The binding of the SBP domain of CRR1 to copper response elements of target promoters in vitro is blocked by Cu(II) or Hg(II), which also deactivate transcription in vivo. Mutagenesis of the SBP domain implicates a conserved His residue in interaction with copper ions, suggesting a mechanism for copper sensing. A metallothionein-like domain at the CRR1 C terminus is implicated in zinc homeostasis.
Abstract
Copper response regulator 1 (CRR1), an SBP-domain transcription factor, is a global regulator of nutritional copper signaling in Chlamydomonas reinhardtii and activates genes necessary during periods of copper deficiency. We localized Chlamydomonas CRR1 to the nucleus in mustard (Sinapis alba) seedlings, a location consistent with its function as a transcription factor. The Zn binding SBP domain of CRR1 binds copper ions in vitro. Cu(I) can replace Zn(II), but the Cu(II) form is unstable. The DNA binding activity is inhibited in vitro by Cu(II) or Hg(II) ions, which also prevent activation of transcription in vivo, but not by Co(II) or Ni(II), which have no effect in vivo. Copper inhibition of DNA binding is reduced by mutation of a conserved His residue. These results implicate the SBP domain in copper sensing. Deletion of a C-terminal metallothionein-like Cys-rich domain impacted neither nutritional copper signaling nor the effect of mercuric supplementation, but rendered CRR1 insensitive to hypoxia and to nickel supplementation, which normally activate the copper deficiency regulon in wild-type cells. Strains carrying the crr1-ΔCys allele upregulate ZRT genes and hyperaccumulate Zn(II), suggesting that the effect of nickel ions may be revealing a role for the C-terminal domain of CRR1 in zinc homeostasis in Chlamydomonas.
INTRODUCTION
Trace metal ions are essential for many types of biological catalysis. Yet, organisms may face metal deficiency in nature because of scarcity or reduced bioavailability. Despite the operation of mechanisms for mobilizing inaccessible forms of required metals, many organisms can remain chronically deficient for an essential micronutrient; accordingly there are pathways for acclimation to these situations. Chlamydomonas reinhardtii is an excellent reference organism for understanding transition metal homeostasis in a photosynthetic eukaryote (Merchant et al., 2006; Burkhead et al., 2009; Hanikenne et al., 2009).
The best-studied response to copper deficiency in Chlamydomonas is the replacement of plastocyanin, the most abundant copper-containing protein in a photosynthetic cell, with cytochrome c6 (Cyt c6), a heme protein (Merchant and Bogorad, 1986). This replacement, which allows for adequate photosynthetic electron transport when copper is limiting, occurs by transcriptional activation of the CYC6 gene (encoding Cyt c6) via copper response elements (CuREs) and a transcription factor, copper response regulator1 (CRR1; Quinn and Merchant, 1995; Kropat et al., 2005). Mutational analysis of CuREs in the promoters of CYC6 and CPX1 (another CRR1 target) identified the sequence GTAC as the core of the CuRE (Quinn et al., 1999, 2000). Mutation of any of the four nucleotides of this core sequence resulted in complete loss of expression during copper deficiency; however, the core sequence by itself was not sufficient for CuRE activity. The same core sequence is found in CuREs from other plants, such as Arabidopsis thaliana and Barbula unguiculata (Nagae et al., 2008; Yamasaki et al., 2009).
CRR1 contains a plant-specific DNA binding domain, called the SBP domain, which was identified originally in an Antirrhinum majus protein that bound the SQUAMOSA promoter (hence the name Squamosa promoter binding protein) (Klein et al., 1996) (Figure 1A). The SBP domain binding site, TNCGTACAA, was suggested initially by sequence alignment of Antirrhinum DEFH84, SQUA, and Arabidopsis AP1 (a SQUA ortholog) promoters (Cardon et al., 1999). Subsequently, in vitro DNA binding experiments identified GTAC as the core sequence of the SBP binding site of Arabidopsis Squamosa promoter binding protein-like (SPL) transcription factors SPL1, SPL3, and SPL8 (Birkenbihl et al., 2005). A more detailed SELEX study of Arabidopsis SPL14 indicated the sequence CCGTAC(A/G) as its DNA binding site (Liang et al., 2008), confirming the recognition of the GTAC core by the SBP domain.
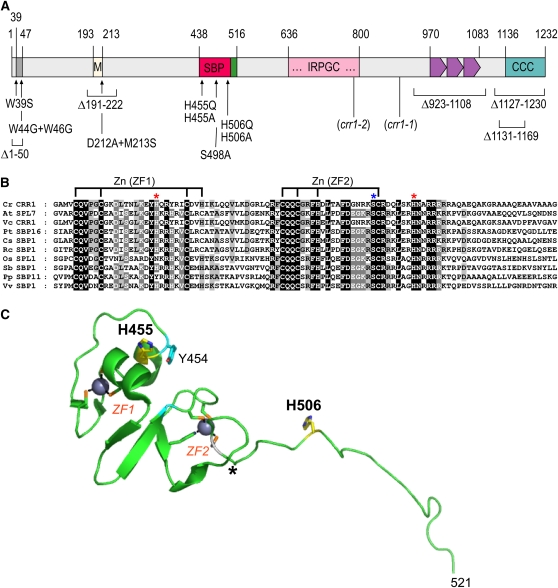
Figure 1.
The CRR1 Protein.
(A) Diagram of CRR1 showing locations of all mutations used in this work. The positions of the frameshift mutations in the crr1 mutants crr1-2 and crr1-1 are marked for reference. Domains from left to right are: dark-gray box = AHA motif; yellow box (M) = Met-rich region; dark pink (SBP) = SBP domain; green box = NLS; pink box (…IRPGC…) = extended SBP region with a highly conserved motif; purple boxes = ankyrin repeats; and turquoise box = Cys-rich region.
(B) Alignment of the SBP domains from CRR1 and SPL7 orthologs. The SBP domains of Chlamydomonas CRR1, a Volvox carteri CRR1 ortholog, Arabidopsis SPL7, and its orthologs in O. sativa, Sorghum bicolor, Populus trichocarpa, Physcomitrella, Ricinus cummunis, Cleome spinosa, and Vitis vinifera. Sequences were aligned using ClustalW. The His residues CRR1 H455 and CRR1 H506 are marked with a red asterisk, and the Ser residue CRR1 S498 is marked with a blue asterisk.
(C) Ribbon diagram of a representative of 20 models deposited as the solution structure of the zinc binding SBP domain of Arabidopsis SPL7 (PDB code: 1UL5). The two zinc coordination complexes are highlighted (ZF1 and ZF2), as are the side chains of the two His residues (His-455 and His-506 in CRR1 residue numbering, as indicated) studied here and the conserved Tyr-454. The region C-terminal to the asterisk is not well defined by the NMR data and is presumably flexible in solution; as can be seen, this is predicted to include His-506 in CRR1.
The ~80–amino acid SBP domains of Arabidopsis SPL4 and SPL7 (the sequences most closely related to the SBP domain of CRR1) have been characterized structurally by NMR spectroscopy. The SBP-DBD contains two consecutive zinc finger-like domains in a single globular domain, each of which adopts a noncanonical fold. Of the 10 potential metal binding ligands conserved in the SBP domain, eight are used to coordinate two Zn ions in N-terminal Cys3His (or Cys4) and C-terminal Cys2HisCys tetrahedral coordination complexes, denoted ZF1 and ZF2, which pack against one another (Yamasaki et al., 2004, 2006). Two highly conserved His residues are not involved in zinc coordination (Figures 1B and 1C). One, His-20 (numbering in the Arabidopsis SPL4 and SPL7 SBP-DBDs; His-455 in CRR1), is conserved in 90% of the SBP proteins (see Supplemental Figure 1A online). It forms part of an aromatic cluster on the surface of ZF1. The other, His-71 (His-506 in CRR1), is absolutely invariant. It is positioned C-terminal to ZF2 within a positively charged tail of the SBP domain, containing the bipartite nuclear localization signal (NLS). His-71 is dynamic in the free SBP domain and has been modeled to participate in some way in DNA binding (Yamasaki et al., 2004). The conformation of this region is unknown in intact CRR1 and may well be less flexible in the context of the entire protein.
In addition to the SBP domain, CRR1 contains an N-terminal AHA motif (for aromatic, hydrophobic, acidic), present also in a subset of SPLs in Arabidopsis, rice (Oryza sativa), and Physcomitrella patens (Riese et al., 2007) (see Supplemental Figure 1B online), an NLS within the SBP domain, ankyrin repeats, and a region of conservation flanking the SBP domain, which we referred to as the extended SBP domain (Kropat et al., 2005) (Figure 1A). The extended SBP domain contains a highly conserved amino acid sequence, IRPGC, but its function is not known. Truncated sequences of Arabidopsis SPL1 and SPL14 containing the AHA motif promoted transcription in yeast, but only one of three AHA-like motifs found in Physcomitrella SBP domain proteins did the same (Stone et al., 2005; Riese et al., 2007). A unique feature in CRR1 among the SPL family is the presence of a Cys-rich C-terminal region, which shows sequence similarity to a metallothionein (MT2) and a recently described putative Cu(I) sensing domain in Drosophila melanogaster metal-responsive factor-1 (MTF-1) (Chen et al., 2008). This domain was of potential interest in metal binding because of the role of CRR1 and its Arabidopsis ortholog, SPL7, in copper homeostasis (Kropat et al., 2005; Yamasaki et al., 2009).
In vivo experiments to test the metal selectivity of CRR1 indicate high specificity for copper ions for downregulating CYC6 expression (Hill et al., 1991; Merchant et al., 1991). In particular, Ag(I), which is an effector of yeast ACE1, was completely ineffective in downregulating CYC6 in vivo despite being taken up by the Chlamydomonas assimilatory Cu(I) transporter (Howe and Merchant, 1992; Page et al., 2009). On the other hand, Hg(II) can turn off CYC6 expression but only at 10- to 20-fold higher concentrations. The response is transient because Hg(II) is rapidly detoxified by coordination to glutathione, and it indicates that Hg(II) interact reversibly in the CRR1-dependent signal transduction pathway (Hill et al., 1991; Howe and Merchant, 1992). Whether Hg(II) mimics copper at the site of the copper sensor is not known. Since Hg(II) can activate dMTF-1–dependent gene expression in flies (albeit not in mammalian cells) (Balamurugan et al., 2004), the presence of a Cys-rich [presumed Hg(II) avid] C-terminal domain in CRR1 was therefore of interest as another candidate metal binding domain.
The Chlamydomonas nutritional copper regulon can also be activated in O2-deprived cells or in medium supplemented with Ni salts in the presence of copper (Quinn et al., 2000, 2002, 2003; Moseley et al., 2002). The responses require CRR1 and the CuRE, indicating that O2 and Ni ions impact the function of a component in the nutritional copper signal transduction pathway. Nickel is not a required nutrient for Chlamydomonas, and it is assumed that Ni ions interfere with the normal operation of some molecule in the copper-sensing pathway. On the other hand, the response to low O2 is physiologically relevant, since crr1 strains fail to grow under hypoxic conditions (Eriksson et al., 2004). In addition, we showed that a CRR1 target gene, CRD1, is required for chlorophyll biosynthesis in hypoxic cells (Quinn et al., 2002).
In this work, we separate the copper-sensing component of the pathway from the nickel- and O2-sensing domain. We also demonstrate that the putative NLS can function to drive a reporter protein to the nucleus, supportive of CRR1 residency in the Chlamydomonas nucleus, and we test the role of the AHA motif, the ankyrin repeats, and alternative translation start sites in CRR1 by site-directed mutagenesis.
RESULTS
CRR1 Is Localized in the Nucleus
In previous work, we established that CRR1 has a DNA binding domain with sequence specificity for the core of the CuRE and that it is required for transcriptional activation of copper deficiency targets in vivo (Kropat et al., 2005). This requires that the protein be located in the nucleus, at least in copper-deficient cells. Since we were unable to generate an effective antibody against CRR1 to test localization of the protein in Chlamydomonas, we constructed an N-terminal tagged version of CRR1 that could be expressed from pEGADxCRR1 in plant cells under the control of the promoter of the 35S RNA of Cauliflower mosaic virus (see Methods). We previously determined that N-terminally tagged (with 6xHA or codon-optimized green fluorescent protein [GFP]) versions of CRR1 do function in vivo based on rescue of the crr1 phenotype (Table 1); however, the low expression of this construct (driven by the CRR1 promoter) precluded visualization by fluorescence microscopy or immunochemical methods, and the heterologous systems were used instead. When the CRR1-containing construct was introduced into mustard (Sinapis alba) seedling hypocotyls (Table 2), we noted nuclear localization of enhanced GFP (EGFP):CRR1 relative to the control construct. By contrast, the EGFP fluorescence was detected in the entire cell, confirming that CRR1 has a functional NLS (potential monopartite NLS residues 68 to 71; bipartite NLS residues 496 to 513) like other SBP domain proteins (Birkenbihl et al., 2005). Because the experiments were conducted in copper-supplemented Murashige and Skoog medium without any effort to initiate copper deficiency, it is likely that the seedlings are copper replete, suggesting that copper deficiency is not required for nuclear localization, at least in the heterologous system.
Table 1.
CRR1 Mutants Characterized in This Study in Vivo
| Rescue Assayed by |
|||||||
| −Cu |
+Ni | −O2 | |||||
| Mutant Type | Location | Residues | Kautsky | Cyt c6 | CYC6 | CYC6 | CYC6 |
| Wild type | Y | Y | Y | Y | Y | ||
| Deletions | |||||||
| ΔAnk | Ankyrin repeats | 923 to 1108 | Y | Y | Y | Y | Y |
| ΔCysN | First half of Cys-rich region | 1131 to 1169 | Y | Y | Y | Y | Y |
| ΔCys | Entire Cys-rich region | 1127 to 1230 | Y | Y | Y | N | N |
| ΔMet | Met-rich region | 191 to 222 | N | N | N | N | |
| ΔInt1 | Deletion of intron 1 | Y | Y | ||||
| ΔSt1 | Frame shift after first ATG | N | N | N | |||
| Tags | |||||||
| GFP | GFP tag at N terminus | Y | Y | ||||
| 6xHA | 6xHA tag at N terminus | Y | Y | ||||
| Chimeras | |||||||
| CRR1-SPL1 | AtSPL1 SBP domain in CRR1 | N | |||||
| CRR1-SPL7 | AtSPL7 SBP domain in CRR1 | N | |||||
| Point mutations | |||||||
| WS | AHA-motif, 39-WAVEDWNWD-47 | W39S | Y | ||||
| GNG | AHA-motif, 39-WAVEDWNWD-47 | W44G/W46G | Y | ||||
| Met-3 | Met-rich region, 210-MPDM-213 | D212A/M213S | Y | Y | |||
| H455Q | Non-Zn coordinating His in SBP ZF1 | H455Q | Y | ||||
| S498A | Possible phosphorylation site in At | S498A | Y | Y | Y | ||
| H506A | Non-Zn coordinating His in SBP tail | H506A | N | ||||
| H506Q | Non-Zn coordinating His in SBP tail | H506Q | N | ||||
For each mutation, the region or residue number(s) deleted or changed in CRR1 is indicated (initiator Met is 1). The column “Rescue Assayed by” indicates whether CRR1 function is recovered upon introduction of the construct into crr1 (Y, yes; N, no). Function was tested in –Cu cells by restoration of photosynthesis (Kautsky curves for fluorescence rise and decay kinetics) or differential gene expression (tested by immunoblots for Cyt c6 abundance and real-time PCR for CYC6 mRNA abundance). For the response to Ni(II) and hypoxia (−O2), differential gene regulation was tested by real-time PCR. See text for other details.
Table 2.
CRR1 Localizes to the Nucleus
| Construct | n | Ratio N/C | sd |
| 35S:EGFP-CRR1 | 10 | 1.02 | 0.53 |
| 35S:EGFP | 6 | 0.16 | 0.11 |
| 35S:DsReda | 10 | 0.16 | 0.06 |
Hypocotyls from mustard seedlings were transiently transformed with a construct expressing chimeric EGFP-CRR1 or the EGFP under the control of the 35S promoter. Pictures from n transformants were taken and analyzed with ImageJ 1.42q software. Mean pixel intensities of background were subtracted from that of whole cells and nuclei. Integrated densities of nuclei alone and of cells without nuclei were calculated, and their ratio (N/C) reflecting distribution of the protein and the standard deviation (sd) were determined.
Another construct using the red fluorescence protein from Discosoma sp (DsRed) confirmed the N/C distribution of the EGFP construct.
For Arabidopsis SPL8, phosphorylation at a Ser residue in the proximity of the NLS was shown to block nuclear localization (Birkenbihl et al., 2005). Mutations that block phosphorylation (Ser to Ala) supported the role of phosphorylation in determining subcellular location. Although SPL8 is unrelated to CRR1, we tested the importance of the analogous Ser residue (Ser-498 in CRR1) for CRR1-dependent expression of CYC6 in vivo (see below). The photosynthetic electron transfer chain is blocked in copper-deficient crr1 at the donor side of photosystem I (PSI) because CYC6 expression (encoding Cyt c6 as a replacement for plastocyanin) cannot be activated in copper-deficient medium. Copper-supplemented crr1 shows normal decay kinetics because it has plastocyanin (Kropat et al., 2005). When CRR1 function is restored (e.g., Cyt c6 is made), the Kautsky curves show normal decay kinetics in copper-deficient medium. We tested a construct carrying a Ser498Ala mutation in CRR1 for its ability to rescue crr1-2.2. The mutated construct did rescue the mutant (Table 2, Figure 2). When we tested whether the Ser498Ala mutation renders CRR1 constitutively active (e.g., by constant nuclear import or by blocking nuclear export in the copper-replete situation), we found that Cyt c6 accumulation remained copper responsive as in the wild-type strain (Table 2). Therefore, we conclude that phosphorylation of a Ser residue in the SBP domain is not likely to be important for CRR1 function.
Figure 2.
In Vivo Functional Analysis of SBP Domain Mutants.
Site-directed mutations in the SBP domain (S498A, H506Q, and H455Q) were introduced into genomic DNA encoding CRR1. The constructs were cotransformed with pArg7.8 into a crr1-2.2arg7 strain. Arg+ prototrophs carrying the test constructs were tested on copper-deficient medium for rescue of crr1 by fluorescence induction and decay kinetics. To normalize the traces, the fluorescence yield at 0.5 s was set to 1. See also Table 1.
[See online article for color version of this figure.]
Mutational Analysis of the Initiator Met, the AHA Motif, and a Met-Rich Domain
The first intron of CRR1 is spliced very slowly, so that it is retained in ~30% of the CRR1 mRNA pool (Kropat et al., 2005). If the intron-retained version of the mRNA is functional, a Met codon downstream of the intron would serve as the initiator instead of the Met upstream of the intron because of the presence of stop codons in the intron. To distinguish the importance of the intron, we tested constructs in which the intron was deleted (to generate a prespliced RNA) and one in which a frame shift was introduced upstream of the intron (to force use of a downstream initiator Met) for their ability to rescue crr1-2.2 by analyzing the restoration of photosynthesis in Cu deficiency. The former construct was completely functional and showed copper-responsive CYC6 expression, whereas the latter was nonfunctional (Table 1), indicating that the slowly spliced intron was not important or regulatory in any way and that the first Met functions as the initiator.
CRR1 contains an AHA motif that is found in several other SBP domain proteins, including SPL7 (Riese et al., 2007), and is proposed to function as a transcriptional activation domain (Stone et al., 2005; Riese et al., 2007; Guo et al., 2008). The AHA motif is a short peptide sequence consisting of aromatic, hydrophobic, and acidic amino acid residues. To test its importance, we generated point mutations in conserved Trp residues of the motif 39WAVEDWNWD47. The Trp residues have been shown to be important for transcriptional activation (Riese et al., 2007). We tested the constructs carrying the mutations for rescue of crr1. Neither the Trp39Ser modification nor the double Trp44Gly/Trp46Gly alteration showed an altered phenotype (Table 1). We concluded that the AHA motif is not required for CRR1 function in nutritional copper signaling.
Within the N-terminal low complexity region, there is a Met-rich region, consisting of the sequence MxMxxM followed by a tandem MxxM, resembling the Mets motif of the CTR family of Cu(I) transporters (Dancis et al., 1994; Puig et al., 2002; Puig and Thiele, 2002). In the Ctr proteins, the Mets motif facilitates Cu(I) transport. Since a synthetic peptide with a single copy of MxxMxxM is sufficient for selective Cu(I) binding (Jiang et al., 2005), the presence of a Mets motif suggested that the domain might be important for Cu(I) binding to CRR1. We deleted 33 residues encompassing the Mets region to generate a derivative, named ΔMet, but this construct failed to rescue crr1-2.2 (Table 1). Since the drastic nature of the mutation might have resulted in loss of protein structure, we introduced point mutations into one of the three Mets motifs. The construct with the alteration Asp212Ala/Met213Ser, named Met3, showed normal CRR1 function in the copper deficiency response and normal fluorescence rise and decay kinetics (Table 1). Although the effect of the mutations in a single Mets motif might be too subtle to distinguish by the Kautsky assay or by CYC6 expression analysis, which varies as a function of copper nutrition and cell densities (Merchant et al., 1991), we note that the Mets motif region is less evident even in the closely related sequence from Volvox and it is absent in Arabidopsis SPL7. Taken together, we suspect that the Mets motif is unlikely to play a part in copper sensing in Chlamydomonas.
The SBP Domain Is Essential for CRR1 Function and May Interact Directly with Copper via a Conserved His Residue
Previously, we demonstrated that the SBP domain binds CuREs in CRR1 targets in a sequence-specific manner (Kropat et al., 2005). Structural studies indicate that this domain binds Zn(II), and we and others showed that Zn(II) binding is required for DNA binding (Birkenbihl et al., 2005; Kropat et al., 2005). The roles of the individual zinc finger (ZF) domains are distinct. Zn(II) binding to ZF1 is required for folding of the SBP domain; by contrast, Zn binding to ZF2 is apparently not required to fold the domain but is essential for maintaining high affinity DNA binding (Yamasaki et al., 2006).
Since only eight of the 10 conserved Cys and His residues are involved in Zn(II) binding (Figure 1B), we investigated whether this domain might be capable of coordinating other metal ions, and in so doing, provide a mechanism for regulation of transcription. SBP purified to homogeneity (≥95%) in the presence of 50 μM Zn(II) added to all buffers (see Methods) was found to contain 4 to 5 mol Zn(II) per mol SBP monomer, as determined by atomic absorbance spectroscopy. Since a molar stoichiometry of 2 Zn(II)/SBP was expected, Zn(II) chelators EDTA (KZn = 8.4 × 1013 M−1 at pH 7.0) and nitrilo-2,2',2"-triacetic acid (NTA; KZn = 9.7 × 107 M−1 at pH 7.0) were added (at 5× molar excess) in an effort to remove any weakly bound Zn(II). Addition of EDTA at lower pH destabilizes the protein, resulting in reversible SBP precipitation; the addition of excess Zn(II) resolubilizes the protein to give a far-UV circular dichroism (CD) spectrum that is indistinguishable from an untreated preparation. SBP incubated in NTA did not immediately precipitate but did so following an overnight incubation at room temperature. Native electrospray ionization mass spectrometry (ESI-MS) in 25 mM ammonium acetate confirms two molar equivalents of Zn(II) bound with high affinity per SBP monomer as expected. SBP purified with a slight modification in which 0.5 mM NTA was used in place of 50 μM Zn(II) resulted in the expected two Zn(II) per SBP monomer stoichiometry (see Methods).
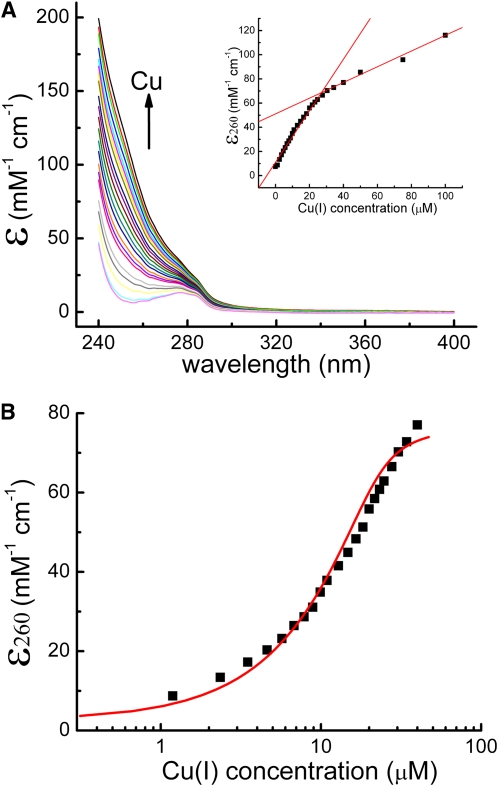
Given the apparently poor solubility of the metal-free apo-SBP (see above), we performed a series of anaerobic Zn(II) displacement experiments to determine if other metals [e.g., Cu(I)] could bind to SBP. Figure 3 shows molar absorption spectra that result upon addition of small aliquots of a concentrated Cu(I) solution to 10 μM Zn(II) SBP. The increased absorbance in the near UV is indicative of Cu(I) → thiolate charge transfer (Liu et al., 2007; Ma et al., 2009) representing coordination to SBP Cys residues. The nearly linear biphasic nature of the binding data (Figure 3A, inset) suggests that the first two Cu(I) ions bind with a similar structure followed by a nonsaturable increase in the absorbance after these two sites are filled. This second phase in the Cu(I) binding curve is consistent with a Cu(I)-induced SBP aggregation as evidenced by strong Cu(I) luminescence measured under the same conditions (see Supplemental Figure 2 online) (Chen et al., 2008).
Figure 3.
Spectroscopic Characterization of Cu(I) Binding to Zn(II)4-SBP under Strictly Anaerobic Conditions.
(A) Molar absorptivity spectra resulting from the addition of small aliquots of a concentrated stock solution of CuCl added into 10 μM Zn(II)4-SBP. Increased absorbance in the near UV is indicative of Cu(I) → thiolate charge transfer. The inset shows molar absorbance at 260 nm as a function of total Cu(I) concentration. The apparent biphasic nature (red lines) suggests two high-affinity Cu(I) sites followed by additional coordination and subsequent structural rearrangement. See text for details.
(B) Preferential binding of Cu(I) over Zn(II). Raw data from (A) were truncated at 50 μM total Cu(I), which corresponds roughly to the first phase of Cu(I) binding from (A). The smooth red line indicates the best fit to a two-site sequential Cu binding model where K1 = 1.8 ± 0.8 × 105 M−1 and K2 = 6 ± 2 × 105 M−1. These analyses reveal that the SBP domain coordinates Cu(I) with a ~400,000-fold preference over Zn(II) (see also Zimmermann et al., 2009).
If one truncates the absorption data following the first phase of the binding curve and performs a nonlinear least squares fitting to a two-site binding model, best fit values of K1 = 1.8 ± 0.8 × 105 M−1 and K2 = 6 ± 2 × 105 M−1 (Kd's of 5.6 and 1.7 μM, respectively) are obtained (Figure 3B). Note that these values report on Cu(I) binding to Zn(II)-loaded SBP and thus effectively represent lower limits for the KCu(I)/KZn affinity ratio; this value of ≈105 for displacement of Zn(II) by Cu(I), meaning that Cu(I) binds 105-fold more tightly than Zn(II), is consistent with that previously measured for a series of Zn(II) binding heavy metal binding domains excised from Arabidopsis zinc transporters (Zimmermann et al., 2009). In any case, such a model requires that Cu(I) will indeed displace Zn(II) upon coordination to the zinc finger domains of SBP.
To test this, we performed the same experiment in the presence of MagFura-2 (MF; KZn = 5.0 × 107 M−1), a common spectroscopic probe for Zn(II) (Figure 4) (Walkup and Imperiali, 1997; VanZile et al., 2002). A 10-min incubation of Zn4-5-SBP with MF did not produce an altered MF2 spectrum (black line), indicating that MF2 is not capable of stripping any Zn(II) from SBP under these conditions; however, this finding does not rule out a kinetically hindered process like removal of Zn(II) from SBP observed for NTA (discussed above). The addition of Cu(I) results in the stochiometric displacement of SBP-bound Zn(II), as evidenced by the appearance of the spectrum corresponding to the Zn(II)-MF complex (Figure 4). Cu(I) substituted SBP is structurally similar to Zn(II) loaded SBP as probed by CD spectroscopy (see Supplemental Figure 3 online). The CD spectrum shows very little secondary structure in the isolated SBP; the ellipticity associated with electronic transitions of aromatic side chains (e.g. Tyr) appear to dominate much of the spectrum (Yamasaki et al., 2004).
Figure 4.
Cu(I) Binding to SBP Displaces Zn(II) under Strictly Anaerobic Conditions.
A total of 5 μM Zn(II)4-SBP was incubated with 30 μM MagFura-2. Cu(I) was added sequentially to the indicated final concentrations; 0 μM (solid black, 0×), 10 μM (red, 2×), 20 μM (green, 4×), and 30 μM (blue, 6×). Addition of 15 μM Cu(I) to MagFura-2 (dashed black line) has a minor effect on the spectrum. Addition of stoichiometric Cu(I) is capable of displacing all SBP-bound Zn(II).
We next tested the isolated SBP domain of CRR1 for binding to a CuRE from the CYC6 promoter by electrophoretic mobility shift assay (EMSA). Zn(II)-loaded SBP shows sequence-specific binding, but increasing amounts of copper [added aerobically as Cu(II)] or mercuric ions (both of which inactivate CRR1-dependent expression in copper-deficient Chlamydomonas cells in vivo) inhibited DNA binding activity (Figure 5A). On the other hand, Ni(II) and Co(II) ions, which are not effective in turning off CYC6 expression in vivo are also ineffective in inhibiting DNA binding in vitro. DNA binding was inhibited completely at a Cu(II):protein ratio of ~2 to 3:1, whereas even the highest tested ratio of Ni(II)/Co(II):protein of 100:1 did not interfere with binding. These results were confirmed by fluorescence anisotropy measurements of protein-DNA binding. When we tested the SBP domain of Arabidopsis SPL7, we observed the same pattern of inhibition of DNA binding by Cu(II) or Hg(II) but not Ni(II) or Co(II) (Figure 5A).
Figure 5.
Copper Impacts the Binding of the Isolated SBP Domain to the CuRE.
(A) Cu(II) and Hg(II) but not Co(II) or Ni(II) inhibit DNA binding of SBP. The SBP domain from CRR1 or SPL7 was incubated with equimolar amounts of fluorescein-labeled CYC6-CuRE double-stranded DNA in the presence of Zn(II) alone or with the indicated molar ratios of Cu(II), Hg(II), Ni(II), or Co(II). Additions of the metal ions to the protein were performed aerobically in the absence of DTT. The filled triangles mark the position of unbound fragment. The shift in mobility of the bound fragment is marked with an open triangle.
(B) His-506 is essential for DNA binding. Wild-type and mutated forms of CRR1-SBP were incubated with fluorescein-labeled double-stranded DNA CYC6-CuRE in the presence of Zn(II) and 1 mM DTT and with or without a 2-fold molar excess of Cu(II) over protein. DTT stabilizes the DNA–protein interaction by reducing Cu(II) to Cu(I).
(C) His-455 mutants are less sensitive to Cu(II) inhibition. Wild-type CRR1-SBP or His-455 substitution mutants were incubated with the indicated Cu(II)/protein molar ratio.
Since SBP domains contain two highly conserved His residues (His-455 and His-506 in CRR1, corresponding to His-20 and His-71 in structurally characterized Arabidopsis SPL4 and SPL7 SBP domains; Yamasaki et al., 2004; Birkenbihl et al., 2005), we mutated them to test their importance for the DNA–protein interaction in the EMSA. Mutation of the C-terminal ZF2 zinc-coordinating Cys residue (Cys64SBP/Cys499CRR1) was used as a non-DNA binding control (Birkenbihl et al., 2005). The absolutely conserved His residue (His71SBP/His506CRR1) is essential for DNA binding (Figure 5B, compare lanes 3 and 5 to 1). This is consistent with structural studies that suggest that the positively charged C-terminal tail containing His-506 is directly involved in a specific protein:DNA interaction (Yamasaki et al., 2006). By contrast, mutation of the highly but not absolutely conserved (in 97 of 120 sequences; Guo et al., 2008) His20SBP/His455CRR1 had no impact on DNA binding (Figure 5B, lanes 7 and 9 compared with 1) as expected from earlier work (Birkenbihl et al., 2005); however, when we tested for Cu(II) inhibition of DNA binding, we noted that the inhibitory effect appeared less pronounced in the H455 mutants (Figure 5C, compare lanes 6 to 10 and 11 to 15 to lanes 1 to 5), revealing the relevance of this residue for the interaction of the SBP domain with Cu ions. Interestingly, when the reducing agent DTT is present in the DNA binding assay (as in Figure 5B), the inhibitory effect of Cu ions is blocked, perhaps because Cu(II) ions are reduced to the Cu(I) form. Note that Cu(II) binding to SBP does not impact the structure (see Supplemental Figure 3 online), and cuprous ions are therefore not inhibitory. On the other hand, binding of Cu(II) to the protein changes the structure. In some experiments, the protein precipitates (similar to the apoprotein; see above). In other experiments, when we could record a spectrum, the Cu(II)-treated protein was clearly structurally different (see Discussion).
DNA Binding Is Required in Vivo
The two noncoordinating His residues in the SBP domain (His-455 and His-506) were mutated in the context of full-length CRR1 to test the impact of the mutations in vivo. The test constructs (or a wild-type control) were introduced into the crr1-2.2 strain and evaluated for recovery of photosynthesis in copper-deficient medium. In copper-replete medium, crr1 mutants are photosynthetically active. CRR1 carrying the His71GlnSBP/His506GlnCRR1 mutation that blocked DNA binding in vitro was unable to activate CYC6 expression in vivo (Table 1) and hence unable to confer photosynthetic growth in copper deficiency (Figure 2). By contrast, the His20GlnSBP/His455GlnCRR1 mutation, which could bind DNA, fully rescued crr1-2.2. We also created chimeric constructs in which the SBP domain of CRR1 was replaced with the SBP domains from SPL7 or SPL1, but these constructs did not function in vivo as assessed by rescue of crr1-2.2. It is possible that there are intraprotein or interprotein interactions with the SBP domain (e.g., with copper delivery factors; see Discussion) that are functionally important in vivo.
The Ankyrin Domain and C-Terminal Cys-Rich Region Distinguish the Hypoxia and Ni Response Region of CRR1 from the Cu Deficiency Response Region
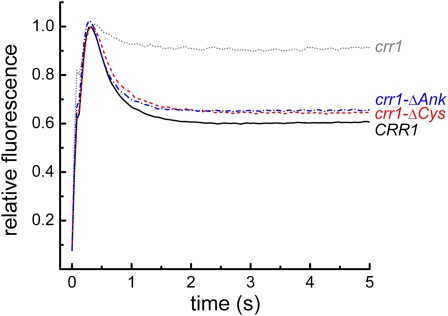
Besides the DNA binding SBP domain, CRR1 has an ankyrin repeat region followed by a C-terminal Cys-rich region that resembles MT2. The ankyrin repeats, which typically function in protein–protein interactions, are found in a subset of SPLs (Riese et al., 2007), and this region was therefore a candidate for functional analysis by mutation. The Cys-rich region was also of interest because of the potential for binding metal ions. Therefore, we deleted the C terminus (residues 1127 to 1230) to generate crr1-ΔCys and made an in-frame deletion of residues 923 to 1108 to generate crr1-ΔAnk, which has an internal deletion of the ankyrin repeats (Figure 1A, Table 1). Constructs carrying these modified versions of CRR1 were able to rescue crr1-2.2 comparable to the constructs encoding wild-type CRR1 with respect to growth and photosynthetic performance in copper-deficient medium (Figure 6). Immunoblot analysis confirmed normal accumulation of Cyt c6 in copper-deficient but not copper-replete medium (Table 1).
Figure 6.
The Ankyrin Repeat Region and the Cys-Rich C-Terminal Region Are Not Involved in the Copper Response.
Constructs encoding wild-type CRR1 or the indicated mutants (see Table 1, Figure 1) were introduced into a crr1-2/arg7 strain. Strains were grown on –Cu solid medium, and fluorescence induction was monitored. Fluorescence induction curves of one representative transformant from each set of constructs are shown. For normalization, the fluorescence at 0.5 s was set to 1.
[See online article for color version of this figure.]
Hg(II) can mimic the action of copper in suppressing the activation of CYC6 expression. This may occur by Hg(II) substitution for copper at a putative copper sensing site in CRR1 (or in other copper sensors) or by Hg(II) binding to a critical thiol, thereby inactivating CRR1 function. The C-terminal Cys-rich region is a candidate site for Hg(II) action as well. Nevertheless, when we tested the crr1-ΔCys construct relative to the wild type in a time-course experiment for downregulation of CYC6 expression, we noted that the response to Hg(II) was unaffected by the deletion (Figure 7). It is therefore more likely that Hg(II) is functioning as a copper mimic in CRR1 (or another copper sensor).
Figure 7.
The Cys-Rich Domain Is Not Required for the Response to Hg(II) Ions.
Wild-type and CRR1ΔCys transformed cells were grown in triplicate under –Cu conditions and supplemented at time t = 0 with 0.5 μM CuCl2, 10 μM HgCl2, or 100 μL water as a control. RNA was collected at the indicated times and analyzed for CYC6 and CBLP by blot hybridization.
When we tested several independent strains carrying the crr1-ΔAnk and -ΔCys encoding constructs for response of CYC6 to hypoxia and nickel supplementation, we noted that the hypoxic response was attenuated for the crr1-ΔAnk construct and greatly diminished for the crr1-ΔCys construct relative to rescue by the construct encoding the wild-type protein (Table 1, Figure 8). Deletion of the ankyrin repeats had only a marginal effect on the CRR1-dependent response of the CYC6 gene to Ni(II) supplementation, but the strain carrying the crr1-ΔCys construct was unable to respond. We conclude that the C-terminal region is not required for nutritional copper signaling but is the target of nickel action, possibly by direct binding of Ni(II) to the Cys-rich region.
Figure 8.
The Cys-Rich C-Terminal Region of CRR1 Mediates the Nickel Response.
CYC6 mRNA abundance was assessed in RNA isolated from individual clones grown in the test condition indicated: –Cu(II) relative to +Cu(II), +Ni(II) relative to –Ni(II), and –O2 relative to +O2 (Schmittgen and Livak, 2008). Four independent transformants were tested for each construct in technical triplicates with a relative standard deviation of 1%. The crr1 strain represents crr1-2.2arg7 rescued with the ARG7 gene. The CRR1 strain represents crr1-2.2arg7 rescued with the ARG7 and CRR1 genes. crr1-ΔAnk and crr1-ΔCys represent crr1-2.2arg7 rescued with ARG7 and mutated CRR1s.
The Cys-Rich Domain Impacts Metal Homeostasis
Since there is no obvious physiological function for nickel in Chlamydomonas, we suspected that the nickel effect might be a clue to a role for the C-terminal region of CRR1 that is involved in regulation of the metabolism of another biologically required transition metal ion. Therefore, we measured the metal content of Chlamydomonas in standard growth conditions or in various stress situations that might impact metal homeostasis (high light, low O2, and high iron) (Long and Merchant, 2008). We found that the zinc content of the ΔCys mutants was ~5-fold greater than that of either the crr1 mutant or the wild-type cells (Figure 9A). This is significant because Chlamydomonas regulates its metal content in wild-type cells tightly (e.g., Merchant et al., 2006; Page et al., 2009) (Figure 10A). The effect of ΔCys is specific for Zn accumulation, and there is no change in Mg, Fe, or Mn accumulation. In the case of copper, the ΔCys mutant is blocked in the hypoxia-dependent increase in Cu content, which is compatible with the domain being involved in hypoxia signaling as well (Figure 9B). The abundance of transcripts encoding putative zinc transporters ZRT1, ZRT2, ZRT3, and ZRT5 is also higher in the ΔCys mutant, as is the abundance of two previously characterized zinc-responsive genes, encoding members of the COG0523 protein family (Haas et al., 2009) (Figure 10B). It is possible that CRR1 plays a role in zinc homeostasis via its C-terminal domain.
Figure 9.
The Cys-Rich Domain Has a Role in Zinc Homeostasis.
The indicated strains (defined in Table 1) were grown in triplicate under the indicated conditions. PFD, photon flux density. Equal numbers of cells were analyzed for zinc (A) and copper (B) content by ICP-MS (see Methods). Bars represent mean values of the biological triplicates ± their standard deviation.
Figure 10.
Increased Zinc Content Correlates with Expression of the Zinc Deficiency Response in crr1-ΔCys Strains.
Independent transformants corresponding to crr1 (triangles apex down), CRR1 (triangles apex up), ΔCys (circles), or the partial ΔCysN (squares) were grown in TAP medium. Cells were collected after reaching a cell density of 8 × 106 per mL and analyzed for metal content or RNA abundance.
(A) Cu and Zn content was measured by ICP-MS. Each symbol represents an independent transformant. The values shown are from one experiment and are within 10% of the values obtained in an independent replication.
(B) Abundance of transcripts encoding candidate Zn transporters (ZRT1, ZRT2, ZRT3, and ZRT5) and members of the COG0523 protein family (protein IDs 117458 and 123019) relative to the CBLP. Mean PCR efficiencies for each gene were determined with LinReg 11x, and transcript abundance was calculated as follows: RTA = 1000 × [mean PCR efficiency for CBLP]CTCBLP × [mean PCR efficiency for ZT]CTZT, where ZT refers to the corresponding gene. Each symbol represents an independent transformant analyzed in technical triplicates. The CT values for each gene tested were within 0.2 cycles within the technical replicates.
DISCUSSION
Nutritional Copper Signaling
A working model for understanding nutritional copper homeostasis is that CRR1, a transcription factor that binds to CuREs, recognizes the GTAC core of these elements via its SBP domain (Quinn et al., 2000). In Arabidopsis, SPL7 is the functional equivalent of CRR1 (Yamasaki et al., 2009). In the moss B. unguiculata, SBP2 is a candidate for the copper response regulator, while in another moss Physcomitrella, we found that SBP11 is the most closely related to CRR1 and SPL7 (Nagae et al., 2008; Figure 1B). Both CRR1 and the CuREs are required for acclimation of Chlamydomonas to nutritional copper deficiency. Here, we show that CRR1 has an NLS (Table 2) and that DNA binding via the SBP domain is one critical component of CRR1 function (Figure 5). The presence of CuREs in genes required for acclimation to copper deficiency, such as those encoding the assimilatory CTR transporters, Cyt c6 replacing plastocyanin in photosynthesis, and other enzymes (e.g., the aerobic oxidative cyclase and ferredoxin 5) allows copper-deficient cells to maintain essential metabolic pathways when the supply of copper in the environment is limiting (Quinn and Merchant, 1995; Moseley et al., 2000; Page et al., 2009; Lambertz et. al., 2010). The presence of CuREs in the 5′ flanking region of these genes provides a mechanism for their enhanced CRR1-dependent expression in copper deficiency. But what prevents the expression of CRR1 targets in a copper-replete cell?
The activity of CRR1 must be regulated by copper, and several mechanistic scenarios are possible. CRR1 may be differently expressed or localized in the copper-replete versus the copper-deficient cell, or its DNA binding activity may be modulated by copper; there are precedents in other systems for either type of regulatory mechanism (Saydam et al., 2001; Brown et al., 2002; Beaudoin et al., 2003; Keller et al., 2005). Blot hybridization indicates that CRR1 expression is not significantly different in copper-replete versus -deficient cells, and the occurrence of Ni(II)-responsive CRR1-dependent regulation of genes in copper-replete cells suggests that the protein is also present in copper-replete cells (Quinn et al., 2003; Kropat et al., 2005). Therefore copper must be regulating CRR1 at the level of activity, perhaps by copper-dependent trafficking of the protein or copper-dependent modification of DNA binding and/or transcriptional activation function(s). The former model is attractive because sequence analysis identifies both an NLS (Figure 1A) and a putative nuclear export signal (residues 574 to 580). However, we were unable to test this directly in Chlamydomonas because of a lack of reagent for visualizing the protein in vivo. Nevertheless, in the heterologous assay, CRR1 was localized to the nucleus even in copper-replete seedlings. Therefore, we presently favor the idea that copper might affect the ability of CRR1 to bind DNA or affect transcriptional activation, either by direct interaction of CRR1 with copper ions or interaction of CRR1 with another copper binding protein.
Two different in vitro experiments support a direct interaction of copper ions with CRR1. In one experiment, we show evidence for the stoichiometric binding of Cu(I) to the purified SBP domain. CD spectra and absorbance spectroscopy confirm that the protein retains structure and that a characteristic metal to thiolate charge transfer is evident (Figures 3 and 4). In a second experiment, we show that the in vitro Zn(II)-dependent DNA binding activity can be abrogated only by metals that also block transcriptional activation in vivo, namely, Cu(II) and Hg(II), but not Co(II) or Ni(II) (Figure 5A). The specificity of this interaction is evident from the fact that a nonliganding His residue in the SBP domain is required (Figure 5C). Interestingly, the effect of copper on inhibiting DNA binding seems to require oxidized Cu(II) ions (Figure 5B). This is consistent with the fact that Cu(I)-SBP formed anaerobically and Zn(II)-SBP appear to adopt similar structures (see Supplemental Figure 2 online).
This leaves open the question of how copper ions become bound to CRR1 in vivo and whether delivery is protein mediated and occurs in the nucleus or cytoplasm. Given the precedent for the involvement of specific protein chaperones for intracellular copper delivery in other cellular compartments, a specific protein for copper delivery to CRR1 in vivo may well be involved (reviewed in Field et al., 2002; Pilon et al., 2006; Banci et al., 2010). Indeed, there are several proteins with Atx1-like copper binding domains encoded in plant genomes whose functions remain to be determined; there is one documented example of the ability of an Atx1-like copper chaperone to delivery Cu(I) specifically to a transcriptional regulator (Cobine et al., 2002). The regulated expression or activity of such a delivery protein might determine the extent of expression of the CRR1 regulon.
There are two candidate protein–protein interaction domains in CRR1: the ankyrin repeats and a potential Leu zipper motif (residues 852 to 894). The ankyrin repeat region is not well conserved between CRR1 and SPL7 (the Arabidopsis ortholog), and based on mutational analysis presented here does not appear to be functionally important for the copper deficiency response (Figure 6, Table 1). One explanation for this result may be that our deletion does not encompass the entire ankyrin domain. Indeed, in more recent analyses, when we used a protein homology/analogy recognition engine that predicts structural similarities (Phyre, http://www.sbg.bio.ic.ac.uk/∼phyre/), all of the top scoring models contained more than seven ankyrin domains. One top hit was a euchromatic histone-Lys N-methyltransferase 1 recognizing histone H3K9 mono- and dimethyl Lys, one of the important epigenetic markers for gene activation/repression. A second interaction domain, a potential Leu zipper motif, is conserved in SPL7 but has not yet been tested for function.
The in vivo importance of Cu(I) versus Cu(II) in the SBP domain function is unclear. Reduced Cu(I) is thought to be the predominant intracellular form of copper due to a high concentration of cellular reductants and the fact that known eukaryotic copper binding regulators, including Ace1, Amt1, Cuf1, and Mac1 of Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Candida glabrata and MTF-1 of Drosophila all bind Cu(I) very tightly, in many cases forming a tetranuclear metal cluster (e.g., Winge, 1998; Brown et al., 2002; Chen et al., 2008). Bacterial copper regulators bind Cu(I) as well, usually to a Cys-containing low coordination number (n ≤ 3) mononuclear Cu(I) site (Changela et al., 2003; Liu et al., 2007; Ma et al., 2009). However, the in vivo and in vitro metal selectivity may be different for CRR1 versus the yeast regulators; in contrast to CRR1, the yeast regulators respond to silver ions (Fürst and Hamer, 1989; Zhou and Thiele, 1991; Casas-Finet et al., 1992; Heredia et al., 2001; Shetty et al., 2004), which is consistent with Ag(I) being a structural surrogate for Cu(I).
Thus, we are open to the possibility that there is a role for Cu(II) ions in nutritional copper signaling in plants. Although it is widely accepted that copper ions are reduced to Cu(I) in vivo by GSH or other low molecular weight thiols, many copper proteins function in electron transfer pathways or other redox reactions in which the Cu(II) oxidation state is stabilized. It seems possible then that in copper-deficient cells, SBP is occupied by Zn(II) and functions as a transcriptional activator. In copper-supplemented cells, the 105 higher affinity for Cu(I) allows substitution of Zn(II) by Cu(I). Oxidation of this Cu(I) to form Cu(II) or alternatively a disulfide bond would change the structure, block DNA binding, and hence transcriptional activation. Another possibility is that Cu(I) initially binds stochiometrically and is isostructural with Zn(II) (see Supplemental Figure 3 online) but at higher molar ratios forms a metal-linked oligomer, which is unlikely to adopt the same structure and thus be nonfunctional for DNA binding. The Cu(I) titrations do not saturate in vitro (see Supplemental Figure 2 online). We cannot distinguish between these models until we can probe the structure of the SBP domain in vivo.
The effect of hypoxia on the CRR1 regulon might be mediated, at least in part, by a decrease in the Cu(II)/Cu(I) ratio under a low partial pressure of O2. The hyperaccumulation of copper in hypoxic cells (Figure 9B) would then result from CRR1- and Cu(I)-dependent upregulation of copper assimilation by CTRs (Page et al., 2009). Since there are several SBP domain proteins in plants, this work raises the question of whether copper nutrition might impact the function of other SPLs as well. While this is possible, it is more likely that copper is delivered to a subset of SPLs (e.g., the ones with the extended SBP domain; Figure 1A) through specific protein–protein interaction.
C-Terminal Cys-Rich Domain
Previously, we observed that the low O2 and nickel responses mediated by CRR1 and the CuREs were not identical to the copper deficiency response, and we therefore did not favor a model in which nickel ions might be interfering with the copper sensing site (Quinn et al., 2000, 2002, 2003). Mutational analysis presented here clearly separates the nickel-responsive domain of CRR1 from the copper-responsive domain (Figures 5A and 8). Arabidopsis SPL7 lacks the C-terminal Cys-rich region and accordingly does not respond to nickel in the same way (Yamasaki et al., 2009), but the Volvox ortholog does include this C-terminal domain (see Supplemental Figure 1B online).
The physiological significance of the nickel response of Chlamydomonas is not known because the alga does not appear to use nickel-containing enzymes (the hydrogenase is an Fe-only type [Happe et al., 1994; Posewitz et al., 2009], and there is no urease [Fernández et al., 2009]), although the genome encodes a candidate nickel transporter (Hanikenne et al., 2009). Nickel is perhaps best viewed as a pharmacological probe of another transition metal ion. The misregulation of zinc content and the ZRT genes (Figures 9 and 10) suggests that CRR1 might be involved also in zinc homeostasis, but the underlying mechanism is, as yet, not clear.
Our present view is that the C-terminal domain is a sensor of zinc status and functions to repress the nutritional zinc regulon. The repression mechanism must require the rest of the CRR1 molecule because crr1 strains remain repressed. For instance, the SBP domain may require an interaction with the C terminus for repressing activity. Deletion of the C-terminal domain relieves repression. The discovery of a molecular connection between copper and zinc homeostasis is potentially significant and might speak to a mechanism for maintaining copper and zinc intracellular ratios. Indeed, recent work suggests that deletion of the Cu(I) chaperone in cyanobacteria results in increased bioavailable zinc, a finding that also suggests crosstalk between zinc and copper homeostasis systems (Dainty et al., 2010). We do know that crr1 strains are unable to grow in zinc-deficient medium, which solidifies the connection between copper and zinc metabolism.
The C-terminal region also impacts (but does not abolish) hypoxic signaling by CRR1 (Figure 8). This might be because the hypoxic response is mediated in part by the redox state of intracellular copper ions (discussed above) and in part by the redox state of the C-terminal thiols on CRR1. More detailed phenotypic comparison of strains carrying the wild-type CRR1 versus the crr1-ΔCys derivative might reveal the importance of hypoxic sensing by the Cys-rich region.
METHODS
Strains and Media
Chlamydomonas reinhardtii strains 2137, CC425, and crr1-2 isolate #2 (referred to subsequently as crr1-2.2) were grown in TAP liquid medium with or without copper (Quinn and Merchant, 1998). If necessary, Arg was added to a concentration of 30 μg/mL. For growth on plates, the media were supplemented with 1.5% EDTA-washed agar. Milli-Q (Millipore) water was used for all media and buffers.
General DNA Manipulations
Bacterial strains used were DH5α for cloning and BL21(DE3) for expression of recombinant proteins. DNA manipulation was according to standard methods (Sambrook et al., 1989) unless stated otherwise. Point mutations and additional endonuclease restriction sites were introduced via a PCR-based method (Sommer et al., 2004). Briefly, the plasmid to be mutagenized was amplified using mutation-carrying sense and antisense oligonucleotides. After PCR, the template was digested with DpnI, and the PCR product was transformed and amplified in Escherichia coli. For manipulations of CRR1, the plasmid CRR1-F1-B1, containing the 6-kb BamHI fragment, which rescues the crr1 mutant, was used (Kropat et al., 2005). All constructs were verified by sequencing. Primer sequences are listed in Supplemental Table 2 online.
Construction of Full-Length cDNA and 35S-EGFP–Tagged Construct
To create a full-length cDNA for CRR1, two fragments of DNA, spanning introns 1 and 2, respectively, were amplified from reverse-transcribed RNA using primer pairs CRR1-50/CRR1-43 and CRR1- 24/CRR1-1 (see Supplemental Table 1 online). These fragments were digested with XbaI/AcuI and AcuI/AscI, respectively, and ligated together with the 1100-bp AcuI fragment from plasmid CRR1-F1-B6 into XbaI/AscI-digested vector pMCS (see below) to yield pCRR1cDNA-Asc. In the next step, a PCR product from CRR1-F1-B6, generated by amplification with primer pair cDNA-Sac/Seq-Ank (see Supplemental Table 1 online) and pCRR1cDNA-Asc, was cut with SacI/AscI and ligated to yield pCRR1cDNA. For construction of an EGFP-tagged CRR1 under the control of the 35S promoter, pCRR1cDNA and pEGAD were digested with BamHI and ligated to yield pEGADxCRR1.
Construction of Deletion and Point Mutants for in Vivo Assay
Plasmid pMCS containing additional restriction sites AatII, MluI, and AscI and lacking the NotI site was created by modification of plasmid pBluescript II KS+ (Stratagene). Primers pKS+MCS-for and -rev (see Supplemental Table 1 online) were annealed, digested with SacII/XbaI, and ligated into SacII/XbaI-digested pBluescript II KS+.
Subclones of plasmid CRR1-F1-B6 (see above) were created by digestion with MluI/XbaI (3504 bp), MluI/AscI (1064 bp), AscI/ClaI (1574 bp), and AatII/ClaI (970 bp) and ligation into pMCS to generate pMCS-MX, pMCS-MA, pMCS-AscC, and pMCS-AatC, respectively. The 1893-bp ApaI and the 928-bp NotI fragments of pMCS-MX were further subcloned in pMCS to generate pMCS-Apa and pMCS-Not, respectively.
To generate the deletion construct of the Cys-rich region pCRRΔCys, first an additional PstI restriction site was introduced in pMCS-AatC by mutagenesis, using primers CYSdel-for and -rev, to generate pMCS-AatC+Pst. The 309-bp PstI fragment in pMCS-AatC+Pst containing the Cys-rich region was then removed by digestion, and the remaining plasmid was religated to yield pMCS-AatC-Pst. Finally, the AatII/ClaI fragment of CRR1-F1-B6 was replaced by the AatII/ClaI fragment of pMCS-AatC-Pst to yield pCRRΔCys.
The upstream region of the Cys-rich domain was deleted by eliminating the 114-bp NotI fragment from pMCS-AatC to yield pMCs-AatC-Not. The 856-bp AatII/ClaI fragment of pMCs-AatC-Not was then used to replace the 970-bp AatII/ClaI fragment of CRR1-F1-B6 to yield pCRR1ΔCysN.
To delete the predicted ankyrin repeats, the 556-bp NcoI fragment of pMCS-AscC was eliminated to yield pMCS-AscC-Nco. The 1019-bp AscI/ClaI fragment of pMCs-AscC-Nco was then used to replace the 1574-bp AscI/ClaI fragment of CRR1-F1-B6 to yield pCRR1ΔAnk.
Deletion of the Met-rich region was accomplished by first introducing an additional NotI site into pMCS-MX by mutagenesis, using primers METdel-for and -rev (see Supplemental Table 1 online) to generate pMCS-MX+Not. Then, the 96-bp NotI fragment containing the Met-rich region of CRR1 was excised from pMCS-MX+Not to yield pMCS-MX-Not. Finally, the 3408-bp MluI/XbaI fragment of pMCS-MX-Not was used to replace the 3504-bp MluI/XbaI fragment of CRR1-F1-B6 to yield pCRR1ΔMet.
The first intron was deleted by introducing consecutively two AgeI sites at the borders of intron1 in pMCS-Apa by mutagenesis using primers INT1del1-for and -rev and INT1del2-for and -rev (see Supplemental Table 1 online) to yield pMCS-Apa+Age. The 175-bp AgeI fragment, corresponding to intron1, was eliminated from pMCS-Apa+Age to generate pMCS-Apa-Age. The 1718-bp ApaI fragment of pMCS-Apa-Age was then used to replace the 1893-bp ApaI fragment of pMCS-MX to generate pMCS-MX-Age, and the MluI/XbaI fragment of pMCS-MXAge was then used to replace the corresponding fragment in CRR1-F1-B6 to yield pCRR1ΔInt1. A frame shift 3′ of the first intron, after the first ATG, was generated by introducing the additional nucleotide A in pMCS-Apa by mutagenesis, using primers STA1del-for and -rev (see Supplemental Table 1 online) to yield pMCS-Apa-St1. The 1894-bp ApaI fragment of pMCS-Apa-St1 was then used to replace the 1893-bp ApaI fragment of pMCS-MX to generate pMCS-MXSt1, and the 3505-bp fragment MluI/XbaI of pMCS-MX-St1 was then used to replace the corresponding 3504-bp fragment in CRR1-F1-B6 to yield pCRR1ΔSt1.
Point mutations in the AHA motif were generated by mutagenesis using the listed primer pairs AHA1-for and -rev or AHA2and3-for and -rev (see Supplemental Table 1 online) and pMCS-Apa as template and cloning the mutagenized fragments back into CRR1-F1-B6, which replaced the original fragments.
Point mutations in the SBP domain were generated by mutagenesis using the listed primer pairs SBPH20Q-for and -rev, SBPH71A-for and –rev, or SBPH71Q-for and -rev (see Supplemental Table 1 online) and pMCS-Not as template and then cloning the mutagenized fragments back into CRR1-F1-B6 replacing the original fragments. Plasmids were purified using Qiagen midiprep columns.
Localization of CRR1-GFP
Onion epidermis cells and mustard hypocotyl epidermis cells were transfected and analyzed as described (Hiltbrunner et al., 2005, 2006; Stolpe et al., 2005).
Fluorescence Induction
Kautsky curves were recorded with a FluorCam 700MF as described (Kropat et al., 2005). In brief, cells were adapted to darkness for 10 min followed by exposure to light. There is a rapid increase in chlorophyll fluorescence representing the reduction of the plastquinone (PQ) pool. The fluorescence decays slowly to a steady state as the PQ pool is reoxidized by PSI. The kinetics of the raise and decay in fluorescence is an indicator of the function of photosystem II (reduces PQ) and PSI (oxidizes PQ).
In Vivo Assay of Mutated CRR1
Mutated constructs were introduced into crr1-2.2 by electroporation. Cells were grown to a density of 3 × 106 cells/mL, collected by centrifugation at 3000g for 5 min. and then treated with isolated autolysin (Harris, 1989) for 0.5 to 1 h. Cell wall lysis was monitored by microscopy by testing for susceptibility to 1% Triton X-100. Cells were collected by centrifugation and resuspended in TAP + 40 mM sucrose to a cell density of 1 × 108 cells/mL. A 250-μL aliquot of these cells was mixed with 0.5 μg pArg7.8, a plasmid containing the argininosuccinate lyase-encoding gene (Debuchy et al., 1989), plus 2 to 3 μg of the DNA construct in a 0.4-cm electroporation cuvette (settings 800 Ω, 25 μF, 800 V, 5.6 to 6.4 ms). Treated cells were incubated overnight in TAP medium in the light, collected by centrifugation, and plated on solid TAP medium to identify Arg prototrophs. After restreaking twice on TAP –Cu plates, cells were tested for their ability to grow on –Cu medium (by visual inspection) and analyzed for photosynthetic electron transfer by fluorescence rise and decay kinetic measurements. crr1 mutants are blocked in the decay component in copper-deficient conditions but show wild-type decay in copper replete medium. To assay the presence of the test DNA in the prototrophs, genomic DNA was isolated from 1.5-mL cultures. Collected cells were resuspended in 500 μL lysis buffer (2% [w/v] CTAB, 100 mM Tris-HCl, pH 8, 1.4 M NaCl, 20 mM EDTA, pH 8, and 2% [v/v] β-mercaptoethanol) and heated for 1 h at 65°C. DNA was then isolated by phenol/chloroform extraction and analyzed by amplification with plasmid specific primers (CRR1 borders screen1-for and -rev and screen2-for and –rev; see Supplemental Table 2 online).
Metal Concentration Determination in Binding Assays
Metal concentrations were determined by atomic absorbance spectroscopy on a Perkin-Elmer AAnalyst 400 using a linear calibration range from 0 to 0.75 ppm Zn(II). The concentration of zinc in protein samples of known concentration (typically 5 to 10 μM quantified by UV absorbance and following an exhaustive dialysis) was determined and compared with a series of standards of known zinc concentration. A final dialysis solution was analyzed in exactly the same way, and the latter determination substracted from the former to obtain protein-associated Zn(II).
ESI-MS
ESI-MS was performed at the Indiana University Mass Spectrometry facility on an Agilent 6130 quadrupole mass spectrometer with liquid chromatography performed on a Waters 1200 HPLC with a C8 stationary phase and 0.1% formic acid mobile phase with a gradient of 5 to 95% acetonitrile. Native ESI-MS was accomplished in 25 mM ammonium acetate buffer at pH 7.
UV-VIS Copper Binding Assays
Copper binding assays were performed anaerobically on a Hewlett Packard 8453 spectrophotometer by monitoring absorbance from 190 to 800 nm at ambient (≈22°C) temperature. Cu(I) solutions were prepared from ultrapure CuCl (Sigma-Aldrich) in 50 mM HEPES and 150 mM NaCl at pH 7.0. Concentration was determined by atomic absorbance spectroscopy as described above. Direct titration absorbance data were plotted as a function of Cu(I) concentration and fit to two sequential site model on DynaFit (Kuzmic, 1996) according to the script provided in Supplemental Figure 4 online. Zn(II) displacement experiments were conducted using MagFura-2 purchased from Invitrogen (Molecular Probes).
Luminescence Copper Binding Assay
Luminescence measurements were performed as described (Chen et al., 2008). The Zn(II)-loaded SBP domain of CRR1 was purified as described and dialyzed to remove residual Zn(II) using buffer L (10 mM HEPES, pH 7.4, 10 μM ZnCl2, and 100 mM NaCl) to exchange the buffer under strictly anaerobic conditions in a glove box. Cu(I) solution was freshly prepared from CuCl in 10 mM MES buffer, pH 6.25, and Cu(I) concentration was determined by ICP-OES (TJA Radial IRIS 1000 inductively coupled plasma optical emission spectrometry). Three milliliters of 10 μM protein in buffer L was titrated under anaerobic conditions with aliquots of Cu(I) solution. Luminescence was monitored with a Photon Technology International Quanta Master Spectrofluorimeter, with excitation at 300 nm and emission at 500 to 675 nm.
CD Spectroscopy
CD spectroscopy was performed on a Jasco J715 spectrometer at the Indiana University Physical Biochemistry Instrumentation Facility using a 0.1-cm cuvette in 25 mM phosphate and 100 mM sodium fluoride at pH 7.0 and 25°C. Samples were prepared anaerobically. The indicated concentration of Cu(I) was incubated with Zn(II)-SBP for 10 min. Spectra are an average of 20 scans.
Construction, Expression, and Purification of SBP Domains
Primers SBPex-for and SBPex-rev (see Supplemental Table 1 online) were used to amplify the SBP domain coding DNA from plasmid CRR1-F1-B6. The product was digested with NdeI and HindIII and ligated into vector pET21b (Novagene) to yield plasmid pET21-WT-SBP, which includes amino acids 424 to 524 of CRR1 with an additional five residues (MKASM) on the N terminus. Mutated versions of the SBP domain H455A, H455Q, C499S, H506Q, and H506A were generated with a PCR-based method (Sommer et al., 2004) using primers SBPH20A-for and -rev, SBPH20Q-for and -rev, SBPC6S-for and -rev, SBPH71Q-for and -rev, and SBPH71A-for and -rev (see Supplemental Table 1 online). SPL7 SBP domains were amplified from cDNA clones provided by Peter Huijser and from plasmid pda01562 (RIKEN). For expression of the proteins, the plasmids were introduced into BL21(DE3) cells, and transformants were grown in Luria-Bertani medium to an OD600 of 0.7. Expression of the protein was initiated by addition of isopropyl β-D-1-thiogalactopyranoside and ZnCl2 to a final concentration of 0.5 mM each, and the cultures were maintained for another 6 h at 37°C. Cells were collected by centrifugation at 5000g and washed with buffer A (20 mM HEPES, 50 mM NaCl, and 50 μM ZnCl2, pH 7.4), and the pellet was resuspended in 5 volumes of buffer A. Complete protease inhibitor without EDTA (Roche) was added, and cells were disrupted by sonication (Sonopuls HD2070, Bandelin; 12 cycles, 30 s/cycle, microtip, 30% output). Cell debris were removed by centrifugation for 30 min at 45,000g, and the resulting supernatant was diluted 2.5-fold with buffer A. The SBP domain was purified by strong cation exchange chromatography on SP-Sepharose (GE Healthcare) using a 10 × 1.6-cm column with a 200-mL linear gradient of buffers A and B (20 mM HEPES, 1 M NaCl, and 50 μM ZnCl2, pH 7.4). Fractions containing the protein of interest were pooled, concentrated, and further purified and depleted of residual zinc ions in the buffer over a 30 × 1-cm Superose12 column (GE Healthcare) with buffer C (10 mM phosphate and 150 mM NaCl, pH 7.4). After purification, proteins were judged to be >95% pure as assessed from Coomassie Brilliant Blue–stained gels. Concentrations of the proteins were determined by spectroscopy using an extinction coefficient of 3355 M−1cm−1 as calculated with ProteinParam (www.expasy.org). To confirm integrity and the reduced state of the cysteines within the proteins, free thiols were assayed with Ellman’s reagent DTNB as described (VanZile et al., 2000). If necessary, Cys residues were reduced in the presence of 50 μM ZnCl2 by addition of 100-fold excess of TCEP for 30 min at room temperature or incubation with 200 mM DTT at 37°C for 4 h. As a final step, reductants and zinc ions were removed from the buffer either by dialysis (against buffer A followed by five rounds against buffer A without ZnCl2) or by gel filtration on the Superose12 column as described above. Only proteins with a free thiol content of >93% were used for experiments. SBP purified in this way contained 4.5 ± 0.5 mol equiv Zn(II) as determined by atomic absorption spectroscopy (see above). A simple modification of the above protocol in which 0.5 mM NTA was used in place of 50 μM ZnCl2 resulted in a preparation containing ≈2.0 mol equiv Zn(II).
EMSA
Appropriate oligonucleotide pairs were mixed at 50 μM each in 10 mM Tris-Cl, 1 mM EDTA, and 10 mM MgCl2, heated to 100°C in a water bath, and annealed by cooling slowly overnight. The indicated amounts of protein and metals were incubated aerobically with 50 pmol double-stranded DNA in binding buffer Z (10 mM Tris-Cl, 0.05% spermidine, 100 mM NaCl, 10 μM ZnCl2, and 4% glycerol, pH 7.5) at 25°C for 10 min and subsequently separated by PAGE (10% monomer) in 0.5× TAE (20 mM Tris, 10 mM acetic acid, and 0.5 mM EDTA, pH 8.5) at 100 V for 45 min. DNA was visualized by UV light for the fluorescein-labeled DNA.
RNA Analysis
RNA abundance was estimated by real-time PCR or by blot hybridization as described previously (Allen et al., 2007). Cells for RNA isolation were collected at a density of 3 to 6 × 106 cells/mL. –Cu cells were grown for three consecutive rounds in –Cu medium, and +Ni cells were collected 6 h after addition of NiCl2 to a final concentration of 50 μM in the medium. −O2 cells were grown for 24 h in 1% air, 2% CO2, and 97% N2 by bubbling before collection. For testing the response to Hg(II) ions, HgCl2 (10 μM) or CuCl2 (0.5 μM) was added to duplicate Cu-deficient cultures. Ten-milliliter samples were collected at the indicated times (before metal addition = 0, 20, 40, 60, and 90 min after addition) for RNA extraction.
Intracellular Metal Content Determination
Cells were grown under the indicated conditions to a cell density of 5 × 106 cells/mL. Sample preparation and metal measurements by ICP-MS were as described (Merchant et al., 2006).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database or the Arabidopsis Genome Initiative database under the following accession numbers: Chlamydomonas CRR1, AAY33924; Volvox carterii CRR1, XP_002948544.1; Arabidopsis SPL7, At5G18830.1; Oryza sativa SPL1, LOC_Os01g18850; Sorghum bicolor SBP1, Sb09g020110; Populus trichocarpa SBP16, XP_002303781.1; Physcomitrella patens SBP11, ABV03806.1; Ricinus cummunis SBP1, XP_002516839.1; Cleome spinosa SBP1, ABD96886.1; and Vitis vinifera SBP1, XP_002270226.1; Chlamydomonas ZRT1, XP_00169290.1; Chlamydomonas ZRT2, XP_001700327.1; Chlamydomonas ZRT3, XP_001693505.1; Chlamydomonas ZRT5, XP_001702078.1; Chlamydomonas 117458, XP_001692771; and Chlamydomonas 123019, XP_001702424.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignments of CRR1 and Other SBP Domain Proteins.
Supplemental Figure 2. Stoichiometric versus Superstoichiometric Binding of Cu(I) to the SBP Domain.
Supplemental Figure 3. Cu(I) Does Not Significantly Distort SBP Structure.
Supplemental Figure 4. Dynafit Script for Two Sequential Site Models.
Supplemental Table 1. Primers Used for DNA Manipulations.
Supplemental Table 2. Primers Used for Functional Analysis.
Acknowledgments
We thank Tim Kunkel at the University of Freiburg for help with mustard transformation and microscopy, Andrew Mason and Chris Mull from the California State University Long Beach for help with ICP-MS, and Peter Huijser for Arabidopsis SPL-encoding DNAs and helpful discussion. This work was supported by the National Institutes of Health (GM42143 to S.S.M., GM042569 to D.P.G., and 1F32GM083562 to D.M.) and Deutsche Forschungsgesellschaft (SO706/1-1 and SO706/2-1 to F.S.).
References
- Allen M.D., del Campo J.A., Kropat J., Merchant S.S. (2007). FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii. Eukaryot. Cell 6: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan K., Egli D., Selvaraj A., Zhang B., Georgiev O., Schaffner W. (2004). Metal-responsive transcription factor (MTF-1) and heavy metal stress response in Drosophila and mammalian cells: A functional comparison. Biol. Chem. 385: 597–603 [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., McGreevy K.S., Rosato A. (2010). Molecular recognition in copper trafficking. Nat. Prod. Rep. 27: 695–710 [DOI] [PubMed] [Google Scholar]
- Beaudoin J., Mercier A., Langlois R., Labbé S. (2003). The Schizosaccharomyces pombe Cuf1 is composed of functional modules from two distinct classes of copper metalloregulatory transcription factors. J. Biol. Chem. 278: 14565–14577 [DOI] [PubMed] [Google Scholar]
- Birkenbihl R.P., Jach G., Saedler H., Huijser P. (2005). Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352: 585–596 [DOI] [PubMed] [Google Scholar]
- Brown K.R., Keller G.L., Pickering I.J., Harris H.H., George G.N., Winge D.R. (2002). Structures of the cuprous-thiolate clusters of the Mac1 and Ace1 transcriptional activators. Biochemistry 41: 6469–6476 [DOI] [PubMed] [Google Scholar]
- Burkhead J.L., Reynolds K.A., Abdel-Ghany S.E., Cohu C.M., Pilon M. (2009). Copper homeostasis. New Phytol. 182: 799–816 [DOI] [PubMed] [Google Scholar]
- Cardon G., Höhmann S., Klein J., Nettesheim K., Saedler H., Huijser P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237: 91–104 [DOI] [PubMed] [Google Scholar]
- Casas-Finet J.R., Hu S., Hamer D., Karpel R.L. (1992). Characterization of the copper- and silver-thiolate clusters in N-terminal fragments of the yeast ACE1 transcription factor capable of binding to its specific DNA recognition sequence. Biochemistry 31: 6617–6626 [DOI] [PubMed] [Google Scholar]
- Changela A., Chen K., Xue Y., Holschen J., Outten C.E., O’Halloran T.V., Mondragón A. (2003). Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301: 1383–1387 [DOI] [PubMed] [Google Scholar]
- Chen X., Hua H., Balamurugan K., Kong X., Zhang L., George G.N., Georgiev O., Schaffner W., Giedroc D.P. (2008). Copper sensing function of Drosophila metal-responsive transcription factor-1 is mediated by a tetranuclear Cu(I) cluster. Nucleic Acids Res. 36: 3128–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobine P.A., George G.N., Jones C.E., Wickramasinghe W.A., Solioz M., Dameron C.T. (2002). Copper transfer from the Cu(I) chaperone, CopZ, to the repressor, Zn(II)CopY: Metal coordination environments and protein interactions. Biochemistry 41: 5822–5829 [DOI] [PubMed] [Google Scholar]
- Dainty S.J., Patterson C.J., Waldron K.J., Robinson N.J. (2010). Interaction between cyanobacterial copper chaperone Atx1 and zinc homeostasis. J. Biol. Inorg. Chem. 15: 77–85 [DOI] [PubMed] [Google Scholar]
- Dancis A., Yuan D.S., Haile D., Askwith C., Eide D., Moehle C., Kaplan J., Klausner R.D. (1994). Molecular characterization of a copper transport protein in S. cerevisiae: An unexpected role for copper in iron transport. Cell 76: 393–402 [DOI] [PubMed] [Google Scholar]
- Debuchy R., Purton S., Rochaix J.-D. (1989). The argininosuccinate lyase gene of Chlamydomonas reinhardtii: An important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8: 2803–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M., Moseley J.L., Tottey S., Del Campo J.A., Quinn J.M., Kim Y., Merchant S. (2004). Genetic dissection of nutritional copper signaling in chlamydomonas distinguishes regulatory and target genes. Genetics 168: 795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández E., Llamas Á., Galván A. (2009). Nitrogen assimilation and its regulation. The Chlamydomonas Sourcebook. Organellar and Metabolic Processes, Stern D., (San Diego, CA: Academic Press; ), pp. 69–113 [Google Scholar]
- Field L.S., Luk E., Culotta V.C. (2002). Copper chaperones: Personal escorts for metal ions. J. Bioenerg. Biomembr. 34: 373–379 [DOI] [PubMed] [Google Scholar]
- Fürst P., Hamer D. (1989). Cooperative activation of a eukaryotic transcription factor: Interaction between Cu(I) and yeast ACE1 protein. Proc. Natl. Acad. Sci. USA 86: 5267–5271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A.Y., Zhu Q.H., Gu X., Ge S., Yang J., Luo J. (2008). Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418: 1–8 [DOI] [PubMed] [Google Scholar]
- Haas C.E., Rodionov D.A., Kropat J., Malasarn D., Merchant S.S., de Crécy-Lagard V. (2009). A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10: 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanikenne M., Merchant S.S., Hamel P.P. (2009). Transition metal nutrition: A balance between deficiency and toxicity. The Chlamydomonas sourcebook, Stern D., (San Diego: Academic Press; ), pp. 333–400 [Google Scholar]
- Happe T., Mosler B., Naber J.D. (1994). Induction, localization and metal content of hydrogenase in the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 222: 769–774 [DOI] [PubMed] [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Source Book. (San Diego: Academic Press; ). [Google Scholar]
- Heredia J., Crooks M., Zhu Z. (2001). Phosphorylation and Cu+ coordination-dependent DNA binding of the transcription factor Mac1p in the regulation of copper transport. J. Biol. Chem. 276: 8793–8797 [DOI] [PubMed] [Google Scholar]
- Hill K.L., Li H.H., Singer J., Merchant S. (1991). Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6. Analysis of the kinetics and metal specificity of its copper-responsive expression. J. Biol. Chem. 266: 15060–15067 [PubMed] [Google Scholar]
- Hiltbrunner A., Tscheuschler A., Viczián A., Kunkel T., Kircher S., Schäfer E. (2006). FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczián A., Bury E., Tscheuschler A., Kircher S., Tóth R., Honsberger A., Nagy F., Fankhauser C., Schäfer E. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15: 2125–2130 [DOI] [PubMed] [Google Scholar]
- Howe G., Merchant S. (1992). Heavy metal-activated synthesis of peptides in Chlamydomonas reinhardtii. Plant Physiol. 98: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Nadas I.A., Kim M.A., Franz K.J. (2005). A Mets motif peptide found in copper transport proteins selectively binds Cu(I) with methionine-only coordination. Inorg. Chem. 44: 9787–9794 [DOI] [PubMed] [Google Scholar]
- Keller G., Bird A., Winge D.R. (2005). Independent metalloregulation of Ace1 and Mac1 in Saccharomyces cerevisiae. Eukaryot. Cell 4: 1863–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J., Saedler H., Huijser P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250: 7–16 [DOI] [PubMed] [Google Scholar]
- Kropat J., Tottey S., Birkenbihl R.P., Depège N., Huijser P., Merchant S. (2005). A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 102: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmic P. (1996). Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal. Biochem. 237: 260–273 [DOI] [PubMed] [Google Scholar]
- Lambertz C., Hemschemeier A., Happe T. (2010). The anaerobic expression of the ferredoxin encoding FDX5 gene of Chlamydomonas reinhardtii is regulated by the Crr1 transcription factor. Eukaryot. Cell 9: 1747–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Nazarenus T.J., Stone J.M. (2008). Identification of a consensus DNA-binding site for the Arabidopsis thaliana SBP domain transcription factor, AtSPL14, and binding kinetics by surface plasmon resonance. Biochemistry 47: 3645–3653 [DOI] [PubMed] [Google Scholar]
- Liu T., Ramesh A., Ma Z., Ward S.K., Zhang L., George G.N., Talaat A.M., Sacchettini J.C., Giedroc D.P. (2007). CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 3: 60–68 [DOI] [PubMed] [Google Scholar]
- Long J.C., Merchant S.S. (2008). Photo-oxidative stress impacts the expression of genes encoding iron metabolism components in Chlamydomonas. Photochem. Photobiol. 84: 1395–1403 [DOI] [PubMed] [Google Scholar]
- Ma Z., Cowart D.M., Scott R.A., Giedroc D.P. (2009). Molecular insights into the metal selectivity of the copper(I)-sensing repressor CsoR from Bacillus subtilis. Biochemistry 48: 3325–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S., Bogorad L. (1986). Rapid degradation of apoplastocyanin in Cu(II)-deficient cells of Chlamydomonas reinhardtii. J. Biol. Chem. 261: 15850–15853 [PubMed] [Google Scholar]
- Merchant S., Hill K., Howe G. (1991). Dynamic interplay between two copper-titrating components in the transcriptional regulation of cyt c6. EMBO J. 10: 1383–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., Allen M.D., Kropat J., Moseley J.L., Long J.C., Tottey S., Terauchi A.M. (2006). Between a rock and a hard place: Trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 1763: 578–594 [DOI] [PubMed] [Google Scholar]
- Moseley J., Quinn J., Eriksson M., Merchant S. (2000). The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19: 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.L., Page M.D., Alder N.P., Eriksson M., Quinn J., Soto F., Theg S.M., Hippler M., Merchant S. (2002). Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14: 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M., Nakata M., Takahashi Y. (2008). Identification of negative cis-acting elements in response to copper in the chloroplastic iron superoxide dismutase gene of the moss Barbula unguiculata. Plant Physiol. 146: 1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M.D., Kropat J., Hamel P.P., Merchant S.S. (2009). Two Chlamydomonas CTR copper transporters with a novel cys-met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell 21: 928–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M., Abdel-Ghany S.E., Cohu C.M., Gogolin K.A., Ye H. (2006). Copper cofactor delivery in plant cells. Curr. Opin. Plant Biol. 9: 256–263 [DOI] [PubMed] [Google Scholar]
- Posewitz M.C., Dubini A., Meuser J.E., Seibert M., Ghirardi M.I. (2009). Hydrogenases, hydrogen production, and anoxia. The Chlamydomonas Sourcebook: Organellar and Metabolic Processes, Stern D., (San Diego, CA: Elsevier; ), pp. 217–255 [Google Scholar]
- Puig S., Lee J., Lau M., Thiele D.J. (2002). Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J. Biol. Chem. 277: 26021–26030 [DOI] [PubMed] [Google Scholar]
- Puig S., Thiele D.J. (2002). Molecular mechanisms of copper uptake and distribution. Curr. Opin. Chem. Biol. 6: 171–180 [DOI] [PubMed] [Google Scholar]
- Quinn J.M., Barraco P., Eriksson M., Merchant S. (2000). Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J. Biol. Chem. 275: 6080–6089 [DOI] [PubMed] [Google Scholar]
- Quinn J.M., Eriksson M., Moseley J.L., Merchant S. (2002). Oxygen deficiency responsive gene expression in Chlamydomonas reinhardtii through a copper-sensing signal transduction pathway. Plant Physiol. 128: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.M., Kropat J., Merchant S. (2003). Copper response element and Crr1-dependent Ni(2+)-responsive promoter for induced, reversible gene expression in Chlamydomonas reinhardtii. Eukaryot. Cell 2: 995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.M., Merchant S. (1995). Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7: 623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.M., Merchant S. (1998). Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 297: 263–279 [DOI] [PubMed] [Google Scholar]
- Quinn J.M., Nakamoto S.S., Merchant S. (1999). Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper response elements and increased translation from a copper deficiency-specific form of the transcript. J. Biol. Chem. 274: 14444–14454 [DOI] [PubMed] [Google Scholar]
- Riese M., Höhmann S., Saedler H., Münster T., Huijser P. (2007). Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene 401: 28–37 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Saydam N., Georgiev O., Nakano M.Y., Greber U.F., Schaffner W. (2001). Nucleo-cytoplasmic trafficking of metal-regulatory transcription factor 1 is regulated by diverse stress signals. J. Biol. Chem. 276: 25487–25495 [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Shetty R.S., Deo S.K., Liu Y., Daunert S. (2004). Fluorescence-based sensing system for copper using genetically engineered living yeast cells. Biotechnol. Bioeng. 88: 664–670 [DOI] [PubMed] [Google Scholar]
- Sommer F., Drepper F., Haehnel W., Hippler M. (2004). The hydrophobic recognition site formed by residues PsaA-Trp651 and PsaB-Trp627 of photosystem I in Chlamydomonas reinhardtii confers distinct selectivity for binding of plastocyanin and cytochrome c6. J. Biol. Chem. 279: 20009–20017 [DOI] [PubMed] [Google Scholar]
- Stolpe T., Süsslin C., Marrocco K., Nick P., Kretsch T., Kircher S. (2005). In planta analysis of protein-protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma 226: 137–146 [DOI] [PubMed] [Google Scholar]
- Stone J.M., Liang X., Nekl E.R., Stiers J.J. (2005). Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 41: 744–754 [DOI] [PubMed] [Google Scholar]
- VanZile M.L., Chen X., Giedroc D.P. (2002). Structural characterization of distinct α3N and α5 metal sites in the cyanobacterial zinc sensor SmtB. Biochemistry 41: 9765–9775 [DOI] [PubMed] [Google Scholar]
- VanZile M.L., Cosper N.J., Scott R.A., Giedroc D.P. (2000). The zinc metalloregulatory protein Synechococcus PCC7942 SmtB binds a single zinc ion per monomer with high affinity in a tetrahedral coordination geometry. Biochemistry 39: 11818–11829 [DOI] [PubMed] [Google Scholar]
- Walkup G.K., Imperiali B. (1997). Fluorescent chemosensors for divalent zinc based on zinc finger domains. Enhanced oxidative stability, metal binding affinity, and structural and functional characterization. J. Am. Chem. Soc. 119: 3443–3450 [Google Scholar]
- Winge D.R. (1998). Copper-regulatory domain involved in gene expression. Prog. Nucleic Acid Res. Mol. Biol. 58: 165–195 [DOI] [PubMed] [Google Scholar]
- Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. (2009). SQUAMOSA Promoter Binding Protein-Like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K., et al. (2004). A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 337: 49–63 [DOI] [PubMed] [Google Scholar]
- Yamasaki K., et al. (2006). An Arabidopsis SBP-domain fragment with a disrupted C-terminal zinc-binding site retains its tertiary structure. FEBS Lett. 580: 2109–2116 [DOI] [PubMed] [Google Scholar]
- Zhou P.B., Thiele D.J. (1991). Isolation of a metal-activated transcription factor gene from Candida glabrata by complementation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88: 6112–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M., Clarke O., Gulbis J.M., Keizer D.W., Jarvis R.S., Cobbett C.S., Hinds M.G., Xiao Z., Wedd A.G. (2009). Metal binding affinities of Arabidopsis zinc and copper transporters: Selectivities match the relative, but not the absolute, affinities of their amino-terminal domains. Biochemistry 48: 11640–11654 [DOI] [PubMed] [Google Scholar]