Caffeic acid O-methyltransferase (COMT) is a key enzyme in the biosynthesis of monolignols, the building blocks of plant lignins. New crystallographic analyses of unliganded and product-bound forms of COMT detail (1) the closed conformational state that productively assembles reactant molecules within the enzyme’s active site and (2) the molecular bases for substrate preferences of the COMTs.
Abstract
Lignin forms from the polymerization of phenylpropanoid-derived building blocks (the monolignols), whose modification through hydroxylation and O-methylation modulates the chemical and physical properties of the lignin polymer. The enzyme caffeic acid O-methyltransferase (COMT) is central to lignin biosynthesis. It is often targeted in attempts to engineer the lignin composition of transgenic plants for improved forage digestibility, pulping efficiency, or utility in biofuel production. Despite intensive investigation, the structural determinants of the regiospecificity and substrate selectivity of COMT remain poorly defined. Reported here are x-ray crystallographic structures of perennial ryegrass (Lolium perenne) COMT (Lp OMT1) in open conformational state, apo- and holoenzyme forms and, most significantly, in a closed conformational state complexed with the products S-adenosyl-l-homocysteine and sinapaldehyde. The product-bound complex reveals the post-methyl-transfer organization of COMT’s catalytic groups with reactant molecules and the fully formed phenolic-ligand binding site. The core scaffold of the phenolic ligand forges a hydrogen-bonding network involving the 4-hydroxy group that anchors the aromatic ring and thereby permits only metahydroxyl groups to be positioned for transmethylation. While distal from the site of transmethylation, the propanoid tail substituent governs the kinetic preference of ryegrass COMT for aldehydes over alcohols and acids due to a single hydrogen bond donor for the C9 oxygenated moiety dictating the preference for an aldehyde.
INTRODUCTION
Lignin is a heterogeneous polymeric macromolecule and a major component of the cell wall in vascular plants. It provides mechanical support for plant tissues (Zhong et al., 1997; Boerjan et al., 2003), protects the plant from pathogen invasion (Hammond-Kosack and Jones, 1996), and enhances the hydrophobicity of the plant vasculature (Kubitzki, 1987). Lignin assembles by dehydrogenative polymerization of monomeric monolignol building blocks (Whetten and Sederoff, 1995). The three prevalent monolignols, p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol, differ only in the degree of methoxylation of the phenolic ring. These give rise to p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) subunits, respectively, in polymerized lignin. The subunit composition of plant lignin is dependent on species, stage of development, tissue type, subcellular localization, and biotic and abiotic factors. Importantly, the relative abundance of the subunit types determines to a large extent the physical properties of the lignin polymer (Boerjan et al., 2003). S-lignin preferentially forms intersubunit ether bond linkages involving the 4-hydroxyl group, whereas G-lignin possesses a greater concentration of less labile β-5, 5-5 biphenyl, and other carbon-carbon intersubunit bonds. The chemical degradation of lignin during pulping occurs more efficiently with wood material possessing a high S/G ratio, whereas forage digestibility is impaired for plant tissue with elevated S-lignin content (reviewed in Li et al., 2008). Therefore, the impact of lignin composition on the usability of plant material for paper manufacturing, forage, and biofuel production is of great economic interest.
p-Coumaric acid derived from the primary metabolite l-Phe serves as the starting material for the monolignols. p-Coumaric acid is transformed serially via hydroxylation and O-methylation of the phenolic ring and also reductions of the oxygenated functionality on the propanoid side chain. These reactions are part of the general phenylpropanoid biosynthetic pathway and are important for the production of not only the monolignols but also other classes of plant compounds, such as the lignans, phenylpropenes, and suberins. In the current view of monolignol biosynthesis, the key O-methylations of hydroxyl groups at the C3 and C5 positions of the phenolic ring of monolignol precursors are catalyzed primarily by distinct S-adenosyl-l-methionine (SAM)–dependent enzymes. Caffeoyl CoA 3-O-methyltransferase (CCoAOMT; EC 2.1.1.104) is primarily responsible for the initial O-methylation of the 3-hydroxyl group and specifically methylates the 3,4-dihydroxy substrate as a CoA-linked thioester.
A second O-methyltransferase termed (historically) caffeic acid O-methyltransferase (COMT; EC 2.1.1.68) catalyzes predominantly the O-methylation of the 5-hydroxyl group of 3-methoxy-4,5-dihydroxy precursors, preferentially as the aldehyde or alcohol (Figure 1). The roles of CCoAOMT and COMT have been inferred largely from the effects on lignin composition of downregulation of these enzymes in transgenic plants. However, in vitro characterization shows that both CCoAOMT and COMT have bifunctional activity in 3- and 5-O-methylation and that COMT accepts as substrates monolignol precursors as free acids, aldehydes, or alcohols (as detailed further below).
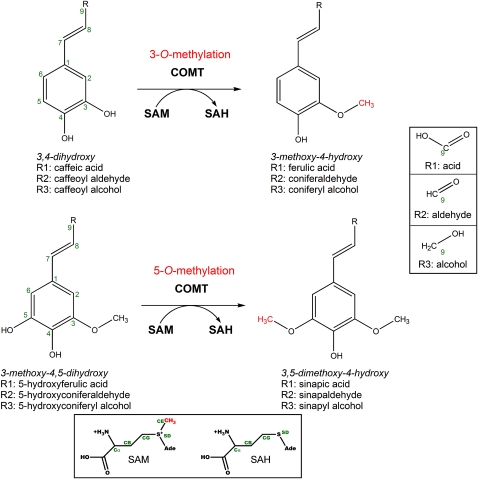
Figure 1.
Potential Monolignol-Precursor Substrates of COMT and the O-Methylation Reaction Catalyzed by COMT Enzymes.
For each of the two hydroxylation/methoxylation patterns of the aromatic ring of the monolignol precursor substrates (3,4-dihydroxy or 3-methoxy-4,5-dihydroxy), the C9 R group of the propene side chain can be the carboxylic acid (R1), aldehyde (R2), or alcohol (R3). The atom numbering of the monolignol carbon skeleton is designated in green (1-9). The methyl group transferred by COMT from SAM is colored red. The box at the bottom of the figure shows the structures and atom nomenclature (green labels) of SAM and SAH. Ade, adenosine; B, G, D, and E represent β, γ, δ, and ε, respectively.
The COMTs belong to the plant type-1 family of SAM-dependent O-methyltransferases (OMTs; the nomenclature of Noel et al. [2003] is used throughout this article). These enzymes are typically ~360 amino acid residues in length, possess an auxiliary N-terminal domain that functions in homodimerization, and do not require metal ions for activity. These properties clearly distinguish the plant type-1 OMTs from a second family, the type-2 OMTs, to which CCoAOMT belongs. The smaller type-2 OMTs contain only the prototypical SAM binding domain (Martin and McMillan, 2002) and are dependent on divalent cations for catalytic activity.
One or more representatives of COMT occur in almost all plant lineages. In vitro, most COMTs generally display a limited degree of permissiveness in substrate acceptance. COMTs from angiosperms such as alfalfa (Medicago sativa), wheat (Triticum aestivum), and aspen (Populus tremuloides) (Li et al., 2000; Parvathi et al., 2001; Zubieta et al., 2002; Ma and Xu, 2008) are kinetically most active against 5-hydroxyconiferaldehyde and then caffeoyl aldehyde, whereas 5-hydroxyconiferyl alcohol and caffeoyl alcohol are slightly less favored, and 5-hydroxyferulic acid and caffeic acid are the poorest substrates (aldehydes > alcohols > acids). In addition, COMTs typically favor O-methylation of the 5-hydroxyl group over the 3-hydroxyl group. However, the COMTs are notable for a complete lack of activity in the O-methylation of the 4-hydroxyl group (Bhuiya and Liu, 2010). Analyses of transgenic alfalfa, maize (Zea mays), and poplar (Populus tremula × Populus alba) plants demonstrate that the most striking effects on lignin composition due to the downregulation of COMT activity are reduced levels of S-subunits without significant changes in G-subunit levels, concomitant with the incorporation of 5-hydroxyconiferyl alcohol into the lignin polymer (Chabbert et al., 1994; Atanassova et al., 1995; Ralph et al., 1997; Zhong et al., 1998; Jouanin et al., 2000; Guo et al., 2001; Ralph et al., 2001). The naturally occurring maize bm3-mutant, which is deficient in COMT activity, shows similar characteristics (Grand et al., 1985; Vignols et al., 1995). Collectively, the available data indicate that the most significant function of COMT in monolignol biosynthesis is the production of syringyl-lignin monomers, preferentially via the O-methylation of the 5-hydroxyconiferaldehyde precursor. Nevertheless, COMTs from a few plant species display disparate substrate specificities. For example, the COMT1 enzyme from tall fescue (Festuca arundinaceae) shows the highest catalytic activity toward caffeoyl aldehyde and 5-hydroxyferulic acid (Chen et al., 2002, 2004). In addition, an OMT from the gymnosperm loblolly pine (Pinus taeda) catalyzes the methylation of monolignol precursors in both free acid and CoA-ester forms (Li et al., 1997).
Earlier crystallographic analyses from this laboratory yielded the structure of COMT from alfalfa (Zubieta et al., 2002), but due to the open conformational state of the enzyme in this structure, the substrate binding pocket was incompletely formed. Therefore, a detailed structural basis for the O-methylation regiospecificity and substrate selectivity of COMT remains to be elucidated. Described here are structure determinations of the perennial ryegrass (Lolium perenne) homolog of COMT (Lp OMT1). A major aim of our studies with perennial ryegrass is the manipulation of lignin composition as a means to improve forage digestibility of transgenic plants (Tu et al., 2010). Of particular relevance is the structure of the closed conformational state of Lp OMT1 in complex with the products S-adenosyl-l-homocysteine and sinapaldehyde occupying a fully assembled phenolic-ligand binding pocket. The atomic resolution view of the amino acid residues involved in forming the substrate binding pocket and the positioning of the methyl-acceptor hydroxyl group in the enzyme’s catalytic site provide a foundation for engineering Lp OMT1 variants with altered regioselectivity. For example, an Lp OMT1 variant with O-methylation activity exclusively for the 3-hydroxyl group of monolignol substrates could potentially be useful for the production of transgenic ryegrass with greatly diminished S-lignin levels and enhanced forage digestibility, or more intriguingly, an Lp OMT1 variant with 4-O-methylation activity may be used to generate novel monolignols with aberrant polymerization properties (Bhuiya and Liu, 2010).
RESULTS
In Vitro Kinetic Analyses of Lp OMT1 Activity
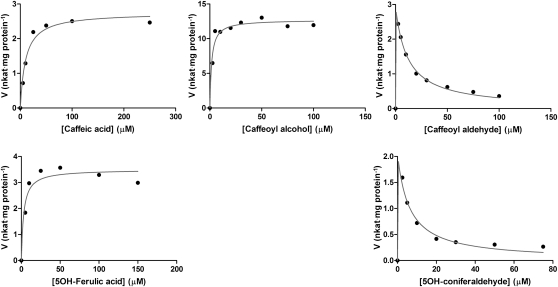
A method based on reversed-phase liquid chromatography coupled with mass spectrometry (LC-MS) was developed for the separation and quantification of substrates and products of Lp OMT1–catalyzed O-methylation reactions. With the chromatographic conditions employed, each of the analyzed substrates (caffeic acid, 5-hydroxyferulic acid, caffeoyl aldehyde, 5-hydroxyconiferaldehyde, and caffeoyl alcohol) was cleanly resolved from the methylated product (the product typically eluted 0.9 min later). Kinetic parameters were determined for OMT1 with each of the three nonaldehyde substrates. Based on measurements with the acid substrates, OMT1 displayed a clear preference for O-methylation of the 5-hydroxyl substituent of a substrate bearing a 3-methoxy substituent (Vmax/Km for 5-hydroxyferulic acid: 3.5 nkat·mg protein−1/3.0 μM), in comparison to 3-O-methylation of a substrate bearing only the single meta substituent (Vmax/Km for caffeic acid: 2.7 nkat·mg protein−1/10.3 μM) (Figure 2). With respect to the C9 substituent, OMT1 showed a strong preference for the alcohol over the acid, as evident from the higher Vmax/Km for caffeoyl alcohol (12.9 nkat·mg protein−1/1.7 μM) versus caffeic acid. In general, these in vitro substrate preference patterns observed with OMT1 are consistent with those reported for the COMTs from aspen (Li et al., 2000), alfalfa (Parvathi et al., 2001; Zubieta et al., 2002), tall fescue (Chen et al., 2004), and wheat (Ma and Xu, 2008), although the Km values measured for Lp OMT1 appear to be consistently lower. Although Lp OMT1 clearly converted the two aldehyde substrates into the corresponding methylated products, a detailed kinetic analysis of these reactions was not possible because severe substrate inhibition was observed above even the lowest substrate concentrations used (1.0 μM) (Figure 2). Previously, Li et al. (2000) observed that 5-hydroxyconiferaldehyde also competitively inhibits the O-methylation of caffeic and 5-hydroxyferulic acids by both aspen COMT and tissue extracts from several other tree species. For aspen COMT (as for the COMTs from most other plant species), 5-hydroxyconiferaldehyde represents the substrate with the greatest binding selectivity (Km = 2.6 μM), a property that may also contribute in part to its potent inhibitory activity (Ki = 0.3 to 2 μM) (Li et al., 2000).
Figure 2.
Enzyme-Kinetics Analyses of Lp OMT1 with Various Monolignol-Precursor Substrates.
In each panel, initial reaction velocity (nkat·mg protein−1) is plotted against substrate concentration (μM), and the displayed curve is derived from nonlinear regression analysis (Prism). The results shown are from a single set of measurements for each substrate.
X-Ray Crystallographic Structures of Distinct Lp OMT1 Catalytic Conformations
The structural analyses of L. perenne OMT1 reported here provide the most complete picture to date of the distinct ligand-bound states of a plant type-1 OMT (Noel et al., 2003). Crystal structures are described for Lp OMT1 in the apoenzyme form, in a form representative of the holoenzyme (bound to S-adenosyl-l-homocysteine [SAH]), and in ternary complexes with SAH and phenolic (sinapaldehyde or coniferaldehyde) products (Table 1). Crystallization trials of Lp OMT1 with SAM consistently yielded structures of the enzyme with bound SAH (indicative of the lability of SAM over the course of the crystallization experiments or low-level transmethylation activity by the enzyme with SAM). The position of the reactive methyl group of SAM can nevertheless be readily modeled from the structure of SAH (as discussed in greater detail below).
Table 1.
Summary of Crystallographic Data Collection and Refinement Statistics for Lp OMT1 Structures
| Structure | OMT1/SAH | OMT1 apo | OMT1/SAH/Sinapaldehyde | OMT1/SAH/Coniferaldehyde |
| Space group | P41212 | P21 | P21 | P21 |
| Unit cell parameters | ||||
| a (Å) | 67.70 | 67.93 | 94.94 | 95.38 |
| b (Å) | 67.70 | 86.54 | 85.23 | 85.06 |
| c (Å) | 249.79 | 131.59 | 98.57 | 98.59 |
| β (°) | 90 | 89.87 | 111.10 | 111.75 |
| Monomers per asymmetric unit | 1 | 4 | 4 | 4 |
| Resolution range (Å)a | 47–1.80 (1.90–1.80) | 60–2.75 (2.90–2.75) | 47–1.85 (1.84–1.85) | 49–2.25 (2.37–2.25) |
| Number of reflections measured | 397,805 | 128,585 | 369,886 | 249,422 |
| Merging R factora | 0.116 (0.454) | 0. 078 (0.364) | 0.085 (0.367) | 0.118 (0.439) |
| Mean (I/σ I)a | 10.4 (2.3) | 11.7 (2.7) | 7.3 (2.1) | 7.1 (2.4) |
| Completenessa | 1.00 (1.00) | 0. 957 (0.946) | 0.957 (0.914) | 0.996 (0.981) |
| Redundancya | 7.2 (3.8) | 3.4 (2.7) | 3.1 (2.4) | 3.6 (2.9) |
| Number of reflections used | 55,209 | 38,072 | 119,372 | 69,341 |
| R factora | 0.212 (0.298) | 0.280 (0.356) | 0.232 (0.404) | 0.232 (0.355) |
| Free R factora | 0.226 (0.315) | 0.334 (0.403) | 0.261 (0.415) | 0.262 (0.362) |
| Number of amino acid residues | 352 | 1,408 | 1,421 | 1,421 |
| Number of water molecules | 313 | 0 | 625 | 314 |
| rmsd from ideality | ||||
| Bond lengths (Å) | 0.005 | 0.010 | 0.006 | 0.007 |
| Bond angles (°) | 1.19 | 1.11 | 1.17 | 1.21 |
| Residues with most favorable conformation (%)b | 98.0 | 93.2 | 97.4 | 97.1 |
| PDB code | 3P9C | See supplemental data onlinec | 3P9I | 3P9K |
Merging R factor = ∑hkl ∑i | Ii(hkl) – <I(hkl)> (denoting average) | ÷ ∑hkl ∑i Ii(hkl).
Values in parentheses describe the highest resolution shell.
As reported by the program MolProbity (Chen et al., 2010).
Supplemental Data Set 1 online: partially refined atomic coordinate set in PDB format (LpCOMT-Apo_refine_10.pdb), which can be displayed with molecular graphics software such as Coot (Emsley and Cowtan, 2004) or PyMol (PyMOL molecular graphics system, version 1.3; Schrödinger). Supplemental Data Set 2 online: crystallographic data (including experimentally measured structure-factor amplitudes and calculated map coefficients; LpCOMT-Apo2_truncated-unique.mtz) that can be input to a crystallographic refinement program such as Phenix (Adams et al., 2010) or displayed with Coot.
Lp OMT1 possesses the same tertiary and quaternary structure as several other plant type-1 OMTs that have been characterized crystallographically (Zubieta et al., 2001, 2002; Liu et al., 2006). Not surprisingly, Lp OMT1 is most structurally similar (a root mean square deviation [rmsd] of 1.51 Å for 332 equivalent residues) to alfalfa COMT (Zubieta et al., 2002), with which it shares 62.7% sequence identity. Slightly higher rmsd values are observed in comparisons with type-1 OMTs having greater sequence divergence: alfalfa chalcone OMT (Zubieta et al., 2001), 2.19 Å rmsd for 312 equivalent residues, with 40.1% sequence identity; alfalfa isoflavone OMT (Zubieta et al., 2001), 1.91 Å rmsd for 310 equivalent residues, with 28.7% sequence identity; and barrel-medic (Medicago truncatula) isoflavonoid OMT (Liu et al., 2006), 3.15 Å rmsd for 265 equivalent residues, with 19.6% sequence identity.
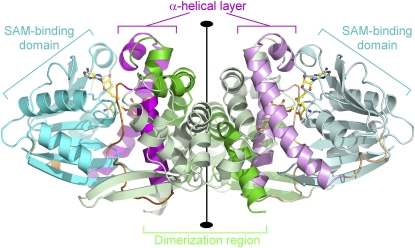
Typical of plant type-1 OMTs, the polypeptide chain fold of Lp OMT1 comprises distinct dimerization and SAM/SAH binding domains, with a α-helical layer sandwiched between the two domains. Two monomers associate to form a symmetric homodimer (Figure 3), in which two tightly intertwined dimerization domains constitute the central core and the SAM binding domains are located peripherally. The active-site cleft in each of the two monomers resides at the interface between the SAM binding domain and the adjacent α-helical layer. Within this layer lies the phenolic substrate binding site, which is lodged in a pocket between two α-helices. Notably, this phenolic binding pocket is also buttressed by α-helical segments from the dimerization region, with one of the helical segments contributed by the opposing monomer.
Figure 3.
Structure of a Homodimer of the Lp OMT1/SAH/Sinapaldehyde Complex.
The homodimer is viewed perpendicular to the twofold rotational axis (vertical) that relates the two monomers. For each of the monomers, the polypeptide chain backbone is color coded by structural domain: dimerization domain (residues 1 to 158), green; SAM/SAH binding domain (residues 199 to 295 and 328 to 360), cyan; α-helical layer involved in phenolic substrate binding (residues 159 to 192 and 304 to 323), magenta; and hinge segments (residues 193 to 198, 296 to 303, and 322 to 327), orange. (One of the monomers is shown with slightly paler shading.) S-adenosyl-homocysteine and sinapaldehyde are shown in ball-and-stick representation and colored according to atom type (carbon, yellow; nitrogen, blue; oxygen, red; and sulfur, gold).
Open and Closed Conformational States of Lp OMT1
Structural comparisons of the various ligand-bound states of OMT1 reveal striking malleability in the positioning of the SAM binding domain. Most conspicuously, relative to the OMT1 holoenzyme, the product-bound OMT1-SAH-sinapaldehyde complex has a substantially more closed conformation, which arises from an approximate 17° inward rotation of the homodimer’s two SAM binding domains with respect to the relatively fixed scaffold formed by the core dimerization region (Figure 4A). These domain movements derive primarily from conformational changes within a hinge segment (residues 296 to 303) in each monomer. The interactions of two α-helices (residues 209 to 219 and 230 to 236) at the top of the SAM binding domain with an extended α-helical segment (residues 159 to 190) in the phenolic-substrate binding region may contribute to stabilizing the closed conformation of OMT1. These interdomain interactions include several hydrogen bonds (between the side chains Asn170-His231, Asn170-Glu235, and Ser181-Thr211) as well as the pairing of the aliphatic faces of two juxtaposed helices. Most significantly, the closed conformational state of OMT1 allows the ribose and Met moieties of the SAM/SAH molecule to associate tightly with the phenolic-substrate binding region (see below).
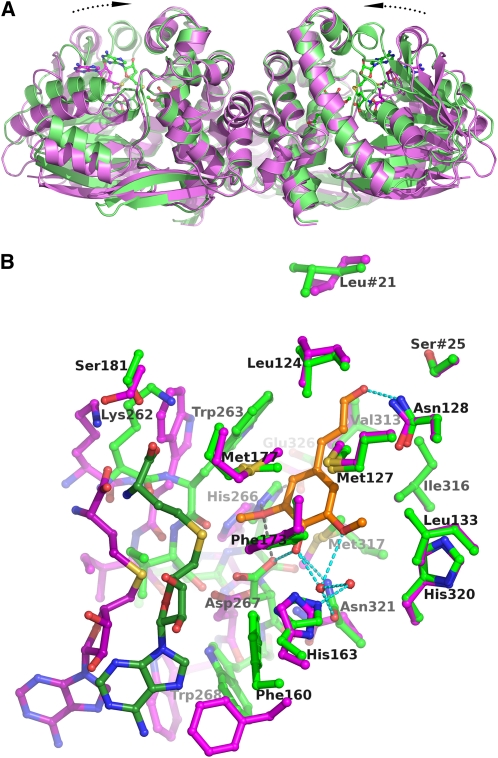
Figure 4.
Comparison of the Open and Closed Conformational States of Lp OMT1.
(A) Comparison of the holoenzyme and holoenzyme/sinapaldehyde forms of the OMT1 homodimer. The structural superposition is based solely on the dimerization domains of the two forms of Lp OMT1. The OMT1 holoenzyme (magenta, with carbon atoms of SAH drawn in darker magenta) exists in an open conformational state, whereas the holoenzyme/sinapaldehyde form (green, with carbon atoms of SAH and sinapaldehyde drawn in darker green) is significantly more closed. The closed conformation of the holoenzyme/sinapaldehyde form arises from an inward rotation (by ~17°) of each SAM binding domain. The orientation of view is identical to that in Figure 3.
(B) Comparison of the active sites in the open and closed conformational states of OMT1. The structures of the OMT1 holoenzyme (open, magenta) and holoenzyme/sinapaldehyde (closed, green) are colored as in (A), except that the carbon atoms of sinapaldehyde are colored orange. With the closed conformational state, the substantial inward shift of the SAM binding domain is evident at the left of the figure (only SAH and residues 262 to 268 and Glu-326, which include the key catalytic residues, are shown). Hydrogen bond interactions involving the sinapaldehyde ligand are shown as dashed lines and are colored as in Figure 5A. The most significant side chain conformational differences between the two forms of OMT1 occur at Phe-160, Ser-181, Trp-263, Met-317, and Asn-321.
In contrast with the marked domain closure observed for the product-bound OMT1 ternary complex, binding of SAM/SAH to OMT1 is associated with only a slight inward movement of the SAM binding domains. Thus, the holoenzyme and apoenzyme forms of OMT1 share essentially the same open conformation, although the SAM binding domains notably appear less ordered overall in electron density maps of apo OMT1. Significantly, in the crystal structures of both the apoenzyme and the ternary complex, the positioning of the SAM binding domain is consistent in the four independent monomers of the two homodimers occurring in the crystallographic asymmetric units.
Binding of SAM/SAH by Lp OMT1
As described for other plant type-1 OMTs, the SAM/SAH molecule within Lp OMT1 adopts an extended conformation (Schubert et al., 2003) and is bound along the C-terminal end of the core β-sheet of the SAM binding domain (Figures 3, 4B, and 5A). In the open conformational state of Lp OMT1, this domain is exclusively responsible for the binding interactions with SAM/SAH. Specifically, the nonpolar side chains of Phe-227, Leu-229, Met-249, Phe-250, and Trp-268 surround the adenine ring of SAM/SAH, which also forms hydrogen bonds with the Asp-248 side chain carboxylate and the backbone amide nitrogen of Met-249. A Gly-rich segment (Gly205-Gly206-Gly207) forms one wall of a channel that is occupied by the ribose ring and the aliphatic portion of the methionine/homocysteine (Met/Hcy) moiety. In addition, hydrogen bond interactions are formed by the Asp-228 side chain carboxylate with both ribose hydroxyl groups, the backbone carbonyl oxygens of Gly-205 and Lys-262 with the α-amino group of the Met/Hcy moiety, and the Lys-262 side chain amino group with the Met/Hcy α-carboxylate group. The ribose ring oxygen also interacts with a water molecule, which in turn forms hydrogen bonds with the side chains of Asp-267 and Trp-268.
Figure 5.
Substrate and Product Binding Sites of Lp OMT1.
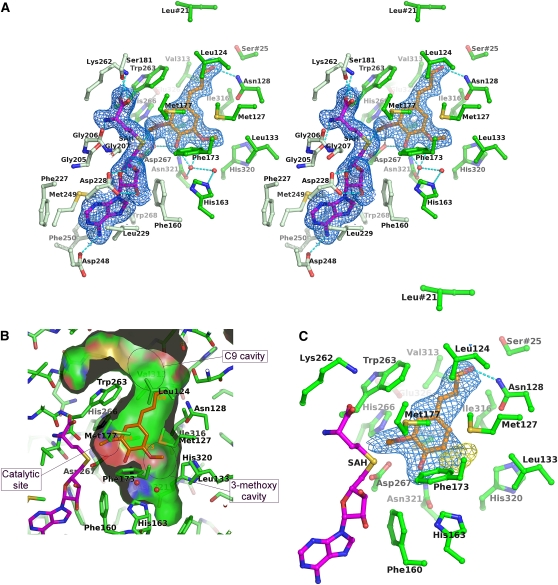
(A) OMT1 amino acid residues that interact with the SAH and sinapaldehyde products (divergent stereoview). The residues that interact with SAH (magenta carbon atoms) are colored light green, whereas those that interact with sinapaldehyde (orange carbon atoms) are colored dark green (only the α-carbon and side chain atoms of each residue are shown, except for the backbone segment Gly205-Gly206-Gly207). Water molecules are represented as small red spheres. Hydrogen bond interactions are shown as cyan dashed lines; those critical to the catalytic reaction (involving His-266, Asp-267, and Glu-326) are shown in dark gray. The substrate binding pocket is capped by the residues Leu-21 and Ser-25, which are contributed by the opposing monomer of the OMT1 homodimer. The blue-colored envelopes surround regions greater than +2.5σ in the omit Fobs-Fcalc electron density map. The atomic model and electron density are shown for the active site of monomer A (primarily, with small contributions from monomer B) of the crystallographic asymmetric unit; the active sites of the other three monomers are essentially indistinguishable.
(B) Surface of the binding pocket for phenolic substrate in the closed conformational state of OMT1. The depicted surface defines the area accessible to a probe sphere with a radius of 1.4 Å. The side of the surface that faces the phenolic substrate binding site is colored according to the identity of the bounding protein atom (carbon, green; nitrogen, blue; oxygen, red; and sulfur, gold); the reverse side of the surface is distinguished by gray coloring. For clarity, the front portion of the surface of the binding pocket has been removed. The positions of bound sinapaldehyde (orange) and two water molecules (red spheres) involved in interactions with the 3-methoxy and 4-hydroxyl groups are shown. The locations of cavities for the 3-methoxy and C9 substituents and the catalytic site are indicated.
(C) Binding of coniferaldehyde by OMT1. OMT1 active-site residues, SAH, and coniferaldehyde are displayed as in (A). The blue-colored envelopes surround regions greater than +1.0 σ in the initial 2Fobs-Fcalc electron density map (calculated prior to inclusion of coniferaldehyde in the atomic model). The yellow-colored envelopes surround regions greater than +2.5σ in the final Fobs-Fcalc electron density map. The difference density lobe indicates a significant fraction of coniferaldehyde molecules are bound with an alternative orientation of the phenyl ring, which transplaces the 3-methoxy group into the OMT1 3-methoxy cavity (as opposed to the catalytic site).
In the closed conformational state of OMT1, these SAM/SAH interactions with the SAM binding domain are retained entirely, but the closer approach of SAM/SAH to the phenolic substrate binding region introduces a number of additional interactions. In particular, the phenyl ring of Phe-160 (which itself adopts a significantly more buried conformation in the closed state of OMT1) together with the side chains of Phe-173 and Met-177 contribute to the binding of the ribose ring and aliphatic portion of the Met/Hcy moiety; and the Ser-181 side chain forms an additional hydrogen bond with the Met/Hcy α-carboxylate group. Notably, all of the amino acid residues that form interactions with SAM/SAH are, in general, highly conserved in all plant type-1 OMTs.
With most OMTs, the SAH product acts as a potent competitive inhibitor of the methylation reaction (Hendricks et al., 2004). This property suggests that SAH and SAM form largely indistinguishable interactions with the enzyme. Indeed, this feature of SAH/SAM binding is particularly evident for the open conformational state of Lp OMT1, in which the additional methyl group of the SAM substrate makes no contacts with the enzyme.
Recognition of Phenolic Substrates/Products by Lp OMT1
The binding mode of phenolic ligands is unequivocally defined in the ternary complex of OMT1/SAH with the sinapaldehyde product, a particularly tightly bound compound in our crystallographic experiments (Figures 5A and 5B). As outlined earlier, the binding pocket is located primarily within the α-helical sandwich layer, with helical segments from the dimerization region lining its internal wall. The SAM/SAH molecule and residues 262 to 268 of the SAM binding domain block the entrance to the pocket in the closed conformation of OMT1. This polypeptide chain segment contributes a number of key residues involved in catalysis and in binding of both the SAM and phenolic substrates.
The precise ligand binding interactions formed by OMT1 are of particular interest, as these not only are integral to the enzyme’s catalytic activity but also confer substrate specificity and regioselectivity for the methylation reaction. In the OMT1-SAH-sinapaldehyde complex, the aromatic and aliphatic core (the phenyl ring and propene side chain) of sinapaldehyde are enveloped by a cluster of nonpolar side chains that include Leu-124, Met-127, Met-177, Trp-263, Val-313, Ile-316, and Met-317. The aldehyde oxygen of the propene side chain is involved in a single hydrogen bond interaction with the amide nitrogen of the Asn-128 side chain. Wang and Pichersky (1999) previously demonstrated that in Clarkia breweri COMT, the residue corresponding to Asn-128, plays a critical role in the recognition of substrates bearing an oxygenated side chain (i.e., the monolignol intermediates). Ala replaces this Asn in OMTs specific for substrates bearing a nonoxygenated, propene side chain (phenylpropenes, such as isoeugenol). The sinapaldehyde 4-hydroxyl group accepts hydrogen bonds from the amide nitrogen of the Asn-321 side chain and a water molecule and is the donor in a hydrogen bond with the Asp-267 carboxylate. The 3-methoxy group occupies a predominantly nonpolar cavity formed by the side chains of Met-127, Leu-133, Phe-173, Ile-316, and His-320. The methoxy-ether oxygen participates in a hydrogen-bonding network with a water molecule that in turn interacts with the side chain of Asn-321, the sinapaldehyde 4-hydroxyl group, and the His-163 side chain imidazole via a bridging water molecule.
Notably, the binding by OMT1 of coniferaldehyde is effectively identical to that of sinapaldehyde (Figure 5C). Furthermore, the residues involved in forming ligand binding interactions, as detailed above for OMT1, are nearly invariant in all well-characterized COMTs, and consequently, substrate recognition is likely achieved identically in all of these COMTs (see Supplemental Figure 1 online). It is therefore intriguing that the disparate binding sites observed for ferulic acid and 5-hydroxyconiferaldehyde in alfalfa COMT (Zubieta et al., 2002) differ significantly from the well-defined site described here for sinapaldehyde in Lp OMT1. The catalytically nonproductive ligand complexes of alfalfa COMT may be representative of prebinding modes that are explored by the incoming substrate molecule when COMT is in the open conformational state and lacks a completely organized phenolic substrate binding pocket and highlight the importance of enzyme dynamics in COMT activity.
DISCUSSION
Open and Closed Conformations of the Lp OMT1 Molecular Architecture
The inward positioning of the SAM binding domains associated with the closed conformational state of OMT1 is intrinsic to the assembly of a fully functional binding pocket for the phenolic substrate. For example, steric contacts with the SAM binding domain induce several side chains (Phe-160, Trp-263, Met-317, and Asn-321) to adopt conformations appropriate for accommodating the phenolic substrate (Figure 4B). More importantly, the OMT1 catalytic residues (His-266, Asp-267, and Glu-326) together with the SAM substrate are brought in immediate proximity of the phenolic substrate. Indeed, the SAM molecule physically closes off the substrate binding pocket and thereby prevents escape of the phenolic substrate and sequesters the activated phenoxide intermediate from bulk solvent. Notably, this role of the SAM substrate in establishing part of the binding pocket for the phenolic substrate is consistent with the order of substrate binding by the OMTs, as binding of the methyl donor SAM precedes that of the phenolic substrate (Huang et al., 2004).
Our structural analyses of Lp OMT1 provide a unique atomic resolution view of distinct conformational states of a single plant type-1 OMT, although considerable variability in the positioning of the SAM binding domain has been previously recognized in these enzymes. For example, in comparison to the structure of (closed-state) isoflavone OMT from alfalfa (Zubieta et al., 2001), the more open conformation of barrel medic isoflavonoid OMT (Liu et al., 2006) was apparent, as evidenced by an outward rotation of the SAM binding domains, a wider active-site cleft, and an inordinately long distance between SAM and the methyl acceptor of the hydroxylated substrate. Similarly, the previously reported structures of ternary complexes of alfalfa COMT (Zubieta et al., 2002) possess conformational states that are intermediate between the open and closed conformations observed for Lp OMT1. Flexibility of the SAM binding domain is evidently an inherent property of the class-I OMTs. This flexibility underlies a critical mechanistic function of these enzymes: the adoption of an initial open state that facilitates binding of SAM first then the hydroxylated substrate and subsequently a closed state in which the substrates and active-site groups are sequestered and appropriately aligned for the catalytic reaction to proceed.
Insight into the Substrate Preferences of COMTs
Collectively, the intricate array of binding interactions between Lp OMT1 and a phenolic ligand (Figure 5A) undoubtedly functions in an enzyme substrate complex to anchor the substrate molecule such that the reactive oxygen atom (the 5-hydroxyl in the case of 5-hydroxyconiferaldehyde, the precursor of sinapaldehyde) forms productive interactions with the enzyme’s catalytic groups and the SAM methyl donor (as discussed further below). As a consequence, COMT enzymes are exclusively selective for O-methylation of a meta (3- or 5-position) hydroxyl group on the substrate phenyl group. In marked contrast, the COMTs never modify the para- or 4-hydroxyl group, which is engaged in hydrogen bonding interactions with a conserved Asn side chain and a water molecule. In isoeugenol and chavicol methyltransferases, which specifically target the 4-hydroxyl group of their phenylpropene substrates, these hydrogen bonding interactions are disrupted by the substitution of the Asn with a nonpolar residue or by the introduction of an additional hydrogen bonding residue (e.g., Glu-165 in C. breweri isoeugenol methyltransferase) at a neighboring site (Wang and Pichersky, 1999; Gang et al., 2002).
Remarkably, the strict regioselective activity of the COMTs is attained despite a moderate degree of promiscuity in substrate acceptance. Our detailed characterization of the binding interactions of the COMTs illuminates the structural basis for this promiscuity. In particular, although the invariant scaffold (the phenyl ring, propene side chain, and 4-hydroxy group) of COMT-accepted substrates forms close interactions with the enzyme, the parts of this scaffold that potentially bear alternative substituents (the 3- and 5-positions of the phenyl ring and the oxygen-bearing C9 of the propene side chain) are situated adjacent to modestly sized cavities within the enzyme’s binding pocket (Figure 5B). These two key features underlie the promiscuity in substrate binding by COMT, which can thereby accommodate an array of distinct but related substrates via a common binding position of the 4-hydroxy phenylpropene scaffold. The interactions formed with the alternative substituents then define the subtleties of the substrate preferences of the COMTs. Thus, in comparison to 5-hydroxyconiferaldehyde, caffeoyl aldehyde would be expected to serve as a less favorable substrate because the COMT cavity that accommodates the 3-methoxy group is occupied by only a hydrogen atom, resulting in the loss of both the nonpolar interactions formed by a methoxy group and the hydrogen bonding network nucleated by a water molecule that interacts with the 3-methoxy and 4-hydroxy oxygens. The proclivity for full occupation of COMT’s 3-methoxy pocket is evidenced by the two distinct orientations observed for the phenyl ring of the coniferaldehyde product in complex with Lp OMT1-SAH (Figure 5C): the first has the lone methoxy group of coniferaldehyde residing as expected in the catalytic site, whereas in the second, this methoxy group instead occupies the opposing 3-methoxy pocket.
The structural basis for aldehyde selectivity (over the less-preferred alcohol and the least-preferred acid forms) is also apparent. Only a single active-site polar group, the carboxamide ND2 group of an Asn (Asn-128 in Lp OMT1), is suitably positioned to interact with the substrate’s C9 oxygenated functionality (Figure 5A). Asn128ND2 serves as a hydrogen bond donor to the C9 aldehyde oxygen in the Lp OMT1-SAH-sinapaldehyde complex. A substrate bearing a C9 alcohol would be less favored than the aldehyde due to the lack of a hydrogen bond acceptor for the hydrogen atom of the C9 hydroxyl group. In addition, although a C9 carboxylate group can be readily accommodated sterically, it would be least favored because of the unavailability of both a hydrogen-bonding partner for the second carboxylate oxygen and a cationic protein group to neutralize the carboxylate’s negative charge.
Our structural characterization of the substrate preferences of Lp OMT1 is consistent with results from enzyme kinetics analyses, as evidenced by the higher O-methylation activity for the 5-hydroxyl over the 3-hydroxyl substituent and for alcohol over acid substrates. Furthermore, because the Lp OMT1 residues that form the phenolic-ligand binding site (as revealed in this work) are absolutely conserved in almost all confirmed COMTs, the substrate preference patterns of all COMTs are expected to be very similar. Therefore, although a full kinetic analysis of Lp OMT1 activity with the aldehyde substrates was not possible, Lp OMT1 very likely resembles other COMTs in that the aldehydes represent the kinetically preferred substrates (Li et al., 2000; Parvathi et al., 2001; Zubieta et al., 2002; Chen et al., 2004; Ma and Xu, 2008). Some support for the high affinity of Lp OMT1 for the phenolic aldehydes is provided by the observation that crystallization of Lp OMT1-SAH–product ternary complexes was effected most readily with the two aldehyde products (coniferaldehyde and sinapaldehyde).
A number of unusual properties pertaining to substrate binding by the COMT enzymes can now be rationalized. For example, the COMT from loblolly pine (Li et al., 1997), the most divergent in sequence of the confirmed COMTs, is active against not only the normal monolignol precursors but also the CoA-thioesters of the acid forms (e.g., caffeoyl CoA). These monolignol CoA-thioesters are more typically methylated by a distinct class of O-methyltransferases, the caffeoyl-CoA OMTs, which possess a more open and expansive ligand binding cleft that serves to accommodate the extended CoA tail of the substrate (Ferrer et al., 2005). Loblolly pine COMT may have the capacity to bind the bulkier esterified substrates due to amino acid substitutions and two small insertions that substantially enlarge the volume of its C9 pocket. Another outstanding question is how certain substrates (the aldehydes in particular) inhibit the activity of Lp OMT1 as well as several other COMTs (Li et al., 2000). One possible mechanism, the entrapment of substrate in exploratory, prebinding modes (as discussed above), seems implausible due to the dependence of inhibition on the identity of the C9 substituent and the absence of specific interactions to this substituent prior to full assembly of the substrate binding site. A more likely mechanism is the adoption by the substrate of a nonproductive binding mode in which the target hydroxyl group occupies the 3-methoxy binding pocket instead of the catalytic site (Figure 5B). Such a flipped binding mode, which closely resembles the catalytically competent enzyme substrate complex, might be expected to be most prevalent for substrates with the greatest affinity overall. The effects of product inhibition, through which regeneration of the active site is impeded by particularly tightly bound product, must also be considered.
In summary, our biochemical and structural studies support a primary role of Lp OMT1 in the 5-O-methylation of 5-hydroxyconiferaldehyde to produce sinapaldehyde, the direct precursor of the monolignol building block (sinapyl alcohol) that is incorporated into syringyl lignin. This result is consistent with functional studies in perennial ryegrass plants, which demonstrate that (1) maximal OMT1 expression coincides with the elevated accumulation of S-lignin in stem tissue (during the early elongation stage of plant development), and (2) transgenic plants with downregulated Lp OMT1 expression have dramatically reduced levels of S-lignin subunits (Tu et al., 2010). Furthermore, similar in planta results have been reported from analogous studies of the COMT enzymes in several other plant species. The preferred use by Lp OMT1 (and most other plant COMTs) of the aldehyde (and to a lesser extent alcohol) forms of the monolignol precursors is consistent with the currently accepted view of the phenylpropanoid biosynthetic pathway in plants (Li et al., 2008).
Additionally, despite the marked preference for 5-O-methylation, Lp OMT1 nevertheless is significantly active in 3-O-methylation, as evidenced here by the conversion in vitro of caffeoyl aldehyde to coniferaldehyde and caffeoyl alcohol to coniferyl alcohol (and also caffeic acid to ferulic acid). This secondary 3-O-methylation activity likely accounts for the slight decreases in levels of G-lignin in OMT1-deficient transgenic ryegrass (Tu et al., 2010). Again, this observed activity of Lp OMT1 is in agreement with the properties of COMTs from other plant species (Maury et al., 1999; Dixon et al., 2001; Parvathi et al., 2001; Do et al., 2007; Ma and Xu, 2008; Guillaumie et al., 2008): in Arabidopsis thaliana, the retained ability to synthesize both G- and S-lignin in CCoAOMT knockouts but not CCoAOMT/COMT double knockouts (Do et al., 2007); and radiochemical labeling studies that demonstrate the incorporation of exogenously supplied caffeoyl alcohol and caffeoyl aldehyde ultimately into both G- and S-subunits of angiosperm lignin (Matsui et al., 2000). Finally, the permissiveness in substrate acceptance by the COMTs intriguingly suggests that developmental adjustments in the in vivo levels of monolignol precursors may be sufficient to modulate the biosynthesis and availability of specific monolignols that are then incorporated through polymerization into lignin.
Inferences on the Catalytic Reaction Mechanism of Lp OMT1
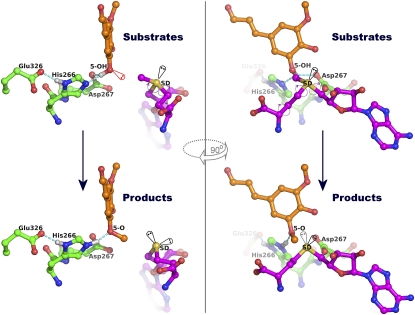
The Lp OMT1-SAH-sinapaldehyde complex is especially illustrative of the mechanistic details of a transmethylation reaction because this complex closely mimics a trapped, postreaction enzyme product state (Figure 6). In particular, the 5-O-methyl group of the sinapaldehyde product remains in van der Waals contact (3.2 Å) with the SAH SD atom (the methyl group carrier); also, the 5-methoxy oxygen atom retains close interactions (2.8 Å) with the side chains of two catalytic residues, His-266 and Asp-267. More significantly, from the structure of this enzyme product state, the arrangement of reactant groups in the prereaction, enzyme substrate complex can be readily inferred (Figure 6). Thus, the labile methyl group resides near the pro-S lone pair of the SAH SD atom, consistent with the required chirality (S-configuration) of the sulfonium moiety of the SAM substrate. In the enzyme substrate complex, the 5-hydroxyl group of the 5-hydroxyconiferaldehyde substrate putatively serves as the donor in a bifurcated hydrogen bond with the His-266 NE2 and Asp-267 OD1 atoms.
Figure 6.
Detailed View of the Lp OMT1–Catalyzed Transmethylation Reaction.
The OMT1 catalytic residues are shown with the inferred positions of the two substrates, SAM and 5-hydroxyconiferaldehyde (top panels), or with the crystallographically observed positions of the SAH and sinapaldehyde products (bottom panels). Atom coloring is as described in Figure 5A. Predicted hydrogen bonds are drawn as cyan dashed lines, with the predicted positions of hydrogen atoms indicated by gray spheres. The conformational change in the Met/Hcy methylene groups of SAM/SAH arises from a rotation about the CG-SD bond, as indicated by the purple curved arrow. Two orthogonal views are shown, approximately perpendicular (left panels) and parallel (right panels) to the vector joining the reactive hydroxyl and methyl groups.
The His-266 imidazole moiety is widely considered to be the catalytic base that deprotonates and thereby activates the nucleophilic hydroxyl group of the substrate. This activation may also involve the Asp-267 carboxylate, either directly or more indirectly by ensuring the appropriate orientation of the hydroxyl proton for abstraction by His-266. Glu-326, which provides the hydrogen bond acceptor in an interaction with the His-266 ND1 nitrogen, functions to establish the His-266 imidazole ring conformation and to promote the basicity of the His-266 NE2 atom for catalytic deprotonation of the phenolic substrate. The geometry of the catalytic and reactant groups appears to be particularly favorable for SN2-nucleophilic attack by the activated 5-hydroxyl group on the methyl group of SAM. Specifically, proton abstraction by His-266 (and Asp-267) occurs roughly in line with the attack on the target methyl group by a lone electron pair from the developing phenoxide; in turn, this nucleophilic attack occurs in line, as expected, with the departure of the sulfur leaving group of the SAH product (Figure 6).
The cleavage of the sulfonium/methyl group bond appears to induce small adjustments in the positions of the Met/Hcy methylene groups (CB and CG) of SAM/SAH (Figure 6). The prereaction conformation of SAM (as observed in the open form of Lp OMT1) projects the labile methyl group directly toward the acceptor hydroxyl group of the substrate. By contrast, the postreaction conformation (observed in the closed, product-bound form of Lp OMT1) effects a change in the orientation of the leaving-group sulfur, thereby rotating the liberated lone-pair electrons away from the methyl group that has been transferred to the product. The accompanying crankshaft movement of the Hcy methylene groups likely arises as a consequence of the anchored positions of the α-amino and carboxylate groups of SAM/SAH and is accommodated within a region of the OMT1 active site immediately adjacent to the highly conserved Gly-Gly-Gly polypeptide segment (see Supplemental Figure 1 online).
An in-depth understanding of the structural framework for the substrate binding and transmethylation activities of Lp OMT1 provides a basis for the rational modification of the enzyme’s regiospecificity and substrate selectivity. For example, amino acid substitutions aimed at occluding the 3-methoxy pocket should prevent the binding of substrates bearing a 3-methoxy group (such as 5-hydroxyconiferyl aldehyde or alcohol) and thus restrict Lp OMT1 activity to the 3-O-methylation of the less bulky substrates (caffeoyl aldehyde or alcohol). Similarly, the introduction of additional hydrogen-bonding groups into the C9 pocket may enhance the binding affinity for phenolic alcohol and acid substrates. Combined substitutions to obstruct the 3-methoxy pocket and disrupt the normal hydrogen-bonding interactions of the 4-hydroxyl group of a phenolic substrate represent a potential strategy for steering the substrate’s 4-hydroxyl group into the catalytic site and thereby create a new O-methyltransferase enzyme that is specific for 4-O-methoxylation (Bhuiya and Liu, 2010). These engineered COMT variants may have applications in the generation of transgenic plants with modified lignins and also in chemo-enzymatic syntheses of novel phenolic compounds.
METHODS
Protein Expression and Purification
The coding sequence of Lolium perenne OMT1 was inserted between the NcoI and BamHI sites of the expression vector pHIS8 (Jez et al., 2000), which under the control of a T7 promoter, yields the target protein fused to an N-terminal octahistidine tag. The Lp OMT1 protein was heterologously overexpressed in the Escherichia coli BL21 (DE3) (Novagen) expression host. E. coli cultures were grown at 37°C in TB medium (Invitrogen) to an optical density (600 nm) of 1.5, induced with 1 mM isopropyl-β-d-thiogalactoside, and allowed to grow for an additional 6 h at 22°C. Bacterial cells were harvested by centrifugation, resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 20 mM imidazole, 1% [v/v] Tween 20, 10% [v/v] glycerol, and 20 mM 2-mercaptoethanol), and lysed by sonication. The Lp OMT1 protein was isolated from the E. coli lysate by affinity chromatography with nickel nitrilotriacetic acid–coupled agarose (Qiagen) and eluted with lysis buffer containing 0.25 M imidazole. The partially purified Lp OMT1 protein was treated with thrombin for cleavage of the octahistidine tag and then further purified by gel exclusion chromatography with a Superdex 200 HR26/60 column (Pharmacia Biosystems).
Lp OMT1 Enzyme Assays
OMT1 O-methylation activity was monitored by LC-MS with an Agilent 1100 series LC-MSD instrument and an Agilent Zorbax Eclipse XDB-C18 (4.6 × 150 mm, 5-mm particle size) reversed-phase column. Chromatographic separations employed a flow rate of 0.5 mL/min and a linear gradient with initial and final mobile phases consisting of 95% (v/v) water, 5% (v/v) acetonitrile, 0.1% (v/v) formic acid, and 5% (v/v) water, 95% (v/v) acetonitrile, and 0.1% (v/v) formic acid, respectively. Monolignol-precursor substrates were obtained commercially (caffeic acid from Indofine Chemical or Sigma-Aldrich) or from R. Dixon (5-hydroxyferulic acid, caffeoyl aldehyde, 5-hydroxyconiferaldehyde, and caffeoyl alcohol). The identities of O-methylation products were confirmed by mass determination and comparison with chromatographic elution times of authentic standards. Quantification was based on absorbance at an appropriate wavelength between 280 and 340 nm.
For enzyme kinetics analyses, the assay mixture (total volume 0.3 mL) contained 0.05 M HEPES, pH 7.5, 10 mM NaCl, and 0.75 mM SAM (Sigma-Aldrich), and eight different initial concentrations of substrate (typically in the range 1 to 150 μM) were used for each fixed quantity of Lp OMT1 (0.9 to 1.5 μg protein). After preincubation of the enzyme with SAM and buffer at 22°C for 10 min, the reaction was initiated by the addition of substrate. Reaction progress was monitored over the course of 25 min by transfer (at timed intervals) of aliquots of the reaction mixture to quench solution (formic acid, 0.4% [v/v] final concentration). The quenched reaction mixtures were prepared for LC-MS analysis by addition of two volumes of methanol, incubation at −20°C for 1 h, and centrifugation to remove particulates. Each substrate enzyme series was assayed in duplicate, and kinetic constants were estimated using nonlinear regression analyses (Prism; GraphPad Software).
Crystallization of Lp OMT1
Crystals of OMT1 were grown by vapor diffusion at 4°C, from 1:1 mixtures of protein solution (10 to 15 mg/mL in 12.5 mM Tris-HCl, pH 7.5, and 50 mM NaCl) and reservoir solution. For cocrystallization experiments, the protein solutions included 2.5 mM SAH (Sigma-Aldrich), SAM (Sigma-Aldrich), or 2.5 mM SAH plus 5 mM sinapaldehyde or coniferaldehyde. The reservoir solution used for both the apoenzyme and holoenzyme forms of OMT1 contained 0.1 M Tris-HCl, pH 8.5, 21% (w/v) polyethylene glycol 8000, 0.2 M calcium acetate, and 2 mM DTT. The reservoir solution for ternary complexes of OMT1 contained 0.1 M sodium succinate, pH 5.5, 28% (w/v) polyethylene glycol monomethyl ether 5000, and 2 mM DTT. Crystal growth typically occurred over a period of 2 to 10 d (to a maximal size of ~0.2 × 0.2 × 0.4 mm) and was sometimes expedited through seeding with finely crushed microcrystals.
X-Ray Diffraction Data
Crystal samples were flash frozen by immersion in liquid nitrogen after a brief incubation in a cryoprotectant solution (consisting of reservoir solution supplemented with 17 to 20% [v/v] ethylene glycol, and where appropriate, SAH and a phenolic aldehyde). X-ray diffraction data were measured from frozen crystals at beamlines 8.2.1 and 8.2.2 of the Advanced Light Source (Lawrence Berkeley National Laboratory) on ADSC Quantum 315 CCD detectors. Diffraction intensities were indexed, integrated, and scaled with the MOSFLM (Leslie, 1992) and SCALA (Evans, 2006) programs.
X-Ray Structure Determination of Lp OMT1
Initial crystallographic phases for the holoenzyme crystal form of Lp OMT1 (complexed with SAH) were determined through molecular replacement (MR) analysis with the Molrep program (Vagin and Teplyakov, 1997). The starting search model for MR was a homology model for a monomer of the Lp OMT1 protein, based on the structure of Medicago sativa COMT (PDB entry 1KYZ) and constructed with the Modeler program (Sali and Blundell, 1993). A refined model of the Lp OMT1 holoenzyme monomer served as a search model for MR solutions for the Lp OMT1 apoenzyme and sinapaldehyde ternary complex structures. The ARP/wARP program (Perrakis et al., 1999) was used for automated rebuilding of initial structural models, using a noncrystallographic symmetry constrained averaged map in the case of the ternary complex. Subsequent structural refinements utilized the CNS (Brunger et al., 1998) and Phenix (Adams et al., 2010) programs. Xfit (McRee, 1999) and Coot (Emsley and Cowtan, 2004) were used for graphical map inspection and manual rebuilding of atomic models. Programs from the CCP4 suite (Collaborative Computational Project Number 4, 1994) were employed for all other crystallographic calculations. Molecular graphics were produced with the program PyMol (the PyMOL molecular graphics system, version 1.3; Schrödinger).
Structural Comparisons of Lp OMT1 and Other OMTs
For structural comparisons of Lp OMT1 with other OMTs, each of the distinct domains of a monomer of the structurally related OMTs was superimposed independently on the corresponding domain of the Lp OMT1 monomer. The SSM program (Krissinel and Henrick, 2004) was used to calculate these superpositions. For structural alignment of the various forms of Lp OMT1, superpositions were based solely on the dimerization domain and were calculated with the CCP4 program LSQKAB (Kabsch, 1976).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: L. perenne Lp OMT1, AAD10253.1; M. sativa Ms COMT, AAB46623.1; Triticum aestivum Ta COMT1, ABP63535.1; Populus tremuloides Pt COMT, AAB61731.1; Festuca arundinaceae Fa COMT, AAK68907.1; Pinus taeda Pt COMT, AAC49708.1; Medicago truncatula (barrel medic) Mt OMT, AAY18581.1; M. sativa Ms Isoflavone, OMT AAC49928.1; M. sativa Ms Chalcone, OMT P93324.1; Clarkia breweri Cb COMT, O23760.1; and C. breweri Cb isoeugenol OMT, O04385.1. Atomic coordinates and x-ray structure factors for the Lp OMT1 structures have been deposited in the Protein Data Bank (http://www.rcsb.org) with the accession codes listed in Table 1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment of Representative Plant COMTs.
Supplemental Data Set 1. Atomic Coordinates for Apo Lp OMT1.
Supplemental Data Set 2. Experimentally Measured Structure-Factor Amplitudes for Apo Lp OMT1.
Acknowledgments
We thank Luciano Martelotto for preparing the E. coli expression vector. This work was supported by National Science Foundation Grants MCB-0645794, MCB-1033033, and MCB-0718064 to J.P.N. Additionally, this material is also based upon work supported by the National Science Foundation under Award EEC-0813570. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. J.P.N. is an investigator with the Howard Hughes Medical Institute. Portions of this research were conducted at the Advanced Light Source, a national user facility operated by Lawrence Berkeley National Laboratory, on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The Berkeley Center for Structural Biology is supported in part by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences. We thank the staff at the Advanced Light Source for assistance with x-ray data collection.
References
- Adams P.D., et al. (2010). PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassova R., Favet N., Martz F., Chabbert B., Tollier M.-T., Monties B., Fritig B., Legrand M. (1995). Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J. 8: 465–477 [Google Scholar]
- Bhuiya M.W., Liu C.J. (2010). Engineering monolignol 4-O-methyltransferases to modulate lignin biosynthesis. J. Biol. Chem. 285: 277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Brünger A.T., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chabbert B., Monties B., Barriere Y., Argillier O. (1994). Biological variability in lignification of maize: expression of the brown midrib bm2 mutation. J. Sci. Food Agric. 64: 455–460 [Google Scholar]
- Chen L., Auh C., Chen F., Cheng X., Aljoe H., Dixon R.A., Wang Z. (2002). Lignin deposition and associated changes in anatomy, enzyme activity, gene expression, and ruminal degradability in stems of tall fescue at different developmental stages. J. Agric. Food Chem. 50: 5558–5565 [DOI] [PubMed] [Google Scholar]
- Chen L., Auh C., Dowling P., Bell J., Lehmann D., Wang Z. (2004). Transgenic down-regulation of caffeic acid O-methyltransferase (COMT) led to improved digestibility in tall fescue (Festuca arundinacea). Funct. Plant Biol. 31: 235–245 [DOI] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. (2010). MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994). The CCP4 suite: Programs for protein crystallography. Acta Crystallog. D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Dixon R.A., Chen F., Guo D., Parvathi K. (2001). The biosynthesis of monolignols: A “metabolic grid”, or independent pathways to guaiacyl and syringyl units? Phytochemistry 57: 1069–1084 [DOI] [PubMed] [Google Scholar]
- Do C.-T., Pollet B., Thévenin J., Sibout R., Denoue D., Barrière Y., Lapierre C., Jouanin L. (2007). Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226: 1117–1129 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Evans P.R. (2006). Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62: 72–82 [DOI] [PubMed] [Google Scholar]
- Ferrer J.-L., Zubieta C., Dixon R.A., Noel J.P. (2005). Crystal structures of alfalfa caffeoyl coenzyme A 3-O-methyltransferase. Plant Physiol. 137: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang D.R., Lavid N., Zubieta C., Chen F., Beuerle T., Lewinsohn E., Noel J.P., Pichersky E. (2002). Characterization of phenylpropene O-methyltransferases from sweet basil: Facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell 14: 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grand C., Parmentier P., Boudet A., Boudet A.M. (1985). Comparison of lignins and of enzymes involved in lignification in normal and brown midrib (bm3) mutant maize seedlings. Physiol. Veg. 23: 905–911 [Google Scholar]
- Guillaumie S., Goffner D., Barbier O., Martinant J.P., Pichon M., Barrière Y. (2008). Expression of cell wall related genes in basal and ear internodes of silking brown-midrib-3, caffeic acid O-methyltransferase (COMT) down-regulated, and normal maize plants. BMC Plant Biol. 8: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D.J., Chen F., Inoue K., Blount J.W., Dixon R.A. (2001). Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa. impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13: 73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack K.E., Jones J.D. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8: 1773–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks C.L., Ross J.R., Pichersky E., Noel J.P., Zhou Z.S. (2004). An enzyme-coupled colorimetric assay for S-adenosylmethionine-dependent methyltransferases. Anal. Biochem. 326: 100–105 [DOI] [PubMed] [Google Scholar]
- Huang T.S., Anzellotti D., Dedaldechamp F., Ibrahim R.K. (2004). Partial purification, kinetic analysis, and amino acid sequence information of a flavonol 3-O-methyltransferase from Serratula tinctoria. Plant Physiol. 134: 1366–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jez J.M., Ferrer J.L., Bowman M.E., Dixon R.A., Noel J.P. (2000). Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry 39: 890–902 [DOI] [PubMed] [Google Scholar]
- Jouanin L., Goujon T., de Nadaï V., Martin M.T., Mila I., Vallet C., Pollet B., Yoshinaga A., Chabbert B., Petit-Conil M., Lapierre C. (2000). Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol. 123: 1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. (1976). A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32: 922–923 [Google Scholar]
- Krissinel E., Henrick K. (2004). Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Kubitzki K.J. (1987). Phenylpropanoid metabolism in relation to land plant origin and diversification. Plant Physiol. 131: 17–24 [Google Scholar]
- Leslie A.G.W. (1992). Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26 [Google Scholar]
- Li L., Popko J.L., Umezawa T., Chiang V.L. (2000). 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J. Biol. Chem. 275: 6537–6545 [DOI] [PubMed] [Google Scholar]
- Li L., Popko J.L., Zhang X.H., Osakabe K., Tsai C.J., Joshi C.P., Chiang V.L. (1997). A novel multifunctional O-methyltransferase implicated in a dual methylation pathway associated with lignin biosynthesis in loblolly pine. Proc. Natl. Acad. Sci. USA 94: 5461–5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Weng J.-K., Chapple C. (2008). Improvement of biomass through lignin modification. Plant J. 54: 569–581 [DOI] [PubMed] [Google Scholar]
- Liu C.J., Deavours B.E., Richard S.B., Ferrer J.L., Blount J.W., Huhman D., Dixon R.A., Noel J.P. (2006). Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses. Plant Cell 18: 3656–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q.H., Xu Y. (2008). Characterization of a caffeic acid 3-O-methyltransferase from wheat and its function in lignin biosynthesis. Biochimie 90: 515–524 [DOI] [PubMed] [Google Scholar]
- Martin J.L., McMillan F.M. (2002). SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12: 783–793 [DOI] [PubMed] [Google Scholar]
- Matsui N., Chen F., Yasuda S., Fukushima K. (2000). Conversion of guaiacyl to syringyl moieties on the cinnamyl alcohol pathway during the biosynthesis of lignin in angiosperms. Planta 210: 831–835 [DOI] [PubMed] [Google Scholar]
- Maury S., Geoffroy P., Legrand M. (1999). Tobacco O-methyltransferases involved in phenylpropanoid metabolism. The different caffeoyl-coenzyme A/5-hydroxyferuloyl-coenzyme A 3/5-O-methyltransferase and caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase classes have distinct substrate specificities and expression patterns. Plant Physiol. 121: 215–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee D.E. (1999). XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125: 156–165 [DOI] [PubMed] [Google Scholar]
- Noel J.P., Dixon R.A., Pickersky E., Zubieta C., Ferrer J.-L. (2003). Structural, functional and evolutionary basis for methylation of plant small molecules. Recent Adv. Phytochem. 37: 37–58 [Google Scholar]
- Parvathi K., Chen F., Guo D.J., Blount J.W., Dixon R.A. (2001). Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J. 25: 193–202 [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris R., Lamzin V.S. (1999). Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Ralph J., Lapierre C., Lu F., Marita J.M., Pilate G., Van Doorsselaere J., Boerjan W., Jouanin L. (2001). NMR evidence for benzodioxane structures resulting from incorporation of 5-hydroxyconiferyl alcohol into lignins of O-methyltransferase-deficient poplars. J. Agric. Food Chem. 49: 86–91 [DOI] [PubMed] [Google Scholar]
- Ralph J., MacKay J.J., Hatfield R.D., O’Malley D.M., Whetten R.W., Sederoff R.R. (1997). Abnormal lignin in a loblolly pine mutant. Science 277: 235–239 [DOI] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Schubert H.L., Blumenthal R.M., Cheng X. (2003). Many paths to methyltransfer: A chronicle of convergence. Trends Biochem. Sci. 28: 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y., Rochfort S., Liu Z., Ran Y., Griffith M., Badenhorst P., Louie G.V., Bowman M.E., Smith K.F, Noel J.P., Mouradov A., Spangenberg G. (2010). Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 22: 3357–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A., Teplyakov A. (1997). MOLREP: An automated program for molecular replacement. J. Appl. Cryst. 30: 1022–1025 [Google Scholar]
- Vignols F., Rigau J., Torres M.A., Capellades M., Puigdomènech P. (1995). The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Pichersky E. (1999). Identification of specific residues involved in substrate discrimination in two plant O-methyltransferases. Arch. Biochem. Biophys. 368: 172–180 [DOI] [PubMed] [Google Scholar]
- Whetten R., Sederoff R. (1995). Lignin biosynthesis. Plant Cell 7: 1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R.Q., Iii W.H., Negrel J., Ye Z.H. (1998). Dual methylation pathways in lignin biosynthesis. Plant Cell 10: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R.Q., Taylor J.J., Ye Z.H. (1997). Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9: 2159–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta C., He X.-Z., Dixon R.A., Noel J.P. (2001). Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Biol. 8: 271–279 [DOI] [PubMed] [Google Scholar]
- Zubieta C., Kota P., Ferrer J.L., Dixon R.A., Noel J.P. (2002). Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]