The interaction of the immune receptor Rx with RanGAP2 is required for stability and balanced partitioning of Rx between the cytoplasm and nucleus, which is essential for activation of the immune response against Potato virus X.
Abstract
The potato (Solanum tuberosum) nucleotide binding–leucine-rich repeat immune receptor Rx confers resistance to Potato virus X (PVX) and requires Ran GTPase-activating protein 2 (RanGAP2) for effective immune signaling. Although Rx does not contain a discernible nuclear localization signal, the protein localizes to both the cytoplasm and nucleus in Nicotiana benthamiana. Transient coexpression of Rx and cytoplasmically localized RanGAP2 sequesters Rx in the cytoplasm. This relocation of the immune receptor appeared to be mediated by the physical interaction between Rx and RanGAP2 and was independent of the concomitant increased GAP activity. Coexpression with RanGAP2 also potentiates Rx-mediated immune signaling, leading to a hypersensitive response (HR) and enhanced resistance to PVX. Besides sequestration, RanGAP2 also stabilizes Rx, a process that likely contributes to enhanced defense signaling. Strikingly, coexpression of Rx with the Rx-interacting WPP domain of RanGAP2 fused to a nuclear localization signal leads to hyperaccumulation of both the WPP domain and Rx in the nucleus. As a consequence, both Rx-mediated resistance to PVX and the HR induced by auto-active Rx mutants are significantly suppressed. These data show that a balanced nucleocytoplasmic partitioning of Rx is required for proper regulation of defense signaling. Furthermore, our data indicate that RanGAP2 regulates this partitioning by serving as a cytoplasmic retention factor for Rx.
INTRODUCTION
Innate immunity of plants protects them against pathogens and shares several features with animal innate immunity (Ausubel, 2005). In plants, immune receptors are encoded by resistance genes, of which the majority are intracellular nucleotide binding–leucine-rich repeat (NB-LRR) proteins containing an NB domain and LRRs (McHale et al., 2006; Tameling and Takken, 2007; Caplan et al., 2008). Upon perception of specific pathogen-derived virulence proteins, so-called effectors, these immune receptors mediate the induction of a strong defense response. This response is referred to as effector-triggered immunity (ETI) and is often associated with a hypersensitive response (HR), a type of programmed cell death at the infection site (Jones and Dangl, 2006). Plant NB-LRRs share structural and functional similarities with the animal nucleotide binding leucine-rich repeat (NLR) family of innate immune receptors (Ye and Ting, 2008) and the metazoan apoptosis factors Apaf-1 and CED-4, which are all classified as STAND (signal transduction ATPases with numerous domains) proteins (Leipe et al., 2004). The NB domain, also referred to as the NB-ARC domain (van der Biezen and Jones, 1998b) consists of the NB, ARC1, and ARC2 subdomains (Albrecht and Takken, 2006; Takken et al., 2006), and together these form an ATPase domain that is proposed to act as a molecular switch, regulating the signaling activity of the receptor by nucleotide-dependent conformational changes (Moffett et al., 2002; Tameling et al., 2002, 2006; Collier and Moffett, 2009; Takken and Tameling, 2009). The C-terminal LRR domain provides recognitional specificity and comprises both a positive and a negative regulatory function (McHale et al., 2006; Collier and Moffett, 2009; Takken and Tameling, 2009). The N-terminal domains of plant NB-LRRs are variable and comprise either a Toll and interleukin-1 receptor-like (TIR) domain or a domain that frequently contains coiled-coil (CC) motifs (Pan et al., 2000; Martin et al., 2003; McHale et al., 2006; Tameling and Takken, 2007). Although a signaling role has been proposed for the TIR domain in some NB-LRRs (Zhang et al., 2004; Michael Weaver et al., 2006; Swiderski et al., 2009), evidence that the CC domain can mediate downstream signaling in NB-LRRs is lacking. Recently it was shown that the NB subdomain can trigger defense signaling when expressed in the absence of the other domains, suggesting that the NB subdomain by itself is responsible for initiating downstream signaling (Rairdan et al., 2008; Collier and Moffett, 2009; Takken and Tameling, 2009).
For some plant NB-LRRs, perception of their cognate effectors is proposed to be mediated by direct binding of the effector (Jia et al., 2000; Deslandes et al., 2003; Dodds et al., 2006; Ueda et al., 2006; Ellis et al., 2007). Other NB-LRRs are thought to act as guards that sense perturbations caused by their cognate effectors of specific host proteins, called guardees, virulence targets, decoys, or baits (Van der Biezen and Jones, 1998a; van der Hoorn and Kamoun, 2008; Collier and Moffett, 2009), which are often bound to the N terminus of NB-LRRs (Collier and Moffett, 2009; Lukasik and Takken, 2009).
NB-LRR activation eventually results in a vast transcriptional reprogramming (Tao et al., 2003; Eulgem, 2005; Tsuda and Katagiri, 2010), but how this is initiated is unclear. The finding that the barley (Hordeum vulgare) NB-LRR protein MLA10 interacts with a WRKY transcriptional repressor in the nucleus upon its activation (Shen et al., 2007) and that the Arabidopsis thaliana NB-LRR RRS1-R is directly fused to a WRKY transcription factor domain (Deslandes et al., 2003) suggest that plant NB-LRRs can directly regulate transcription, perhaps similar to the human NLR protein CIITA (Wright and Ting, 2006; García and Parker, 2009). Previously, nuclear localization of NB-LRRs was not expected, as discernible canonical nuclear localization signals (NLSs) were generally not predicted in these proteins. Later, using more powerful prediction programs, it was found that various NB-LRRs do contain canonical NLSs (Shen and Schulze-Lefert, 2007; Caplan et al., 2008; Liu and Coaker, 2008). However, experimental proof that these motifs are functional has so far only been provided for RPS4 (Wirthmueller et al., 2007). Classical nuclear import involves binding of the NLS by importin-α, which together with other factors (e.g., importin-β, Ran, and Ran GTPase-activating protein [RanGAP]) mediates active transport of the protein through the nuclear pores into the nucleus (Lange et al., 2007; Meier, 2007; Stewart, 2007). To date, several NB-LRRs have been detected in the nucleus, of which indeed some are likely imported directly via importin-α. However, others, such as MLA10 and MLA1, are potentially imported by an alternative mechanism, as canonical NLS motifs are not detected in their sequences (Deslandes et al., 2003; Burch-Smith et al., 2007; Shen et al., 2007; Shen and Schulze-Lefert, 2007; Wirthmueller et al., 2007; Caplan et al., 2008; Liu and Coaker, 2008; Cheng et al., 2009). This is conceivable, as 43% of the nuclear proteins in yeast do not contain an NLS (Lange et al., 2007).
Potato (Solanum tuberosum) Rx is a CC-NB-LRR that confers durable resistance to most strains of Potato virus X (PVX) by recognition of its coat protein (CP) through the C-terminal part of its LRR domain (Bendahmane et al., 1995, 1999; García-Arenal and McDonald, 2003; Rairdan and Moffett, 2006). Interestingly, Rx is also able to detect the CP of other Potexviruses, and through a single amino acid substitution in the LRR it even recognizes a Carlavirus (Farnham and Baulcombe, 2006; Baurès et al., 2008; Candresse et al., 2010). The PVX resistance phenotype is also displayed in other Solanaceous species transgenic for Rx, such as Nicotiana benthamiana (Bendahmane et al., 1999; Candresse et al., 2010). Activation of Rx by CP suppresses virus replication early after infection without triggering an HR, a response referred to as extreme resistance (Tozzini et al., 1991; Kohm et al., 1993; Bendahmane et al., 1999). However, when CP is overexpressed or Rx function is weakened, an Rx-dependent HR is induced (Bendahmane et al., 1999; Sacco et al., 2007; Tameling and Baulcombe, 2007), indicating that Rx is not functionally different from other NB-LRRs. Indeed, Rx-triggered HR requires Hsp90 and SGT1, proteins that are also required for functionality of other NB-LRR proteins (Peart et al., 2002b; Lu et al., 2003; Shirasu, 2009).
In studies aimed at identifying proteins that are required for Rx function, a RanGAP was found to copurify with Rx from N. benthamiana (Sacco et al., 2007; Tameling and Baulcombe, 2007). Together with the small GTPase Ran, RanGAPs are essential for regulation of nucleocytoplasmic trafficking of macromolecules through the nuclear pores in eukaryotic cells (Meier, 2007; Stewart, 2007; Meier and Brkljacic, 2009). Ran hydrolyzes GTP with the aid of RanGAP in the cytoplasm (Bischoff et al., 1994), and upon binding to the Ran-specific nuclear transport factor NTF2, Ran-GDP is recycled back to the nucleus where Ran is reloaded with GTP (Quimby et al., 2000). In plants and animals, part of the cytoplasmic RanGAP pool is concentrated at the outside of the nuclear envelope (Görlich and Kutay, 1999; Rose and Meier, 2001; Pay et al., 2002). In plants, this is mediated by the plant-specific N-terminal tryptophan-proline-proline (WPP) domain of RanGAP that binds to anchor proteins at the nuclear envelope (Rose and Meier, 2001; Pay et al., 2002; Xu et al., 2007; Zhao et al., 2008). The WPP domain of RanGAP2 and the CC domain of Rx are required and sufficient for RanGAP2–Rx interaction (Sacco et al., 2007; Tameling and Baulcombe, 2007). RanGAP2 also binds to the nearly identical CC domain of the potato immune receptor GPA2 but does not bind to the more distantly related CC domain of the pepper (Capsicum annuum) immune receptor Bs2. Silencing of RanGAP2 revealed its requirement for Rx-mediated resistance to PVX but not for N-mediated resistance to Tobacco mosaic virus (Sacco et al., 2007; Tameling and Baulcombe, 2007). What role RanGAP2 plays in Rx signaling has remained elusive to date. Here, we report that, despite the lack of a discernible canonical NLS in Rx, this immune receptor localizes both to the cytoplasm and the nucleus. This was observed upon transient expression of Rx, followed by noninvasive confocal microscopy and was confirmed by stable expression of Rx driven by its native promoter and subsequent biochemical fractionation. Coexpression of Rx with RanGAP2, or domains thereof, dramatically changed the nucleocytoplasmic partitioning of Rx. This effect is likely achieved by physical sequestration of Rx by RanGAP2, as this requires the WPP domain and is independent of its GAP activity. In addition, silencing of RanGAP2 alone or both RanGAPs simultaneously affected the Rx partitioning in the opposite way. This indicates that RanGAP2 facilitates the nucleocytoplasmic partitioning of Rx by serving as a cytoplasmic retention factor. Coexpression of Rx with RanGAP2 potentiates the Rx-mediated defense response, and in addition to physical sequestration, it also results in stabilization of the protein, a process that likely contributes to this enhanced activity. Interestingly, coexpression of Rx with the RanGAP2-WPP domain fused to the SV40 NLS results in hyperaccumulation of both proteins in the nucleus and leads to a suppression of Rx signaling. Thus, the nucleocytoplasmic partitioning of Rx dictates its functionality and is facilitated by RanGAP2.
RESULTS
Rx Localizes Both to the Cytoplasm and the Nucleus
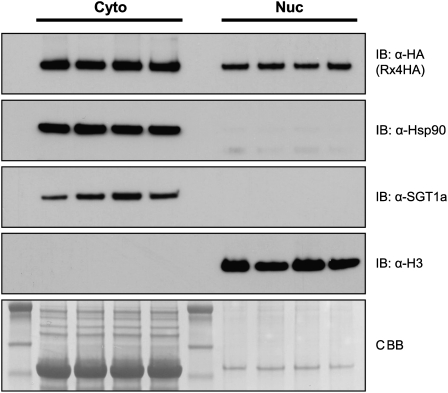
To study the subcellular localization of Rx in plants stably expressing the protein, nucleocytoplasmic fractionation studies were performed using transgenic N. benthamiana:Rx4HA showing extreme resistance to PVX (Lu et al., 2003; Tameling and Baulcombe, 2007). In these plants, expression of Rx, fused to four copies of the hemagglutinin (HA) epitope tag, is controlled by its native regulatory sequences. Surprisingly, despite the absence of a discernible NLS in the Rx protein sequence, fractionation revealed that Rx-4HA was indeed present in both the nuclear and in the nuclei-depleted fraction (Figure 1). Hsp90 and SGT1 are part of a chaperone complex that stabilizes NB-LRRs in a potentially signaling-competent state (Shirasu, 2009). While these two proteins were only detected in the nuclei-depleted fraction, the nuclear protein histone H3 was indeed only detected in the nuclear extract (Figure 1). So, under native conditions, Rx localizes both to the cytoplasm and the nucleus.
Figure 1.
Rx Localizes to the Cytoplasm and Nucleus in Transgenic N. benthamiana:Rx4HA.
Immunoblot showing the presence of Rx-4HA in nuclei-depleted soluble (Cyto) and nuclear (Nuc) fractions of transgenic N. benthamiana:Rx4HA plants, which express Rx-4HA under control of the Rx native regulatory sequences. Fractionation was performed in four replicates. The nuclei-enriched fraction was loaded in a relatively 10-fold higher concentration. Hsp90, SGT1, and histone H3, detected by immunoblotting, were used as fractionation markers. Coomassie blue (CBB) staining of the blot was used as loading control.
Rx and RanGAP2 Coexpression Alters Nucleocytoplasmic Partitioning of Rx
Knowing the subcellular localization of Rx, we set out to perform microscopy-based colocalization studies of Rx and its interactor RanGAP2 in N. benthamiana to study the role of RanGAP2 in Rx-mediated resistance to PVX. Therefore, Rx and domains thereof were fused to green fluorescent protein (GFP) and either coexpressed with a negative control or in combination with RanGAP2 in N. benthamiana. Although expression was driven by the strong 35S promoter of Cauliflower mosaic virus (CaMV), most GFP fusion proteins were only just detectable by confocal microscopy, indicating that they are not overexpressed. Only the CC domain showed a higher signal (Figure 2). The GFP-Rx construct is fully functional as it provides extreme resistance to PVX in transgenic potato and mediates HR in N. benthamiana when coexpressed with the CP of PVX (PVX-CP) (Slootweg et al., 2010). Functionality of the Rx-CC-GFP and Rx-NB-ARC-GFP fusions was demonstrated by transcomplementation assays, in which the domains lacking in each fusion were coexpressed in trans in N. benthamiana (Moffett et al., 2002). For both fusions, this coexpression resulted in resistance to PVX (Slootweg et al., 2010). Functionality of the GFP-Rx-CC-NB fusion was shown by coexpression with an NB-ARC-LRR-Myc fusion, in combination with PVX-CP, which induced an HR (see Supplemental Figure 1 online).
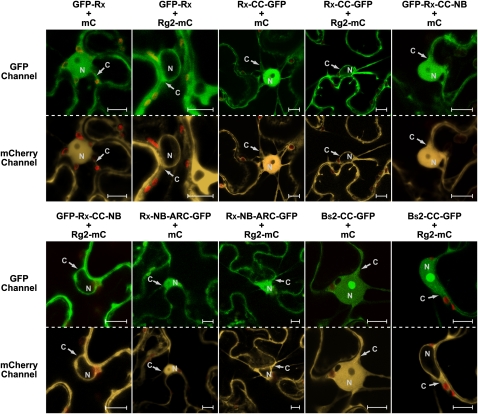
Figure 2.
RanGAP2 Coexpression with Rx Alters the Nucleocytoplasmic Partitioning of Rx through the Rx CC Domain.
Confocal images of N. benthamiana leaf epidermal cells transiently coexpressing GFP-Rx, Rx-CC-GFP, GFP-Rx-CC-NB, Rx-NB-ARC-GFP, or Bs2-CC-GFP (detected by the GFP channel) with Rg2-mC or mC alone (detected by the mCherry channel). Red structures are chloroplasts. C, cytoplasm; N, nucleus. Bars = 10 μm.
As expected, GFP-Rx localized both to the nucleus and the cytoplasm when coexpressed with the free monomeric RFP derivative mCherry (mC) (Shaner et al., 2004), serving as a negative control (Figure 2). Free mC and GFP also localize to both cellular compartments, as their molecular mass is below the size exclusion limit (~40 kD) for passive diffusion through the nuclear pores (Merkle, 2003). However, GFP-Rx, with a predicted molecular mass of 140 kD, largely exceeds this exclusion limit and must therefore be actively imported into the nucleus. We also studied the localization of the CC, CC-NB (only containing the NB subdomain and lacking the ARC1 and ARC2 subdomains), and the NB-ARC domains of Rx. All these fusions, when coexpressed with mC, showed a localization similar to full-length Rx (Figure 2), but unlike for the latter, passive diffusion might contribute to their nuclear accumulation. Furthermore, Rx-CC-GFP accumulated relatively stronger in the nucleus, compared with the other fusions (Figure 2).
To analyze the subcellular localization of RanGAP2, the protein was fused to mC (Rg2-mC) and also transiently expressed in N. benthamiana. Rg2-mC localizes solely to the cytoplasm (Figure 2), similar to the two Arabidopsis RanGAPs (Rose and Meier, 2001; Pay et al., 2002). The latter show a concentration at the nuclear envelope, a feature that we occasionally observed in cells that expressed RanGAP2 to low levels (see Supplemental Figure 2 online). In most cells, potential nuclear envelope localization is likely masked by equal fluorescence intensities in the cytoplasm and at the nuclear envelope.
Interestingly, when Rg2-mC and GFP-Rx were coexpressed, a dramatic change was observed in the localization of Rx, as GFP-Rx was now only detected in the cytoplasm (Figure 2). Rx-CC-GFP and GFP-Rx-CC-NB showed a similar change in localization, whereas Rx-NB-ARC-GFP localization was not affected upon coexpression with Rg2-mC, indicating that the Rx-CC domain is required for relocation. The localization of Bs2-CC-GFP, which does not bind to RanGAP2 (Tameling and Baulcombe, 2007), but also localizes to the cytoplasm and nucleus, is not changed when coexpressed with Rg2-mC (Figure 2). All the evoked relocation effects with the various RanGAP2 fusions presented in this article are highly robust and are observed in virtually all cells in the infiltrated area as assessed by quick laser scanning of the leaf sample and by imaging of ~10 cells per combination within a single experiment. Each coexpression was at least tested in three independent experiments. The cytoplasmic localization of Rg2-mC itself was not influenced by GFP-Rx coexpression (see Supplemental Figure 3 online). Together, these data show that coexpression of Rx with RanGAP2 affects Rx localization, suggesting that RanGAP2 facilitates the nucleocytoplasmic partitioning of this immune receptor.
Rx Also Interacts with RanGAP1
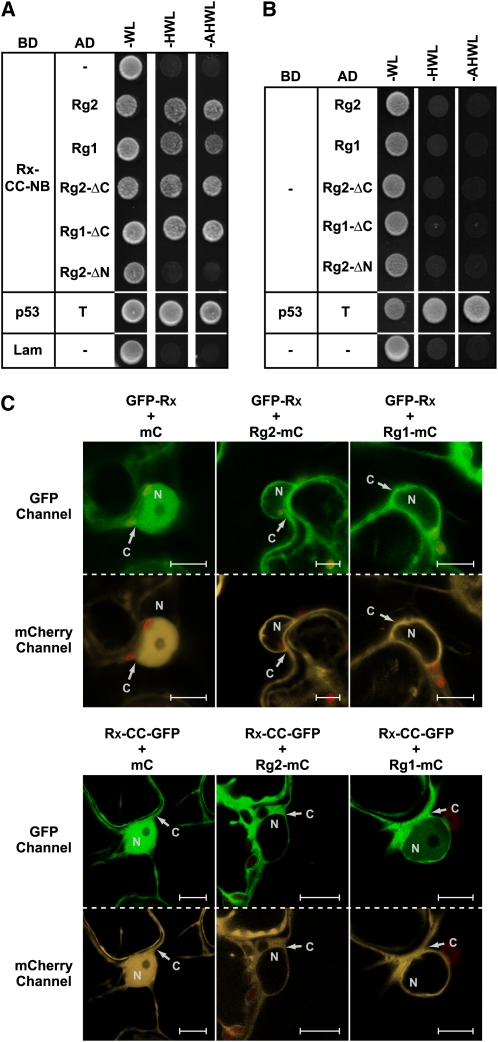
In previous studies, where the interaction between Rx and RanGAP2 was analyzed by coimmunoprecipitation (co-IP) experiments in N. benthamiana, an interaction with the paralog RanGAP1 (Rg1) was not detected (Sacco et al., 2007; Tameling and Baulcombe, 2007). To study possible interaction in another system, we performed yeast two-hybrid assays for which two Rx bait constructs were made comprising either both the CC and the NB subdomain or the NB subdomain alone. No yeast growth was observed on selective plates when either Rx-NB or the empty bait vector was cotransformed with any of the RanGAP constructs (Figures 3A and 3B). This was expected, as RanGAP2 specifically binds to the Rx-CC domain (Sacco et al., 2007; Tameling and Baulcombe, 2007). Indeed, the Rx-CC-NB construct did result in growth when cotransformed with either Rg2 or Rg2-ΔC, but not with Rg2-ΔN that lacks the Rx-CC–interacting WPP domain. Interestingly, the combination with Rg1 or Rg1-ΔC also resulted in growth (Figure 3A), showing that in yeast not only RanGAP2 but also RanGAP1 interacts with Rx and that this interaction is based on binding of the Rx-CC to the WPP domain of the RanGAPs.
Figure 3.
Both RanGAP1 and RanGAP2 Bind to Rx and Affect Its Nucleocytoplasmic Partitioning When Coexpressed with the Immune Receptor.
(A) and (B) Growth of yeast cotransformed with the Rx-CC-NB bait construct (A) (fused to the GAL4 DNA binding domain [BD]) or the empty bait vector (B) with the indicated RanGAP prey constructs (fused to the GAL4 activation domain [AD]) or empty prey vector (−). Growth on –WL plates indicates that bait and prey constructs are both present. Growth on –HWL and –AHWL plates indicates interaction between bait and prey proteins. Plates were photographed after 2 d of incubation. Cotransformation of murine p53 (p53) with the SV40 large T-antigen (T) and human lamin (Lam) or empty bait vector with the empty prey vector (−) served as positive and negative controls, respectively.
(C) Confocal images of N. benthamiana leaf epidermal cells transiently coexpressing GFP-Rx or Rx-CC-GFP (detected by the GFP-channel) and Rg2-mC, Rg1-mC, or mC alone (detected by the mCherry channel). Red structures are chloroplasts. C, cytoplasm; N, nucleus. Bars = 10 μm.
We subsequently checked whether RanGAP1 coexpression also relocates Rx. Indeed, similar to Rg2-mC, Rg1-mC coexpression also leads to a primarily cytoplasmic localization of GFP-Rx and Rx-CC-GFP (Figure 3C), suggesting that RanGAP1 also interacts with Rx in planta. From the imaging of many cells it seems that RanGAP1 is less efficient in relocating Rx-CC compared with RanGAP2, while both RanGAP proteins accumulated to similar levels (Figure 3C) (Tameling and Baulcombe, 2007). For example, this is visible in Figure 3C in the bottom panel, where in contrast with coexpression with Rg2-mC, coexpression with Rg1-mC does not result in full depletion of Rx-CC-GFP from the nucleus. Probably RanGAP1 has a lower binding affinity for Rx, which explains why this interaction was not detected in previous co-IP experiments that involve extensive washing steps (Sacco et al., 2007; Tameling and Baulcombe, 2007). These data show that in addition to RanGAP2, RanGAP1 also binds to Rx in yeast and that this interaction is very likely to occur in the plant.
GAP Activity of RanGAP2 Is Not Required for Rx Relocation
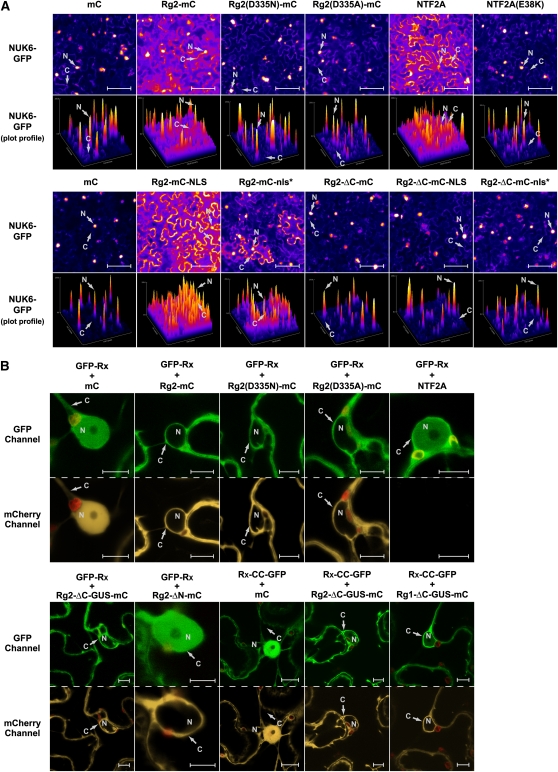
To investigate whether the effect of RanGAP2 coexpression on the nucleocytoplasmic partitioning of Rx is specific for this protein or is a more general phenomenon caused by increased GAP-activity, we also studied possible relocation of two nuclear marker proteins. For this we used the maize (Zea mays) transcription factor R (Shieh et al., 1993; Zhao et al., 2006) and the Phytophthora infestans effector NUK6, fused to GFP (Kanneganti et al., 2007). Both proteins contain NLS motifs and are imported via the classical nuclear import mechanism (Meier, 2007). Moreover, nuclear import of NUK6-GFP in N. benthamiana cells has been shown to depend on the importin receptors Impα1 and Impα2 (Kanneganti et al., 2007). First, both proteins were transiently expressed and multiple cells were visualized by wide-field fluorescence microscopy. The GFP fluorescence is displayed by pseudocolors using the Fire lookup table (LUT) in the ImageJ software to improve visualization of the different GFP signal intensity levels. Finally, the intensities of the whole field of view covering multiple cells were plotted in an intensity profile of representative images. Each coexpression was analyzed by performing at least three independent experiments, using four to five images per combination, which displayed highly reproducible effects. Indeed, both R-GFP and NUK6-GFP localized strongly to the nucleus when expressed in combination with the negative control, which was mC (Figure 4A, top panel; see Supplemental Figure 4 online). To our surprise, although there was still clear accumulation of both proteins in the nucleus, there was a marked decrease in nuclear import when Rg2-mC was coexpressed with the R or NUK6 proteins (Figure 4A, top panel; see Supplemental Figure 4 online). However, nuclear import was much more diminished when Arabidopsis NTF2A was coexpressed. NTF2A specifically imports Ran-GDP into the nucleus, which is essential for proper functioning of the nucleocytoplasmic trafficking machinery (Merkle, 2003). NTF2A overexpression in N. benthamiana was previously shown to block nuclear import of R-GFP (Zhao et al., 2006). Indeed, NUK6-GFP primarily localized to the cytoplasm when coexpressed with NTF2A, whereas coexpression with the loss-of-function mutant NTF2A(E38K) (Zhao et al., 2006) did not have this effect (Figure 4A, top panel). These data show that similar to NTF2A overexpression, RanGAP2 overexpression affects nuclear protein import in general, albeit to a much lesser extent. In Drosophila melanogaster, it was shown that RanGAP overexpression leads to an increased nuclear pool of RanGAP, which has an inhibitory effect on nucleocytoplasmic trafficking, most likely by depleting the nuclear Ran-GTP pool due to its GAP activity (Kusano et al., 2003). This likely also occurs in N. benthamiana because fusion of RanGAP2 to the SV40 NLS (Rg2-mC-NLS), thereby directing it to the nucleus, resulted in a much more pronounced increase of coexpressed NUK6-GFP in the cytoplasm, as when Rg2-mC was used (Figure 4A, bottom panel). Fusion with a mutated nonfunctional SV40 NLS (Rg2-mC-nls*) had an effect on NUK6-GFP localization that was similar to coexpression with non-NLS-fused RanGAP2 (Rg2-mC) (Figure 4A, bottom and top panels, respectively). Stronger nuclear accumulation of Rg2-mC-NLS, but not of Rg2-mC-nls*, compared with Rg2-mC, was indeed detected (see Supplemental Figure 5 online).
Figure 4.
Nucleocytoplasmic Partitioning of Rx Is Altered as a Result of Physical Sequestration by RanGAP2 in the Cytoplasm and Not by an Increased GAP Activity.
(A) NUK6-GFP (a nuclear marker protein) was transiently coexpressed in N. benthamiana leaf epidermal cells with the indicated constructs. The localization of NUK6-GFP was assessed by bright-field fluorescence microscopy. The GFP signal was converted to the Fire LUT to display the different intensities. With the same color representation, intensity profiles (plot profiles) are displayed below each image to indicate the local relative GFP intensities in the cytoplasm and nucleus. Bars = 100 μm.
(B) Confocal images of N. benthamiana leaf epidermal cells transiently coexpressing GFP-Rx or Rx-CC-GFP (detected by the GFP-channel) with NTF2A (not fused to mC) or the indicated constructs of RanGAP2 (Rg2) or RanGAP1 (Rg1), fused to mC or mC alone (detected by the mCherry channel). Red structures are chloroplasts. C, cytoplasm; N, nucleus. Bars = 10 μm.
In RanGAPs, the region of the LRR domain that contacts Ran and is highly conserved in RanGAPs of eukaryotes is essential for its GAP activity on Ran (Hillig et al., 1999). To determine whether the GAP activity of RanGAP2 is required for Rx relocation when coexpressed, a highly conserved Asp residue at position 335 in the LRR domain of RanGAP2 was changed to either Asn or Ala (see Supplemental Figure 6 online), as analogous substitutions in RanGAPs of Saccharomyces cerevisiae, Schizosaccharomyces pombe, and human RanGAP strongly inhibit GAP activity (Haberland and Gerke, 1999). Rg2(D335N)-mC and Rg2(D335A)-mC localize and accumulate similar to wild-type Rg2-mC (Figure 4B, top panel, mCherry channel). However, when NUK6-GFP or R-GFP was coexpressed with these mutants, increased accumulation of the protein in the cytoplasm, as observed when coexpressed with Rg2-mC, was no longer observed (Figure 4A, top panel; see Supplemental Figure 4 online). This indicates that the GAP activity in these mutants is indeed abolished, and that it is the increased GAP activity in the nucleus upon coexpression with wild-type RanGAP2 that inhibits nuclear import of NUK6-GFP and R-GFP. Furthermore, we also deleted the complete LRR domain of RanGAP2, leaving only the N-terminal WPP domain (Rg2-ΔC-mC), which does not exert GAP activity. When coexpressed, this fusion does not affect NUK6-GFP localization and neither do Rg2-ΔC-mC-NLS or Rg2-ΔC-mC-nls* fusions (Figure 4A, bottom panel).
To investigate whether altered nucleocytoplasmic partitioning of Rx upon RanGAP coexpression can also be ascribed to the increase in nuclear GAP activity, GFP-Rx was also coexpressed with Rg2(D335N)-mC and Rg2(D335A)-mC. While these GAP activity mutants lost the ability to relocate NUK6-GFP or R-GFP (Figure 4A, top panel; see Supplemental Figure 4 online), they still relocated GFP-Rx to the cytoplasm (Figure 4B, top panel). We also fused the WPP domain of Rg2 to both β-glucuronidase (GUS) and mC (Rg2-ΔC-GUS-mC), thereby generating a recombinant protein that due to its size does not passively diffuse into the nucleus (Figure 4B, bottom panel). Similar to coexpression with Rg2-mC (Figure 4B, top panel), coexpression with the Rg2-ΔC-GUS-mC fusion protein, and also with Rg1-ΔC-GUS-mC, gave rise to increased amounts of GFP-Rx, as well as Rx-CC-GFP, in the cytoplasm (Figure 4B, bottom panel). By contrast, deletion of the N-terminal WPP domain from Rg2-mC, leaving only the GAP activity–harboring LRR domain (Rg2-ΔN-mC), did not alter Rx localization when this fusion protein was coexpressed with GFP-Rx, while Rg2-ΔN-mC accumulated to similar levels as Rg2-mC (Figure 4B, bottom and top panels, respectively). Coexpression of GFP-Rx with NTF2A, which strongly suppresses nuclear import of NUK6-GFP (Figure 4A, top panel), did not affect GFP-Rx localization (Figure 4B, top panel). Together, these data show that the inhibitory effect on the classical nuclear import pathway, either by increasing GAP activity in the cell (through overexpression of RanGAP2) or by overexpression of NTF2A, has no influence on the nucleocytoplasmic partitioning of Rx. Moreover, it is very likely that depletion of Rx from the nucleus is caused by sequestration of the protein in the cytoplasm as a result of physical interaction of the WPP domain of RanGAP2 with the CC domain of Rx. This suggests that RanGAP2 serves as a cytoplasmic retention factor regulating nucleocytoplasmic partitioning of Rx.
RanGAP2 Functions as a Cytoplasmic Retention Factor Balancing Rx Nucleocytoplasmic Partitioning
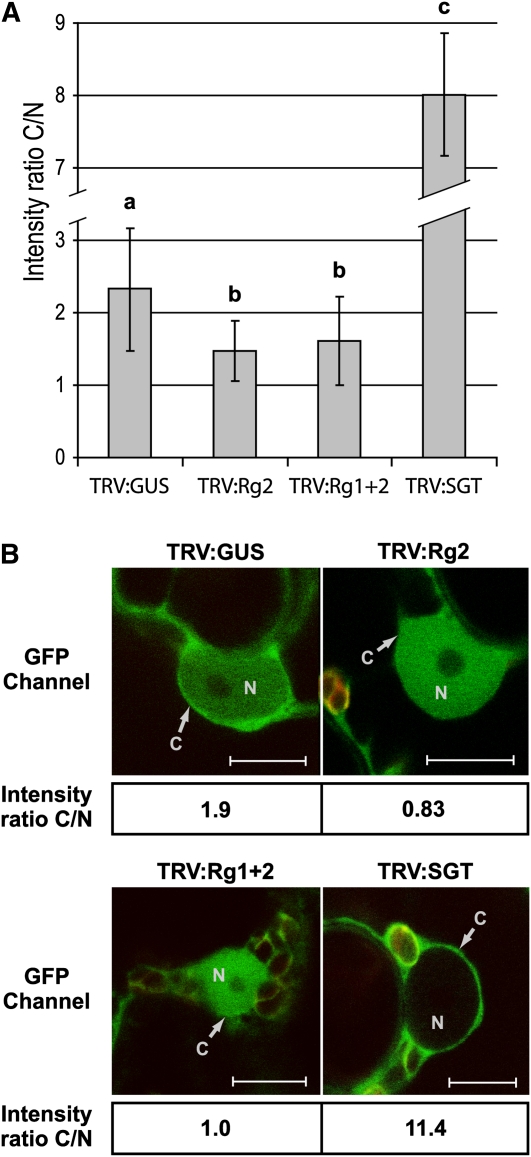
To test whether RanGAP2 indeed could serve as a cytoplasmic retention factor, GFP-Rx localization was studied in N. benthamiana in which RanGAP2 alone, or both RanGAPs simultaneously, were silenced by virus-induced gene silencing (VIGS) using recombinant Tobacco rattle virus (TRV). The intensity of GFP-Rx in the cytoplasm and nucleus was quantified based on 38 to 55 imaged cells per TRV construct from different plants, and the average ratio between the intensity in the cytoplasm and the nucleus (C/N) was plotted (Figure 5A). Both inoculation with TRV:Rg2 and TRV:Rg1+2 VIGS constructs resulted in a significantly reduced C/N ratio of GFP-Rx when compared with TRV:GUS, which served as a negative control. By contrast, inoculation with TRV:SGT, which silences SGT1, caused a dramatic increase in the C/N ratio, a phenomenon also observed by Slootweg et al. (2010). Representative images are shown in Figure 5B. These data strongly support the hypothesis that RanGAP2 serves as a cytoplasmic retention factor for the Rx immune receptor.
Figure 5.
Silencing of RanGAP2 Alone or Both RanGAPs Simultaneously Leads to Increased Rx Accumulation in the Nucleus.
Wild-type N. benthamiana seedlings were inoculated with the indicated recombinant TRV constructs to induce VIGS. TRV:GUS serves as a negative control. TRV:Rg2 targets RanGAP2 alone and TRV:Rg1+2 targets both RanGAPs. TRV:SGT silences SGT1. Three weeks after TRV inoculation, GFP-Rx was transiently expressed, and the localization of GFP-Rx was determined by confocal microscopy 3 d later. From TRV:GUS, TRV:Rg2, TRV:Rg1+2, and TRV:SGT, in total 38, 51, 55, and 4 cells were imaged, respectively. For the first three TRV constructs, six different plants (two leaves for each plant) were sampled. The intensities of the GFP signal in the cytoplasm (C) and nucleus (N) were quantified using ImageJ software.
(A) Histogram showing the mean C/N intensity ratios. Bars represent the average C/N intensity ratios for Rx and their standard deviations for the various treatments. Different letters above the data points indicate significant (P < 0.01) differences between the means (as determined by the Bonferroni and Dunnett t test).
(B) Representative images used for quantification of the C/N ratios. Calculated C/N intensity ratios are provided below each image. Red structures are chloroplasts. Bars = 10 μm.
Coexpression of Rx with RanGAP2 Results in Enhanced Defense Signaling
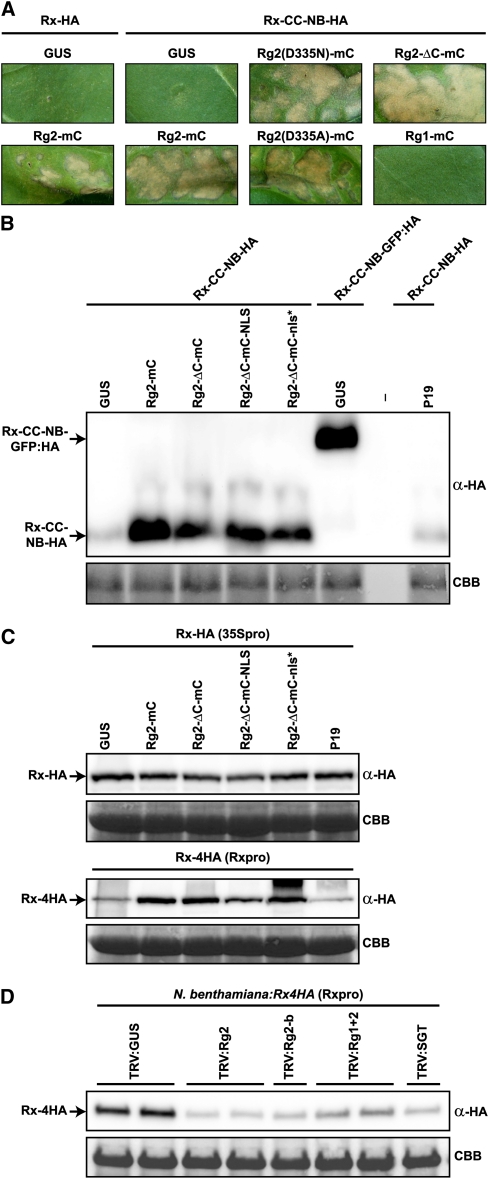
Previously it was shown that in tobacco (Nicotiana tabacum) Rx-HA has weak auto-activity when expressed under control of the CaMV 35S promoter, as it induces a mild HR in the absence of PVX-CP in young leaves after agroinfiltration. This auto-activity is enhanced when parts of the C terminus of Rx are deleted (Bendahmane et al., 2002). It was concluded that the LRR domain and the ARC subdomains have both a positive and a negative regulatory function on downstream signaling initiated by Rx. In mature fully expanded tobacco leaves, neither expression of Rx-HA nor Rx-CC-NB-HA resulted in visible cell death (Figure 6), an observation also reported by others (Sacco et al., 2007). However, coexpression with Rg2-mC led to a weak HR in combination with Rx-HA and a strong HR in combination with Rx-CC-NB-HA (Figure 6). So, the very weak auto-activity of these Rx constructs is enhanced by coexpression with RanGAP2, a phenomenon that was also reported by Sacco et al. (2007). To investigate whether the influence of RanGAP2 overexpression on nucleocytoplasmic trafficking (Figure 4A) plays a role in potentiating the Rx-mediated HR, the GAP activity mutants Rg2(D335N)-mC and Rg2(D335A)-mC were coexpressed with Rx-CC-NB-HA. The mutants caused an HR with an intensity that was similar to the HR induced by Rg2-mC (Figure 6), indicating that the general inhibition of nuclear import does not play a role in this process. This is further confirmed by the finding that Rg2-ΔC-mC, lacking GAP activity and only containing the WPP domain, also provides this phenotype (Figure 6). Although Rg2-ΔC-mC is not fused to GUS and therefore is sufficiently small to passively diffuse into the nucleus, in addition to potentiating Rx signaling, its expression also results in a relative increase in GFP-Rx accumulation in the cytoplasm (Figure 7A). As mentioned above, this is very likely mediated by physical sequestration of Rx in the cytoplasm. Note that an Rg2-ΔC-mC/GFP-Rx complex formed in the cytoplasm cannot passively diffuse into the nucleus. Coexpression of Rx-CC-NB-HA with Rg1-mC, which likely only binds weakly to Rx (see above), did not result in an HR (Figure 6A). These data suggest that the relative increase of Rx and Rx-CC-NB in the cytoplasm causes enhanced Rx-mediated defense signaling. In subsequent analyses, we checked the Rx-CC-NB-HA and Rx-HA levels in total extracts after coexpression with GUS (as a negative control), Rg2-mC, Rg2-ΔC-mC, Rg2-ΔC-mC-NLS, or Rg2-ΔC-mC-nls* by immunoblotting (Figure 6B). Besides their effect on the nucleocytoplasmic partitioning and defense signaling of Rx, all these RanGAP2 fusions also caused significant stabilization of Rx-CC-NB-HA, resulting in levels similar to the Rx-CC-NB-GFP:HA fusion (Figure 6B). The stabilizing effect of GFP on this fusion was reported previously by Rairdan et al. (2008) and put forward as the reason why, unlike Rx-CC-NB-HA, the Rx-CC-NB-GFP:HA fusion leads to HR when expressed in tobacco or N. benthamiana. However, the stabilizing effect was not apparent with the Rx-HA fusion (Figure 6C). It could be that this effect is too small to detect above the variation in CaMV 35S-expressed Rx-HA protein levels in the different extracts and that the levels of Rx have already reached a plateau. To test this, Rx-4HA, under control of its own promoter, was transiently coexpressed with the same constructs (Figure 6C). The Rx promoter leads to much lower expression levels than the CaMV 35S promoter (Bendahmane et al., 2002). Indeed, all coexpressed RanGAP2 constructs now displayed a clear stabilizing effect on Rx-4HA, which supports the idea that in leaf tissue in which Rx-HA expression is driven by the 35S promoter, Rx protein levels cannot increase further. In accordance with this finding, silencing of RanGAP2 alone or both RanGAPs simultaneously leads to a destabilization of Rx-4HA in transgenic N. benthamiana:Rx4HA plants (Figure 6D). Silencing of SGT1 also leads to destabilization of Rx-4HA in these plants, which has been shown previously (Azevedo et al., 2006). In summary, these data show that coexpression of the Rg2 fusions that physically interact with Rx not only has an effect on Rx localization and stability but also enhances defense signaling of Rx-HA and Rx-CC-NB-HA leading to HR.
Figure 6.
Coexpression of Rx with Different RanGAP2 Fusions and Silencing of RanGAP2 Affect Rx Protein Stability and Rx-Mediated Defense Signaling Activity.
(A) Rx-HA or Rx-CC-NB-HA was transiently coexpressed with GUS as a negative control or the indicated RanGAP constructs in N. tabacum. Leaves were photographed at 4 d after infiltration to visualize the HR resulting from enhanced auto-activity of the two Rx constructs. The Rx constructs were agroinfiltrated at OD600 = 0.5.
(B) Rx-CC-NB-HA, Rx-CC-NB-GFP:HA, or Rx-HA were transiently coexpressed in N. benthamiana with GUS, silencing suppressor P19, or the indicated RanGAP2 (Rg2) constructs, and total extracts were analyzed by immunoblotting.
(C) Similar to (B), except that now full-length Rx was expressed; either Rx-HA under control of the CaMV 35S promoter (35Spro; top panel) or Rx-4HA under control of its endogenous promoter (Rxpro; bottom panel). Note that Rxpro-driven expression results in at least 30-fold lower Rx protein levels.
(D) The indicated TRV constructs were used for VIGS in N. benthamiana:Rx4HA plants, in which Rx-4HA expression is also under control of its native regulatory sequences. TRV:GUS served as a negative control. TRV:Rg2 and TRV:Rg2-b target RanGAP2 alone, while TRV:Rg1+2 targets both RanGAPs simultaneously. TRV:SGT silences SGT1. Leaves were harvested for immunoblot analysis at 24 d after TRV inoculation.
(B) to (D) Total protein extracts were analyzed by immunoblotting using α-HA. Coomassie blue staining (CBB) of the blots is used as loading control.
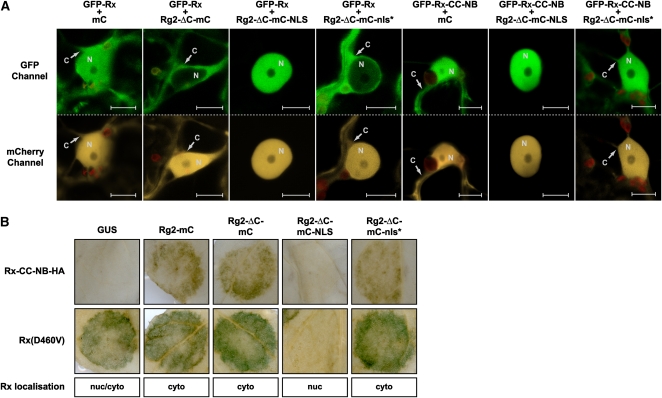
Figure 7.
Hyperaccumulation of Rx in the Nucleus Blocks Its Auto-Activity.
(A) Confocal images of N. benthamiana leaf epidermal cells transiently coexpressing GFP-Rx or GFP-Rx-CC-NB (detected by the GFP channel) with the indicated RanGAP2 (Rg2) constructs fused to mC, or mC alone (detected by the mCherry channel). Red structures are chloroplasts. C, cytoplasm; N, nucleus. Bars = 10 μm.
(B) The weak and strong auto-active Rx variants Rx-CC-NB-HA and Rx(D460V), respectively, were transiently coexpressed in N. benthamiana (at OD600 = 0.3 and 0.1, respectively) with the indicated Rg2 constructs or GUS in circular areas. Chlorophyll was removed to visualize the HR, which appears as a dark spot. The subcellular localization of Rx is indicated below the pictures.
Hyperaccumulation of Rx in the Nucleus Blocks the HR
When the WPP domain of RanGAP2 is fused to an NLS, the resulting Rg2-ΔC-mC-NLS protein exclusively localizes to the nucleus (Figure 7A). As expected, the control Rg2 fusion, containing a nonfunctional mutated NLS (Rg2-ΔC-mC-nls*), localizes, similar to Rg2-ΔC-mC, both to the cytoplasm and the nucleus (Figure 7A). When GFP-Rx was coexpressed with Rg2-ΔC-mC-NLS, hyperaccumulation of Rx in the nucleus was observed (Figure 7A), whereas coexpression with Rg2-ΔC-mC-nls* or Rg2-ΔC-mC resulted in the expected opposite phenotype, which is a relative increase of GFP-Rx in the cytoplasm (Figure 7A). Similar results were obtained when GFP-Rx-CC-NB was coexpressed with the various RanGAP2 fusions (Figure 7A; data not shown for Rg2-ΔC-mC). The massive accumulation of Rx in the nucleus is most likely caused by the nuclear import of the protein through physical association with the SV40 NLS-equipped WPP domain of RanGAP2.
To investigate what effect Rx hyperaccumulation in the nucleus has on Rx-mediated defense signaling, the slightly auto-active Rx-CC-NB-HA fusion was coexpressed with the different RanGAP2 fusions in N. benthamiana. Just as observed in tobacco (Figure 6A), Rx-CC-NB-HA did not cause HR in combination with GUS, but when coexpressed with Rg2-mC or Rg2-ΔC-mC, a clear HR was mounted (Figure 7B, top row). Strikingly, when Rx-CC-NB-HA was coexpressed with Rg2-ΔC-mC-NLS, no HR was observed (Figure 7B, top row), while total Rx-CC-NB-HA protein levels were increased to similar degrees as with Rg2-ΔC-mC and Rg2-ΔC-mC-nls* (Figure 6B). Coexpression with the control protein Rg2-ΔC-mC-nls* again gave HR, similar to expression with Rg2-ΔC-mC (Figures 6B and 7B, top row). This shows that the increased nuclear accumulation of Rx-CC-NB-HA suppresses Rx function. In a random mutagenesis screen, Rx mutants were identified that have an auto-active phenotype when agroinfiltrated in tobacco or N. benthamiana (Bendahmane et al., 2002). To further substantiate our findings, we subjected the respectively strong and weak auto-active Rx(D460V) and Rx(E400K) mutants to coexpression studies. Unlike Rx-CC-NB-HA, the mutants already triggered an HR when coexpressed with GUS (Figure 7B, bottom row; see Supplemental Figure 7 online). This auto-activity was not significantly potentiated by coexpression with Rg2-mC, Rg2-ΔC-mC, or Rg2-ΔC-mC-nls*, as observed for Rx-CC-NB-HA (Figure 7B, top row; see Supplemental Figure 7 online). However, the HR was fully suppressed when the Rx mutants were coexpressed with Rg2-ΔC-mC-NLS (Figure 7B, bottom row; see Supplemental Figure 7 online). We also included auto-active domain-swap constructs between Rx and the GPA2 immune receptor in our experiments (see Supplemental Figure 7 online). GPA2 provides resistance to the potato cyst nematode Globodera pallida and has an amino acid sequence that is over 88% identical to that of Rx (van der Vossen et al., 2000). In the R1G23R45 swap construct, the complete NB-ARC domain of Rx has been swapped with that of GPA2, which results in auto-activity (see Supplemental Figure 7 online). The same phenotype is observed when the LRR domain of GPA2 is swapped with that of Rx (swap G13R45; see Supplemental Figure 7 online) (Rairdan and Moffett, 2006). When coexpressed with Rg2-ΔC-mC-NLS, the HR triggered by these Rx-GPA2 swaps is completely suppressed, confirming that indeed the immune receptor cannot trigger defense signaling when solely localized to the nucleus, whereas signaling activity is potentiated when it accumulates and is stabilized in the cytoplasm. Since the CC domain of GPA2 also interacts with RanGAP2 (Sacco et al., 2007), and because we show that the auto-activity of the G13R45 swap construct, containing the GPA2-CC domain, is also suppressed when coexpressed with Rg2-ΔC-mC-NLS (see Supplemental Figure 7 online), our data strongly suggest that in addition to what is the case for Rx, nuclear hyperaccumulation of GPA2 will also block its ability to initiate defense signaling.
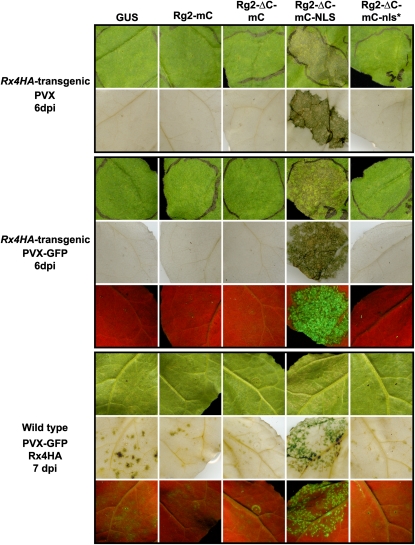
Hyperaccumulation of Rx in the Nucleus Causes Loss of Resistance to PVX
Suppression of the HR upon Rx hyperaccumulation in the nucleus prompted us to test whether altered nucleocytoplasmic partitioning of Rx also affects the actual resistance to PVX. RanGAP2 silencing in N. benthamiana:Rx4HA plants leads to partially compromised resistance to PVX, resulting in spread of the virus accompanied by local and systemic lesions (Tameling and Baulcombe, 2007). These lesions are the result of a trailing Rx-mediated HR, triggered by increased amounts of PVX compared with fully resistant plants. Such lesions are not observed in wild type susceptible, PVX-infected N. benthamiana leaves. We agroinfiltrated a binary PVX construct together with GUS, Rg2-mC, Rg2-ΔC-mC, or Rg2-ΔC-mC-nls* in N. benthamiana:Rx4HA. This did not affect extreme resistance, since lesions were not detected (Figure 8, top panel). However, when Rg2-ΔC-mC-NLS was expressed in combination with PVX, many lesions appeared in the infiltrated area (Figure 8, top panel), indicating partial loss of resistance similar to the situation when RanGAP2 is silenced. As reported previously, these lesions are indeed associated with increased virus titers, since clear GFP foci were detected under UV light when PVX-GFP was agroinfiltrated (Figure 8, middle panel). As expected, these GFP foci became even more intense when the partially resistant transgenic N. benthamiana:Rx(D399V) line (Sacco et al., 2007) was used in this assay (see Supplemental Figure 8 online), indicating that Rg2-ΔC-mC-NLS coexpression here results in a near complete loss of resistance. These results were confirmed in wild-type N. benthamiana, in which both the Rx-4HA and the RanGAP2 constructs were transiently coexpressed, in combination with PVX-GFP. The amount of agrobacteria carrying the Rx-4HA construct was chosen in such a way that partial resistance occurred, whereby a few PVX-induced lesions already appeared when coexpressed with GUS (Figure 8, bottom panel). Indeed, many more lesions appeared when Rx-4HA was coexpressed with Rg2-ΔC-mC-NLS. By contrast, coexpression of Rx-4HA with Rg2-ΔC-mC or Rg2-ΔC-mC-nls* and to a lesser extent also with Rg2-mC led to a reduced lesion phenotype compared with the GUS control (Figure 8, bottom panel). This increased level of resistance is likely caused by enhanced Rx-mediated defense signaling, which is reflected by the increased level of auto-activity induced by Rx-HA and by the increased Rx-4HA protein levels upon coexpression with these Rg2 fusions (Figures 6A and 6C). In summary, these data clearly show that hyperaccumulation of Rx in the nucleus not only suppresses the HR triggered by auto-active forms of Rx but also compromises the actual Rx-mediated resistance to PVX. Conversely, when Rx accumulates and is stabilized in the cytoplasm, defense against PVX is enhanced.
Figure 8.
Hyperaccumulation of Rx in the Nucleus Causes Loss of Resistance to PVX.
The indicated constructs were transiently expressed in transgenic N. benthamiana:Rx4HA (top and middle panels) or transiently coexpressed with Rx-4HA (from a binary vector) in wild-type N. benthamiana (bottom panel) in the marked areas on one leaf. Simultaneously, PVX or PVX-GFP inoculation was performed by coinfiltrating a dilute suspension of agrobacterium carrying a binary vector encoding infectious PVX or PVX-GFP. The appearance of spot lesions (Rx-mediated trailing HR) visible under daylight and GFP foci visible under UV light indicate partial loss of resistance. To be able to study both compromised and enhanced resistance, Rx-4HA was expressed transiently in wild-type N. benthamiana (bottom panel), where the induced resistance was to a certain extent already partial in the negative control (GUS). At 6 or 7 d after inoculation (dpi), leaves were photographed under daylight (first row of panels) or UV light (third row of middle and bottom panels), the latter to visualize the GFP fluorescent foci. After that, chlorophyll was removed and leaves were photographed under daylight to visualize the spot lesions (second row of panels).
DISCUSSION
Localization of Rx in Both the Cytoplasm and Nucleus Is Required for Defense Signaling
Although the NB-LRR protein Rx does not contain discernible canonical NLS motifs, it does localize to both the cytoplasm and nucleus (Figures 1 and 2). Furthermore, we found that a forced disequilibrium of the nucleocytoplasmic partitioning of Rx toward nuclear accumulation compromises defense signaling (Figures 7 and 8; see Supplemental Figures 7 and 8 online), showing that a balanced nucleocytoplasmic partitioning is essential for effective Rx-mediated immune signaling. Coexpression of Rx with RanGAP2, or particular domains thereof, led to drastic changes in the nucleocytoplasmic partitioning of Rx (Figures 2, 4, and 7). This was not the result of increased cellular GAP activity, as the loss-of-GAP-activity mutants, Rg2(D335N)-mC and Rg2(D335A)-mC, caused the same Rx relocation as the wild-type protein (Figure 4). Interestingly, coexpression with Rg2-ΔC-GUS-mC or Rg2-ΔC-mC, in which the GAP activity–harboring LRR domain is deleted, had the same effect, showing that the WPP domain only is responsible for Rx relocation. As RanGAP2 binds to the CC domain of Rx through its WPP domain, this indicates that most likely Rx is physically sequestered by the cytoplasmically localized RanGAP2 fusions. The observation that coexpression of Rx with Rg2-ΔC-mC-NLS, which is actively imported into the nucleus, results in nuclear hyperaccumulation of Rx is also in favor of sequestration of Rx by RanGAP2 (Figure 7). This suggests that RanGAP2 regulates the nucleocytoplasmic partitioning of Rx by functioning as a cytoplasmic retention factor, a mechanism that is common for nuclear proteins (see below; Xu and Massagué, 2004; Kaminaka et al., 2006; García and Parker, 2009; Seo et al., 2010). Increased levels of such a cytoplasmic retention factor are expected to result in a change in nucleocytoplasmic partitioning toward accumulation in the cytoplasm and decreased levels in a change toward accumulation in the nucleus. These phenomena were indeed observed for Rx, as a change toward cytoplasmic accumulation occurred when Rx was coexpressed with RanGAP2, and a change toward nuclear accumulation took place when RanGAP2 was silenced alone or both RanGAPs were silenced simultaneously (Figures 5A and 5B).
In previous co-IP studies, no interaction between Rx and RanGAP1 was detected (Sacco et al., 2007; Tameling and Baulcombe, 2007). However, here, we show that RanGAP1 is not only able to bind to Rx in yeast (Figures 3A and 3B) but also in plants, as RanGAP1 also physically sequesters Rx in the cytoplasm (Figure 3C). RanGAP1 probably has a lower binding affinity for Rx than RanGAP2, as the relocation of Rx-CC-GFP by RanGAP1 was less dramatic (Figure 3C), which explains why this interaction was not detected in previous co-IP experiments that involve extensive washing steps (Sacco et al., 2007; Tameling and Baulcombe, 2007). If RanGAP1 also contributes to Rx function, it is probably not substantial, as specific RanGAP2 silencing led to partial loss of resistance to PVX, whereas specific RanGAP1 silencing did not (Tameling and Baulcombe, 2007). Furthermore, we found that while coexpression of Rg2-mC enhanced the Rx-CC-NB-HA auto-activity, coexpression with Rg1-mC did not (Figure 6A).
The RAR1-SGT1-HSP90 chaperone complex is known to stabilize NB-LRRs, including Rx, in a potentially signaling-competent state in the cytoplasm (Hubert et al., 2003; Holt et al., 2005; Azevedo et al., 2006; Mestre and Baulcombe, 2006; Botër et al., 2007). Such a stabilizing mechanism is perhaps not required in the nucleus, as no SGT1 signal was observed in the nuclei-enriched fraction (Figure 1). This is in contrast with a previous report that describes the presence of a small pool of SGT1 in the nucleus (Noël et al., 2007). Indeed, a very faint HSP90 signal was observed in the nuclear-enriched fraction (Figure 1), which indicates that very low levels of HSP90 and perhaps additional cochaperones also mediate the stabilization of Rx in the nucleus.
RanGAP2 fusions that sequester Rx in the cytoplasm promote Rx-mediated defense signaling; vice versa, the RanGAP2 fusion (Rg2-ΔC-mC-NLS) that sequesters Rx in the nucleus suppresses Rx function (Figures 2 and 6 to 8; see Supplemental Figures 7 and 8 online). This phenomenon is both observed in assays in which the HR is induced by auto-active Rx mutants (Figures 6 and 7; see Supplemental Figure 7 online) and in PVX resistance assays (Figure 8; see Supplemental Figure 8 online). The observation that auto-active Rx mutants do not trigger an HR when they are depleted from the cytoplasm and hyperaccumulate in the nucleus indicates that postrecognition signaling cannot be initiated when Rx resides solely in the nucleus. Thus, for Rx to be able to initiate defense signaling, a balanced partitioning of the immune receptor between the cytoplasm and nucleus is required.
Coexpression of Rx with the various RanGAP2 fusions does not only alter the localization of Rx but also stabilizes the protein, thereby resulting in higher Rx levels. Although the effect was not detected when Rx-HA expression was driven by the CaMV 35S promoter, we could observe a clear stabilization when Rx-4HA expression was driven by its own promoter (Figure 6C). The stabilizing effect was also clear on the Rx-CC-NB-HA fusion (Figure 6B), a phenomenon that was also observed by others (P. Moffett, personal communication). In agreement with this finding, silencing of RanGAP2 alone, or both RanGAPs simultaneously, resulted in lower levels of Rx in N. benthamiana:Rx4HA plants (Figure 6D), indicating that binding to RanGAP2 is required to maintain a certain steady state level of Rx. As a moderately enhanced defense signaling is observed when full-length Rx is coexpressed with the RanGAP2 fusions (Figures 6A and 8), the question arises whether this is caused by Rx relocation, by increased Rx protein stability, or both. Interestingly, fusing full-length Rx to a nuclear export signal (NES) also leads to strong accumulation of Rx in the cytoplasm. However, instead of promoting resistance to PVX, this resulted in compromised resistance to the virus (Slootweg et al., 2010). This indicates that although Rx-RanGAP2 coexpression also causes relocation of Rx to the cytoplasm, in this case, the stabilizing effect on Rx, leading to higher Rx protein levels, overrules the relocation effect as observed with Rx-NES by Slootweg et al. (2010) and results in enhanced Rx-mediated defense signaling. This phenomenon is supported by the following observation; silencing of SGT1 leads to strong accumulation of Rx in the cytoplasm (Figure 5), which is similar to coexpression of Rx with RanGAP2 (Figure 2) but does not cause an increase in total Rx protein levels (Figure 6D). In agreement with this finding, SGT1 silencing leads to decreased instead of increased Rx-mediated defense signaling (Peart et al., 2002b; Tameling and Baulcombe, 2007). Furthermore, the previously observed partial loss-of-resistance phenotype caused by RanGAP2 silencing (Tameling and Baulcombe, 2007) is probably the result of enhanced nuclear accumulation of Rx, in combination with decreased total Rx protein levels (Figures 5 and 6D).
Redirection to the cytoplasm of nuclear-resident NB-LRRs, such as MLA10, N, RPS4, and snc1, by fusion to a NES compromises their ability to activate defense signaling (Burch-Smith et al., 2007; Shen et al., 2007; Wirthmueller et al., 2007; Cheng et al., 2009). This suggests that the requirement of a particular equilibrium in the nucleocytoplasmic partitioning of NB-LRRs for a proper regulation of the defense response is conserved in plants. However, there are no reports on the effect on defense signaling activation when these NB-LRR proteins are depleted from the cytoplasm and are forced to strongly accumulate in the nucleus. Here, we show that nuclear hyperaccumulation of Rx suppresses Rx function (Figures 7 and 8; see Supplemental Figures 7 and 8 online). This indicates that for proper functioning, NB-LRRs should be present in the cytoplasm as well as in the nucleus. This was confirmed by Slootweg et al. (2010), who found that fusion of Rx directly to the SV40 NLS leads to nuclear hyperaccumulation and inhibition of resistance to PVX. Upon its activation, the barley NB-LRR, MLA10, is proposed to relieve the repression of defense gene transcription mediated by two transcriptional regulators in the nucleus by binding to them. This subsequently results in activation of ETI (Shen et al., 2007). It would be interesting to test whether an auto-active mutant of MLA10 fused to an NLS triggers a stronger defense response than the nonfused mutant or whether such a fusion compromises defense signaling, similar to what we found for Rx. Such analyses should reveal whether this phenomenon is specific for Rx or whether for other NB-LRRs nuclear accumulation also inhibits defense signaling. Furthermore, activation of MLA1 appears to result in moderately increased levels of MLA1 in the nucleus (Shen et al., 2007). To study whether activation of Rx also affects its nucleocytoplasmic partitioning, agrobacteria containing binary constructs expressing either infectious PVX or PVX-CP were infiltrated in N. benthamiana:Rx4HA plants. Leaves were subsequently subjected to nucleocytoplasmic fractionation (see Supplemental Figure 10 online). No clear changes in Rx partitioning were observed, indicating that activation of Rx does, at least not detectably, affect its nucleocytoplasmic partitioning. However, it cannot be excluded that the rate of nucleocytoplasmic shuttling is equally affected in both directions. In summary, our data point out that a balanced partitioning between the cytoplasm and the nucleus is required for proper functioning of the immune receptor Rx.
Potential Roles of Rx in the Nucleus and Cytoplasm
Rx might play a role in the nucleus in transcriptional reprogramming that is common for induction of ETI and systemic acquired resistance (Tao et al., 2003; Eulgem, 2005; Wang et al., 2006; Vlot et al., 2008; Tsuda and Katagiri, 2010). However, in contrast with a proposed stimulatory effect for the MLA immune receptors (Shen et al., 2007), Rx might exert an inhibitory effect in the nucleus on defense gene transcription leading to ETI. This is supported by our finding that nuclear hyperaccumulation of auto-active Rx inhibits defense signaling. However, since the Rx-mediated resistance response is very rapid (Kohm et al., 1993), Rx might also directly activate an antiviral mechanism in the cytoplasm, which is the location where PVX replicates and where the virus is detected by Rx (Slootweg et al., 2010). A recent report shows that inhibition of PVX replication probably involves translational inhibition of viral RNA via an argonaute protein that might target a particular viral secondary RNA structure without using PVX-derived small RNAs as guide strands (Bhattacharjee et al., 2009). Thus, it is possible that upon activation, Rx immediately initiates this antiviral response in the cytoplasm and that additionally ETI (including systemic acquired resistance) is also induced in the cytoplasm. Alternatively, as described above, the inhibitory effect of Rx on ETI in the nucleus might be relieved by relocating a transcriptional repressor to the cytoplasm. These possibilities need to be explored in future research.
Regulation of Nucleocytoplasmic Partitioning of Rx and Possible Roles of RanGAP in This Process and in Initiating Defense Signaling
Since Rx does not contain a discernible canonical NLS motif, it remains to be determined how Rx is imported into the nucleus. Rx might either contain a complex NLS that is not readily distinguishable from its primary sequence, or it uses a piggyback mechanism that involves binding to an NLS-containing carrier protein (Lange et al., 2007; Genoud et al., 2008). However, we found that overexpression of NTF2A, which has a dominant inhibitory effect on nucleocytoplasmic trafficking (Zhao et al., 2006), decreases nuclear import of the NLS-containing protein NUK6, but not of Rx (Figures 4A and 4B). This suggests that Rx is imported into the nucleus in an importin-independent manner. Several examples of importin-independent nuclear import exist, and for these proteins import occurs by nucleoporin proteins present in the nuclear pore complex (Xu and Massagué, 2004). It would be interesting to test whether Rx indeed binds directly to nucleoporin proteins. As mentioned above, we hypothesize that RanGAP2 regulates the dynamic nucleocytoplasmic partitioning of Rx by serving as a retention factor that sequesters a certain amount of Rx in the cytoplasm. This option seems to be a common theme for nuclear proteins (Xu and Massagué, 2004; Kaminaka et al., 2006; Garcia and Parker, 2009; Seo et al., 2010). An illustrative example of a cytoplasmic retention factor is Lesions Simulating Disease resistance 1 that retains part of the pool of Arabidopsis bZIP10, which is a transcription factor that is a positive regulator of ETI and PTI, in the cytoplasm to antagonize plant immune signaling (Kaminaka et al., 2006).
Apart from the scenario in which RanGAP2 only regulates nucleocytoplasmic partitioning of Rx, it is also possible, but not mutually exclusive, that it is involved in PVX-CP perception. In this case, both RanGAP1 and RanGAP2 would act as guardees or baits that are targeted by PVX-CP. Manipulation of the RanGAPs is subsequently sensed by Rx, which thereby activates ETI (Van der Biezen and Jones, 1998a; Collier and Moffett, 2009). In the bait and switch model of Collier and Moffett (2009), it is proposed that RanGAP2 (bait) facilitates the interaction of PVX-CP with the LRR domain of Rx, meaning that PVX-CP is expected to directly interact with both RanGAP and Rx simultaneously. Similarly, a mammalian picornavirus targets Ran GTPase, thereby shutting down nucleocytoplasmic trafficking and facilitating virus replication in the cytoplasm (Porter et al., 2006). To date, no interaction has been found between PVX-CP and RanGAP2 or Rx (Rairdan and Moffett, 2006; Tameling and Baulcombe, 2007). Possibly, the interaction is too weak and/or too transient to capture in co-IP or yeast two-hybrid assays, or perhaps the RanGAPs do not serve as guardees/baits. Specific RanGAP2 silencing does not affect PVX replication and movement in susceptible wild-type N. benthamiana plants, which argues against RanGAP2 being a virulence target of PVX-CP (Tameling and Baulcombe, 2007). However, RanGAP1, which was not targeted by the used VIGS construct, could also be manipulated by PVX-CP. Unlike silencing of either RanGAP1 or RanGAP2, silencing of both RanGAPs simultaneously led to severe morphological phenotypes, including strongly deformed leaves and severe reduction in root biomass (see Supplemental Figure 9A online). Nevertheless, replication and movement of PVX-GFP was not affected (see Supplemental Figure 9B online). So, either both RanGAPs are not required for viral replication and/or movement or the residual amounts of these proteins that are present upon silencing of the encoding genes are still sufficient to aid these processes. Since a stronger downregulation of the RanGAP transcripts would most likely be lethal, it will be hard to obtain conclusive evidence for the RanGAPs being virulence targets of PVX-CP.
Besides RanGAP2, several additional nuclear transport components are important for plant innate immunity. Two Arabidopsis nucleoporin proteins (Nups) that are part of the nuclear pore complex, Nup96 and Nup88, are required for resistance to virulent and avirulent bacteria and the autoimmune response of the auto-active NB-LRR snc1 (Zhang and Li, 2005; Cheng et al., 2009). Interestingly, a mutation in Nup88 caused a reduction in nuclear accumulation of snc1-GFP (Cheng et al., 2009). A suppressor mutation in Arabidopsis importin α3 was also found to be important for the snc1-mediated autoimmune response (Palma et al., 2005). Future studies should provide more information how Rx and other NB-LRRs that lack canonical NLS motifs are imported into the nucleus and what roles these NB-LRRs play in the cytoplasm and nucleus to regulate plant innate immunity.
METHODS
Plant Material
Wild-type Nicotiana benthamiana, N. benthamiana:Rx4HA (Lu et al., 2003), N. benthamiana:Rx(D399V) (Sacco et al., 2007), and Nicotiana tabacum cv SR1 were grown in the greenhouse under 16 h light at 21°C and 8 h darkness at 19°C at a relative humidity of ~75%.
Plasmid Construction
All oligonucleotide sequences can be found in Supplemental Table 1 online. The pBIN+-GFP-Rx-CC-NB construct (SOL172) encodes amino acids 1 to 293 of Rx. This region was amplified by PCR with oligonucleotides wo192 and wo193 by which a 5′ BamHI site, a Gly-Ser linker, a stop-codon, and a 3′ SalI site were introduced. The PCR product was subcloned in the pGEM T-easy vector (Promega; SOL169). The Rx-CC-NB fragment was excised from SOL169 with BamHI and SalI and cloned in BamHI/SalI-digested pRAP-GFP-Rx1 (Slootweg et al., 2010), by which full-length Rx was exchanged with the CC-NB fragment, resulting in pRAP-GFP-Rx-CC-NB. The pBIN+-GFP-Bs2-CC construct encodes amino acids 1 to 151 of Bs2 from pepper (Capsicum annuum; Tai et al., 1999). This region was amplified by PCR with oligonucleotides eo1 and eo2, introducing a 5′ NcoI and a 3′ BamHI site. The fragment was digested with NcoI and BamHI and ligated in NcoI/BamHI-digested pRAP:CC-GFP (Slootweg et al., 2010), thereby replacing the Rx-CC with the Bs2-CC fragment. The expression cassettes comprising 35Spro:GFP-Rx-CC-NB:Tnos or 35Spro:Bs2-CC-GFP:Tnos were cloned in AscI/PacI-digested pBIN+ (van Engelen et al., 1995).
mC was PCR amplified from the plasmid pRSET-B mCherry (Shaner et al., 2004) with oligonucleotides wo125 and wo129, introducing a 5′ BamHI and a 3′ XmaI site, and subcloned in the pGEM T-easy vector resulting in pGEM T-mCherry (SOL2). Amplification of the full-length cDNAs from RanGAP2 (Rg2), RanGAP1 (Rg1), and the RanGAP2-ΔC (encoding amino acids 1 to 112) and RanGAP2-ΔN (encoding amino acids 107 to 541) fragments has been described previously (Tameling and Baulcombe, 2007). These fragments were subcloned previously in the PCR II blunt Topo vector (Invitrogen). All fragments were excised with SalI/BamHI and cloned in the binary pC/SBPc vector digested with the same enzymes, of which the multiple cloning site was adapted by first ligating an oligonucleotide linker created with oligonucleotides wo88 and wo89 in XbaI/SalI-digested pC/SBPc (Tameling and Baulcombe, 2007). This resulted in the Rg2-csBP, Rg1-csBP, Rg2-ΔC-csBP, and Rg2-ΔN-csBP constructs, in which the csBP-tag was replaced with mCherry. For this, the mCherry fragment was excised from SOL2 with BamHI/XmaI and ligated in the BamHI/XmaI-digested csBP fusion constructs. This resulted in the pBIN61-Rg2-mC (SOL6), pBIN61-Rg1-mC (SOL9), pBIN61-Rg2-ΔC-mC (SOL15), and pBIN61-Rg2-ΔN-mC (SOL12) constructs, which include the CaMV 35S promoter and nos terminator (this is applicable to all pBIN61-based plasmids described here). For pBIN61-mC (SOL39), the mCherry fragment was released with SalI and XmaI from SOL2 and ligated in SalI/XmaI-digested pC/SBPc, thereby exchanging csBP with mCherry. Full-length GUS was amplified with oligonucleotides wo177 and wo178, introducing a 5′ XhoI site and a 3′ XmaI site and subcloned in the pGEM T-easy vector. The GUS fragment was excised with XhoI and XmaI and ligated in XhoI/XmaI-digested SOL15, resulting in the pBIN61-Rg2-ΔC-mC-GUS (SOL122) construct.
Site-directed mutagenesis was performed by overlap extension PCR. For the D335N mutation in RanGAP2, the mismatch oligonucleotides wo155 and wo156 were used. For the D335A mutation in RanGAP2, the mismatch oligonucleotides wo157 and wo158 were used. The mutated RanGAP2 fragments were subcloned in pGEM T-easy, excised with SalI and BamHI, and ligated in SalI/BamHI-digested SOL9, thereby replacing wild-type RanGAP1, resulting in the pBIN61-Rg2(D335N)-mC (SOL30) and pBIN61-Rg2(D335A)-mC (SOL31) constructs. For the introduction of the SV40 NLS (Haasen et al., 1999), an oligonucleotide linker was designed with oligonucleotides wo169 and wo170 and for the mutated nonfunctional SV40 NLS (Haasen et al., 1999), an oligonucleotide linker was created with oligonucleotides wo171 and wo172. The linkers were ligated in XhoI/XmaI-digested SOL2, resulting in pGEM T-mCherry-NLS and pGEM T-mCherry-nls*, respectively. The mCherry-NLS and mCherry-nls* fragments were excised with BamHI and XmaI and ligated in BamHI/XmaI-digested Rg2-csBP and Rg2-ΔC-csBP constructs, resulting in pBIN61-Rg2-mC-NLS (SOL120), pBIN61-Rg2-mC-nls* (SOL121), pBIN61-Rg2-ΔC-mC-NLS (SOL85), and pBIN61-Rg2-ΔC-mC-nls* (SOL86). The RanGAP1-ΔC fragment (encoding amino acids 1 to 107) was amplified by PCR with oligonucleotides wo111 and wo114, introducing a 5′ SalI and 3′ BamHI site, subcloned in pGEM T-easy, excised with SalI and BamHI, and ligated in SalI/BamHI-digested SOL122, resulting in pBIN61-Rg1-ΔC-mC-GUS (SOL168).
For the pGBKT7-Rx-CC-NB yeast two-hybrid bait plasmid (SOL1001), the Rx-CC-NB fragment (encoding amino acids 1 to 293) was amplified by PCR with oligonucleotides wo190 and wo191, introducing a 5′ EcoRI and 3′ BamHI site. For the pGBKT7-Rx-NB bait plasmid (SOL1005), the Rx-NB fragment (encoding amino acids 139 to 293) was amplified by PCR with oligonucleotides wo198 and wo199, introducing a 5′ EcoRI and 3′ BamHI site. Fragments were subcloned in pGEM T-easy, excised with EcoRI and BamHI, and subsequently ligated in EcoRI/BamHI-digested pGBKT7 (Clontech). The multiple cloning site of the prey vector pGADT7 (Clontech) was modified by ligating an oligonucleotide linker generated with wo200 and wo201 in the EcoRI/XhoI-digested vector. The RanGAP fragments that were subcloned in PCR II Blunt TOPO or pGEM T-easy were excised with SalI and BamHI and ligated in the SalI/BamHI-digested modified pGADT7 vector, resulting in pGADT7-Rg2 (SOL1007), pGADT7-Rg1 (SOL1010), pGADT7-Rg2-ΔC (SOL1008), pGADT7-Rg2-ΔN (SOL1009), and pGADT7-Rg1-ΔC (SOL1017).
The pBIN+-GFP-Rx, pBIN+-Rx-CC-GFP, and pBIN+-Rx-NB-ARC-GFP constructs (containing the CaMV 35S promoter and nos terminator) are described by Slootweg et al. (2010). pBIN61-Rx-HA, pBIN61-Rx-CC-NB-HA (encoding amino acids 1 to 293), and pB1-Rx(D460V) (containing the 5′ and 3′ Rx regulatory sequences) are described by Bendahmane et al. (2002). pBIN61-Rx-CC-NB-GFP:HA is described by Rairdan et al. (2008). pBIN61-P19 is described by Voinnet et al. (2003). The binary plasmids pH2GW7-AtNTF2a, pH2GW7-AtNTF2a(E38K), and pGWB5-R-GFP all contain the CaMV 35S promoter and are described by Zhao et al. (2006). NUK6-GFP in the pGD binary vector containing the CaMV 35S promoter is described by Kanneganti et al. (2007). For construction of plasmids used in the supplemental data, see Supplemental Methods online.
Agroinfiltration
Binary plasmids were introduced in Agrobacterium tumefaciens strain C58C1, carrying the helper plasmid pCH32, or MOG101. Transient expression was performed as described previously (Mestre and Baulcombe, 2006), with some modifications. Bacteria were resuspended in MMA containing 200 μM acetosyringone (Van der Hoorn et al., 2000). Unless indicated, the suspensions carrying Rx constructs or NUK6-GFP were infiltrated at OD600 = 0.3 and suspensions carrying RanGAP constructs GUS, NTF2A, or NTF2A(E38K) at OD600 = 0.5. VIGS assay is described in Supplemental Methods online.
HR and PVX Resistance Assays
The HR induced by auto-activation of Rx fusion proteins was visualized by removing chlorophyll from leaves with 100% ethanol. Resistance to PVX was tested by agroinfiltration with a suspension of Agrobacterium (OD600 of 0.001) carrying the binary PVX plasmid pGR106 (Lu et al., 2003) or the binary PVX-GFP plasmid pGR208 (Peart et al., 2002a). These were mixed with the additional Agrobacterium suspensions (e.g., to express GUS or RanGAP2) and simultaneously agroinfiltrated. In wild-type N. benthamiana, resistance was provided by including Agrobacterium carrying the binary plasmid pB1-Rx4HA (Lu et al., 2003). This suspension was infiltrated at OD600 = 0.5 because this results in partial resistance.
Confocal and Bright-Field Fluorescence Microscopy
Confocal microscopy was performed on N. benthamiana epidermal cells using a Zeiss LSM 510 confocal microscope (Carl-Zeiss) with a ×40 1.2 numerical aperture water-corrected objective. The argon laser was used to excite at 488 nm for GFP and chlorophyll, and the HeNe laser at 543 nm to excite mCherry. GFP and chlorophyll emission were detected through a band-pass filter of 505 to 550 nm and through a 650-nm long-pass filter, respectively. mCherry emission was detected through a band-pass filter of 600 to 650 nm.
Bright-field fluorescence microscopy was performed with a Nikon 90i epifluorescence microscope equipped with a monochromatic camera (Nikon Ds-Qi.1Mc), using a ×4 0.13 numerical aperture objective. GFP and mCherry emission was examined using the GFP filter cube (GFP-B, EX 460 to 500, DM 505, BA 510 to 560) or the TRITC filter cube (EX540/25, DM 565, BA 605/55), respectively. The GFP fluorescence intensities were converted to the Fire LUT in ImageJ software (Abramoff et al., 2004).
Statistical analysis on the intensity measurements using ImageJ software was performed in SPSS15.0 using one-way analysis of variance (P < 0.01), followed by Bonferroni and Dunnett t (two-sided) posthoc multiple comparisons.
Protein Extraction and Immunoblotting
The subcellular fractionation, as shown in Figure 1, was performed according to Shen et al. (2007). For total protein extracts, two fresh leaf discs (1 cm in diameter) were ground in 8 M urea, 1% SDS, and 100 mM DTT or in 2× sample buffer. Samples were boiled in the presence of SDS loading buffer, centrifuged, and subsequently SDS-PAGE was performed.
HA-tag proteins were detected by immunoblotting (Moffett et al., 2002) using anti-HA-peroxidase (clone 3F10; Roche). HSP90 and SGT1 were detected using polyclonal antibodies raised against SGSa from Arabidopsis thaliana SGT1a or barley (Hordeum vulgare) HSP90-NTD, respectively, which were described previously (Takahashi et al., 2003; Azevedo et al., 2006). Histone H3 and PEPC were detected using polyclonal anti-H3 (acetyl K18) (ab1191; Abcam) and polyclonal anti-PEPC (100-4163; Rockland), respectively. Coomassie Brilliant Blue R 250 staining of the membranes after protein gel blot analysis was used to demonstrate equal loading.
Yeast Two-Hybrid Analysis
Interaction studies were performed in yeast strain PJ69-4a using the Matchmaker GAL4 two-hybrid system according to the manufacturer’s protocol. From the transformation plates, 10 colonies were picked per combination and transferred to new SD/-WL and SD/-AHWL plates and incubated for 7 d at 30°C to confirm the growth. Of each transformation combination, a colony was used to inoculate a 4-mL SD/-WL culture. After 18 h of growth, a 10× dilution range of these cultures was made from OD600 = 1 to OD600 = 1 × 10−5. Of each dilution, 5 μL was spotted on SD/-WL and SD/-AHWL plates and on plates lacking His, Leu, and Trp, containing 2.5 mM 3-amino-1,2,4-triazole (SD/-HWL + 2.5 mM 3-amino-1,2,3-triazole).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under accession numbers EF396238 (RanGAP1), EF396237 (RanGAP2), and AJ011801 (Rx).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The GFP-Rx-CC-NB Fusion Protein Is Functional in Activating Defense Signaling in a Transcomplementation Assay.
Supplemental Figure 2. Nuclear Rim Localization Is Occasionally Visible in Cells That Express Rg2-mC at Low Levels.
Supplemental Figure 3. GFP-Rx Expression Does Not Affect the Localization of RanGAP2.
Supplemental Figure 4. Wild-Type RanGAP2 Also Changes the Nucleocytoplasmic Partitioning of R-GFP.
Supplemental Figure 5. Fusion of RanGAP2 with the SV40 NLS Leads to a Strong Accumulation of RanGAP2 in the Nucleus.
Supplemental Figure 6. Multiple Sequence Alignment of Part of the LRR Domain of N. benthamiana RanGAP2 and the Saccharomyces pombe rna1p RanGAP Protein.
Supplemental Figure 7. Hyperaccumulation of Rx and Rx-GPA2 Swaps in the Nucleus Blocks Auto-Activity Resulting in the Absence of an HR.
Supplemental Figure 8. Targeting of Rx to the Nucleus Causes Near Complete Loss of Resistance to PVX in Transgenic N. benthamiana:Rx(D399V) Plants.
Supplemental Figure 9. Virus-Induced Gene Silencing of RanGAP1 and RanGAP2 Simultaneously Has No Detectable Effect on PVX-GFP Replication and Movement.
Supplemental Figure 10. Activation of Rx Does Not Provoke a Change in Its Nucleocytoplasmic Partitioning in N. benthamiana:Rx4HA Plants.
Supplemental Table 1. Primers Used in Plasmid Construction.
Acknowledgments
We thank the European Union–funded Integrated Project Bioexploit, the Netherlands Organization for Scientific Research (NWO; VENI Grant 863.08.018 to W.I.L.T.), and the Centre for BioSystems Genomics, which is part of the Netherlands Genomics Initiative and NWO, for support. We thank Peter Moffett for sharing unpublished results and materials. Frank Takken is acknowledged for critical reading of the manuscript. We also thank Sophien Kamoun and Iris Meier for sharing materials.
References
- Abramoff M.D., Magalhaes P.J., Ram S.J. (2004). Image processing with ImageJ. Biophotonics International 11: 36–41 [Google Scholar]
- Albrecht M., Takken F.L. (2006). Update on the domain architectures of NLRs and R proteins. Biochem. Biophys. Res. Commun. 339: 459–462 [DOI] [PubMed] [Google Scholar]
- Ausubel F.M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6: 973–979 [DOI] [PubMed] [Google Scholar]
- Azevedo C., Betsuyaku S., Peart J., Takahashi A., Noël L., Sadanandom A., Casais C., Parker J., Shirasu K. (2006). Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 25: 2007–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurès I., Candresse T., Leveau A., Bendahmane A., Sturbois B. (2008). The Rx gene confers resistance to a range of Potexviruses in transgenic Nicotiana plants. Mol. Plant Microbe Interact. 21: 1154–1164 [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Farnham G., Moffett P., Baulcombe D.C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32: 195–204 [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Kanyuka K., Baulcombe D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A., Köhn B.A., Dedi C., Baulcombe D.C. (1995). The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J. 8: 933–941 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S., Zamora A., Azhar M.T., Sacco M.A., Lambert L.H., Moffett P. (2009). Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control. Plant J. 58: 940–951 [DOI] [PubMed] [Google Scholar]
- Bischoff F.R., Klebe C., Kretschmer J., Wittinghofer A., Ponstingl H. (1994). RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA 91: 2587–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botër M., Amigues B., Peart J., Breuer C., Kadota Y., Casais C., Moore G., Kleanthous C., Ochsenbein F., Shirasu K., Guerois R. (2007). Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell 19: 3791–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith T.M., Schiff M., Caplan J.L., Tsao J., Czymmek K., Dinesh-Kumar S.P. (2007). A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candresse T., Marais A., Faure C., Dubrana M.P., Gombert J., Bendahmane A. (2010). Multiple coat protein mutations abolish recognition of Pepino mosaic potexvirus (PepMV) by the potato Rx resistance gene in transgenic tomatoes. Mol. Plant Microbe Interact. 23: 376–383 [DOI] [PubMed] [Google Scholar]
- Caplan J., Padmanabhan M., Dinesh-Kumar S.P. (2008). Plant NB-LRR immune receptors: From recognition to transcriptional reprogramming. Cell Host Microbe 3: 126–135 [DOI] [PubMed] [Google Scholar]
- Cheng Y.T., Germain H., Wiermer M., Bi D., Xu F., García A.V., Wirthmueller L., Després C., Parker J.E., Zhang Y., Li X. (2009). Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21: 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S.M., Moffett P. (2009). NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14: 521–529 [DOI] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Peeters N., Feng D.X., Khounlotham M., Boucher C., Somssich I., Genin S., Marco Y. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Catanzariti A.M., Teh T., Wang C.I., Ayliffe M.A., Kobe B., Ellis J.G. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103: 8888–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.G., Dodds P.N., Lawrence G.J. (2007). Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu. Rev. Phytopathol. 45: 289–306 [DOI] [PubMed] [Google Scholar]
- Eulgem T. (2005). Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10: 71–78 [DOI] [PubMed] [Google Scholar]
- Farnham G., Baulcombe D.C. (2006). Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proc. Natl. Acad. Sci. USA 103: 18828–18833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A.V., Parker J.E. (2009). Heaven’s Gate: Nuclear accessibility and activities of plant immune regulators. Trends Plant Sci. 14: 479–487 [DOI] [PubMed] [Google Scholar]
- García-Arenal F., McDonald B.A. (2003). An analysis of the durability of resistance to plant viruses. Phytopathology 93: 941–952 [DOI] [PubMed] [Google Scholar]
- Genoud T., Schweizer F., Tscheuschler A., Debrieux D., Casal J.J., Schäfer E., Hiltbrunner A., Fankhauser C. (2008). FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Kutay U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Haasen D., Köhler C., Neuhaus G., Merkle T. (1999). Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 20: 695–705 [DOI] [PubMed] [Google Scholar]
- Haberland J., Gerke V. (1999). Conserved charged residues in the leucine-rich repeat domain of the Ran GTPase activating protein are required for Ran binding and GTPase activation. Biochem. J. 343: 653–662 [PMC free article] [PubMed] [Google Scholar]
- Hillig R.C., Renault L., Vetter I.R., Drell T., IV, Wittinghofer A., Becker J. (1999). The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol. Cell 3: 781–791 [DOI] [PubMed] [Google Scholar]
- Holt B.F., III, Belkhadir Y., Dangl J.L. (2005). Antagonistic control of disease resistance protein stability in the plant immune system. Science 309: 929–932 [DOI] [PubMed] [Google Scholar]
- Hubert D.A., Tornero P., Belkhadir Y., Krishna P., Takahashi A., Shirasu K., Dangl J.L. (2003). Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22: 5679–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., McAdams S.A., Bryan G.T., Hershey H.P., Valent B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kaminaka H., Näke C., Epple P., Dittgen J., Schütze K., Chaban C., Holt B.F., III, Merkle T., Schäfer E., Harter K., Dangl J.L. (2006). bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 25: 4400–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti T.D., Bai X., Tsai C.W., Win J., Meulia T., Goodin M., Kamoun S., Hogenhout S.A. (2007). A functional genetic assay for nuclear trafficking in plants. Plant J. 50: 149–158 [DOI] [PubMed] [Google Scholar]
- Kohm B.A., Goulden M.G., Gilbert J.E., Kavanagh T.A., Baulcombe D.C. (1993). A potato virus X resistance gene mediates an induced, nonspecific resistance in protoplasts. Plant Cell 5: 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano A., Staber C., Chan H.Y., Ganetzky B. (2003). Closing the (Ran)GAP on segregation distortion in Drosophila. Bioessays 25: 108–115 [DOI] [PubMed] [Google Scholar]
- Lange A., Mills R.E., Lange C.J., Stewart M., Devine S.E., Corbett A.H. (2007). Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 282: 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe D.D., Koonin E.V., Aravind L. (2004). STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 343: 1–28 [DOI] [PubMed] [Google Scholar]
- Liu J., Coaker G. (2008). Nuclear trafficking during plant innate immunity. Mol. Plant 1: 411–422 [DOI] [PubMed] [Google Scholar]
- Lu R., Malcuit I., Moffett P., Ruiz M.T., Peart J., Wu A.J., Rathjen J.P., Bendahmane A., Day L., Baulcombe D.C. (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22: 5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik E., Takken F.L. (2009). STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 12: 427–436 [DOI] [PubMed] [Google Scholar]
- Martin G.B., Bogdanove A.J., Sessa G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54: 23–61 [DOI] [PubMed] [Google Scholar]
- McHale L., Tan X., Koehl P., Michelmore R.W. (2006). Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 7: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I. (2007). Composition of the plant nuclear envelope: Theme and variations. J. Exp. Bot. 58: 27–34 [DOI] [PubMed] [Google Scholar]
- Meier I., Brkljacic J. (2009). Adding pieces to the puzzling plant nuclear envelope. Curr. Opin. Plant Biol. 12: 752–759 [DOI] [PubMed] [Google Scholar]
- Merkle T. (2003). Nucleo-cytoplasmic partitioning of proteins in plants: Implications for the regulation of environmental and developmental signalling. Curr. Genet. 44: 231–260 [DOI] [PubMed] [Google Scholar]
- Mestre P., Baulcombe D.C. (2006). Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 18: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael Weaver L., Swiderski M.R., Li Y., Jones J.D. (2006). The Arabidopsis thaliana TIR-NB-LRR R-protein, RPP1A; protein localization and constitutive activation of defence by truncated alleles in tobacco and Arabidopsis. Plant J. 47: 829–840 [DOI] [PubMed] [Google Scholar]
- Moffett P., Farnham G., Peart J., Baulcombe D.C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël L.D., Cagna G., Stuttmann J., Wirthmüller L., Betsuyaku S., Witte C.P., Bhat R., Pochon N., Colby T., Parker J.E. (2007). Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19: 4061–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma K., Zhang Y., Li X. (2005). An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 15: 1129–1135 [DOI] [PubMed] [Google Scholar]
- Pan Q., Wendel J., Fluhr R. (2000). Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol. 50: 203–213 [DOI] [PubMed] [Google Scholar]
- Pay A., Resch K., Frohnmeyer H., Fejes E., Nagy F., Nick P. (2002). Plant RanGAPs are localized at the nuclear envelope in interphase and associated with microtubules in mitotic cells. Plant J. 30: 699–709 [DOI] [PubMed] [Google Scholar]
- Peart J.R., Cook G., Feys B.J., Parker J.E., Baulcombe D.C. (2002a). An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J. 29: 569–579 [DOI] [PubMed] [Google Scholar]
- Peart J.R., et al. (2002b). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99: 10865–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter F.W., Bochkov Y.A., Albee A.J., Wiese C., Palmenberg A.C. (2006). A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 103: 12417–12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimby B.B., Lamitina T., L’Hernault S.W., Corbett A.H. (2000). The mechanism of ran import into the nucleus by nuclear transport factor 2. J. Biol. Chem. 275: 28575–28582 [DOI] [PubMed] [Google Scholar]
- Rairdan G.J., Collier S.M., Sacco M.A., Baldwin T.T., Boettrich T., Moffett P. (2008). The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20: 739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan G.J., Moffett P. (2006). Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A., Meier I. (2001). A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc. Natl. Acad. Sci. USA 98: 15377–15382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco M.A., Mansoor S., Moffett P. (2007). A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J. 52: 82–93 [DOI] [PubMed] [Google Scholar]
- Seo P.J., Kim M.J., Park J.Y., Kim S.Y., Jeon J., Lee Y.H., Kim J., Park C.M. (2010). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61: 661–671 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Shen Q.H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ülker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shen Q.H., Schulze-Lefert P. (2007). Rumble in the nuclear jungle: Compartmentalization, trafficking, and nuclear action of plant immune receptors. EMBO J. 26: 4293–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh M.W., Wessler S.R., Raikhel N.V. (1993). Nuclear targeting of the maize R protein requires two nuclear localization sequences. Plant Physiol. 101: 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K. (2009). The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60: 139–164 [DOI] [PubMed] [Google Scholar]
- Slootweg E., et al. (2010). Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22: 4195–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. (2007). Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8: 195–208 [DOI] [PubMed] [Google Scholar]
- Swiderski M.R., Birker D., Jones J.D. (2009). The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol. Plant Microbe Interact. 22: 157–165 [DOI] [PubMed] [Google Scholar]
- Tai T.H., Dahlbeck D., Clark E.T., Gajiwala P., Pasion R., Whalen M.C., Stall R.E., Staskawicz B.J. (1999). Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA 96: 14153–14158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A., Casais C., Ichimura K., Shirasu K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 11777–11782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken F.L.W., Albrecht M., Tameling W.I.L. (2006). Resistance proteins: Molecular switches of plant defence. Curr. Opin. Plant Biol. 9: 383–390 [DOI] [PubMed] [Google Scholar]
- Takken F.L.W., Tameling W.I.L. (2009). To nibble at plant resistance proteins. Science 324: 744–746 [DOI] [PubMed] [Google Scholar]
- Tameling W.I.L., Baulcombe D.C. (2007). Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 19: 1682–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling W.I.L., Elzinga S.D., Darmin P.S., Vossen J.H., Takken F.L.W., Haring M.A., Cornelissen B.J.C. (2002). The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14: 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling W.I.L., Takken F.L.W. (2007). Resistance proteins: Scouts of the plant innate immune system. Eur. J. Plant Pathol. 121: 243–255 [Google Scholar]
- Tameling W.I.L., Vossen J.H., Albrecht M., Lengauer T., Berden J.A., Haring M.A., Cornelissen B.J.C., Takken F.L.W. (2006). Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140: 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Xie Z., Chen W., Glazebrook J., Chang H.S., Han B., Zhu T., Zou G., Katagiri F. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozzini A.C., Ceriani M.F., Saladrigas M.V., Hopp H.E. (1991). Extreme resistance to infection by Potato virus X in genotypes of wild tuber-bearing Solanum species. Potato Res. 34: 317–324 [Google Scholar]
- Tsuda K., Katagiri F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13: 459–465 [DOI] [PubMed] [Google Scholar]
- Ueda H., Yamaguchi Y., Sano H. (2006). Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol. Biol. 61: 31–45 [DOI] [PubMed] [Google Scholar]
- Van der Biezen E.A., Jones J.D. (1998a). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23: 454–456 [DOI] [PubMed] [Google Scholar]
- van der Biezen E.A., Jones J.D.G. (1998b). The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8: R226–R227 [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A.L., Laurent F., Roth R., De Wit P.J.G.M. (2000). Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13: 439–446 [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A.L., Kamoun S. (2008). From Guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen E.A., van der Voort J.N., Kanyuka K., Bendahmane A., Sandbrink H., Baulcombe D.C., Bakker J., Stiekema W.J., Klein-Lankhorst R.M. (2000). Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: A virus and a nematode. Plant J. 23: 567–576 [DOI] [PubMed] [Google Scholar]
- van Engelen F.A., Molthoff J.W., Conner A.J., Nap J.P., Pereira A., Stiekema W.J. (1995). pBINPLUS: An improved plant transformation vector based on pBIN19. Transgenic Res. 4: 288–290 [DOI] [PubMed] [Google Scholar]
- Vlot A.C., Klessig D.F., Park S.W. (2008). Systemic acquired resistance: The elusive signal(s). Curr. Opin. Plant Biol. 11: 436–442 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller L., Zhang Y., Jones J.D.G., Parker J.E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Wright K.L., Ting J.P. (2006). Epigenetic regulation of MHC-II and CIITA genes. Trends Immunol. 27: 405–412 [DOI] [PubMed] [Google Scholar]
- Xu L., Massagué J. (2004). Nucleocytoplasmic shuttling of signal transducers. Nat. Rev. Mol. Cell Biol. 5: 209–219 [DOI] [PubMed] [Google Scholar]
- Xu X.M., Meulia T., Meier I. (2007). Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr. Biol. 17: 1157–1163 [DOI] [PubMed] [Google Scholar]
- Ye Z., Ting J.P. (2008). NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Curr. Opin. Immunol. 20: 3–9 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Dorey S., Swiderski M., Jones J.D. (2004). Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 40: 213–224 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Li X. (2005). A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell 17: 1306–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Brkljacic J., Meier I. (2008). Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 20: 1639–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Leung S., Corbett A.H., Meier I. (2006). Identification and characterization of the Arabidopsis orthologs of nuclear transport factor 2, the nuclear import factor of ran. Plant Physiol. 140: 869–878 [DOI] [PMC free article] [PubMed] [Google Scholar]