Although it is one of central importance to plant reproductive success, the structure of sporopollenin, the main constituent of exine in pollen, is still poorly understood. This work shows that two Arabidopsis oxidoreductases are involved in a biosynthetic pathway leading to sporopollenin that may have been a key determinant in the evolution of land plants.
Abstract
The precise structure of the sporopollenin polymer that is the major constituent of exine, the outer pollen wall, remains poorly understood. Recently, characterization of Arabidopsis thaliana genes and corresponding enzymes involved in exine formation has demonstrated the role of fatty acid derivatives as precursors of sporopollenin building units. Fatty acyl-CoA esters synthesized by ACYL-COA SYNTHETASE5 (ACOS5) are condensed with malonyl-CoA by POLYKETIDE SYNTHASE A (PKSA) and PKSB to yield α-pyrone polyketides required for exine formation. Here, we show that two closely related genes encoding oxidoreductases are specifically and transiently expressed in tapetal cells during microspore development in Arabidopsis anthers. Mutants compromised in expression of the reductases displayed a range of pollen exine layer defects, depending on the mutant allele. Phylogenetic studies indicated that the two reductases belong to a large reductase/dehydrogenase gene family and cluster in two distinct clades with putative orthologs from several angiosperm lineages and the moss Physcomitrella patens. Recombinant proteins produced in bacteria reduced the carbonyl function of tetraketide α-pyrone compounds synthesized by PKSA/B, and the proteins were therefore named TETRAKETIDE α-PYRONE REDUCTASE1 (TKPR1) and TKPR2 (previously called DRL1 and CCRL6, respectively). TKPR activities, together with those of ACOS5 and PKSA/B, identify a conserved biosynthetic pathway leading to hydroxylated α-pyrone compounds that were previously unknown to be sporopollenin precursors.
INTRODUCTION
In flowering plants, pollen grains are formed within the anther tissues of the stamen, the male reproductive organ. Molecular and genetic studies have identified a number of genes that are expressed during stamen and pollen development (Ma, 2005). Many of them regulate anther cell differentiation, tapetum function, and microsporocyte development into pollen grains. In particular, there have been a number of studies that describe Arabidopsis thaliana mutants showing abnormal pollen structure resulting in male-sterile or partially sterile phenotypes.
The different stages of pollen development have been carefully documented (Owen and Makaroff, 1995; Sanders et al., 1999; Scott et al., 2004; Blackmore et al., 2007). Meiosis in anthers gives rise to haploid microsporocytes arranged in tetrads surrounded by a callose wall. Then, callose is hydrolyzed and the free microsporocytes are released in the anther locule. At this stage, cellulosic primexine and pectocellulosic intine are produced by the developing microspores and constitute the inner layers of the pollen wall. The outer layer of pollen wall, called the exine, is composed primarily of sporopollenin, a polymer of phenylpropanoid and lipidic monomers covalently coupled by ether and ester linkages (Rozema et al., 2001; Bubert et al., 2002; Ahlers et al., 2003). Sporopollenin precursors are produced in the sporophyte tapetal cell layer surrounding the anther locule, then secreted and deposited on the pollen surface. The sporopollenin polymer confers on the exine unparalleled physical strength, chemical inertness, and elasticity. However, the high level of resistance of sporopollenin to chemical degradation makes it particularly difficult to analyze by chemical methods (Bubert et al., 2002; Ahlers et al., 2003).
Significant progress in our understanding of exine formation processes has been made recently, thanks to genetic and molecular studies of Arabidopsis mutants exhibiting defects in exine structure and deposition (Aarts et al., 1997; Paxson-Sowders et al., 2001; Ariizumi et al., 2003, 2004; Ito et al., 2007; Morant et al., 2007; Yang et al., 2007; Guan et al., 2008; Suzuki et al., 2008; de Azevedo Souza et al., 2009; Dobritsa et al., 2009a; Tang et al., 2009). Unfortunately, in most cases, annotations of the mutated Arabidopsis genes found to be responsible for pollen phenotypes are primarily based on sequence similarity, and the exact functions of the corresponding proteins in pollen development remain unknown. For example, the Male sterility2 (ms2) mutant exhibits a lack of exine, and MS2 shows sequence similarity with fatty acyl-CoA reductases (Aarts et al., 1997). In agreement with this observation, bacteria transformed with a MS2 cDNA produce fatty alcohols from uncharacterized precursors (Doan et al., 2009). A loss-of-function mutation in the FACELESS POLLEN1/YORE-YORE/WAX2 gene that impacts exine structure and wax synthesis (Ariizumi et al., 2003; Chen et al., 2003; Kurata et al., 2003) was reported recently to be allelic to CER3, a gene previously described to fulfill an unknown function in wax synthesis (Rowland et al., 2007). DIHYDROFLAVONOL 4-REDUCTASE-LIKE1 (DRL1), another gene required for pollen development and male fertility (Tang et al., 2009), shares sequence similarity with DIHYDROFLAVONOL REDUCTASE (DFR), a gene implicated in the biosynthesis of anthocyanins and proanthocyanidins (Shirley et al., 1992), but the DRL1 enzyme is also of unknown biochemical function.
Recently acos5, a male-sterile mutant, and dex2, a male-semisterile mutant, have both been shown to be impaired in exine formation (Morant et al., 2007; de Azevedo Souza et al., 2009). For both ACYL-COA SYNTHETASE5 (ACOS5) and DEX2, the catalytic activities of the encoded proteins were characterized biochemically, providing evidence for specific roles in sporopollenin biosynthesis. ACOS5, originally annotated as a 4-coumarate:CoA ligase-like protein, displays fatty acyl-CoA synthetase activity with medium- to long-chain fatty acids (C8-C18 fatty acids) (de Azevedo Souza et al., 2009), and DEX2 encodes CYP703A2, which has been shown to in-chain hydroxylate lauric acid (C12:0 fatty acid) (Morant et al., 2007). Recently, CYP704B1 has been demonstrated also to be implicated in exine formation and to catalyze the ω-hydroxylation of long-chain fatty acids (Dobritsa et al., 2009b). These studies unequivocally demonstrate that fatty acids are essential precursors of sporopollenin. At late stages of pollen grain development, lipophilic tryphine is deposited on the surface and within the chambers of exine and constitutes the pollen coat (Scott et al., 2004; Blackmore et al., 2007; Grienenberger et al., 2009), but this is a process distinct from exine formation and probably involves a distinct biosynthetic pathway or pathways.
We recently demonstrated that two Arabidopsis polyketide synthases, POLYKETIDE SYNTHASE A (PKSA) and PKSB, play critical roles in sporopollenin biosynthesis, acting downstream of ACOS5. pksa pksb double mutants are completely male sterile and lack an exine. In vitro, both proteins accept fatty acyl-CoA esters synthesized by ACOS5 and condense them to malonyl-CoA to yield triketide and tetraketide α-pyrones as reaction products (Kim et al., 2010). Here, we show by in situ hybridization of mRNAs and by immunolocalization of corresponding proteins that two Arabidopsis oxidoreductases, one encoded by the DRL1 gene previously described by Tang et al. (2009) and the other annotated as CINNAMOYL COA REDUCTASE-LIKE6 (CCRL6) (Hamberger et al., 2007), are coexpressed with ACOS5, PKSA, and PKSB in anther tapetum cells. We show that recombinant enzymes produced in bacteria accept the tetraketide α-pyrones produced by PKSA and PKSB as substrates to reduce the carbonyl function on the tetraketide alkyl chain to a secondary alcohol function. Phylogenetic studies showed that the oxidoreductases belong to a gene family conserved from mosses to flowering plants. Since similar gene conservation holds true for PKSA, PKSB, and ACOS5 genes, it appears that the whole biosynthetic pathway leading from medium or long-chain fatty acids to sporopollenin units is highly conserved and may have been a key determinant in the evolution of land plants.
RESULTS
Genes Involved in Pollen Cell Wall Formation Are Tightly Coregulated
Pollen wall formation requires exquisite and coordinated spatio-temporal regulation of numerous biosynthetic genes by specific transcriptional regulators (Alves-Ferreira et al., 2007; Ito et al., 2007; Yang et al., 2007). In particular, many genes involved in exine biosynthesis in Arabidopsis have been shown to be repressed by the MS1 transcription factor in wild-type plants and to be overexpressed in the ms1 mutant (Ito et al., 2007; Yang et al., 2007). Examination of Arabidopsis microarray data showed that, during development of wild-type and ms1 anthers, several uncharacterized genes are coregulated with genes involved in exine formation. These genes may therefore encode unknown players of sporopollenin biosynthesis (http://bbc.botany.utoronto.ca/ntools/cgi-bin/ntools_expression_angler.cgi; Toufighi et al., 2005). Among the genes coexpressed during pollen development, two were annotated as CHALCONE SYNTHASE-LIKE and identified in the companion article to this one (Kim et al., 2010) as encoding PKSA and PKSB. PKSA and PKSB catalyze the condensation of fatty acyl-CoA esters produced by ACOS5 with malonyl-CoA to yield tri- and tetraketide α-pyrone compounds as reaction products (Kim et al., 2010). Several other tightly coregulated genes were annotated as oxidoreductases/dehydrogenases that all have unknown physiological substrates (http://www.Arabidopsis.org/). Among them, the proteins encoded by At4g35420 (DRL1; Tang et al., 2009) and At1g68540 (CCRL6; Hamberger et al., 2007) contain putative NAD(P)H binding domains and share 45% identity (see Supplemental Figure 1 online). Both proteins display sequence similarity with two well-characterized plant oxidoreductases: DFR, an enzyme of anthocyanin synthesis (35 and 29% identity; 52 and 43% similarity, respectively) (Shirley et al., 1992), and cinnamoyl-CoA reductase (CCR) that is involved in lignin biosynthesis (~35% identity and 53% similarity for both At4g35420 and At1g68540 encoded proteins) (Lacombe et al., 1997). These plant enzymes belong to a superfamily whose members are also encountered in microbial and mammalian kingdoms and share a conserved N-terminal sequence that is likely involved in the interactions with NAD(P)H (see Supplemental Figure 1 online) (Baker and Blasco, 1992; Lacombe et al., 1997). DRL1 has been shown to be required for male fertility (Tang et al., 2009), but neither the DRL1 expression pattern nor its exact role in pollen wall formation has been described in detail.
At4g35420 and At1g68540 Expression Profiles during Flower Development
We explored publicly available microarray databases, such as Genevestigator (https://www.genevestigator.com/gv/index.jsp) (Hruz et al., 2008) and the Arabidopsis eFP browser (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi) (Winter et al., 2007), to determine the tissue expression patterns of At4g35420 (DRL1) and At1g68540 (CCRL6). The results of this search indicated that both were preferentially expressed in young flower buds, in accordance with their high coregulation scores, similar to the expression patterns reported for PKSA, PKSB, and ACOS5 (de Azevedo Souza et al., 2009; Kim et al., 2010). To facilitate their designation in the following paragraphs, genes corresponding to At4g35420 (DRL1) and At1g68540 (CCRL6) were named TETRAKETIDE α-PYRONE REDUCTASE1 (TKPR1) and TKPR2, respectively, in anticipation of the enzymatic activities described below for the two corresponding proteins.
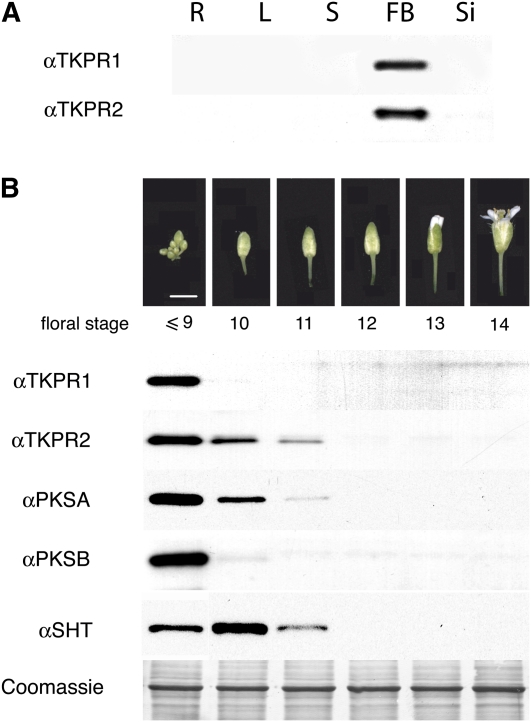
Measurements of relative mRNA abundance by quantitative RT-PCR in RNA preparations from various organs confirmed the flower-specific expression of TKPR1 and TKPR2 in contrast with At1g25460, a close homolog (Figure 10) that displayed a strikingly different expression pattern and is expressed mainly in young seedlings (see Supplemental Figure 2 online). Relative protein levels were also investigated by immunoblotting protein extracts with specific polyclonal antibodies raised against the recombinant proteins (Figure 1). The analysis of various organs confirmed that both TKPR proteins specifically accumulated in flowers, and no signal could be detected in extracts from other organs (Figure 1A).
Figure 10.
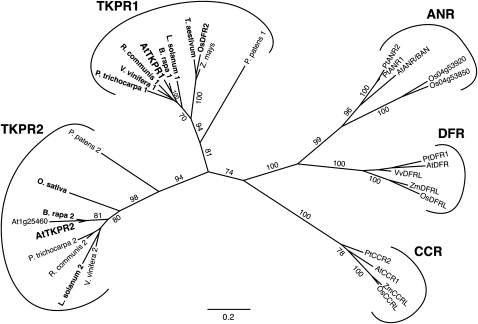
Phylogenetic Analysis of Selected TKPR, CCR, ANR, and DFR Protein Sequences.
Sequences from Arabidopsis, poplar, rice, Physcomitrella, Vitis, wheat (Triticum aestivum), maize (Zea mays), soybean (Glycine max), Nicotiana, tomato (Solanum lycopersicum), Ricinus, Brassica, and Sorghum were aligned and used to construct an unrooted maximum likelihood tree using PhyML 3.0 and FigTree v.1.2.3. Bootstrap values (1000 replicates) above 70% are given on branches. Proteins that are expressed in flower tissues (as inferred from EST libraries or published data) are indicated in bold. Genes and expression data used in this analysis are given in Supplemental Table 2 online, and the sequence alignment is available in Supplemental Data Set 1 online. Bar = 0.2 amino acid substitutions.
Figure 1.
Developmental Expression of TKPR1 and TKPR2 Proteins.
(A) Protein extracts from different plant tissues were immunoblotted with polyclonal antibodies raised against each recombinant protein. Expression levels in roots (R), leaves (L), stems (S), flower buds (FB), and siliques (Si) were compared. Six micrograms of total protein was loaded per lane.
(B) Comparison of TKPR, PKS, and SHT protein accumulation at the different stages of flower development illustrated in the top photographs. Approximate flower developmental stages are shown as defined by Smyth et al. (1990). Equal loading (6 μg per lane) of protein is shown by Coomassie blue staining of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase.
[See online article for color version of this figure.]
Protein extracts from flower buds at different stages of development were analyzed next, and distinct kinetics of accumulation were found for the two proteins (Figure 1B). Both proteins were most highly abundant in the youngest buds tested, but the TKPR1 protein disappeared rapidly in more mature buds, whereas TKPR2 levels decreased more slowly. These differential kinetics of accumulation are comparable to those observed for PKSA and PKSB (Kim et al., 2010) that are shown here for comparison. This suggests that TKPR1 and PKSB protein levels are tightly coregulated, as are TKPR2 and PKSA levels. SHT, an acyltransferase participating in pollen development, was taken as an internal marker in the same bud extracts and displayed a maximum accumulation at a later stage of flower development, in accordance with its role in pollen coat formation (Figure 1B) (Grienenberger et al., 2009).
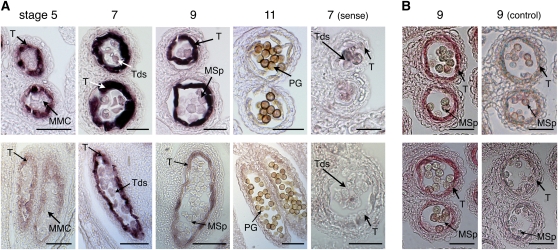
To determine the precise sites of TKPR1 and TKPR2 expression in flower tissues, in situ hybridization and immunolocalization experiments were performed and showed the specific accumulation of both transcripts and proteins in the anthers (Figure 2). In situ hybridization experiments at different stages of anther development demonstrated the tapetum-specific expression of both TKPR genes. The two genes displayed similar but distinct temporal expression patterns. The highest hybridization signal for both was found at stage 7 of anther development (Figure 2A); however, the TKPR1 expression pattern was broader over developmental time. Consistent with this, antibodies raised against each recombinant protein detected TKPR protein accumulation specifically in tapetum cells at a later stage (stage 9) of anther development (Figure 2B).
Figure 2.
Tapetum-Specific Expression of TKPR1 and TKPR2.
Top panels: TKPR1 expression; bottom panels: TKPR2 expression.
(A) TKPR1 and TKPR2 mRNAs were localized by in situ hybridization of gene-specific antisense probes to sections of wild-type (Col-0) flowers. Sense probes were used for controls. Stages of anther development are according to Sanders et al. (1999). Dark precipitates indicate hybridization of the probe. Stage 7 shows highest hybridization signals for both of TKPR1 and TKPR2 in the tapetum, but TKPR2 expression was more restricted temporally.
(B) Specific antibodies detected TKPR1 and TKPR2 protein accumulation in the tapetum of anthers at stage 9 of development. Controls were performed using preimmune sera.
MMC, microspore mother cells; Tds, tetrads; T, tapetum; MSp, microspores; PG, pollen grain. Bars = 70 μm.
Disruption of Oxidoreductase Genes Differentially Affects Pollen Development
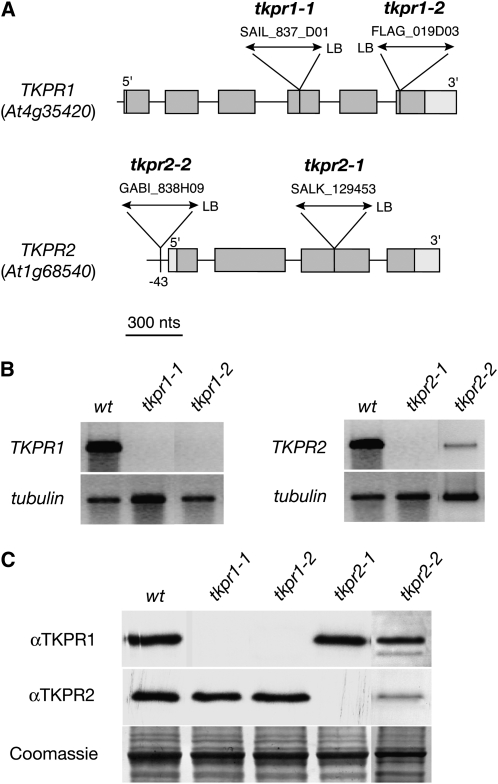
T-DNA insertion alleles in At4g35420/TKPR1 and At1g68540/TKPR2 were obtained from public collections (Samson et al., 2002; Alonso et al., 2003), and homozygous populations were generated. Two tkpr1 alleles and two tkpr2 alleles were isolated and characterized (Figure 3). We assayed gene expression by RT-PCR with RNA from wild-type and mutant flower buds as templates. Using primers flanking the sites of T-DNA insertions (primer sequences are given in Supplemental Table 1 online), no transcript could be detected for three of the mutant lines (Figure 3B). Moreover, in those insertion lines, no protein accumulation could be detected by immunoblotting protein extracts with specific antibodies (Figure 3C). These data indicate that tkpr1-1, tkpr1-2, and tkpr2-1 mutant lines are null alleles of At4g35420 and At1g68540, respectively. In the case of the tkpr2-2 mutant, T-DNA insertion in the promoter sequence (Figure 3A) strongly reduced gene expression, as shown by low transcript and protein levels detected by RT-PCR and immunoblotting, respectively (Figures 3B and 3C).
Figure 3.
Molecular Characterization of TKPR1 and TKPR2 Insertion Alleles.
(A) Positions of T-DNA insertions in the different mutant lines are shown, and for each the position of the left border (LB) is indicated. Boxes denote exons and thin horizontal lines denote introns.
(B) RT-PCR analysis of gene expression in wild-type and mutant flower buds. No amplicon was detectable in tkpr1-1, tkpr1-2, and tkpr2-1 samples, whereas a weak signal was recorded in tkpr2-2 buds, using primers shown in Supplemental Table 1 online. TUBULIN transcripts were amplified as a positive control.
(C) Protein accumulation in wild-type and mutant bud extracts was evaluated by immunoblotting with specific antibodies raised against recombinant proteins. No protein accumulation was detected in tkpr1-1, tkpr1-2, and tkpr2-1 lines, confirming that gene expression was knocked out. A faint band was detected in the tkpr2-2 sample, indicating that T-DNA insertion in the promoter sequence (A) knocked down gene expression. Equal loading of protein is shown by Coomassie blue staining of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase.
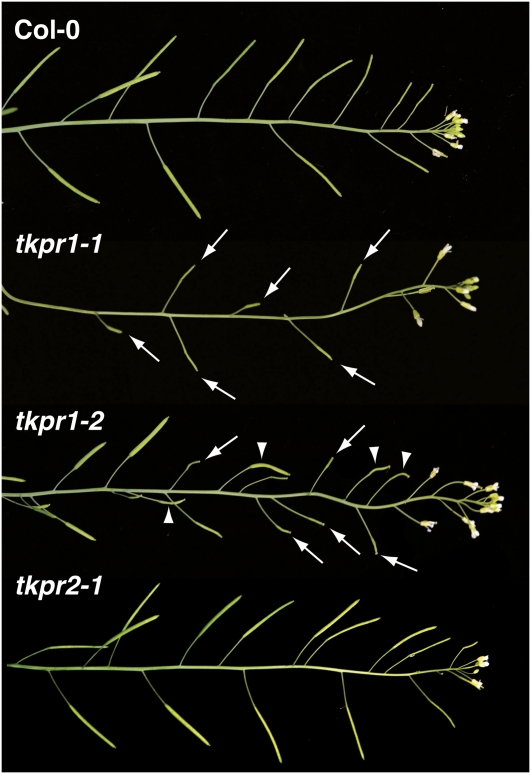
Figure 4 illustrates the inflorescence phenotypes of homozygous mutants, showing that the disruption of TKPR1 and TKPR2 differently affected plant fertility. The tkpr1-1 line was sterile and produced no pollen, and its siliques had no seeds, similar to the results reported by Tang et al. (2009). By contrast, tkpr1-2 displayed a semisterile phenotype; it produced little pollen, and its inflorescences bore three types of siliques: a few that were filled with seeds like the wild type, some that were empty (arrows in Figure 4), and a third category that was bent and contained only a few seeds (arrowheads in Figure 4). Mutations in TKPR2 did not affect fertility, as illustrated for tkpr2-1 in Figure 4.
Figure 4.
Differential Impacts of tkpr Mutations on Plant Fertility.
The inflorescences of different Arabidopsis genotypes are shown. Compared with Col-0 wild-type siliques that are filled with seeds, those of tkpr1-1 are empty (arrows), and this line is sterile. By contrast, tkpr1-2 inflorescences bear three types of siliques: some are filled with seeds, some are completely empty (arrows), and a third category contains only a few seeds (arrow heads), thus revealing a semisterile phenotype. Mutations in TKPR2 did not affect fertility, as shown here for tkpr2-1.
[See online article for color version of this figure.]
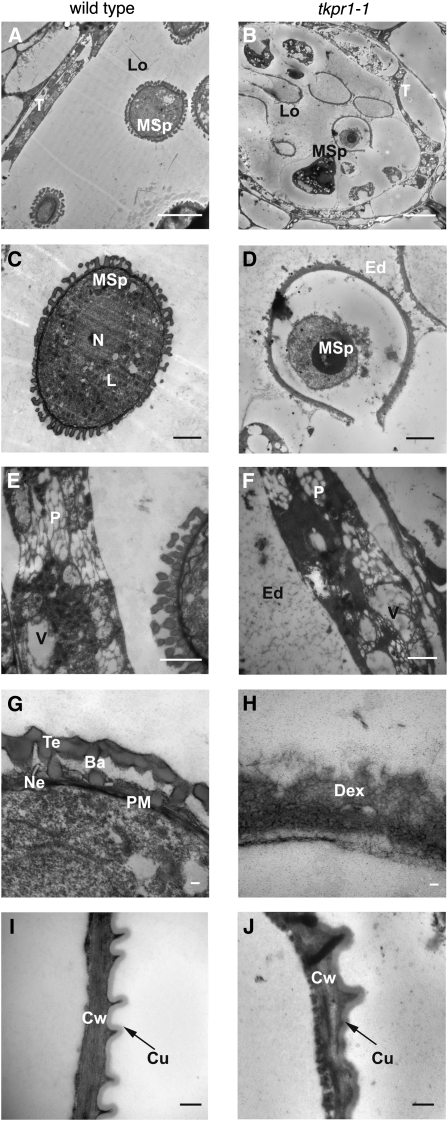
tkpr1-1 anthers and microspores were examined in detail and compared with the wild type at stage 9 of development (Sanders et al., 1999) by transmission electron microscopy (TEM). Figure 5 shows that in wild-type anthers, a thick reticulate exine with distinct baculae and tecta had formed around free, uninucleate microspores. In mutant flowers, however, the tapetum was highly vacuolated (Figure 5B), and microspores were profoundly affected, with an exine structure that appeared very thin (Figure 5D) and completely disorganized without baculae and tecta (Figure 5H). In contrast with the cytoplasmicaly dense microspores in wild-type anthers (Figure 5C), tkpr1-1 microspores at this stage were disorganized, largely devoid of cytoplasm, and showed signs of rupture. Finally, tkpr1-1 locules contained a fibrillar, electron-dense network (Figure 5F) that was not observed in wild-type anthers and could represent unpolymerized sporopollenin precursors. By contrast, cell walls of wild-type and mutant anthers were similar, with visible superficial cuticle layer (Figures 5I and 5J).
Figure 5.
Exine Formation Is Impaired in tkpr1-1 Anthers.
TEM analysis of sections of anthers and microspores from wild-type ([A], [C], [E], [G], and [I]) or tkpr1-1 ([B], [D], [F], [H], and [J]) plants at stage 9 of anther development (Sanders et al., 1999). Details are shown for anther locule ([A] and [B]), microspore ([C] and [D]), tapetum ([E] and [F]), exine ([G] and [H]), and anther wall ([I] and [J]).
Ba, baculae; Cu, cuticle; Cw, cell wall; Dex, defective exine structure; Ed, electron-dense material; L, lipid droplets; Lo, locule; MSp, microspore; N, nucleus; Ne, nexine; P, plastid filled with plastoglobuli; PM, plasma membrane; T, tapetal cell; Te, tectum; V, vacuole containing electron-dense material. Bars = 10 μm in (A) and (B), 2 μm in (C) to (F), 500 nm in (I) and (J), and 100 nm in (G) and (H).
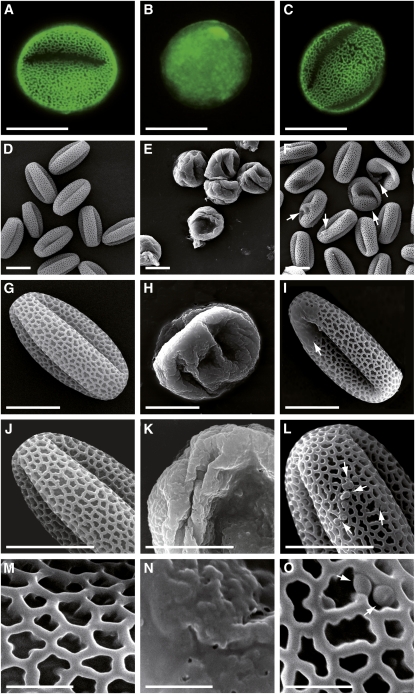
Pollen grains from the other genotypes were examined by fluorescence microscopy after staining with auramine O, a fluorescent dye that reveals exine patterns (Dobritsa et al., 2009a, 2009b). Fluorescence of the few tkpr1-2 mutant pollen grains that were produced (Figure 6B) was strongly attenuated compared with the wild-type pattern (Figure 6A), whereas only subtle alterations were seen in the exine patterns of tkpr2-1 pollen whose overall fluorescence intensity was slightly reduced (Figure 6C).
Figure 6.
Comparison of Exine Architecture in Wild-Type, tkpr1-2, and tkpr2-1 Pollen.
(A), (D), (G), (J), and (M) Wild-type pollen.
(B), (E), (H), (K), and (N) tkpr1-2 pollen.
(C), (F), (I), (L), and (O) tkpr2-1 pollen; arrows indicate exine defects.
(A) to (C) Epifluorescence microscope images of wild-type and mutant pollen. Pollen was stained with the fluorescent dye auramine O and visualized using fluorescein isothiocyanate settings.
(D) to (O) Scanning electron micrographs of wild-type and mutant pollen grains.
Bars = 10 μm in (A) to (L) and 2 μm in (M) to (O).
[See online article for color version of this figure.]
We also examined tkpr1-2 and tkpr2-1 pollen grains in detail by scanning electron microscopy (Figures 6D to 6O). Many tkpr1-2 pollen grains were severely distorted (Figures 6E and 6H), showing large areas where the reticulate exine pattern was no longer apparent (Figures 6K and 6N), and these pollen grains tended to aggregate in comparison to the well-separated wild-type grains (Figures 6E and 6D, respectively). More subtle defects were detected in tkpr2-1 (arrows in Figure 6F), and higher magnification images showed a few limited areas without a visible exine network (arrow in Figure 6I). At even higher magnification (Figures 6L and 6O), a few spherical protrusions were visible inside and outside the mutant exine tecta. Similar defects were observed in tkpr2-2 pollen (see Supplemental Figure 3 online).
Enzymatic Activities of TKPR1 and TKPR2 Proteins Produced in Escherichia coli
The coding regions of the two putative reductase genes were cloned in a vector that introduced an N-terminal glutathione S-transferase tag and were expressed in E. coli. Each recombinant protein was purified by affinity chromatography on glutathione beads (see Supplemental Figure 4 online), used to raise polyclonal antisera, and tested for catalytic activity.
Sequence analysis of TKPR1 (At4g35420) and TKPR2 (At1g68540) predicted proteins showed that they both possess consensus NADP/NAD binding motifs (see Supplemental Figure 1 online). Therefore, considering the fact that these two putative oxidoreductase genes are tightly coexpressed with ACOS5 and the polyketide synthases PKSA and PKSB in tapetal cells during pollen development (Figures 1 and 2) (de Azevedo Souza et al., 2009; Kim et al., 2010), we examined the possibility that these enzyme activities could function in a common biochemical pathway. Particularly, the tri- and tetraketide compounds produced by PKSA and PKSB through condensation of fatty acyl-CoAs with malonyl-CoA (Kim et al., 2010) bear several chemical functions that are potential targets for reduction reactions.
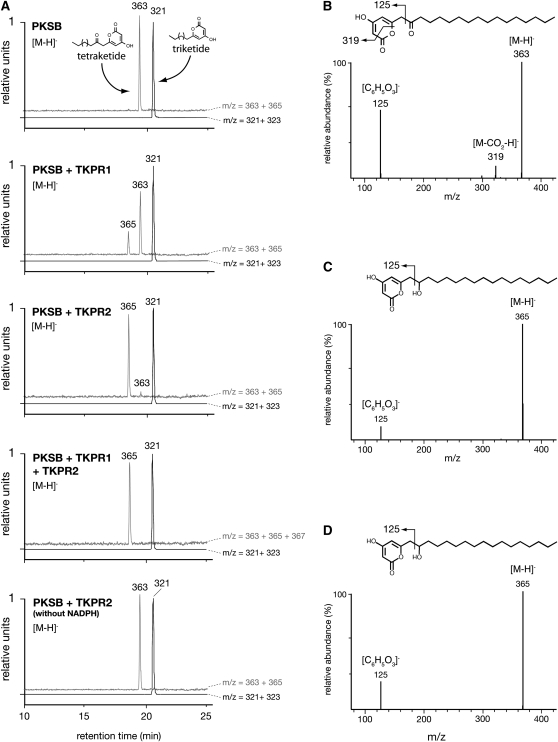
As shown in Figure 7A (top profile), incubation of PKSB in the presence of palmitoyl-CoA and malonyl-CoA produced tri- and tetraketide α-pyrones with mass-to-charge (m/z) values of 321 and 363, respectively. When recombinant TKPR1 protein and NADPH were added to the PKS incubation medium, the peak of the tetraketide product decreased and a new peak appeared with a m/z value of 365 that is 2 atomic mass units higher than that of the tetraketide substrate (m/z = 363), indicating that TKPR1 had catalyzed reduction of the tetraketide α-pyrone. The triketide peak (m/z =321) remained unchanged, and its putative reduced product (m/z =323) was not detected, thus indicating that the triketide α-pyrone is not a substrate for the reductase.
Figure 7.
Identification of TKPR Reaction Products by LC-MS/MS.
Tri- and tetraketide compounds were synthesized by PKSB in the presence of palmitoyl-CoA and malonyl-CoA and incubated with recombinant TKPR enzymes. Reaction products were separated by UPLC and identified by negative electrospray ionization (ESI) mass spectrometry.
(A) Compounds were detected by their m/z values. Black curves show ions corresponding to the triketide compound and its putative reduction product (m/z of 321 and 323, respectively); gray curves show ions corresponding to the tetraketide compound and its putative reduction product (m/z of 363 and 365, respectively). The 321 (triketide) and 363 (tetraketide) peaks of the control incubated without NADPH (bottom profile) have been normalized to 1. Only the tetraketide compound was substrate for TKPR1 and TKPR2. No product corresponding to a double reduction of the tetraketide substrate (m/z = 367) was detected when TKPR1 and TKPR2 were incubated together.
(B) Fragmentation pattern of [M-H]− ion of the tetraketide substrate (m/z = 363) and putative fragmentation scheme of the molecule.
(C) Fragmentation pattern of [M-H]− ion of the reduced product obtained after incubation with TKPR1 and its putative fragmentation scheme.
(D) Fragmentation pattern of [M-H]− ion of the reaction product of TKPR2 is identical to that of TKPR1 reaction product (C), thus indicating that both enzymes catalyze the reduction of the carbonyl function borne by the aliphatic chain of the tetraketide compound.
The TKPR2 protein proved even more active, since almost all of the tetraketide α-pyrone had disappeared and was supplanted by the reduced compound of 365 m/z value (Figure 7A) after incubation with enzyme. These results demonstrate that TKPR1 and TKPR2 are reductases active with tetraketide but not triketide α-pyrones as substrates. Moreover, when TKPR1 and TKPR2 were incubated together, no new peak resulting from a double reduction (m/z = 367) was recorded. This suggests that both enzymes target the same chemical function in vitro. The occurrence of a carbonyl function borne by the alkyl chain of tetraketide compounds constitutes the major structural difference from triketide products (cf. structures in top profile of Figure 7A). This makes the carbonyl function the most likely target of the reduction reaction that affects only the tetraketide compounds. Moreover, in collision-induced dissociation experiments (Figures 7B to 7D), we observed the presence of fragments at the 125 m/z value in the mass spectrum of reduction reaction products (Figures 7C and 7D) and in the tetraketide substrate (Figure 7B), which is diagnostic for the lactone ring of the tetraketide α-pyrones (Saxena et al., 2003; Rubin-Pitel et al., 2008). Thus, these results suggest that the lactone ring remained unaffected by the reduction reaction. Taken together, these data indicate that both TKPR1 and TKPR2 reduced the carbonyl function of the tetraketide α-pyrones to a secondary alcohol function. Therefore, the proteins encoded by At4g35420/DRL1 and At1g68540/CCRL6 were named TKPR1 and TKPR2, respectively.
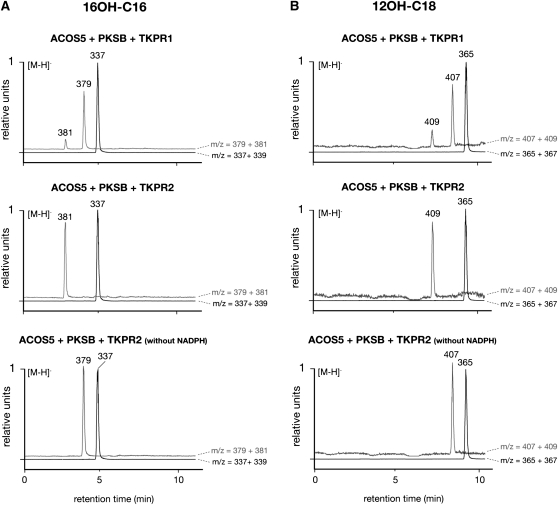
Because the role of CYP450 hydroxylases in modification of fatty acid sporopollenin precursors has been demonstrated previously (Morant et al., 2007; Dobritsa et al., 2009b), we tested the ability of TKPR to reduce hydroxylated tetraketide α-pyrone compounds. These were generated by first incubating 16-OH palmitic or 12-OH stearic acids with ACOS5, ATP, and CoA. The corresponding CoA esters were then condensed to malonyl CoA using recombinant PKSB (Kim et al., 2010) to yield the hydroxylated tetraketide compounds, which were assayed as TKPR1 and TKPR2 substrates (Figures 8A and 8B). In controls incubated in the absence of NADPH, tri- and tetraketide compounds were produced by PKS enzyme (bottom profiles in Figures 8A and 8B). When TKPR incubations were performed in the presence of NADPH, new peaks were detected that corresponded to reduced tetraketides derived from 16-OH palmitic and 12-OH stearic acids (species of 381 and 409 m/z values, respectively). It is noteworthy that TKPR2 catalyzed the complete reduction of the tetraketide substrates, whereas TKPR1 was less active, similar to results obtained using palmitoyl-CoA as substrate (Figure 7A). Altogether, these results show that ACOS5, PKS, and TKPR activities catalyze a series of biosynthetic reactions yielding hydroxylated tetraketide compounds that serve as sporopollenin precursors.
Figure 8.
ACOS5, PKS, and TKPR Activities Define a Sequence of Biosynthetic Reactions.
The 16-hydroxy palmitic (A) and 12-hydroxy stearic (B) acids were incubated successively with ACOS5, PKSB, and TKPR1 or TKPR2 recombinant enzymes (see Methods). Putative reaction products were detected by their m/z values after separation by UPLC and negative electrospray ionization (ESI) mass spectrometry. Black curves show ion species corresponding to triketides, and their putative reduction products and gray curves show ion species from tetraketide substrates and products. Controls for the reduction reaction were run without NADPH and gave rise to tri- and tetraketide ions that were normalized to 1.
TKPR Proteins Localize to Different Cellular Compartments
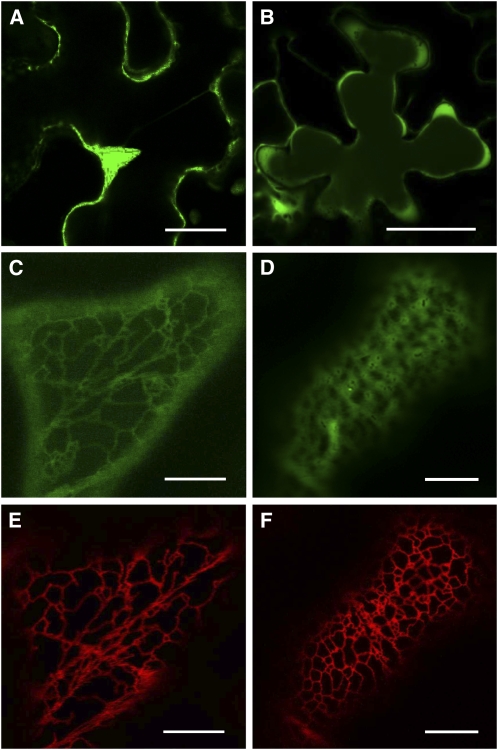
To investigate the cellular localization of TKPR1 and TKPR2 proteins, translational fusions upstream to enhanced green fluorescent protein (eGFP) and placed under the control of the cauliflower mosaic virus 35S promoter (p35S:TKPR1/2-eGFP) were generated. The constructs were transiently expressed in Nicotiana benthamiana leaves, together with a reporter construct of an endoplasmic reticulum (ER) marker (mRFP-HDEL). As illustrated in Figure 9, observations by confocal microscopy showed that the two TKPR fusions localize to different cellular compartments. The TKPR1-eGFP fluorescence (Figure 9A) delineates the periphery of epidermal cells. At higher magnification, most of the TKPR1-eGFP fluorescence pattern (Figure 9C) coincides with the reticulate network of mRFP-HDEL that is typical of ER localization (Figure 9E) (Robinson et al., 2007). These data indicate that TKPR1 enzyme is, at least in part, associated with ER. By contrast, TKPR2-eGFP fluorescence that is also located in the cell periphery (Figure 9B) appears, at higher magnification, clearly distinct from the typical reticulate pattern of the ER marker (cf. green and red fluorescence patterns in Figures 9D and 9F, respectively), with TKPR2-eGFP diffuse fluorescence (Figure 9D) being reminiscent of a cytoplasm labeling. Therefore, TKPR2 enzyme is primarily cytosolic and not associated with ER as TKPR1.
Figure 9.
Expression of Fluorescent Fusion Proteins Reveals Distinct Subcellular Localizations for TKPR1 and TKPR2.
TKPR1/2-eGFP and mRFP-HDEL (ER marker protein) constructs were transiently expressed in N. benthamiana leaves by agrotransformation and observed by confocal laser scanning microscopy. eGFP fluorescence ([A] to [D]); mRFP fluorescence ([E] and [F]). TKPR1-eGFP fluorescence is detected at the periphery of epidermal cells (A). At higher magnification, a considerable part of TKPR1-eGFP fluorescence pattern (C) coincides with the reticulate network of the ER marker (E), thus revealing that TKPR1 is associated with ER. TKPR2-eGFP fluorescence (B) is located in cell cytoplasm and, at higher magnification (D), is clearly distinct from the reticulate pattern of the ER marker (F). Thus, it is typical of a cytosolic localization. Bars = 40 μm in (A) and (B) and 10 μm in (C) to (F).
Phylogenetic Analysis of TKPR Genes
TKPR1 and TKPR2 belong to a gene superfamily that includes members of mammalian, bacterial, and plant origins (see Supplemental Figure 1 online) (Baker et al., 1990; Baker and Blasco, 1992; Lacombe et al., 1997). Plant family members whose functions are unknown have been referred to as DRL (Tang et al., 2009) and CCR-like (Hamberger et al., 2007) since the functions of DFR/TT3 in anthocyanin synthesis and CCR in lignin biosynthesis had been characterized several years ago (Shirley et al., 1992; Lacombe et al., 1997). ANTHOCYANIDIN REDUCTASE (ANR), encoding another flavonoid biosynthetic enzyme (Xie et al., 2003), constitutes another clade of the family (Figure 10).
Phylogenetic analysis showed that TKPR1 and TKPR2 define two new clades of plant reductase sequences, formerly annotated as DRL and CCRL genes, distinct from the DFR, ANR, and CCR clades (Figure 10). These two new clades include homologs from several plant species, ranging from the moss Physcomitrella patens to various angiosperms. Strikingly, most of the species examined possess a single homolog in each clade. This indicates that these genes appeared early in evolution and were likely present in a common land plant ancestor (Figure 10; see the list of genes in Supplemental Table 2 online). As previously reported, Arabidopsis ACOS5, PKSA, and PKSB genes show similar patterns of phylogenetic conservation (de Azevedo Souza et al., 2009; Kim et al., 2010), and, like ACOS5, PKSA, and PKSB homologs, TKPR homologs are expressed in male organs of poplar (Populus spp) and rice (Oryza sativa). These results suggest a conservation of the biosynthetic pathway involving ACOS5, PKS, and TKPR activities that leads to sporopollenin precursors in land plants.
DISCUSSION
Previously Unknown Gene Functions Required for Pollen Cell Wall Biogenesis
Although it is one of the most robust plant biopolymers and of central importance to plant reproductive success, the structure of sporopollenin, the main constituent of exine, is still poorly understood. Chemical analysis (Rozema et al., 2001; Bubert et al., 2002; Ahlers et al., 2003) and genetic and biochemical studies (Morant et al., 2007; de Azevedo Souza et al., 2009) have demonstrated the presence of lipidic and phenolic units in the sporopollenin polymer. The isolation of numerous mutants affected in exine synthesis has shown, on the other hand, that a large number of genes expressed in the anther tissues participate in the formation of the pollen cell wall, thus suggesting a high degree of complexity of the underlying processes. Here and in the companion article (Kim et al., 2010), we show the contribution of enzymes encoded by four metabolic genes acting downstream of the fatty acyl-CoA synthetase ACOS5 that produces fatty acyl-CoA esters as central intermediates in sporopollenin biosynthesis (de Azevedo Souza et al., 2009). Two polyketide synthases encoded by PKSA and PKSB catalyze the synthesis of α-pyrone products by decarboxylative condensation of the fatty acyl-CoA esters with malonyl-CoA (Kim et al., 2010), and two reductases (DRL1/TKPR1 and CCRL6/TKPR2) were found to be active on the tetraketide products produced by PKSA and PKSB activities (Figures 7 and 8). TKPR activity creates a new alcohol function whose biosynthetic origin is thus distinct from those of the hydroxyl groups introduced in ω- and in-chain positions of fatty acids by CYP450 enzymes (Morant et al., 2007; Dobritsa et al., 2009b). Our data indicate that polyhydroxylated long-chain α-pyrones are building units of sporopollenin, thus adding a new function to the polyketide class of natural compounds that exhibits an amazing functional diversity (Hertweck, 2009).
Defects in genes that are directly associated with sporopollenin synthesis have strikingly different impacts on pollen development: the acos5 mutant fails to produce viable pollen (de Azevedo Souza et al., 2009) similar to the tkpr1-1 mutant (Figure 5; Tang et al., 2009). Another allele, tkpr1-2, produced pollen severely affected in exine formation (Figure 6), whereas tkpr2-1 and tkpr2-2 lines were fertile and produced pollen grains exhibiting only slightly modified exine patterns (Figure 6; see Supplemental Figure 3 online). The differential impacts of TKPR1 (DRL1) and TKPR2 inactivation on pollen development could not have been anticipated from biochemical data since both TKPR1 and TKPR2 proteins have similar reduction activities in vitro on the tetraketide α-pyrones produced by PKSA and PKSB, with TKPR2 being the most active enzyme (Figures 7 and 8). These results suggest that in vivo, the two reductases do not fulfill exactly the same functions. The distinct subcellular localizations of TKPR1 and TKPR2 that have been evidenced in Figure 9 may support these conclusions. TKPR1 appeared preferentially localized to the ER, as also shown for PKSA and PKSB (Kim et al., 2010). With CYP703A2 and CYP704B1 hydroxylases being putatively localized to the ER, they may form, with PKSs and TKPR1, metabolons located in the ER and yielding polyhydroxylated α-pyrones. In this model, cytoplasmic TKPR2 enzyme might be preferentially active on distinct substrates released in the cytosol. In situ hybridization experiments showed that TKPR1 and TKPR2 have similar temporal and spatial patterns of gene expression but that TKPR1 expression is initiated at an earlier stage of anther development (Figure 2). Immunodetection of the proteins with specific antibodies in flower tissues at various stages of development (Figure 1) showed distinct kinetics of TKPR protein accumulation, with TKPR1 accumulating more transiently than TKPR2. The severe phenotype of tkpr1 pollen, fully sterile or poorly fertile depending on the allele, suggests that TKPR1-catalyzed polyketide reduction is essential at an early step of exine formation, possibly for initiating the reticulated exine pattern. It is noteworthy that tapetal cells of tkpr1-1 mutant appeared highly vacuolated compared with the wild type (Figure 5), thus suggesting a role of TKPR1 expression in normal tapetum development. We were unable to demonstrate a clear differential accumulation of TKPR1 and TKPR2 proteins at early stages of anther development due to too weak immunological signals (data not shown).
It is interesting that, in parallel with these apparent in vivo differences in TKPR function, the two PKS enzymes that supply TKPR substrates may also have slightly different in vivo functions, despite their identical in vitro activities (Kim et al., 2010). Our results are in agreement with the hypothesis that, in vivo, pairs of PKS and TKPR enzymes work together to specify particular structural attributes of the sporopollenin polymer, and with the model whereby the TKPR1/PKSB pair plays an essential role in polyhydroxylated long-chain α-pyrones biosynthesis at a critical early stage of exine deposition.
It is noteworthy here that the sequential action of ACOS5 (fatty acid acyl-CoA synthase), PKS, and TKPR enzymes on fatty acid substrates ends up with the chemical reduction of the carboxylic function of the initial fatty acid into a secondary alcohol function in the tetraketide α-pyrone molecule. This new hydroxyl function is then prone to form ether or ester linkages in the highly complex sporopollenin polymer as proposed for hydroxyl groups introduced by the P450 hydroxylases (Morant et al., 2007; Dobritsa et al., 2009b). Since hydroxy fatty acids have been demonstrated to be substrates of ACOS5, PKS, and TKPR activities acting sequentially to synthesize sporopollenin units (Figure 8), the network of ester and ether interlinkages in the sporopollenin polymer could be particularly dense, resulting in an unparalleled resistance to degradation.
The Sporopollenin Biosynthetic Pathway Is Conserved in Land Plants
It is believed that the high level of resistance of the pollen cell wall to biotic and abiotic stresses has played a pivotal role in the conquest of land by plants (Rozema et al., 2001; Blackmore et al., 2007). Indeed, the widespread occurrence of sporopollenin in pollen grains and spores of land plant lineages indicates the conservation of an ancient biosynthetic pathway during plant evolution. In agreement with this assumption, it has been noticed that genes involved in exine formation, such as ACOS5, CYP703A2, CYP704B1, and PKSA/PKSB, are conserved from moss to gymnosperms and angiosperms (Morant et al., 2007; de Azevedo Souza et al., 2009; Dobritsa et al., 2009b; Kim et al., 2010). In the case of PKSA and PKSB, several putative orthologs are specifically expressed in anthers or tapetal cells of various plant species (Kim et al., 2010).
TKPR1 (DRL1) and TKPR2 belong to a superfamily of reductases/dehydrogenases that harbors an N-terminal putative NAD(P)H binding domain called a Rossmann fold and includes DFR, ANR, and CCR involved in flavonoid and lignin biosynthesis (Shirley et al., 1992; Lacombe et al., 1997; Yau et al., 2005; Tang et al., 2009). The reductase/dehydrogenase gene family appears widespread in living organisms since it also includes cholesterol dehydrogenases and UDP-galactose epimerases from bacteria and 3β-hydroxysteroid dehydrogenases of mammalian origin (see Supplemental Figure 1 online) (Baker et al., 1990; Baker and Blasco, 1992). Moreover, the overall structural resemblance of the gene family members indicates that they may be derived from a common ancestor (Baker et al., 1990; Baker and Blasco, 1992; Lacombe et al., 1997; Yau et al., 2005; Tang et al., 2009).
Our results show that TKPR2, a newly discovered reductase, and TKPR1/DRL1 (Tang et al., 2009) are coexpressed in Arabidopsis anthers (Figure 2) and belong to the same reductase/dehydrogenase family. Phylogenetic analysis showed that TKPR1 and TKPR2 genes cluster in distinct clades with homologs from various plant species (Figure 10). Besides Arabidopsis, poplar, rice, grape (Vitis vinifera), sorghum (Sorghum bicolor), and Physcomitrella possess gene members clustering in each clade. As observed for PKSA and PKSB genes (Kim et al., 2010), these TKPR orthologs appear to be specifically expressed during anther development. In the case of Arabidopsis (this work), poplar (Wilkins et al., 2009), and rice (Yau et al., 2005), a member of each clade was found to be expressed in male reproductive organs, thus suggesting that the functions of the encoded reductases are not fully redundant, even though those of Arabidopsis have been shown here to catalyze the same reaction in vitro. Thus, our work shows that, like the Arabidopsis ACOS5, CYP703A2, CYP704B1, and PKSA/PKSB genes, TKPR1 and TKPR2 represent ancient conserved genes present in all land plants surveyed to date. The functions of these enzymes revealed by this and other work (Morant et al., 2007; de Azevedo Souza et al., 2009; Dobritsa et al., 2009b; Kim et al., 2010) strongly suggest that they participate in a conserved biochemical pathway that generates sporopollenin precursors.
A Model for Sporopollenin α-Pyrone Monomer Synthesis
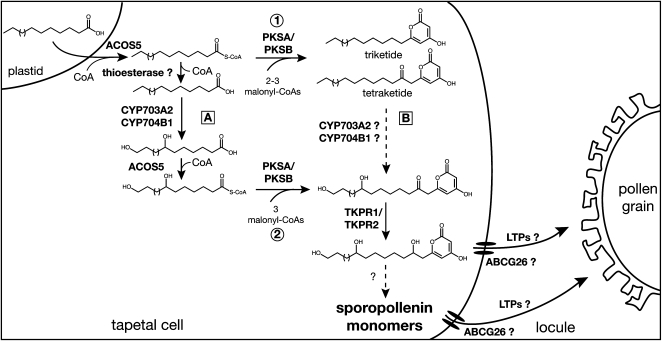
The results described here and in the companion article (Kim et al., 2010), provide compelling new data illuminating a previously undiscovered, conserved metabolic pathway involving fatty acyl-CoA extension and reduction steps catalyzed by PKS and TKPR enzymes, respectively, which leads to polyketide α-pyrone compounds that are shown here to be essential sporopollenin precursors. In Figure 11, we propose a working model that includes different possibilities for the sequential actions of the enzymes involved in sporopollenin unit synthesis. After CoA ester formation by ACOS5, PKSs can catalyze acyl-CoA extension to tri- and tetraketide α-pyrones (Figure 11, route 1). CYP450 hydroxylases have been shown to be active on free fatty acids (Morant et al., 2007; Dobritsa et al., 2009b), thus implicating putative thioesterase and CoA ester regeneration steps upstream and downstream of the hydroxylation step as shown on the left side of the scheme (Figure 11, route A). We have shown that hydroxy fatty acyl-CoAs are the most efficient substrates for PKSA and PKSB, consistent with an ER localization (Kim et al., 2010), and can yield various hydroxy polyketides (route 2). These latter compounds might also arise from the hydroxylation of the alkyl α-pyrones by CYP450s (route B), but this possibility remains to be examined experimentally. Finally, reduction of the carbonyl function of the hydroxy tetraketide α-pyrones by TKPRs gives rise to an additional hydroxyl function in the alkyl chain of the polyketide products, generating more highly hydroxylated polyketides that are proposed constituents of a sporopollenin polymer highly cross-linked by ester and ether bonds. The association of TKPR1 with ER, where hydroxylation by P450s takes place, may be important in this respect. It remains to be determined whether these hydroxylated polyketides are direct sporopollenin monomers exported into the locule by the possible actions of ABCG26, a putative ATP binding cassette transporter of sporopollenin precursors in Arabidopsis (Quilichini et al., 2010), and lipid transfer proteins (as has been suggested in rice; Zhang et al., 2010) or if other unknown modifications occur before export and polymerization in the pollen cell wall (Figure 11). Examination of the metabolic changes in the sporopollenin mutants that we have characterized should shed more light on the complex molecular network leading to sporopollenin biosynthesis.
Figure 11.
A Putative Scheme of Sporopollenin Biosynthesis.
Fatty acids synthesized in plastids are esterified to CoA by ACOS5 and then extended to triketide and tetraketide products by PKSs that catalyze the condensation to malonyl CoA. Extension of fatty acyl-CoAs by PKSs may take place either directly (route 1) or after hydroxylation by CYP450s (route 2). CYP450 hydroxylases have been shown to have free fatty acids as substrates, thus implicating CoA ester hydrolysis by a putative thioesterase and regeneration of the CoA esters by ACOS5 upstream and downstream of the hydroxylation step, respectively (route A). However, hydroxyl groups might be putatively introduced by CYP450s at the tetraketide level (route B) before the reduction of the carbonyl function by TKPRs. Resulting polyhydroxylated tetraketide compounds may then be exported by ABCG26 and lipid transfer proteins (LTPs) to the locule as sporopollenin building units or further processed before transport and polymerization in the pollen wall.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana Columbia (Col-0) and Wassilewskija ecotypes were grown as follows. For segregation analysis and transformant selection, seeds were surface sterilized, and after a cold treatment (2 d at 4°C in the dark) were sown on Murashige and Skoog medium supplemented with adequate antibiotic and germinated at 20°C under 12 h 70 μmol m−2 s−1 fluorescent lighting. Twelve days later, the plants were subcultured on soil and transferred to growth chambers with light/dark cycles of 16 h/8 h for 2 weeks and then to the greenhouse. Homozygous seeds were sown and grown directly on soil. Nicotiana benthamiana plants were grown in the greenhouse under the same conditions. T-DNA insertion mutants were obtained from SALK, SAIL (Alonso et al., 2003), and Gabi-Kat (Rosso et al., 2003) collections via the Nottingham Arabidopsis Stock Centre and from FLAGdb/FST at Institut National de la Recherche Agronomique (INRA) Versailles (Samson et al., 2002). The lines used in this study were SAIL_837_D01 and FLAG_019D03 for tkpr1 and GABI_838H09 and SALK_129453 for tkpr2. In progeny, homozygous insertion lines were identified by PCR using gene-specific and vector-specific primers (see Supplemental Table 1 online).
Phylogenetic Studies
Putative TKPR orthologs were searched using the TBLASTN algorithm (blosum62 matrix) (Altschul et al., 1990) in plant EST databases (the National Center for Biotechnology Information, http://blast.ncbi.nlm.nih.gov/Blast.cgi, and Dana-Farber Cancer Institute Plant Gene Indice, http://compbio.dfci.harvard.edu/tgi/plant.html). Translated sequences of candidate genes were aligned using the MUSCLE 3.7 program (Edgar, 2004) with a maximum number of 16 iterations and then curated by removing gap positions. The resulting multiple sequence alignment is available as Supplemental Data Set 1 online. Sources of amino acid sequences used to generate these alignments are given in Supplemental Table 2 online. A bootstrap maximum likelihood tree was constructed using PhyML 3.0 (Guindon and Gascuel, 2003) with the WGA amino acid substitution model. The tree was edited using FigTree v.1.2.3. Bootstrap values are based on 1000 iterations.
Microscopy
For fluorescence microscopy, pollen was stained with 0.01% auramine O in water for 5 min, washed twice with water, and examined with a Nikon E800 microscope using fluorescein isothiocyanate settings. For electron scanning microscopy, pollen grains were coated with gold particles (S150A sputter coater; Edwards High Vacuum) for 3 min at 50 mV and viewed with a Philips XL30 ESEM (FEI) under high vacuum conditions and at 20 to 30 kV. The microscope was equiped with a Thornley-Everhart secondary electron detector and a PGT Spirit EDX microanalysis system (Princeton Gamma Tech). For TEM, tissue fixation, embedding, sectioning, and visualization were performed in the UBC Bioimaging Facility as described by Kim et al. (2010). For subcellular localization of TKPR-eGFP fusions, the cDNA of TKPR1/2 was PCR amplified using the primer pairs given in Supplemental Table 1 online and inserted into the expression vector pB7FWG2 (VIB) using the Gateway cloning system (Invitrogen) to obtain C-terminal fusions with eGFP. The mRFP fluorescent protein with the C-terminal extension-HDEL served as an ER marker protein (Robinson et al., 2007). Both constructs allow the in planta expression of proteins under the control of the constitutive cauliflower mosaic virus double 35S promoter. Leaves of 4-week-old N. benthamiana plants were agroinfiltrated and subsequently placed in a growth chamber for 96 h. Fluorescence microscopy was performed using a LSM 700 confocal laser microscope (Carl Zeiss). Two lasers were used as the excitation sources at 488 and 555 nm, and light emission was detected with the short-pass filter at 613 nm for RFP constructs and short-pass filter at 515 nm for GFP constructs. Images were recorded and processed using ZEN 2009 (Carl Zeiss).
RT-PCR
Reverse transcription and quantitative RT-PCR analysis of TKPR expression was performed as described for PKS genes (Kim et al., 2010) using forward and reverse primers shown in Supplemental Table 1 online. In the case of quantitative RT-PCR, ACTIN2 (At3g18780) was used as a reference gene. For knockout mutant validation, TUBULIN3 (At5g62700) was used as a positive control of RT-PCR.
Production of Recombinant Proteins and Specific Antibodies and Immunological Techniques
The methods and procedures were as described in the companion article (Kim et al., 2010). Briefly, full coding sequences were cloned in pGEX-KG and expressed in E. coli. Recombinant proteins were purified on glutathione agarose and analyzed on SDS-PAGE gels as illustrated in Supplemental Figure 4 online. Purified preparations were used to raise polyclonal antibodies in rabbits. Antibody specificity for each TKPR protein was enhanced by preincubation with an acetonic powder of Arabidopsis leaves and then with the other recombinant TKPR protein. Antibody specificity was checked against purified recombinant proteins on immunoblots (see Supplemental Figure 5 online). Bud tissues were fixed, dehydrated in an ethanol series, and embedded in paraplast. Ten-micrometer-thick sections were incubated with a 200-fold dilution of the serum. After washing and incubation with phosphatase-labeled goat anti-rabbit antibody, phosphatase activity was revealed with FastRed. Sections were observed under a Nikon E800 microscope.
In Situ Hybridization
Methods used for probe synthesis, tissue fixation and hybridization were as described by Kim et al. (2010). Digoxigenin-labeled probes were synthesized by PCR using gene-specific primers (see Supplemental Table 1 online). Probe specificity was checked by hybridization to PCR-amplified cDNA of TKPR1 and TKPR2 as shown in Supplemental Figure 6 online.
Chemical Synthesis of Fatty Acyl-CoA Esters
Palmitoyl-CoA and stearoyl-CoA esters were chemically prepared from palmitic and stearic acids according to published procedures (Funa et al., 2006). CoA ester preparations were purified by reverse-phase HPLC and characterized by liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis.
Enzyme Activity Assays
ACOS5 Activity
In a volume of 100 μL, 5 μg of recombinant protein was incubated in 60 mM sodium phosphate buffer containing 10 mM MgCl2, 5 mM ATP, 2.5 mM DDT, 1 mM CoASH, and 0.1 mM fatty acid (from a 1 mM stock solution in 0.1% Triton X-100). The mixture was incubated at 30°C. For ACOS5 activity measurements, the reaction was stopped by addition of 10 μL of 1 n HCl and, after extraction of residual fatty acid with ethyl acetate, CoA esters were analyzed by LC-MS/MS. ACOS5 activity was used for the biosynthesis of various fatty acyl-CoA esters that were tested as PKS substrates.
PKS Activity
When chemically synthesized CoA esters were used, 5 μg of enzyme was incubated in 100 μL volume containing 100 mM sodium phosphate, pH 7.0, 0.1 mM fatty acyl-CoA ester, and 0.1 mM malonyl-CoA. PKS activity was evaluated as described by Kim et al. (2010) and used to synthesize the substrates of TKPRs. PKS activity was also assayed with various fatty acyl-CoA esters synthesized by ACOS5. To that aim, after 15 min ACOS5 incubation, 5 μg of PKSA or PKSB protein were added to the incubation medium together with malonyl-CoA and incubated for an additional hour. Activity was measured as for chemically synthesized fatty acyl-CoA substrates. A variety of tetraketide compounds synthesized by PKS were tested as substrates of TKPRs.
TKPR Activity
To evaluate reductase activity on PKS reaction products, PKSs and TKPRs (5 μg each), 0.1 mM malonyl-CoA, and 1 mM NADPH were added to the reaction mixture at the end of 15 min of ACOS5 incubation, and incubation was carried out for one additional hour. The reaction was stopped with 10 μL 1 n HCl, and products were extracted with 500 μL ethyl acetate. Solvent was evaporated under nitrogen stream, and the reaction products were dissolved in 100 μL methanol and analyzed by LC-MS/MS.
Characterization of Reaction Products by LC-MS/MS
Reaction products were resolved and identified as described by Kim et al. (2010). For analysis, an Acquity UPLC system (Waters) coupled to a Quattro Premier XE triple quadrupole MS system (Waters Micromass) was used. Full-scan, selected ion recording, daughter scan, and multiple reaction monitoring modes were used for mass analysis.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At4g35420 (TKPR1), At1g68540 (TKPR2), At1g02050 (PKSA), At4g34850 (PKSB), At1g62940 (ACOS5), At3g18780 (ACTIN2), and At5g62700 (TUBULIN3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Multiple Alignment of Oxidoreductase Predicted Protein Sequences Obtained with ClustalW.
Supplemental Figure 2. Developmental Expression Profiles of TKPR1, TKPR2, and At1g25460.
Supplemental Figure 3. Comparison of Exine Architecture in Wild-Type and tkpr2-2 Pollen.
Supplemental Figure 4. Analysis of Recombinant Protein Preparations at Different Steps of Purification.
Supplemental Figure 5. Characterization of Antibodies Raised against TKPR Proteins.
Supplemental Figure 6. Specificity of TKPR Nucleotidic Probes.
Supplemental Table 1. Primers Used in Cloning, Genotyping, and RT-PCR Experiments.
Supplemental Table 2. Genes and Expression Data Used for Constructing the Phylogenetic Tree of Figure 10.
Supplemental Data Set 1. Sequence Alignments Used for Constructing the Phylogenetic Tree Shown in Figure 10.
Supplementary Material
Acknowledgments
The assistance of D. Meyer (Institut de Biologie Moléculaire des Plantes) in histochemical analysis, M. Alioua (Institut de Biologie Moléculaire des Plantes) in DNA sequencing, and M. Erhardt (Institut de Biologie Moléculaire des Plantes) for TEM is gratefully acknowledged. We thank J.-H. Lignot (Institut Pluridisciplinaire Hubert Curien, Strasbourg) for kind help with scanning electron microscopy, C. Ritzenthaler (Institut de Biologie Moléculaire des Plantes) for help with confocal microscopy, and D. Debayle (Institut de Biologie Moléculaire des Plantes) for assistance in LC-MS/MS analysis at the initial stages of the work. We also thank Michael Friedmann (University of British Columbia) for helpful comments and advice and the University of British Columbia Bioimaging Facility for technical assistance and advice. We are grateful to the Salk Genomic Analysis Laboratory (La Jolla, CA), INRA Versailles (France), and Bielefeld University (Germany) for providing the T-DNA mutants and to the Nottingham Arabidopsis Stock Centre for distributing the seeds. The UPLC-MS/MS system was cofinanced by the Centre National de la Recherche Scientifique, the Université de Strasbourg, the Région Alsace, INRA, and Tepral Company. This work was supported by doctoral fellowships of the Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche to E.G. and B.L, and by a Natural Sciences and Engineering Research Council Discovery grant to C.J.D.
References
- Aarts M.G., Hodge R., Kalantidis K., Florack D., Wilson Z.A., Mulligan B.J., Stiekema W.J., Scott R., Pereira A. (1997). The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Ahlers F., Lambert J., Wiermann R. (2003). Acetylation and silylation of piperidine solubilized sporopollenin from pollen of Typha angustifolia L. Z. Naturforsch., C, J. Biosci. 58: 807–811 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Alves-Ferreira M., Wellmer F., Banhara A., Kumar V., Riechmann J.L., Meyerowitz E.M. (2007). Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol. 145: 747–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T., Hatakeyama K., Hinata K., Inatsugi R., Nishida I., Sato S., Kato T., Tabata S., Toriyama K. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39: 170–181 [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Hatakeyama K., Hinata K., Sato S., Kato T., Tabata S., Toriyama K. (2003). A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol. Biol. 53: 107–116 [DOI] [PubMed] [Google Scholar]
- Baker M.E., Blasco R. (1992). Expansion of the mammalian 3 beta-hydroxysteroid dehydrogenase/plant dihydroflavonol reductase superfamily to include a bacterial cholesterol dehydrogenase, a bacterial UDP-galactose-4-epimerase, and open reading frames in vaccinia virus and fish lymphocystis disease virus. FEBS Lett. 301: 89–93 [DOI] [PubMed] [Google Scholar]
- Baker M.E., Luu-The Y., Simard J., Labrie F. (1990). A common ancestor for mammalian 3 beta-hydroxysteroid dehydrogenase and plant dihydroflavonol reductase. Biochem. J. 269: 558–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore S., Wortley A.H., Skvarla J.J., Rowley J.R. (2007). Pollen wall development in flowering plants. New Phytol. 174: 483–498 [DOI] [PubMed] [Google Scholar]
- Bubert H., Lambert J., Steuernagel S., Ahlers F., Wiermann R. (2002). Continuous decomposition of sporopollenin from pollen of Typha angustifolia L. by acidic methanolysis. Z. Naturforsch., C, J. Biosci. 57: 1035–1041 [DOI] [PubMed] [Google Scholar]
- Chen X., Goodwin S.M., Boroff V.L., Liu X., Jenks M.A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo Souza C., Kim S.S., Koch S., Kienow L., Schneider K., McKim S.M., Haughn G.W., Kombrink E., Douglas C.J. (2009). A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T.T., Carlsson A.S., Hamberg M., Bülow L., Stymne S., Olsson P. (2009). Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J. Plant Physiol. 166: 787–796 [DOI] [PubMed] [Google Scholar]
- Dobritsa A.A., Nishikawa S.-I., Preuss D., Urbanczyk-Wochniak E., Sumner L.W., Hammond A., Carlson A.L., Swanson R.J. (2009a). LAP3, a novel plant protein required for pollen development, is essential for proper exine formation. Sex. Plant Reprod. 22: 167–177 [DOI] [PubMed] [Google Scholar]
- Dobritsa A.A., Shrestha J., Morant M., Pinot F., Matsuno M., Swanson R., Møller B.L., Preuss D. (2009b). CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 151: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funa N., Ozawa H., Hirata A., Horinouchi S. (2006). Phenolic lipid synthesis by type III polyketide synthases is essential for cyst formation in Azotobacter vinelandii. Proc. Natl. Acad. Sci. USA 103: 6356–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E., Besseau S., Geoffroy P., Debayle D., Heintz D., Lapierre C., Pollet B., Heitz T., Legrand M. (2009). A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J. 58: 246–259 [DOI] [PubMed] [Google Scholar]
- Guan Y.F., Huang X.Y., Zhu J., Gao J.F., Zhang H.X., Yang Z.N. (2008). RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 147: 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Hamberger B., Ellis M., Friedmann M., de Azevedo Souza C., Barbazuc B., Douglas C.J. (2007). Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of angiosperm gene families. Can. J. Bot. 85: 1182–1201 [Google Scholar]
- Hertweck C. (2009). The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 48: 4688–4716 [DOI] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Nagata N., Yoshiba Y., Ohme-Takagi M., Ma H., Shinozaki K. (2007). Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19: 3549–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.S., et al. (2010). LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22: 4045–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T., Kawabata-Awai C., Sakuradani E., Shimizu S., Okada K., Wada T. (2003). The YORE-YORE gene regulates multiple aspects of epidermal cell differentiation in Arabidopsis. Plant J. 36: 55–66 [DOI] [PubMed] [Google Scholar]
- Lacombe E., Hawkins S., Van Doorsselaere J., Piquemal J., Goffner D., Poeydomenge O., Boudet A.M., Grima-Pettenati J. (1997). Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: Cloning, expression and phylogenetic relationships. Plant J. 11: 429–441 [DOI] [PubMed] [Google Scholar]
- Ma H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Morant M., Jørgensen K., Schaller H., Pinot F., Møller B.L., Werck-Reichhart D., Bak S. (2007). CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen H.A., Makaroff C.A. (1995). Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185: 7–21 [Google Scholar]
- Paxson-Sowders D.M., Dodrill C.H., Owen H.A., Makaroff C.A. (2001). DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol. 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- Quilichini T.D., Friedmann M.C., Samuels A.L., Douglas C.J. (2010). ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol. 154: 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Herranz M.-C., Bubeck J., Pepperkok R., Ritzenthaler C. (2007). Membrane dynamics in the early secretory pathway. Crit. Rev. Plant Sci. 26: 199–225 [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Rowland O., Lee R., Franke R., Schreiber L., Kunst L. (2007). The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett. 581: 3538–3544 [DOI] [PubMed] [Google Scholar]
- Rozema J., Broekman R.A., Blokker P., Meijkamp B.B., de Bakker N., van de Staaij J., van Beem A., Ariese F., Kars S.M. (2001). UV-B absorbance and UV-B absorbing compounds (para-coumaric acid) in pollen and sporopollenin: the perspective to track historic UV-B levels. J. Photochem. Photobiol. B 62: 108–117 [DOI] [PubMed] [Google Scholar]
- Rubin-Pitel S.B., Zhang H., Vu T., Brunzelle J.S., Zhao H., Nair S.K. (2008). Distinct structural elements dictate the specificity of the type III pentaketide synthase from Neurospora crassa. Chem. Biol. 15: 1079–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson F., Brunaud V., Balzergue S., Dubreucq B., Lepiniec L., Pelletier G., Caboche M., Lecharny A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y.-C., Lee P.Y., Truong M.T., Beals T.P., Goldberg R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322 [Google Scholar]
- Saxena P., Yadav G., Mohanty D., Gokhale R.S. (2003). A new family of type III polyketide synthases in Mycobacterium tuberculosis. J. Biol. Chem. 278: 44780–44790 [DOI] [PubMed] [Google Scholar]
- Scott R.J., Spielman M., Dickinson H.G. (2004). Stamen structure and function. Plant Cell 16(Suppl): S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley B.W., Hanley S., Goodman H.M. (1992). Effects of ionizing radiation on a plant genome: Analysis of two Arabidopsis transparent testa mutations. Plant Cell 4: 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Masaoka K., Nishi M., Nakamura K., Ishiguro S. (2008). Identification of kaonashi mutants showing abnormal pollen exine structure in Arabidopsis thaliana. Plant Cell Physiol. 49: 1465–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L.K., Chu H., Yip W.K., Yeung E.C., Lo C. (2009). An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New Phytol. 181: 576–587 [DOI] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Wilkins O., Nahal H., Foong J., Provart N.J., Campbell M.M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149: 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.Y., Sharma S.B., Paiva N.L., Ferreira D., Dixon R.A. (2003). Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]
- Yang C., Vizcay-Barrena G., Conner K., Wilson Z.A. (2007). MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19: 3530–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau C.P., Zhuang C.X., Zee S.Y., Yip W.K. (2005). Expression of a microsporocyte-specific gene encoding DIHYDROFLAVONOL 4-REDUCTASE-LIKE protein is developmentally regulated during early microsporogenesis in rice. Sex. Plant Reprod. 18: 65–74 [Google Scholar]
- Zhang D., Liang W., Yin C., Zong J., Gu F., Zhang D. (2010). OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 154: 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.