained with Coomassie blue (BioSafe Coomassie; Bio-Rad) except for one lane containing raw extract that was subjected to GXCP (Figure 3). The stained gel was dried on filter paper and sent for MS/MS de novo sequencing.
The sample preparation for electrospray ionization (ESI)-MS and the measurements were conducted by OMX (GmbH) using the following protocol. All organic solvents and chemicals used for sample preparation and ESI-MS were of analytical grade. Formic acid was purchased from Baker. DTT and iodoacetamide were obtained from Sigma-Aldrich and trypsin (NB sequencing grade modified trypsin, catalog no. 37283) from Serva.
An aliquot of each gel spot was crushed into small particles and destained using 50 mM ammonium bicarbonate in 50% acetonitrile (ACN). The samples were reduced with 10 mM DTT for 45 min at 56°C, and SH groups were subsequently alkylated with 55 mM iodoacetamide. Gel particles were dehydrated in ACN, swelled in digestion buffer containing 12.5 ng/μL trypsin in 50 mM ammonium bicarbonate, and incubated for 2 h at 50°C. Peptides were extracted into the digestion buffer during the incubation time. The supernatant was collected, acidified with a final concentration of 1% formic acid, and stored at 8°C for 24 h maximum.
Static nanoESI-MS was performed on a Q-TOF Premier tandem mass spectrometer (Waters) as described before with minor modifications (Granvogl et al., 2007). Peptide MS spectra were scanned in a mass range of 400 to 1500 m/z. Corresponding data were collected at a scan rate of 2 s. For external calibration, the fragmentation pattern from 500 fmol/μL [Glu1]-Fibrinopeptide B (Sigma-Aldrich) in 50% ACN and 0.1% formic acid was used. The signal at 421.759, which resulted from autolysis of trypsin, was used for internal calibration of MS spectra and to monitor enzyme activity. Fragment ion spectra were recorded at a scan time of 3 s for at least 1 min or until a fragmentation pattern appeared that was adequate for de novo sequencing of peptides. Manual interpretation of MS/MS spectra was assisted by signal deconvolution using the MaxEnt3 algorithm of the MassLynx 4.1 software.
Herbivore Performance in the Field
The experiments in the United States were performed on an irrigated field plot at the field station in the Lytle Ranch Preserve, Utah. We randomly planted 20 pairs of wild-type plants and transgenic plants for all three lines (60 pairs in total) and estimated total leaf damage separately for each feeding guild on May 5, 14, and 23 and June 1, 2010. Estimations were performed without knowing the identity of the scored individual. The damage data were analyzed by paired t tests.
The field site in Germany was situated near Dornburg on a former agricultural field. We planted 20 pairs of wild-type plants and irSPI2a+b line 1 in each of the two experiments (June 6 and July 15, 2005). The plants were watered when necessary, and the flower buds were removed on all genotypes before opening. We quantified leaf damage as described by Schmidt and Baldwin (2006b). To summarize shortly, the herbivore damage of each individual leaf was categorized according to damage classes, and then a mean damage level was calculated for each plant. After arcsine transformation, the data were tested with a paired t test.
Herbivore Performance in the Glasshouse
For M. sexta on stably silenced plants (Figure 7), M. sexta eggs were obtained from our in-house culture. Freshly hatched larvae were reared on uninduced leaf tissue, separated by genotypes in boxes (three boxes per genotype). The leaf material was exchanged daily, and 2 d after hatching, 30 larvae per genotype and treatment were weighed and separated in individual containers containing two leaf discs (25 mm), from either untreated or MeJA-induced plants (treated on day of hatching), on moist filter paper. Four leaf discs were cut from a single fully expanded leaf, supplying two caterpillars with material from one plant (15 plants per genotype and treatment). The boxes were randomized and kept at 23 to 25°C in a shaded area in the glasshouse. Leaf discs were exchanged daily and the larval mass was recorded on days 2, 4, 6, 8, and 10. The leaf discs were scanned and the consumed leaf area determined using ImageJ 1.38 (http://rsb.info.nih.gov/ij/index.html). As a measure of consumption, we related the consumed leaf area to larval mass at day 2. To estimate food conversion efficiency the mass gain between days 2 and 4 was related to the leaf area consumed in this period. From each silencing construct, only one line was included to make the experiment more manageable. The experiment was repeated three times with different lines and in some cases also allowing larvae to feed directly on the plants, with similar results. The statistical analysis (repeated measures ANOVA) is explained in detail in Supplemental Tables 1 and 2 online.
For M. sexta on VIGS plants (see Supplemental Figure 7 online), the experiment was performed as above but with 30 caterpillars per genotype and VIGS vector. All larvae were transferred to MeJA-induced material in individual boxes on day 4. The experiment was repeated two times with similar results.
For Spodoptera exigua performance on stably silenced lines (Figure 8D), neonates, freshly hatched from eggs supplied by the Plant Protection Center of Bayer, were reared communally in boxes on uninduced leaf tissue at room temperature, as described above. After 3 d, we transferred 21 larvae per genotype and treatment to individual containers (three larvae per plant replicate). From day 6 on, half of the caterpillars were fed on MeJA-induced leaf material, and after 9 d they reached a size that allowed weighing. irSPI2a+b line 2 and irSPI1/2ab line 2 were not included in the setup with uninduced material to make the experiment more manageable. Two repetitions of the experiment had to be aborted prematurely because of high mortality, in spite of a higher replicate number of 30 individuals. For statistical analysis, see Supplemental Table 3 online.
The experiments with Spodoptera littoralis on stably silenced lines were performed exactly as with M. sexta, with 28 larvae per genotype for control plants and 32 larvae per genotype for MeJA-treated plants. For statistics, see Supplemental Table 2 online.
For S. littoralis performance on VIGS-silenced plants (see Supplemental Figures 7B to 7E online), eggs were obtained from the Plant Protection Center of Bayer. The experiments were performed as with M. sexta, transferring 12 larvae per genotype and vector to individual boxes on day 6, but only supplying them with uninduced tissue throughout the experiment. The experiment was repeated two times with similar results. To measure the consumption early in the larval development, we repeated the experiment with larvae feeding only on wild-type plants inoculated with CV or vSPI2c (see Supplemental Figures 7C to 7E online). The larvae were weighed after hatching and then transferred to individual containers containing leaf discs. Larval mass was recorded on alternate days using an analytical balance with a readability of 0.01 mg, and the consumed leaf area was determined as described above.
Statistics
The software package SPSS 17.0 for Windows was used for all statistical analyses where appropriate. For the linear mixed-effects model fitted to the M. sexta performance data (see Supplemental Figure 7A online), we used R 2.9.0 in combination with the package lme4 (R Development Core Team; http://www.R-project.org). If necessary, data were transformed to meet the assumption of homogeneity of variances, as specified for each test. Further details of the tests and applied models are described in the figure legends and in the supplemental material.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GU133368 (Sn SPI1a), GU133369 (Sn SPI1b), GU133370 (Sn SPI2a, gDNA), GU133371 (Sn SPI2a mRNA), GU133372 (Sn SPI2c-R2), and GU133373 (Sn SPI2c-R3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Copy Number of SPI Genes in S. nigrum.
Supplemental Figure 2. Number of T-DNA Inserts in the Genomes of the Transgenic Lines with Constructs irSPI2a+b and irSPI1/2a+b.
Supplemental Figure 3. Plant Height, Flower Diameter, and Number of Viable and Nonviable Seeds in Wild-Type and SPI-Silenced S. nigrum Plants.
Supplemental Figure 4. Multiple Sequence Alignment (ClustalW) of Translated cDNA Sequences from All Three SPI2 Genes Containing Two Sequence Repeats.
Supplemental Figure 5. Characterization of Transgenic Lines with Construct irSPI2a+b+c.
Supplemental Figure 6. Natural Herbivory in Utah and Germany.
Supplemental Figure 7. Herbivore Performance after VIGS of SPI2c.
Supplemental Figure 8. TPI Profiles of Different Solanum spp Using GXCP.
Supplemental Table 1. Statistical Analysis for Data in Figure 7A.
Supplemental Table 2. Statistical Analysis for Data in Figure 8A.
Supplemental Table 3. Statistical Analysis for Data in Figure 8D.
Supplemental Table 4. Primer Sequences.
Supplementary Material
Acknowledgments
We thank H. Merker, A. Prehl, S. Lindermann, A. Hackett, A. Rauch, and T. Mentrup for assistance in the lab and in the field, A. van Doorn for countless helpful discussions, K. Gase, A. Wissgott, W. Kröber, and S. Kutschbach for the design and generation of silencing constructs and for plant transformation, D. Kessler, A. Berg, and D. Rosenberger for insect culture, D. Kessler, C. Diezel, and the 2010 Utah field crew for performing the field herbivory assays, M. Kallenbach for help with volatile analysis, J. Schumacher for statistical advice, J. Kellmann and K. Groten for managing the legal and practical obstacles to conducting field experiments with transgenic plants in Germany, Brigham Young University for the use of the Lytle Ranch Preserve field station, German and U.S. authorities for constructive regulatory oversight, the Max Planck Society for funding, and four anonymous reviewers for their helpful comments and suggestions.
References
- Abdeen A., Virgós A., Olivella E., Villanueva J., Avilés X., Gabarra R., Prat S. (2005). Multiple insect resistance in transgenic tomato plants over-expressing two families of plant proteinase inhibitors. Plant Mol. Biol. 57: 189–202 [DOI] [PubMed] [Google Scholar]
- Atkinson A.H., Heath R.L., Simpson R.J., Clarke A.E., Anderson M.A. (1993). Proteinase inhibitors in Nicotiana alata stigmas are derived from a precursor protein which is processed into five homologous inhibitors. Plant Cell 5: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayés A., Comellas-Bigler M., Rodríguez de la Vega M., Maskos K., Bode W., Aviles F.X., Jongsma M.A., Beekwilder J., Vendrell J. (2005). Structural basis of the resistance of an insect carboxypeptidase to plant protease inhibitors. Proc. Natl. Acad. Sci. USA 102: 16602–16607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayés A., de la Vega M.R., Vendrell J., Aviles F.X., Jongsma M.A., Beekwilder J. (2006). Response of the digestive system of Helicoverpa zea to ingestion of potato carboxypeptidase inhibitor and characterization of an uninhibited carboxypeptidase B. Insect Biochem. Mol. Biol. 36: 654–664 [DOI] [PubMed] [Google Scholar]
- Beekwilder J., Schipper B., Bakker P., Bosch D., Jongsma M. (2000). Characterization of potato proteinase inhibitor II reactive site mutants. Eur. J. Biochem. 267: 1975–1984 [DOI] [PubMed] [Google Scholar]
- Beuning L.L., Spriggs T.W., Christeller J.T. (1994). Evolution of the proteinase inhibitor I family and apparent lack of hypervariability in the proteinase contact loop. J. Mol. Evol. 39: 644–654 [DOI] [PubMed] [Google Scholar]
- Bezzi S., Kessler D., Diezel C., Muck A., Anssour S., Baldwin I.T. (2010). Silencing NaTPI expression increases nectar germin, nectarins, and hydrogen peroxide levels and inhibits nectar removal from plants in nature. Plant Physiol. 152: 2232–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown D.P., Wilkinson H.S., Gatehouse J.A. (1997). Differentially regulated inhibitor-sensitive and insensitive protease genes from the phytophagous insect pest, Helicoverpa armigera, are members of complex multigene families. Insect Biochem. Mol. Biol. 27: 625–638 [DOI] [PubMed] [Google Scholar]
- Brioschi D., Nadalini L.D., Bengtson M.H., Sogayar M.C., Moura D.S., Silva-Filho M.C. (2007). General up regulation of Spodoptera frugiperda trypsins and chymotrypsins allows its adaptation to soybean proteinase inhibitor. Insect Biochem. Mol. Biol. 37: 1283–1290 [DOI] [PubMed] [Google Scholar]
- Brown E.S., Dewhurst C.F. (1975). Genus Spodoptera (Lepidoptera, Noctuidae) in Africa and Near East. Bull. Entomol. Res. 65: 221–226 [Google Scholar]
- Brown T. (1993). Hybridization analysis of DNA blots. Curr. Protoc. Mol. Biol. 21: 2.10.11–2.10.16 [DOI] [PubMed] [Google Scholar]
- Brown T. (1999). Southern blotting. Curr. Protoc. Mol. Biol. 68: 2.9.1–2.9.20 [DOI] [PubMed] [Google Scholar]
- Bubner B., Gase K., Baldwin I.T. (2004). Two-fold differences are the detection limit for determining transgene copy numbers in plants by real-time PCR. BMC Biotechnol. 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubner B., Gase K., Berger B., Link D., Baldwin I.T. (2006). Occurrence of tetraploidy in Nicotiana attenuata plants after Agrobacterium-mediated transformation is genotype specific but independent of polysomaty of explant tissue. Plant Cell Rep. 25: 668–675 [DOI] [PubMed] [Google Scholar]
- Christeller J.T. (2005). Evolutionary mechanisms acting on proteinase inhibitor variability. FEBS J. 272: 5710–5722 [DOI] [PubMed] [Google Scholar]
- Christou P., Capell T., Kohli A., Gatehouse J.A., Gatehouse A.M.R. (2006). Recent developments and future prospects in insect pest control in transgenic crops. Trends Plant Sci. 11: 302–308 [DOI] [PubMed] [Google Scholar]
- De Leo F., Bonade-Bottino M.A., Ceci L.R., Gallerani R., Jouanin L. (1998). Opposite effects on spodoptera littoralis larvae of high expression level of a trypsin proteinase inhibitor in transgenic plants. Plant Physiol. 118: 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leo F., Gallerani R. (2002). The mustard trypsin inhibitor 2 affects the fertility of Spodoptera littoralis larvae fed on transgenic plants. Insect Biochem. Mol. Biol. 32: 489–496 [DOI] [PubMed] [Google Scholar]
- Dunse K.M., Kaas Q., Guarino R.F., Barton P.A., Craik D.J., Anderson M.A. (2010a). Molecular basis for the resistance of an insect chymotrypsin to a potato type II proteinase inhibitor. Proc. Natl. Acad. Sci. USA 107: 15016–15021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunse K.M., Stevens J.A., Lay F.T., Gaspar Y.M., Heath R.L., Anderson M.A. (2010b). Coexpression of potato type I and II proteinase inhibitors gives cotton plants protection against insect damage in the field. Proc. Natl. Acad. Sci. USA 107: 15011–15015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds J.M., Chweya J.A. (1997). Black Nightshades. Solanum nigrum L. and Related Species. (Gatersleben, Germany: Institute of Plant Genetics and Crop Plant Research/Rome: International Plant Genetic Resources Institute; ). [Google Scholar]
- Gatehouse J.A. (2008). Biotechnological prospects for engineering insect-resistant plants. Plant Physiol. 146: 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C., Le Métayer M., Bonadé-Bottino M., Pham-Delègue M.-H., Jouanin L. (1998). High level of resistance to proteinase inhibitors may be conferred by proteolytic cleavage in beetle larvae. Insect Biochem. Mol. Biol. 28: 229–237 [DOI] [PubMed] [Google Scholar]
- Giri A.P., Harsulkar A.M., Deshpande V.V., Sainani M.N., Gupta V.S., Ranjekar P.K. (1998). Chickpea defensive proteinase inhibitors can be inactivated by podborer gut proteinases. Plant Physiol. 116: 393–401 [Google Scholar]
- Granvogl B., Gruber P., Eichacker L.A. (2007). Standardisation of rapid in-gel digestion by mass spectrometry. Proteomics 7: 642–654 [DOI] [PubMed] [Google Scholar]
- Green T.R., Ryan C.A. (1972). Wound-induced proteinase inhibitor in plant leaves: A possible defense mechanism against insects. Science 175: 776–777 [DOI] [PubMed] [Google Scholar]
- Halitschke R., Baldwin I.T. (2003). Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J. 36: 794–807 [DOI] [PubMed] [Google Scholar]
- Hartl M., Merker H., Schmidt D.D., Baldwin I.T. (2008). Optimized virus-induced gene silencing in Solanum nigrum reveals the defensive function of leucine aminopeptidase against herbivores and the shortcomings of empty vector controls. New Phytol. 179: 356–365 [DOI] [PubMed] [Google Scholar]
- Horn M., Patankar A.G., Zavala J.A., Wu J., Dolecková-Maresová L., Vujtechová M., Mares M., Baldwin I.T. (2005). Differential elicitation of two processing proteases controls the processing pattern of the trypsin proteinase inhibitor precursor in Nicotiana attenuata. Plant Physiol. 139: 375–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma M.A., Bakker P.L., Peters J., Bosch D., Stiekema W.J. (1995). Adaptation of Spodoptera exigua larvae to plant proteinase inhibitors by induction of gut proteinase activity insensitive to inhibition. Proc. Natl. Acad. Sci. USA 92: 8041–8045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma M.A., Bakker P.L., Stiekema W.J. (1993). Quantitative determination of serine proteinase inhibitor activity using a radial diffusion assay. Anal. Biochem. 212: 79–84 [DOI] [PubMed] [Google Scholar]
- Jongsma M.A., Bolter C. (1997). The adaptation of insects to plant protease inhibitors. J. Insect Physiol. 43: 885–895 [DOI] [PubMed] [Google Scholar]
- Kant M.R., Baldwin I.T. (2007). The ecogenetics and ecogenomics of plant-herbivore interactions: Rapid progress on a slippery road. Curr. Opin. Genet. Dev. 17: 519–524 [DOI] [PubMed] [Google Scholar]
- Kessler A., Baldwin I.T. (2001). Defensive function of herbivore-induced plant volatile emissions in nature. Science 291: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Kessler A., Halitschke R., Baldwin I.T. (2004). Silencing the jasmonate cascade: Induced plant defenses and insect populations. Science 305: 665–668 [DOI] [PubMed] [Google Scholar]
- Kong L., Ranganathan S. (2008). Tandem duplication, circular permutation, molecular adaptation: How Solanaceae resist pests via inhibitors. BMC Bioinformatics 9(suppl. 1): S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A., Pieterse C.M.J. (2008). Cross talk in defense signaling. Plant Physiol. 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krügel T., Lim M., Gase K., Halitschke R., Baldwin I.T. (2002). Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12: 177–183 [Google Scholar]
- Liu J., Xia K.-F., Zhu J.-C., Deng Y.-G., Huang X.-L., Hu B.-L., Xu X., Xu Z.-F. (2006). The nightshade proteinase inhibitor IIb gene is constitutively expressed in glandular trichomes. Plant Cell Physiol. 47: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Marathe R., Dinesh-Kumar S.P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Luo M., Wang Z.Y., Li H.P., Xia K.F., Cai Y.P., Xu Z.F. (2009). Overexpression of a weed (Solanum americanum) proteinase inhibitor in transgenic tobacco results in increased glandular trichome density and enhanced resistance to Helicoverpa armigera and Spodoptera litura. Int. J. Mol. Sci. 10: 1896–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M., Cambra I., Carrillo L., Diaz-Mendoza M., Diaz I. (2009). Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiol. 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins T., Oliveira L., Garcia P. (2005). Larval mortality factors of Spodoptera littoralis in the Azores. Biocontrol 50: 761–770 [Google Scholar]
- Mosolov V.V., Valueva T.A. (2005). Proteinase inhibitors and their function in plants: A review. Appl. Biochem. Microbiol. 41: 227–246 [PubMed] [Google Scholar]
- Mosolov V.V., Valueva T.A. (2008). Proteinase inhibitors in plant biotechnology: A review. Appl. Biochem. Microbiol. 44: 233–240 [PubMed] [Google Scholar]
- Pautot V., Holzer F.M., Walling L.L. (1991). Differential expression of tomato proteinase inhibitor I and II genes during bacterial pathogen invasion and wounding. Mol. Plant Microbe Interact. 4: 284–292 [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W., Horgan G.W., Dempfle L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe R.N., Ralph S.G., Külheim C., Jancsik S.I., Bohlmann J. (2009). Poplar defense against insects: genome analysis, full-length cDNA cloning, and transcriptome and protein analysis of the poplar Kunitz-type protease inhibitor family. New Phytol. 184: 865–884 [DOI] [PubMed] [Google Scholar]
- Pichare M.M., Kachole M.S. (1994). Detection of electrophoretically separated protease inhibitors using X-ray film. J. Biochem. Biophys. Methods 28: 215–224 [DOI] [PubMed] [Google Scholar]
- Rawlings N.D., Morton F.R., Kok C.Y., Kong J., Barrett A.J. (2008). MEROPS: The peptidase database. Nucleic Acids Res. 36(Database issue): D320–D325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H.J. (2000). Primer3 on the WWW for general users and for biologist programmers. Bioinformatics Methods and Protocols: Methods in Molecular Biology, Misener S., Krawetz S.A., (Totowa, NJ: Humana Press; ), pp. 365–386 [DOI] [PubMed] [Google Scholar]
- Ryan C.A. (1973). Proteolytic enzymes and their inhibitors in plants. Annu. Rev. Plant Physiol. 24: 173–196 [Google Scholar]
- Ryan C.A. (1990). Protease inhibitors in plants - Genes for improving defenses against insects and pathogens. Annu. Rev. Phytopathol. 28: 425–449 [Google Scholar]
- Schmidt D.D., Baldwin I.T. (2006a). Transcriptional responses of Solanum nigrum to methyl jasmonate and competition: A glasshouse and field study. Funct. Ecol. 20: 500–508 [Google Scholar]
- Schmidt D.D., Kessler A., Kessler D., Schmidt S., Lim M., Gase K., Baldwin I.T. (2004). Solanum nigrum: a model ecological expression system and its tools. Mol. Ecol. 13: 981–995 [DOI] [PubMed] [Google Scholar]
- Schmidt S., Baldwin I.T. (2006b). Systemin in Solanum nigrum. The tomato-homologous polypeptide does not mediate direct defense responses. Plant Physiol. 142: 1751–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto-Lavino E., Du G., Frohman M.A. (2007). 3′ End cDNA amplification using classic RACE. Nat. Protoc. 1: 2742–2745 [DOI] [PubMed] [Google Scholar]
- Sin S.-F., Yeung E.C., Chye M.-L. (2006). Downregulation of Solanum americanum genes encoding proteinase inhibitor II causes defective seed development. Plant J. 45: 58–70 [DOI] [PubMed] [Google Scholar]
- Steppuhn A., Baldwin I.T. (2007). Resistance management in a native plant: Nicotine prevents herbivores from compensating for plant protease inhibitors. Ecol. Lett. 10: 499–511 [DOI] [PubMed] [Google Scholar]
- Steppuhn A., Schuman M.C., Baldwin I.T. (2008). Silencing jasmonate signalling and jasmonate-mediated defences reveals different survival strategies between two Nicotiana attenuata accessions. Mol. Ecol. 17: 3717–3732 [DOI] [PubMed] [Google Scholar]
- Tamhane V.A., Giri A.P., Kumar P., Gupta V.S. (2009). Spatial and temporal expression patterns of diverse Pin-II proteinase inhibitor genes in Capsicum annuum Linn. Gene 442: 88–98 [DOI] [PubMed] [Google Scholar]
- Tamhane V.A., Giri A.P., Sainani M.N., Gupta V.S. (2007). Diverse forms of Pin-II family proteinase inhibitors from Capsicum annuum adversely affect the growth and development of Helicoverpa armigera. Gene 403: 29–38 [DOI] [PubMed] [Google Scholar]
- van Dam N.M., Horn M., Mares M., Baldwin I.T. (2001). Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J. Chem. Ecol. 27: 547–568 [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A.L. (2008). Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 59: 191–223 [DOI] [PubMed] [Google Scholar]
- Winterer J., Bergelson J. (2001). Diamondback moth compensatory consumption of protease inhibitor-transformed plants. Mol. Ecol. 10: 1069–1074 [DOI] [PubMed] [Google Scholar]
- Xie J., Ouyang X.Z., Xia K.F., Huang Y.F., Pan W.B., Cai Y.P., Xu X.P., Li B.J., Xu Z.F. (2007). Chloroplast-like organelles were found in enucleate sieve elements of transgenic plants overexpressing a proteinase inhibitor. Biosci. Biotechnol. Biochem. 71: 2759–2765 [DOI] [PubMed] [Google Scholar]
- Xu Z.-F., Qi W.-Q., Ouyang X.-Z., Yeung E., Chye M.-L. (2001). A proteinase inhibitor II of Solanum americanum is expressed in phloem. Plant Mol. Biol. 47: 727–738 [DOI] [PubMed] [Google Scholar]
- Zavala J.A., Giri A.P., Jongsma M.A., Baldwin I.T. (2008). Digestive duet: Midgut digestive proteinases of Manduca sexta ingesting Nicotiana attenuata with manipulated trypsin proteinase inhibitor expression. PLoS One 3: e2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala J.A., Patankar A.G., Gase K., Baldwin I.T. (2004a). Constitutive and inducible trypsin proteinase inhibitor production incurs large fitness costs in Nicotiana attenuata. Proc. Natl. Acad. Sci. USA 101: 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala J.A., Patankar A.G., Gase K., Hui D.Q., Baldwin I.T. (2004b). Manipulation of endogenous trypsin proteinase inhibitor production in Nicotiana attenuata demonstrates their function as antiherbivore defenses. Plant Physiol. 134: 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu-Salzman K., Koiwa H., Salzman R.A., Shade R.E., Ahn J.-E. (2003). Cowpea bruchid Callosobruchus maculatus uses a three-component strategy to overcome a plant defensive cysteine protease inhibitor. Insect Mol. Biol. 12: 135–145 [DOI] [PubMed] [Google Scholar]
- Zhu-Salzman K., Luthe D.S., Felton G.W. (2008). Arthropod-inducible proteins: Broad spectrum defenses against multiple herbivores. Plant Physiol. 146: 852–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.