In animals, sorting nexin (SNX) proteins associate with vacuolar protein sorting (VPS) proteins to form the retromer complex, which is involved in protein sorting and a variety of developmental processes. By contrast, this study shows that plant SNXs are dispensable for VPS assembly and functions, suggesting that the two retromer subcomplexes should be seen as molecular modules.
Abstract
Sorting nexins (SNXs) are conserved eukaryotic proteins that associate with three types of vacuolar protein sorting (VPS) proteins to form the retromer complex. How SNXs act in this complex and whether they might work independently of the retromer remains elusive. Here, we show by genetic and cell imaging approaches that the Arabidopsis thaliana SNX1 protein recruits SNX2 at the endosomal membrane, a process required for SNX1-SNX2 dimer activity. We report that, in contrast with the mammalian retromer, SNXs are dispensable for membrane binding and function of the retromer complex. We also show that VPS retromer components can work with or independently of SNXs in the trafficking of seed storage proteins, which reveals distinct functions for subcomplexes of the plant retromer. Finally, we provide compelling evidence that the combined loss of function of SNXs and VPS29 leads to embryo or seedling lethality, underlining the essential role of these proteins in development.
INTRODUCTION
The retromer is a multiprotein complex, first identified in yeast, that is strongly conserved among eukaryotes (Attar and Cullen, 2010). The retromer is involved in the recycling of transmembrane receptors, known in yeast as vacuolar sorting receptors, which mediate the transport of vacuolar/lysosomal hydrolases. Interestingly, new functions of this complex have been recently established in multicellular organisms. Indeed, the retromer complex is involved in trafficking of the sorting receptor Wntless and the secretion of its ligand Wnt in Caenorhabditis elegans, Drosophila melanogaster, and human cells (Eaton, 2008). In mammals, the retromer complex is composed of two subcomplexes, one consisting of dimers of SNX1, SNX2, SNX5, and SNX6 (Wassmer et al., 2009) and the other, also known as the core retromer, containing VPS26, VPS29, and VPS35 (Attar and Cullen, 2010). SNXs are characterized by the presence of a PHOX (NADPH Phagocyte Oxidase) homology (PX) domain, which is involved in the interaction with various phosphoinositides (PIPs) of the endosomal membranes (Teasdale et al., 2001; Seaman and Williams, 2002). SNXs also possess a BAR domain, which mediates dimerization and binding to highly curved membranes (Carlton et al., 2004). The depletion of SNXs causes membrane dissociation of VPS proteins and hence leads to the loss of retromer function, underlining the crucial role played by SNXs in the assembly and function of the entire retromer (Rojas et al., 2007). SNXs bind to endosomal membranes independently of VPS proteins, generate membrane curvature, and are required for the recruitment of the core retromer (Carlton et al., 2004; Rojas et al., 2007). However, complete VPS subcomplex attachment to the endosomal membrane is achieved by the action of the small GTPase Rab7 (Rojas et al., 2008; Seaman et al., 2009). SNXs are conserved proteins found in all eukaryotic systems and are involved in intracellular sorting and trafficking of membrane proteins (Carlton et al., 2005). We identified the first plant SNX in Brassica oleracea as an interactor of various receptor kinases (Vanoosthuyse et al., 2003). Strikingly, the Arabidopsis thaliana genome contains only three SNX genes versus 10 in yeast and 33 in mammals. In a previous work, we described snx1 null mutants, which exhibit a semidwarf phenotype associated with some other subtle developmental defects (Jaillais et al., 2006). This weak phenotype suggested a possible redundancy between SNX1 and the other two SNX proteins in Arabidopsis, SNX2a and SNX2b. Functional redundancy has already been reported for the mouse SNX1 and SNX2, which are close homologs of the plant SNXs. In mice, whereas snx1 or snx2 single mutants are viable and fertile, snx1 snx2 double mutants are embryonic lethal (Schwarz et al., 2002). A similar lethality phenotype was described in mice lacking a functional VPS26 gene, in which the retromer function is altered (Lee et al., 1992). These data underline the absolute requirement of the two retromer subcomplexes for proper embryo development in mice. Here, we address the function of SNX proteins in plant development through the analysis of single and combinatorial snx loss-of-function mutants in Arabidopsis. We show that, contrary to the yeast and mammalian retromer, SNX dimers are not required for core retromer recruitment on the endosomal membrane and are not essential for core retromer function. However, we found that both SNXs and VPS29 are required for proper Arabidopsis development, as plants impaired simultaneously in SNX and VPS29 functions do not survive. Finally, we also demonstrate that SNX2a and SNX2b, although seemingly redundant, have distinct functions in trafficking of storage proteins and plant development.
RESULTS
SNX1 and SNX2 Act Redundantly in Seedling Growth and Development
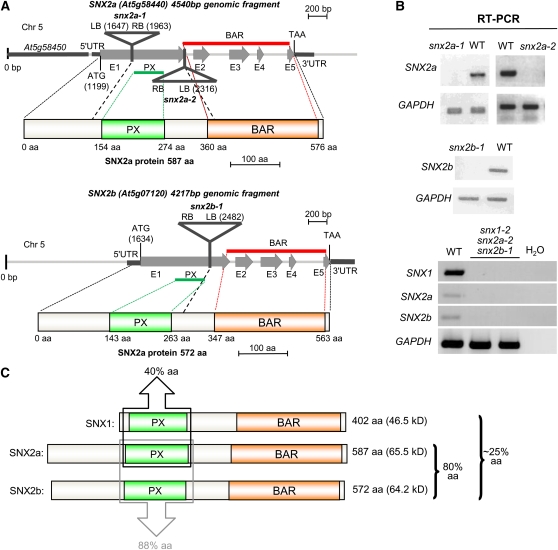
Homozygous T-DNA insertion alleles of SNX2a (i.e., snx2a-1 and snx2a-2) and SNX2b (snx2b-1) genes obtained from public collections were isolated and used in crossing with the previously described snx1-2 line (Jaillais et al., 2006) to generate snx double and triple mutant lines. The absence of full-length transcripts in the respective mutant lines was then demonstrated by RT-PCR analysis (Figures 1A and 1B). All T-DNA insertions in SNX2 genes occurred in exon 1, which encodes the N terminus, the PX domain, and the very beginning of the BAR domain of SNX2 proteins (Figure 1A). Comparison of the respective amino acid sequences of SNXs shows the high overall percentage identity between SNX2a and SNX2b sequences (80% identity), whereas SNX1 shares only 25% identity with SNX2s (Figure 1C).
Figure 1.
Identification of snx Mutants.
(A) Genomic structure of the SNX2a (At5g58440) and SNX2b (At5g07120) genes and position of the T-DNA insertions identified in the snx2a and snx2b mutant lines. Gray arrows indicate exons labeled from E1 to E5. The predicted proteins are shown below each gene, with the PX and BAR domains labeled. aa, amino acids; LB, left border; RB, right border; UTR, untranslated region.
(B) RT-PCR analyses reveal that snx2a-1, snx2a-2, and snx2b-1 single mutants (top panels) and the snx1-2 snx2a-2 snx2b-1 triple mutant (bottom panel) do not express the corresponding full-length mRNAs. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a positive control and water as a negative control. WT, wild-type plant.
(C) Comparison of amino acid sequences and domain localization of SNX1, SNX2a, and SNX2b proteins. Percentages indicate amino acid identity (UniProtKB) between the full-length proteins or in the boxed regions as indicated. SNX1 shares 24% amino acid identity with SNX2a and 25% with SNX2b. The black box indicates the ~40% identity between any SNX2 PX domain and the SNX1 PX domain. The gray box indicates the amino acid identity between SNX2 PX domains.
[See online article for color version of this figure.]
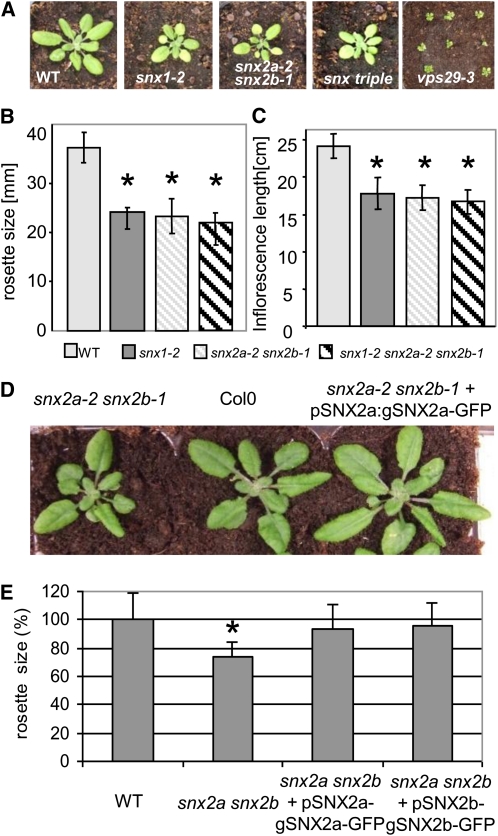
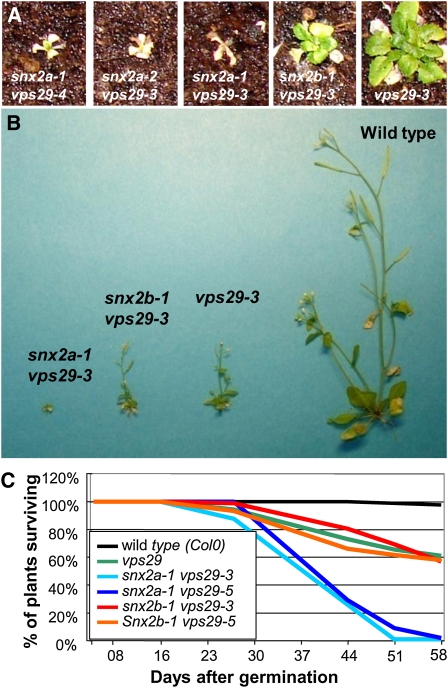
To uncover the function of Arabidopsis SNX proteins, we then examined the phenotypes of single, double, and triple mutants disrupted in SNX genes. Whereas the snx2a or snx2b single mutants were indistinguishable from wild-type plants, the snx2a-1 snx2b-1 and snx2a-2 snx2b-1 double mutants displayed phenotypes similar to the snx1 single mutants, with reduced rosette size and inflorescence length, as well as root gravitropism defects (Jaillais et al., 2006) (Figures 2A to 2C; see Supplemental Figure 1 online). Strikingly, in double mutants impaired in both SNX1 and SNX2 functions (i.e., snx1 snx2a or snx1 snx2b) as well as in triple mutants (snx1-2 snx2a-1 snx2b-1), no additional morphological defects were observed (Figures 2A to 2C; see Supplemental Figure 1 online). This result suggests that SNX1 with either of the two SNX2 proteins is required for proper Arabidopsis growth. To determine whether SNX2a and SNX2b are interchangeable, we introduced a genomic fragment of either SNX2a or SNX2b fused to the green fluorescent protein (GFP) sequence into the snx2a-2 snx2b-1 double mutant. Either construct complemented the mutant growth phenotype, indicating a common function for the two SNX2s (Figures 2D and 2E; see Supplemental Figure 2 online). Surprisingly, Arabidopsis plants interrupted in all three SNX genes exhibited rather weak developmental phenotypes compared with the strong defects found in vps29 single mutants (Figure 2A; Jaillais et al., 2007). Altogether, our data suggest that SNXs might not play an essential role in retromer function and that SNX2a and SNX2b are redundant and work together with SNX1 in seedling growth. We previously reported strong genetic interactions between SNX1 and VPS29 (Jaillais et al., 2007). Thus, we next looked for possible genetic interactions between SNX2 and VPS29 genes. Surprisingly, whereas we observed no marked effect of the loss of SNX2b function in the vps29 background, we found a drastic developmental defect caused by nonfunctional SNX2a leading to lethality of snx2a vps29 plants 51 to 58 d after germination (Figures 3A to 3C). This observation implies that SNX2s and VPS29 have distinct roles, which nonetheless overlap in plant growth and development, and that SNX2a and SNX2b do not strictly share identical functions during development.
Figure 2.
Phenotypes of snx Mutants and Complementation Test.
(A) Rosette phenotype of wild-type (WT), snx1 single, snx2a snx2b double, snx1 snx2a snx2b triple, and vps29 single mutant plants after 4 weeks of growth in soil.
(B) Comparison of the rosette size of 4-week old plants between wild-type and snx1-2 single, snx2a-2 snx2b-1 double, and snx1-2 snx2a-2 snx2b-1 triple mutant plants.
(C) Comparison of the inflorescence length of 7-week-old plants between wild-type and snx1-2 single, snx2a-2 snx2b-1 double, and snx1-2 snx2a-2 snx2b-1 triple mutant plants.
(D) Rosette phenotype of wild-type plant and snx2a-2 snx2b-1 double mutants nontransformed or transformed with the pSNX2a:gSNX2a-GFP genomic construct after 4 weeks of growth in soil.
(E) pSNX2a:SNX2a-GFP and pSNX2b:SNX2b-GFP genomic constructs rescue the rosette phenotype of 21-d-old snx2a-2 snx2b-1 double mutant plants.
In (B), (C), and (E), bars represent sd, the number of plants analyzed was 26, and asterisks indicate that mutant lines are statistically different from the wild type (P values < 0.001).
[See online article for color version of this figure.]
Figure 3.
SNX1, SNX2, and VPS29 Genetically Interact.
(A) Rosette phenotype of various allelic combinations of snx2a vps29 and snx2b-1 vps29-3 double mutants and vps29-3 single mutant plants after 4 weeks of growth in soil shows the differential genetic interactions between SNX2a, SNX2b, and VPS29 genes.
(B) Phenotype of snx2a-1 vps29-3 and snx2b-1 vps29-3 double mutants, vps29-3 single mutant, and wild-type plants after 50 d of growth in soil.
(C) Survival of wild-type and vps29-3 single, snx2a-1 vps29-1, snx2a-1 vps29-3, snx2b-1 vps29-1, and snx2b-1 vps29-3 double mutant plants at up to 58 d of growth in soil.
SNXs Localize to the Same Endosome and Interact to Form Dimers in Vivo and in Planta
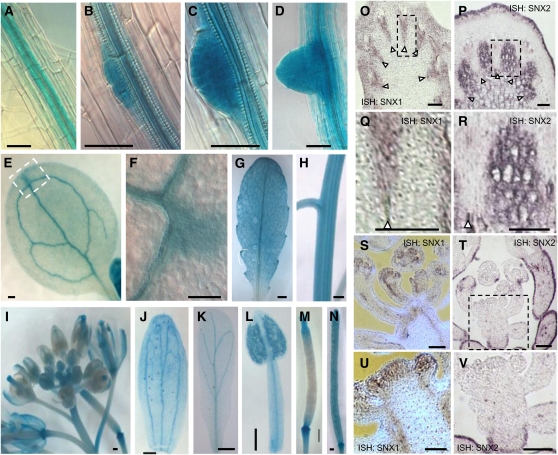
We first investigated the expression patterns of SNX genes to determine whether regions of coexpression existed between these genes. For this purpose, we generated constructs based on a genomic sequence of either SNX1, SNX2a, or SNX2b fused in frame with the β-glucuronidase (GUS) reporter gene and introduced them into the relevant snx mutant plants. GUS staining, which allows detection of the respective SNX-GUS fusion proteins, revealed that the transgenes were active in all plant organs analyzed, including cotyledons, hypocotyls, roots, stems, floral buds, flowers, and leaves (Figures 4A to 4N; see Supplemental Figure 3 online). Vascular tissues of all organs and lateral root primordia showed strong GUS staining (Figures 4A to 4D). The ubiquitous expression of SNXs was confirmed by RT-PCR analysis (see Supplemental Figure 4 online) and in situ hybridization on sections (Figures 4O to 4V). Interestingly, this latter approach revealed some subtle differences between the expression of SNX1 and SNX2 genes, with SNX2a and SNX2b probes giving similar hybridization signals. In vascular tissues of inflorescence stems, SNX1 was expressed exclusively in the xylem pole, whereas SNX2 mRNA was found in the pith and interfascicular regions (Figures 4O to 4R). A differential expression pattern was also observed in floral apical meristems, in which only SNX1 transcripts were detected (Figures 4S to 4V). SNX2 genes were also found to be strongly expressed in the stem epidermis compared with SNX1 (Figures 4P and 4T).
Figure 4.
Expression Pattern of SNXs.
(A) to (N) SNX-GUS–expressing lines reveal that SNX proteins display a highly similar expression pattern throughout the plant. Similar expression patterns were observed for the three At-SNX lines. Results presented are from SNX1-GUS. In vegetative tissues, SNX-GUS proteins (visualized by GUS activity generating blue color) are expressed mostly in the vascular tissue of the root (A), in young root primordia ([B] to [D]), cotyledons ([E] and [F]), leaves (G), and stem (H). SNX-GUS proteins are also highly expressed in inflorescences (I), in the vascular tissues of sepals (J) and petals (K), as well as in anthers (L) and siliques ([M] and [N]).
(O) to (V) In situ hybridization was performed using probes specific for SNX1 or SNX2. SNX2a and SNX2b probes gave identical hybridization signals, here referred to as ISH:SNX2. In the inflorescence stem ([O] to [R]), SNX1 is expressed mainly in cortical layers and in the xylem poles ([O] and [Q]). SNX2s show a complementary expression pattern restricted to the interfascicular regions and pith ([P] and [R]) of the stem. SNX1 ([S] and [U]) and SNX2 ([T] and [V]) expression in inflorescence meristems of Col-0 wild-type plants. Only SNX1 is expressed in the inflorescence meristem (S) and in very young floral buds (U).
Bars = 50 μm in (A) to (F) and (O) to (R), 250 μm in (G) to (N), and 100 μm in (S) to (V). Arrowheads point to xylem poles. Dashed-line rectangles in (E), (O), (P), and (T) highlight the areas enlarged in (F), (Q), (R), and (V), respectively.
Next, to check whether SNXs localize to the same subcellular compartments, we used the previously described Arabidopsis transgenic lines stably expressing SNXs tagged with either the GFP or the monomeric red fluorescent protein (mRFP) (Jaillais et al., 2006). By confocal microscopy analysis of root tip cells, we found all SNXs to be present in the cytosol and also to colocalize to punctate intracellular compartments (Figures 5A and 5B; see Supplemental Figure 5 online). After crossing the different SNX2-fluorescent protein lines with lines expressing markers of the prevacuolar compartment (RABF2b-GFP and SNX1-mRFP), trans-Golgi network (TGN; VHA-a1-GFP), or recycling endosome (GNOM-GFP), we detected a good colocalization of SNX2 proteins with SNX1 and RABF2b (Figure 5A; see Supplemental Figures 5A to 5C online) as deduced from the Pearson’s coefficient (r) (Bolte and Cordelières, 2006). By contrast, only a few compartments exhibited colabeling of SNX2-mRFP with the other two markers (see Supplemental Figures 5D to 5G online). This observation is in agreement with our previous data showing that SNX1 and VPS29 colocalize to the same endosomal compartments, also RABF2b positive, which we defined as sorting endosomes/prevacuolar compartment (PVC) (Jaillais et al., 2008), and with the localization of VPS35 to the PVC (Yamazaki et al., 2008). These SNX-VPS29–containing compartments exhibited sensitivity to two drugs altering endosomes, Brefeldin A and Wortmannin (Wm) (Jaillais et al., 2008; see Supplemental Figures 5H to 5M online). Although we cannot exclude partial colocalization between SNX proteins and the TGN, it is worth noting that we previously reported that Wm has no effect on the TGN labeled with VHA-a1-GFP in Arabidopsis root cells, supporting the idea that SNX proteins predominantly localized to the PVC (Jaillais et al., 2008). Surprisingly, SNXs and VPS29 have recently been localized to the TGN in a study based on the expression of retromer components in tobacco (Nicotiana tabacum) BY2 cells and Arabidopsis protoplasts, as well as on the immunodetection of SNX2a or VPS29 in root cells (Niemes et al., 2010). Although the reason for such a discrepancy remains unclear, it is likely to reflect the differences in the procedure used to study retromer localization. In our experiments, we used only live imaging of root tissues expressing functional retromer-fluorescent proteins under the control of their endogenous promoter.
Figure 5.
SNXs Colocalize and Interact in Vivo and in Planta.
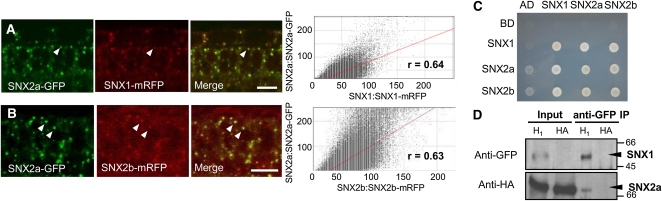
(A) and (B) SNX2a-GFP fusion proteins are found both in the cytosol and in punctate compartments and colocalize with SNX1-mRFP (A) and SNX2b-mRFP (B). Arrowheads indicate colocalizing SNX2a-GFP and SNX-labeled endosomes. The scatterplots at right correspond to the colocalization events for each pair of fusion proteins. r, Pearson’s coefficient. Bars = 10 μm.
(C) Y2H assays reveal that SNXs interact to form homodimers and heterodimers. Yeast was grown on a medium lacking Trp, His, and Leu. Growth on this medium indicates protein interaction. AD, Gal4 activation domain; BD, Gal4 binding domain.
(D) IP experiments followed by protein gel blot analysis demonstrate that SNX1-SNX2a complexes form in planta. Leaf extracts from Col-0 plants coexpressing SNX1-GFP and HA-SNX2a (H1) or expressing only HA-SNX2a (HA), used as a control for the anti-GFP antibody specificity, were treated with an anti-GFP antibody (anti-GFP IP). The respective immunoprecipitates were analyzed by protein gel blotting with either an anti-GFP (top panel) or anti-HA (bottom panel) antibody. Input lanes correspond to leaf extracts before IP. Molecular mass markers are indicated on the right in kilodaltons.
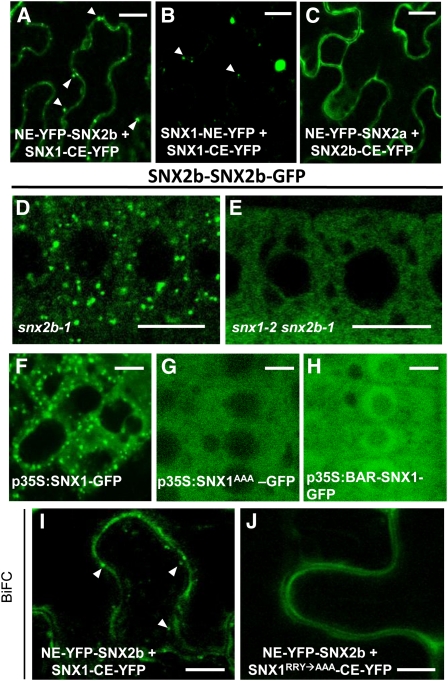
As SNXs are known to associate to form homo- or heterodimers in yeast and mammalian cells (Zhong et al., 2002), we next investigated the possible interactions between SNXs by yeast two-hybrid (Y2H) assays. We found that all SNXs were able to form homo- and heterodimers (Figure 5C). Since SNX proteins are characterized by the presence of a PX and a BAR domain, we investigated the role of these domains in SNX homo/heterodimerization. We cloned the PX and BAR domains of SNX1 and SNX2b for Y2H assays, respectively. We also generated mutated forms of SNXs by replacing the strongly conserved motif RRY of their PX domain by AAA. This mutation was shown to inhibit the PIP binding properties of the PX domain of hSNX1 and to be essential for membrane association (Zhong et al., 2002). Whereas the PX2b domain alone was not capable of interacting with any BAR domain or entire SNX sequence, we generally found interactions between the BAR domains and wild type or mutated sequences of SNXs (see Supplemental Figures 6A to 6F online). These results indicate that the presence of the BAR domain is necessary for the homo- and heterodimerization of SNXs. The mutated SNX1AAA was unable to interact with itself, wild-type SNX1, or the SNX1 BAR domain, underlining the importance of the PX domain for SNX1 homodimerization (see Supplemental Figures 6A and 6D online). To confirm the formation of SNX homo- and heterodimers in planta, we performed an immunoprecipitation (IP) analysis on leaf protein extracts from Arabidopsis Columbia-0 (Col-0) transgenic plants that coexpressed SNX proteins tagged with hemagglutinin (HA) or GFP under the control of the relevant SNX promoters. Immunoprecipitates obtained with either anti-HA or anti-GFP antibody were analyzed by protein gel blot. Only SNX1-SNX2a or SNX1-SNX2b heterocomplexes were detected (Figure 5D; see Supplemental Figures 6G and 6H online), while we failed to isolate any SNX homodimers or SNX2a-SNX2b heterodimers (see Supplemental Figures 6I to 6L online). Finally, to validate these interactions and determine the cytosolic or endomembrane localization of SNX dimers, we performed bimolecular fluorescence complementation (BiFC) assays in tobacco leaf epidermal cells (Desprez et al., 2007). We found that SNX1 formed heterodimers with SNX2a or SNX2b, which localized both in the cytosol and to punctate compartments (Figure 6A; see Supplemental Figure 7A online). These compartments labeled by SNX1 are likely to correspond to endosomes. We also found SNX1 homodimers to localize exclusively to endomembrane compartments, whereas SNX2 homodimers or SNX2a-SNX2b heterodimers were observed only in the cytosol (Figures 6B and 6C; see Supplemental Figures 7B and 7C online). This observation suggests that SNX1 could play an important role in the endosomal localization of SNX1-SNX2 complexes.
Figure 6.
SNX Membrane Association.
(A) to (C) BiFC experiments in tobacco epidermal leaf cells show that only SNX1-containing dimers can associate with endosomes (A) and (B), whereas SNX2 dimers remain cytosolic (C). White arrowheads indicate endosomes. NE-YFP, N-terminal end of yellow fluorescent protein; CE-YFP, C-terminal end of yellow fluorescent protein. Bars = 10 μm.
(D) to (E) In Arabidopsis root cells, SNX2b-GFP localizes to endosomes in a snx2b mutant background (D) but is cytosolic in snx1 snx2b double mutants (E).
(F) to (H) The PX domain is required for endosomal localization of SNX1. Unlike SNX1-GFP (F), SNX1RRY→AAA-GFP or the BAR domain of SNX1 fused to GFP is unable to bind to endosomes in root tip cells ([G] and [H]).
(I) and (J) SNX1 is required for the recruitment of SNX2 proteins to endosomes. In BiFC experiments, wild-type forms of SNX1 and SNX2 interact both in endosomes (white arrowheads) and in the cytosol (I), whereas SNX1RRY→AAA interacts with SNX2 only in the cytosol in tobacco epidermal cells (J). Bars = 10 μm.
SNX1 Endosomal Membrane Localization Is Dependent on a PX–PIP Interaction and Is Required for the Recruitment of SNX2 Proteins to Endosomes
To confirm a role for SNX1 in membrane association of SNX1-SNX2 complexes in Arabidopsis root cells, we looked at SNX2b-GFP localization in snx2b single or snx1 snx2b double mutants. SNX2b-GFP was diffuse in the cytosol in snx1 snx2b mutants, whereas SNX2b-GFP localized to endosomes in the control snx2b mutant (Figures 6D and 6E). This demonstrates that SNX1 is required for membrane association of SNX2b.
To gain insight into how the endosomal binding of SNX1 occurs, we first investigated the importance of phosphatidylinositol-3-phosphate (PI3P) in SNX1 membrane association. PI3P is a major phosphoinositide of endosomal membranes that interacts with the PX domain of yeast and mammalian SNXs (Burda et al., 2002; Zhong et al., 2002). Wm is a potent inhibitor of the activity of PI 3-kinase, the enzyme catalyzing PI3P production. When mammalian cells are treated with Wm, SNX1 is released from the endosomal membrane and becomes cytosolic (Zhong et al., 2002). We have previously shown that long Wm treatment (60 min) did not alter SNX localization but affected endosome homeostasis, leading to abnormally enlarged endosomes (Jaillais et al., 2006, 2008; see Supplemental Figures 5L and 5O online). Interestingly, we found that short-term Wm treatment (15 min) displaced SNX1-GFP from endosomes, leading to its complete solubilization (see Supplemental Figures 7D to 7G online). By contrast, other endosomal-resident proteins, such as RABF2b, were not relocated upon short Wm treatment (see Supplemental Figures 7H and 7I online). This result suggests that PI3P is important for the endosomal localization of SNX1. To determine the relevance of the RRY motif in membrane association of SNX1, we generated SNX1RRY→AAA-GFP–expressing transgenic plants. The mutated protein was found only in the cytosol, indicating that the localization of SNX1 to the endosomal membrane is dependent on a PX domain–mediated interaction with phosphoinositides (Figures 6F and 6G). Consistent with this, when we overexpressed only the BAR domain of SNX1 tagged with GFP, we found it to be cytosolic (Figure 6H).
We used the SNX1RRY→AAA mutated form in BiFC assays to see the impact of the mutation on SNX1-SNX2 complex formation and localization. Whereas the mutated SNX1 could still form heterodimers with SNX2a or SNX2b, these heterocomplexes were detected only in the cytosol of tobacco epidermal leaf cells (Figures 6I and 6J). Altogether, our data indicate that SNX1 is required for the recruitment of SNX2 proteins to endosomes and that the formation of SNX1-SNX2a or SNX1-SNX2b complexes at the endosomal membrane is essential for SNX functions in plant development.
SNX Proteins Are Not Essential for VPS29 Localization and Function
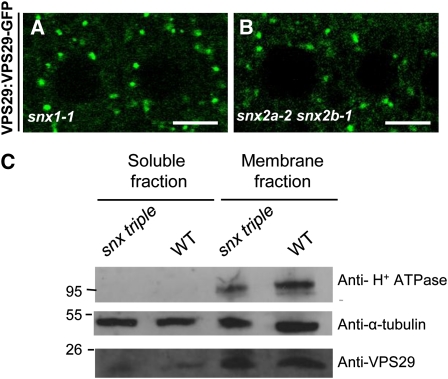
To determine whether SNX1 or SNX2 proteins are important for VPS29 membrane localization, we examined the distribution of VPS29-GFP (Jaillais et al., 2007) in snx1 or snx2a snx2b backgrounds. We found VPS29-GFP to remain associated with endosomes in the snx mutant backgrounds (Figures 7A and 7B). This observation was confirmed by subcellular fractionation and protein gel blot analysis, which revealed that endogenous membrane-bound VPS29 levels did not decrease in the snx1-2 snx2a-2 snx2b-1 triple mutant background (Figure 7C). These results indicate that SNXs are not required for VPS29 association with endosomal membranes. This cannot rule out the possibility that SNXs and VPS29 might physically interact. To clarify this point, we searched for physical associations between these proteins, but we were unable to detect any interactions using Y2H, BiFC assays, or in planta IP performed on plants coexpressing VPS29-GFP and HA-SNXs and using either anti-HA or anti-GFP antibodies for IP (data not shown).
Figure 7.
SNX Proteins Are Not Essential for VPS29 Membrane Localization.
(A) and (B) VPS29-GFP localizes to endosomes and cytosol in root cells of snx1 single (A) or snx2a snx2b double (B) mutants. Bars = 10 μm.
(C) Protein extracts from 15-d-old wild-type and triple snx1-2 snx2a-2 snx2b-1 mutant seedlings were subjected to membrane fractionation and analyzed by immunoblotting using an anti-H+ATPase antibody (top panel) for control of membrane purity and equal loading, an anti-α-tubulin antibody (middle panel) for control of equal loading, and an anti-VPS29 antibody (bottom panel). Molecular mass (in kilodaltons) markers are indicated on the left. WT, wild type Col-0.
SNX Proteins Are Involved in 12S Globulin but Not in 2S Albumin Maturation
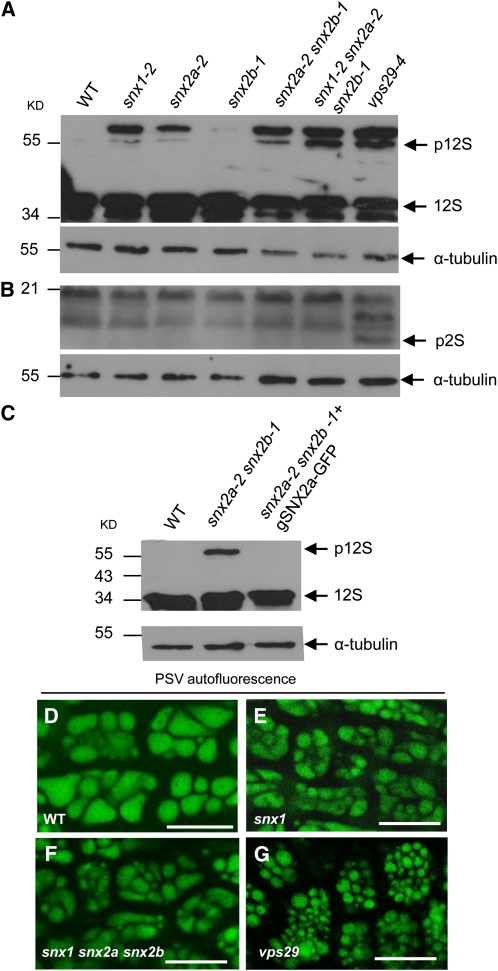
In embryo cells of mature seeds, the loss of function of VPS29 or VPS35 was shown to cause mis-sorting of the major seed storage proteins 12S globulin and 2S albumin, with a portion of them being secreted to the extracellular space instead of being routed to the protein storage vacuoles (PSVs) (Yamazaki et al., 2008). This defect is accompanied by accumulation of storage protein precursors and reduced sizes of PSVs. To determine whether the loss of function of SNX proteins leads to comparable effects in mature seeds, we first compared by immunoblot analysis the storage protein patterns of a variety of snx mutant seeds with those of wild-type and vps29 seeds. Remarkably, whereas vps29 seeds accumulated abnormally high levels of the storage protein precursors pro12S globulin (p12S) and pro2S albumin (p2S), seeds from snx1 and snx2a single, snx2a snx2b double, and snx1 snx2a snx2b triple mutants accumulated only p12S (Figures 8A and 8B). Surprisingly, snx2b seeds exhibited a wild-type-like pattern. This latter result has to be reconciled with our previous observation that loss of function of SNX2b has a weaker effect than loss of function of SNX2a in the vps29 background. These data clearly demonstrate that SNX2a and SNX2b do not strictly share the same functions in plant development. Introduction of the pSNX2a:gSNX2a-GFP construct in a snx2a snx2b double mutant fully complemented the maturation defect of 12S globulin (Figure 8C). Our results indicate that SNXs work with the core retromer for p12S globulin trafficking but are dispensable for 2S albumin maturation, with the latter relying only on a functional core retromer. Finally, the confocal microscope study of the autofluorescent PSVs in mature dry seeds revealed that snx1 and snx1 snx2a snx2b seeds had PSVs of similar sizes, smaller than those of the wild type but larger than vps29 PSVs (Figures 8D to 8G). These observations indicate that impaired SNX functions cause defects in storage protein maturation associated with a slight alteration in PSV size. When the core retromer subcomplex is altered, a stronger phenotype is observed, probably because of drastic alterations of protein trafficking pathways.
Figure 8.
Distinct Role of SNXs and VPS29 in Mediating Maturation of Seed Storage Proteins.
(A) and (B) Pattern of seed storage proteins showing abnormal accumulation of precursor forms of 12S globulin (A) and 2S albumin (B) after immnoblot analysis of wild-type (WT), snx1-2, snx2a-2, snx2b-1 single, snx2a-2 snx2b-1 double, snx1-2 snx2a-2 snx2b-1 triple, and vps29-4 single mutant seeds. Similar protein loading was confirmed by α-tubulin detection. p12S and p2S indicate precursor forms of 12S globulin and 2S albumin, respectively, whereas 12S and 2S indicate mature 12S globulin and 2S albumin, respectively. Molecular mass markers are indicated on the right in kilodaltons.
(C) Abnormal accumulation of precursor forms of 12S globulin in snx2a-2 snx2b-1 double mutant seeds is rescued by transformation with the pSNX2a:gSNX2a-GFP construct. Seed proteins were subjected to immunoblotting using an anti-12S globulin antibody. Similar protein loading was confirmed by α-tubulin. p12S and 12S indicate precursor and mature forms of 12S globulin, respectively. Molecular mass markers are indicated on the left in kilodaltons.
(D) to (G) Compared with the wild type (D), the PSVs are weakly fragmented in the snx1 and snx triple mutants ([E] and [F]) but to a lesser extent than in the vps29 mutant (G). For each confocal microscopy image, several embryo cells (approximately four to eight) from dry mature seeds are shown. Bars = 10 μm.
[See online article for color version of this figure.]
DISCUSSION
SNXs Are Dispensable for the Core Retromer Functions
Altogether, our phenotypic analyses of null mutants, genetic interaction studies, electrophoretic and cellular analyses of mature seeds indicate that SNXs are not essential components for the plant retromer membrane association and functions. This finding is quite distinct from the key role played by the mammalian SNX1 and SNX2 in the proper retromer endosomal localization and function (Rojas et al., 2007). This raises the question of whether SNX proteins are actual components of the plant retromer complex. Our genetic analysis clearly demonstrates that SNX and VPS proteins can function in the same developmental pathways, as exemplified by the embryonic or plantlet lethality observed in some combinations of SNX and VPS29 loss-of-function mutants compared with the weaker phenotypes detected in single null mutants. It is noteworthy that similar genetic interactions have been described in C. elegans between the worm SNX1 homolog VPS5 and the VPS26 or VPS35 genes. Impaired functions for both VPS5 and VPS26 or VPS35 lead to embryonic lethality in the worm, whereas single null mutants are viable (Coudreuse et al., 2006).
A Key Role for SNX1 in SNX Subcomplex Assembly and Function
Our work also shows that SNX function requires heterodimerization between SNX1 and SNX2a or SNX2b, as well as endosomal membrane association of these heterocomplexes. Hence, SNX1 appears to be a key factor in the establishment of a functional SNX complex, as its presence is essential for the recruitment of the other two SNX2 proteins to the endosomal membrane. Although the three plant SNXs possess a PX domain, which is a putative domain for interaction with membrane PI3Ps, our data demonstrate that only SNX1 has the ability to associate with endomembranes in planta. Our earlier phylogenetic analysis of plant SNXs clearly indicated the existence of two classes of SNXs with particular structural features (Vanoosthuyse et al., 2003). Overall, SNX2 sequences share 80% amino acid identity with each other but only 25% with SNX1. Besides, while the PX domains of SNX2s are 88% identical, they share only 40% amino acid identity with that of SNX1. In addition, SNX1 has a short (around 30 amino acids) N-terminal sequence before the PX domain, whereas the two SNX2s exhibit an ~150–amino acid long N-terminal domain (Figure 1C). It is likely that these structural differences play a crucial role in the distinct membrane binding capabilities of SNXs. Thus, although SNX2a and SNX2b possess a PX domain, these domains do not seem sufficient to mediate autonomous membrane binding in planta. The necessity of SNX1 for SNX2 endosome recruitment remains unclear, in particular with regard to previous data showing that the SNX2b PX domain is capable of binding PI3P in vitro (Phan et al., 2008). It is possible that the binding ability of SNX2 proteins to PIPs is only weak and would require oligomerization with SNX1 to ensure endosome binding in planta. Such an increase in lipid association has already been described for animal dynamins, whose weak binding to PIPs is enhanced upon oligomerization (Klein et al., 1998).
Mammalian SNX1 has been described to tubulate membranes and to sense membrane curvature (Carlton et al., 2004). In yeast, such a function has not been reported, and there is no example to date that plant SNXs are involved in membrane tubulation. Because we have shown that plant SNXs are distinguishable from mammalian SNXs regarding core retromer association with the endosomal membrane, we may question whether plant SNXs have conserved a role in membrane tubulation. Further studies are needed to understand better how SNX1 associates with the membrane and recruits SNX2a and SNX2b proteins.
SNX and VPS Subcomplexes Act as Molecular Modules
Although SNX2a and SNX2b seem to act redundantly in plant development, we found that, contrary to SNX2a, SNX2b is dispensable for the proper maturation of 12S globulin storage proteins in seeds. This specific role of SNX2a in seeds is comparable to the distinct functional divergence recently described between the three VPS35 isoforms (Hashiguchi et al., 2010). These data leave open the possibility that SNX and VPS complexes consisting of various isoforms might have specific functions in certain tissues, developmental stages, or environmental conditions. This assumption is supported by the differential expression patterns we found for SNX1 and SNX2 in the vascular tissues of inflorescence stems, epidermal cells, and the shoot apical meristem, as well as by the earlier observation that VPS29 is not expressed in the shoot apical meristem (Jaillais et al., 2007). Finally, our finding that the core retromer can act independently of the SNX complex sustains the idea that both complexes might have distinct cellular and physiological functions. Interestingly, in mammalian cells, SNX1 is involved in the trafficking of G protein–coupled receptors independently of the core retromer (Gullapalli et al., 2006). Altogether, our work and these latter data reveal that the two retromer subcomplexes, as well as their single components, should be seen as molecular modules that can work in concert or separately in mediating different trafficking pathways or developmental processes.
METHODS
Plant Material
The Arabidopsis thaliana Col-0 accession was used as the wild-type plant. The Arabidopsis T-DNA insertion lines snx2a-1 (GABI274A06) and snx2b-1 (GABI105E07) were obtained from Bernd Weisshaar (Max Planck Institute for Plant Breeding Research, Cologne, Germany) and snx2a-2 (SALK127971) from the SALK Institute (Alonso et al., 2003). The snx1-1, snx1-2 (Jaillais et al., 2006), vps29-3, vps29-4, and vps29-5 mutants (Jaillais et al., 2007), as well as the SNX1-GFP, GFP-RABF2b (Jaillais et al., 2006), VPS29-mRFP, VPS29-GFP (Jaillais et al., 2007), GNOM-GFP (Geldner et al., 2003), and VHA-a1-GFP (Dettmer et al., 2006) lines were previously described.
Genotyping of snx Mutants
Genotyping mutant alleles of SNX1 was described previously (Jaillais et al., 2006, 2007). Genotyping wild-type allele of SNX2a was performed with Nex2a-4F/Nex2a-R2 primers and mutant alleles with either Nex2a-R2/LB2SALK for snx2a-2 or Nex2a-4F/LBGABI for snx2a-1. Genotyping wild-type and mutant alleles of SNX2b was performed with the primer pairs Nex2b-F5/Nex2b-R2 and Nex2b-F5/LBGABI, respectively. Primer sequences are given in Supplemental Table 1 online. The absence of SNX expression in mutant seedlings was confirmed by RT-PCR using Nex12F/Nex13 for snx1, Nex2a1/Nex2a5R for snx2a, and Nex2bF4/Nex2bR2 for snx2b (see Supplemental Table 1 online) and a high number (35) of amplification cycles.
Root Gravitropic Response of snx Mutants
Root gravitropism was analyzed as previously described by Jaillais et al. (2006). Briefly, seedlings were grown vertically for 7 d in Petri dishes and turned by 135°. Realignment to the gravity vector was recorded after 40 h. Each root was assigned 1 of 12 30° sectors.
Generation of Expression Vectors and Plant Transformation
The genomic DNA fragments containing the SNX2a and SNX2b genes were amplified by PCR (Phusion Taq; Fermentas) using specific Gateway primers and recombined in pDONR207 (Invitrogen). These fragments were then subcloned in the pMDC107 (Curtis and Grossniklaus, 2003) vector for C-terminal GFP6 fusion and in the pGIInK:mRFP vector for C-terminal mRFP fusion (Rotman et al., 2005).
cDNAs corresponding to the full-length SNX1 sequence and to the BAR domain of SNX1 were amplified by RT-PCR and recombined in the pDONR207 vector (Invitrogen). Site-directed mutagenesis for SNX1RRY→AAA and SNX2bRRF→AAA was performed on residues Arg-66, Arg-67, and Tyr-68, and Arg-189, Arg-190, and Phe-191, respectively. Mutations were created by PCR using specific primers designed with the Stratagen QuikChange Primer Design Program (http://www.stratagene.com/qcprimerdesign). All fragments were then subcloned in pK7FGW2, for C-terminal enhanced GFP fusion (Karimi et al., 2002). The three full-length cDNA sequences of SNXs were cloned in the pAlligator2 vector to generate HA N-terminal fusion proteins (Bensmihen et al., 2004). These tagged constructs were introduced into Col-0 accession as well as into various snx mutants as previously described (Jaillais et al., 2006). For each line, plants with a single T-DNA insertion were chosen for further experiments, and at least five independent transformants were analyzed.
For Y2H, cDNAs corresponding to full-length SNX1 and SNX2 sequences and to the BAR and PX domains of SNX1 and SNX2b were amplified by RT-PCR and recombined in the pDONR207 vector (Invitrogen). These cDNAs, as well as the mutated versions of SNX1 and SNX2b (SNX1RRY→AAA and SNX2bRRF→AAA), were subcloned in the pPC97 vector for Gal4-BD fusions and in the pPC86CYH2 for Gal4-AD fusions (Walhout and Vidal, 2001). All primer pairs used are presented in Supplemental Table 1 online.
Expression Analysis of SNX Genes
To generate SNX-GUS lines, SNX1, SNX2a, and SNX2b genes were cloned in pMDC163 vector (Curtis and Grossniklaus, 2003). These constructs were used to transform snx1-1, snx2a-1, and snx2b-1 mutants, respectively. GUS staining was performed as described by Geldner et al. (2004).
In situ hybridization on sections was performed as described (Zluvova et al., 2006). To produce the probes, we cloned in pGEMTeasy (Promega) the entire open reading frame (ORF) of SNX1 (amplified with Nex12F/Nex39R) and the N-terminal region upstream of the PX domain of SNX2a (amplified with Nex2a1/Nex2aR3) and SNX2b ORF (amplified with Nex2bF4/Nex2bR3), respectively. Primer sequences are given in Supplemental Table 1 online. We used these clones to generate antisense and sense probes.
For the RT-PCR analysis, RNA was isolated from various organs of adult Arabidopsis Col-0 plants. Pairs of primers were designed to specifically amplify each SNX cDNA, and primer pairs were as follows: Nex14/Nex15 for SNX1, Nex2a1/Nex2a2 for SNX2a, and Nex2b1/Nex2b2 for SNX2b. PCR products were sequenced to ensure the selectivity of the amplification. As a control, we used the ubiquitously expressed ACTIN gene (primers Act1 and Act2). All primer pairs used are presented in Supplemental Table 1 online.
Y2H Assays, Immunoprecipitation, and Protein Analysis
Y2H assays were performed as described previously (Jaillais et al., 2007). Immunoprecipitation and protein gel blot analysis were performed as described using anti-GFP (Roche) or anti-HA11 (Covance) antibodies (Jaillais et al., 2007). Total proteins were extracted from 100 seeds of each line, and 10 μg of proteins were separated and analyzed by immunoblotting using anti-2S albumin (1:1000), anti-12S globulin (1:10000) (Shimada et al., 2003), and monoclonal anti-α tubulin (1:2000; Sigma-Aldrich; clone B-5-1-2) antibodies.
For cell fractionation analysis, 300 mg of 15-d-old seedlings were ground in liquid nitrogen, and the powder obtained was resuspended in 1 mL of microsome extraction buffer (50 mM HEPES, pH 7.4, 500 mM sucrose, 1 mM DTT, 5 mM EDTA, and 5 mM EGTA; Roche protease inhibitor cocktail). Extracts were centrifuged at 13,000g for 20 min at 4°C, and supernatants centrifuged again at 13,000g for 10 min at 4°C. Supernatants were then subjected to ultracentrifugation (TLA 100.3 rotor; Beckman) at 100,000g for 90 min at 4°C. Supernatants (soluble fraction) and pellet samples (membrane fraction) were analyzed by immunoblot using anti-α-tubulin (1:2000 dilution; Sigma-Aldrich), anti-H+-ATPase (1:1000 dilution; Agrisera), and anti-VPS29 (Jaillais et al., 2007; 1:1000 dilution) antibodies.
BiFC
SNX cDNAs (ORF from ATG to end with and without STOP) were cloned by Gateway recombination in pBiFP vectors for fusion with either the N-terminal end or C-terminal end of yellow fluorescent protein (Desprez et al., 2007) and transformed in C58 pMP90 Agrobacterium tumefasciens strain. Nicotiana benthamiana leaves were infiltrated as described (Voinnet et al., 2003). After 4 d, infected leaves were analyzed by confocal microscopy. Each experiment was repeated at least three times independently, and more than five infected leaves were analyzed per experiment.
Confocal Microscopy Analysis
Arabidopsis roots of 7-d-old seedlings grown on Murashige and Skoog medium were placed in LM medium and analyzed on an LSM-510 confocal laser scanning microscope (Zeiss) as described (Jaillais et al., 2006). Each experiment was repeated at least three times independently, and >10 plants were analyzed per experiment.
Quantitative Estimation of Colocalization
Confocal images were analyzed with the ImageJ software using the JACoP plugin (Bolte and Cordelières, 2006) to obtain the Pearson's coefficient. A minimum of 15 cells per line was analyzed.
Microscopy and Drug Treatments
Brefeldin A (100 μM in DMSO/ethanol) and Wm (33 μM in DMSO) treatments were performed as described previously (Jaillais et al., 2006, 2007). Autofluorescence of PSVs was analyzed by confocal microscopy after mounting dry seeds in glycerol 87% and squeezing them between a glass slide and cover slip. Each experiment was repeated at least three times independently, and >10 seeds were analyzed per experiment.
Statistical Analysis of Results
All experiments were repeated at least three times independently. All calculations were performed using Microsoft Excel 2003. P values were obtained using a two-sided Student’s test assuming unequal variances.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At5g06140 and Q9FG38 (UniProtKB) for SNX1 gene and protein, respectively; At5g58440 and Q8L5Z7 (UniProtKB) for SNX2a gene and protein, respectively; At5g07120 and B9DFS6 (UniProtKB) for SNX2b gene and protein, respectively; andAt3g47810 and Q9STT2 (UniProtKB) for VPS29 gene and protein, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypes of snx Mutants.
Supplemental Figure 2. Complementation Test of the snx2a-2 snx2b-1 Mutant.
Supplemental Figure 3. Expression Pattern of SNX2a-GUS and SNX2b-GUS Fusion Proteins.
Supplemental Figure 4. Expression Pattern of SNXs.
Supplemental Figure 5. SNXs Colocalize to an Endosomal Compartment.
Supplemental Figure 6. SNXs Interact in Vivo and in Planta.
Supplemental Figure 7. SNX Dimerization and Membrane Association.
Supplemental Table 1. Primers Used in This Work.
Supplementary Material
Acknowledgments
We thank the Max Planck Institute and the SALK Institute for providing the insertion mutant lines. We also thank I. Hara-Nishimura for the gift of anti-2S albumin and anti-12S globulin antibodies, K. Schumacher for the VHA-a1-GFP line, G. Jürgens for the GNOM-GFP line, M. Vidal for pPC97-based and pPC86-CYH2S-based vectors, F. Parcy for pBiFP and pAlligator2 vectors, O. Voinnet for p19 vector, C. Lionnet, C. Chamot, and F. Simian for technical assistance at the Platim IFR128, and V. Bayle for helpful discussions. This work was supported by the Agence Nationale de la Recherche (ANR) BLANC RETROMER project (Grant ANR-08-BLAN-0142). N.T. was supported by a PhD grant from Cluster 9 Région Rhône-Alpes, France.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Attar N., Cullen P.J. (2010). The retromer complex. Adv. Enzyme Regul. 50: 216–236 [DOI] [PubMed] [Google Scholar]
- Bensmihen S., To A., Lambert G., Kroj T., Giraudat J., Parcy F. (2004). Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 561: 127–131 [DOI] [PubMed] [Google Scholar]
- Bolte S., Cordelières F.P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224: 213–232 [DOI] [PubMed] [Google Scholar]
- Burda P., Padilla S.M., Sarkar S., Emr S.D. (2002). Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 115: 3889–3900 [DOI] [PubMed] [Google Scholar]
- Carlton J., Bujny M., Peter B.J., Oorschot V.M., Rutherford A., Mellor H., Klumperman J., McMahon H.T., Cullen P.J. (2004). Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr. Biol. 14: 1791–1800 [DOI] [PubMed] [Google Scholar]
- Carlton J., Bujny M., Rutherford A., Cullen P. (2005). Sorting nexins—Unifying trends and new perspectives. Traffic 6: 75–82 [DOI] [PubMed] [Google Scholar]
- Coudreuse D.Y., Roël G., Betist M.C., Destrée O., Korswagen H.C. (2006). Wnt gradient formation requires retromer function in Wnt-producing cells. Science 312: 921–924 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E.F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S. (2008). Retromer retrieves wntless. Dev. Cell 14: 4–6 [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N., Richter S., Vieten A., Marquardt S., Torres-Ruiz R.A., Mayer U., Jürgens G. (2004). Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131: 389–400 [DOI] [PubMed] [Google Scholar]
- Gullapalli A., Wolfe B.L., Griffin C.T., Magnuson T., Trejo J. (2006). An essential role for SNX1 in lysosomal sorting of protease-activated Receptor-1: Evidence for retromer-, Hrs-, and Tsg101-independent functions of sorting nexins. Mol. Biol. Cell 17: 1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi Y., Niihama M., Takahashi T., Saito C., Nakano A., Tasaka M., Morita M.T. (2010). Loss-of-function mutations of retromer large subunit genes suppress the phenotype of an Arabidopsis zig mutant that lacks Qb-SNARE VTI11. Plant Cell 22: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miège C., Gaude T. (2008). Evidence for a sorting endosome in Arabidopsis root cells. Plant J. 53: 237–247 [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miège C., Rollin C., Gaude T. (2006). AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109 [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Santambrogio M., Rozier F., Fobis-Loisy I., Miège C., Gaude T. (2007). The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell 130: 1057–1070 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Klein D.E., Lee A., Frank D.W., Marks M.S., Lemmon M.A. (1998). The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide binding. J. Biol. Chem. 273: 27725–27733 [DOI] [PubMed] [Google Scholar]
- Lee J.J., Radice G., Perkins C.P., Costantini F. (1992). Identification and characterization of a novel, evolutionarily conserved gene disrupted by the murine H beta 58 embryonic lethal transgene insertion. Development 115: 277–288 [DOI] [PubMed] [Google Scholar]
- Niemes S., Langhans M., Viotti C., Scheuring D., San Wan Yan M., Jiang L., Hillmer S., Robinson D.G., Pimpl P. (2010). Retromer recycles vacuolar sorting receptors from the trans-Golgi network. Plant J. 61: 107–121 [DOI] [PubMed] [Google Scholar]
- Phan N.Q., Kim S.J., Bassham D.C. (2008). Overexpression of Arabidopsis sorting nexin AtSNX2b inhibits endocytic trafficking to the vacuole. Mol. Plant 1: 961–976 [DOI] [PubMed] [Google Scholar]
- Rojas R., Kametaka S., Haft C.R., Bonifacino J.S. (2007). Interchangeable but essential functions of SNX1 and SNX2 in the association of retromer with endosomes and the trafficking of mannose 6-phosphate receptors. Mol. Cell. Biol. 27: 1112–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R., van Vlijmen T., Mardones G.A., Prabhu Y., Rojas A.L., Mohammed S., Heck A.J., Raposo G., van der Sluijs P., Bonifacino J.S. (2008). Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J. Cell Biol. 183: 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman N., Durbarry A., Wardle A., Yang W.C., Chaboud A., Faure J.E., Berger F., Twell D. (2005). A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15: 244–248 [DOI] [PubMed] [Google Scholar]
- Schwarz D.G., Griffin C.T., Schneider E.A., Yee D., Magnuson T. (2002). Genetic analysis of sorting nexins 1 and 2 reveals a redundant and essential function in mice. Mol. Biol. Cell 13: 3588–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N., Harbour M.E., Tattersall D., Read E., Bright N. (2009). Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J. Cell Sci. 122: 2371–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N., Williams H.P. (2002). Identification of the functional domains of yeast sorting nexins Vps5p and Vps17p. Mol. Biol. Cell 13: 2826–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Fuji K., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2003). Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 16095–16100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale R.D., Loci D., Houghton F., Karlsson L., Gleeson P.A. (2001). A large family of endosome-localized proteins related to sorting nexin 1. Biochem. J. 358: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V., Tichtinsky G., Dumas C., Gaude T., Cock J.M. (2003). Interaction of calmodulin, a sorting nexin and kinase-associated protein phosphatase with the Brassica oleracea S locus receptor kinase. Plant Physiol. 133: 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Walhout A.J., Vidal M. (2001). High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24: 297–306 [DOI] [PubMed] [Google Scholar]
- Wassmer T., Attar N., Harterink M., van Weering J.R., Traer C.J., Oakley J., Goud B., Stephens D.J., Verkade P., Korswagen H.C., Cullen P.J. (2009). The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev. Cell 17: 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M., Shimada T., Takahashi H., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2008). Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant Cell Physiol. 49: 142–156 [DOI] [PubMed] [Google Scholar]
- Zhong Q., Lazar C.S., Tronchère H., Sato T., Meerloo T., Yeo M., Songyang Z., Emr S.D., Gill G.N. (2002). Endosomal localization and function of sorting nexin 1. Proc. Natl. Acad. Sci. USA 99: 6767–6772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zluvova J., Nicolas M., Berger A., Negrutiu I., Monéger F. (2006). Premature arrest of the male flower meristem precedes sexual dimorphism in the dioecious plant Silene latifolia. Proc. Natl. Acad. Sci. USA 103: 18854–18859 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.