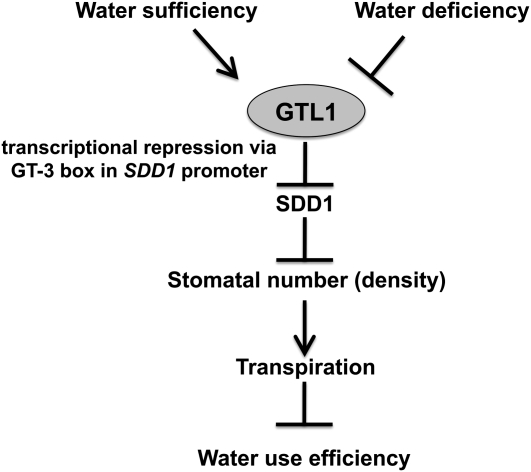
This work provides evidence that Arabidopsis GTL1 functions as a focal regulator of water use efficiency and water stress tolerance. The results establish a potential paradigm for how the environment influences stomatal development to reduce transpiration under low water availability conditions.
Abstract
A goal of modern agriculture is to improve plant drought tolerance and production per amount of water used, referred to as water use efficiency (WUE). Although stomatal density has been linked to WUE, the causal molecular mechanisms have yet to be determined. Arabidopsis thaliana GT-2 LIKE 1 (GTL1) loss-of-function mutations result in increased water deficit tolerance and higher integrated WUE by reducing daytime transpiration without a demonstrable reduction in biomass accumulation. gtl1 plants had higher instantaneous WUE that was attributable to ~25% lower transpiration and stomatal conductance but equivalent CO2 assimilation. Lower transpiration was associated with higher STOMATAL DENSITY AND DISTRIBUTION1 (SDD1) expression and an ~25% reduction in abaxial stomatal density. GTL1 expression occurred in abaxial epidermal cells where the protein was localized to the nucleus, and its expression was downregulated by water stress. Chromatin immunoprecipitation analysis indicated that GTL1 interacts with a region of the SDD1 promoter that contains a GT3 box. An electrophoretic mobility shift assay was used to determine that the GT3 box is necessary for the interaction between GTL1 and the SDD1 promoter. These results establish that GTL1 negatively regulates WUE by modulating stomatal density via transrepression of SDD1.
INTRODUCTION
Drought causes water deficit that limits plant growth and survival because root water uptake from the soil is insufficient to meet the transpirational requirements of the plant (Blum, 1996). Water deficit reduces leaf cell turgor, restricting cell expansion, canopy area development, and photosynthetic source size, thus negatively affecting biomass accumulation and yield (Chaves et al., 2003). An effective plant drought acclimation or adaptation strategy is used to reduce transpirational water loss, which conserves soil moisture and allows plants to maintain an adequate water status to sustain critical physiological and biochemical processes (Nobel, 1999; Chaves et al., 2003). However, a reduction in transpirational water loss often leads to a decline in biomass accumulation because carbon assimilation is also reduced (Sinclair et al., 1984; Udayakumar et al., 1998).
Transpiration and CO2 uptake occur primarily through stomata, the pores bordered by a pair of guard cells (Hetherington and Woodward, 2003). Conductance through these pores regulates transpirational flux and water use (Bacon, 2004; Chaerle et al., 2005; Morison et al., 2008; Huang et al., 2009; Song and Matsuoka, 2009) and is modulated by stomatal movements (opening and closing) and/or density (Hetherington and Woodward, 2003; Yoo et al., 2009; Casson and Hetherington, 2010; Kim et al., 2010). Alteration of stomatal aperture in response to the environment is a well-understood process that has been linked functionally to drought tolerance and water use efficiency (WUE; Chaerle et al., 2005; Nilson and Assmann, 2007; Kim et al., 2010). Likewise, there is substantial understanding of stomatal development determinants (Bergmann and Sack, 2007; Casson and Hetherington, 2010), and it is known that stomatal density is regulated by environmental factors such light, CO2, temperature, humidity, and drought (Lake et al., 2001; Bergmann, 2004; Casson and Gray, 2008; Casson and Hetherington, 2010). However, it is unclear how these environment factors regulate the developmental determinants and what the consequences of altered stomatal density are on drought tolerance and WUE.
Stomata develop predominantly in the leaf epidermis but do exist in other organs (Bergmann and Sack, 2007; Dong and Bergmann, 2010). Asymmetric division of a protodermal meristemoid mother cell forms a meristemoid and a larger stomatal lineage ground cell, which differentiates into an epidermal pavement cell or another meristemoid. The triangular-shaped meristemoid differentiates into a guard mother cell (GMC) that undergoes a symmetric division, forming a pair of guard cells. Then, morphogenesis of the stoma occurs (Bergmann and Sack, 2007; Dong and Bergmann, 2010). This basal pathway of stomatal lineage is regulated by well-characterized genetic determinants. BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE and SPEECHLESS (SPCH) are necessary for asymmetric division of a meristemoid mother cell to produce a meristemoid (MacAlister et al., 2007; Dong et al., 2009). MUTE facilitates meristemoid differentiation to a GMC (MacAlister et al., 2007; Pillitteri et al., 2007). FAMA then regulates differentiation of guard cells (Ohashi-Ito and Bergmann, 2006). INDUCER OF CBF EXPRESSION (also annotated as SCREAM) 1 and 2 physically interact with SPCH, MUTE, and FAMA and regulate the basal pathway (Kanaoka et al., 2008).
A current model for Arabidopsis thaliana stomatal development includes a signaling pathway that negatively regulates the basal pathway of stomatal lineage, which is necessary to achieve a balance between pavement and guard cells in the leaf epidermis (Bergmann and Sack, 2007; Casson and Hetherington, 2010). Determinants of this negative signal regulatory pathway include the leucine-rich repeat receptor-like protein TOO MANY MOUTHS (TMM) that is presumed to interact with the ERECTA (ER) family members of leucine-rich repeat receptor-like kinases (Shpak et al., 2005). STOMATAL DENSITY AND DISTRIBUTION1 (SDD1) encodes a subtilisin-like Ser protease that likely processes propeptides into ligands that activate the TMM-ER complex (Berger and Altmann, 2000; von Groll et al., 2002). Ligand interaction with the receptor is presumed to activate a mitogen-activated protein kinase (MAPK) cascade that includes YODA (YDA; MAPKKK), MKK4/5 (MAPKK), and MPK3/6 (MAPK). Then, SPCH is phosphorylated by MPK3/6, which leads to its inactivation and repression of the basal pathway (Bergmann et al., 2004; Wang et al., 2007; Lampard et al., 2008). Recently, the secretory peptides EPIDERMAL PATTERNING FACTOR1 (EPF1), EPF2, and EPF-like 9 (also annotated as STOMAGEN) have been implicated as ligands of TMM that regulate the MAPK cascade, independently of SDD1 (Hara et al., 2007; Hunt and Gray, 2009; Hunt et al., 2010; Sugano et al., 2010).
The GT-2 transcription factor family proteins contain two trihelix DNA binding domains that interact with GT cis-acting elements (GT elements) during transcriptional regulation (Zhou, 1999). GT elements were identified initially in the promoters of light-regulated genes, such as pea (Pisum sativum) RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE SMALL SUBUNIT 3A and rice (Oryza sativa) PHYTOCHROME A (PHYA) (Green et al., 1987; Dehesh et al., 1990; Zhou, 1999). GT-2 transcription factor proteins have been implicated in other processes, including endoreduplication, petal development, and abiotic stress tolerance in Arabidopsis and soybean (Glycine max; Brewer et al., 2004; Breuer et al., 2009; Xie et al., 2009). In this report, evidence is presented that GT-2 LIKE 1 (GTL1) functions as a focal regulator of water stress tolerance and WUE through a mechanism that involves transcriptional repression of SDD1 and regulation of stomatal density and transpiration. GTL1 expression is downregulated by dehydration, establishing a potential paradigm for how the environment influences stomatal development to reduce transpiration under low water availability conditions.
RESULTS
GTL1 Is Involved in Plant Water Stress Responses
Two independent alleles harboring T-DNA insertions in the first exon and first intron of Arabidopsis GTL1 were annotated as gtl1-4 (SALK_005972) and gtl1-5 (SALK_044308) (Figure 1A), respectively, because three other T-DNA insertion alleles (gtl1-1, gtl1-2, and gtl1-3) were described previously (Breuer et al., 2009). The gtl1-4 T-DNA insertion location is the same as gtl1-3, based on sequence data analysis (http://signal.salk.edu; Salk Institute Genomic Analysis Laboratory). Homozygous gtl1-4 and gtl1-5 plants were identified by PCR amplification using allele-specific primers (Figure 1B; see Supplemental Table 1 online). GTL1 transcript was undetectable in gtl1-4 plants but could be detected at very low abundance in gtl1-5 plants relative to the wild type, which suggests that these mutations may cause loss of function and reduced function of GTL1, respectively (Figure 1C).
Figure 1.
gtl1 T-DNA Insertional Mutations Enhance Survival and Maintenance of Leaf RWC under Water Deficit Stress.
(A) The schematic illustrates T-DNA locations in the first exon (black bar) and the first intron (black line) in GTL1 for gtl1-4 and gtl1-5, respectively. Hatched bars indicate 5′- and 3′-untranslated regions. Arrows indicate the positions of primers used in (B).
(B) Homozygosity of the T-DNA insertion in glt1-4 and gtl1-5 was determined by PCR analysis of the GTL1 genomic fragment using left primers (LP) and right primers (RP) and the T-DNA insertion using a T-DNA–specific left border primer (LB) with LP for gtl1-4 and LB with RP for gtl1-5. Col-0 is the wild type.
(C) GTL1 expression level in wild-type, gtl1-4, and gtl1-5 plants was determined by RT-PCR analysis with forward and reverse primers (GTL1F and GTL1R) and ACTIN2 (ACT2; reference standard).
(D) and (E) Plant water stress responses were analyzed in 3-week-old wild-type and gtl1 (gtl1-4 and gtl1-5) plants grown under a long-day photoperiod (16 h light/8 h dark, 30% relative humidity). Five containers of each genotype (20 plants/container) were evaluated in three independent experiments. Relative SWC is the soil water relative to the soil water at day 0 of withholding water and is the average of five containers (see Supplemental Figure 1B online). The photograph in (D) illustrates results of one replicate from one experiment. Plant survival (E) was determined 4 d after rewatering (mean ± se, n = 5).
(F) In a separate experiment, 4-week-old wild-type and gtl1 plants were grown under a short-day photoperiod (8 h light/16 h dark, 60% relative humidity) and exposed to water deficit stress by withholding water. Leaf RWC (mean ± se, n = 3 to 4) and relative SWC (see Supplemental Figure 1C online) were determined. In (E) and (F), mean values of gtl1-4 and gtl1-5 plants are significantly different from the wild type at *P < 0.05 and **P < 0.01.
gtl1-4 and gtl1-5 plants were better able to survive low relative soil water content (SWC) than were wild-type plants (Figure 1D). More than 80% of gtl1-4 and gtl1-5 and <10% of wild-type plants survived SWC of 15% ± 1.4% (Figure 1E). Most of the wild-type plants wilted at 15% ± 1.4% SWC, while most gtl1-4 and gtl1-5 plants exhibited less severe leaf wilting symptoms (see Supplemental Figure 1A online). Increased water deficit survival of gtl1 plants was associated with the capacity to maintain higher leaf relative water content (RWC) than the wild type at 13% ± 1.0% SWC (Figure 1F).
GTL1 transcript was abundant in whole shoots of well-watered 4-week-old Columbia-0 (Col-0) plants but was less abundant in those of plants exposed to water deficit stress caused by withholding irrigation for 11 d (see Supplemental Figure 2A online). This treatment induced expression of the dehydration-responsive COR15a gene (see Supplemental Figure 2A online; Baker et al., 1994), indicating the plants were experiencing water deficit. Dehydration stress of detached shoots also caused a reduction in GTL1 expression (see Supplemental Figure 2B online). Results deposited in the Genevestigator database (https://www.genevestigator.com; Zimmermann et al., 2004; Perera et al., 2008) also indicate that GTL1 is downregulated in response to water deficit stress and was confirmed by the induction of DREB2A expression (see Supplemental Figure 2C online; Liu et al., 1998). These results indicate that GTL1 is expressed when plants have sufficient available water but is downregulated by water deficit.
gtl1 Mutations Improve WUE by Reducing Transpiration
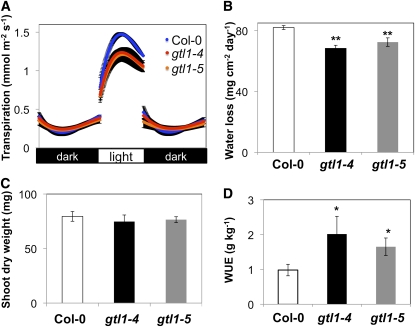
To understand the physiological mechanisms by which gtl1 plants are more water stress tolerant through maintenance of higher leaf water status under low soil moisture conditions, transpiration rates were assessed by gravimetric analyses over diurnal light/dark periods. gtl1 plants exhibited lower light period (but not dark period) transpiration rates than wild-type plants when grown under both water-sufficient (Figure 2A) and water deficit (see Supplemental Figure 3 online) conditions. Lower transpiration rates resulted in reduced daily water loss from gtl1 plants (Figure 2B), which likely enhanced the capacity of gtl1 plants to maintain higher leaf RWC (Figure 1F) and tolerate water deficit stress (Figures 1D and 1E). The significant reduction in transpiration (Figures 2A and 2B) was not associated with decreased shoot dry weight (Figure 2C) or total leaf area (see Supplemental Figures 4A and 4B online), suggesting that reduced stomatal conductance, which led to reduced water loss, did not result in a concomitant reduction in biomass accumulation. Consequently, gtl1 plants had higher integrated WUE (biomass/water use) than did the wild type (Figure 2D). These results indicate that GTL1 is a negative regulator of WUE.
Figure 2.
gtl1 Plants Have Reduced Transpiration and Improved Integrated WUE.
(A) to (C) Diurnal transpiration rate (A) and light period water loss (B) of 5-week-old wild-type (Col-0) and gtl1 plants grown under a 10-h diurnal photoperiod was gravimetrically determined (mean ± se, n = 4). Shoot dry weight (C) was measured at the completion of the experiment (mean ± se, n = 4).
(D) Integrated WUE of wild-type and gtl1 plants under water-sufficient conditions was calculated from gravimetric measurements of water loss and shoot dry weight over a period of 6 weeks (mean ± se, n = 12 to 16). In (B) to (D), asterisks indicate that mean values of gtl1 plants are significantly different from the wild type at *P < 0.05 or **P < 0.01.
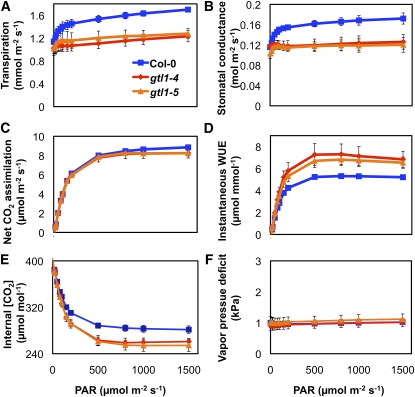
To test the hypothesis that gtl1 mutations reduced transpiration to a greater extent than CO2 assimilation, gas exchange (water and CO2) of fully expanded leaves of wild-type and gtl1 plants was determined using an infrared gas analyzer. Leaf transpiration of gtl1 plants was 26.0% ± 1.9% lower than that of wild-type plants at saturating light levels (Figure 3A). Stomatal conductance of gtl1 plants was also lower than in the wild type (Figure 3B), indicating that the reduced transpiration of gtl1 plants was due to decreased water loss through stomata. Net CO2 assimilation rates of gtl1 plants and the wild type were not significantly different (Figure 3C). Consequently, gtl1 plants had higher instantaneous WUE (CO2 assimilation/transpiration) (Figure 3D), which was attributable to reduced transpiration (Figure 3A). Vapor pressure deficit (VPD) was similar for all measurements (Figure 3F), indicating that the lower transpiration and stomatal conductance in gtl1 plants were not due to different VPD.
Figure 3.
gtl1 Plants Exhibit Higher Instantaneous WUE Due to Lower Transpiration and Stomatal Conductance.
Leaf transpiration, stomatal conductance, net CO2 assimilation, instantaneous WUE, internal CO2 concentration, and VPD were determined on individual leaves of 8-week-old wild-type (Col-0) and gtl1 plants (8-h diurnal photoperiod) using a Li-Cor 6400 gas exchange system (mean ± se, n = 4).
Quantum efficiency of gtl1 and wild-type plants was similar (see Supplemental Figure 5A online), indicating there was no difference in the capacity to use photons to fix carbon. In addition, dark respiration and the light compensation point (light intensity at which CO2 assimilation is equal to respiration) of gtl1 plants and the wild type were similar (see Supplemental Figures 5B and 5C online). Internal CO2 concentration (ci) in leaves of gtl1 plants was lower than in those of the wild type (Figure 3E), possibly due to a reduced CO2 flux from the air to the substomatal cavity because of the lower stomatal conductance in in gtl1 plants. These results indicate that gtl1 and wild-type plants have equivalent CO2 assimilation and respiration rates. Furthermore, gtl1 plants have higher instantaneous WUE, due primarily to reduced transpiration without an appreciable reduction in net CO2 assimilation and biomass accumulation under our experimental conditions.
GTL1 Regulates Stomatal Density but Does Not Affect Stomatal Aperture or Opening/Closing
gtl1 mutations reduced leaf transpiration because of decreased stomatal conductance (Figures 3A and 3B), which may be caused by effects on stomatal aperture or stomatal density (Hetherington and Woodward, 2003; Yoo et al., 2009). There was no difference in leaf abaxial stomatal aperture of gtl1 and wild-type plants under water-sufficient conditions (see Supplemental Figure 6A online). Abscisic acid (ABA) treatment caused an equivalent reduction in transpiration rates of gtl1 and wild-type plants (see Supplemental Figures 6B and 6C online), indicating that both had equivalent stomatal responsiveness to ABA. Seed germination and seedling root growth of gtl1 plants and the wild type also responded to ABA similarly (see Supplemental Figures 6D and 6E online). These data indicate that GTL1 does not regulate ABA responsiveness, and the lower transpiration rates of gtl1 plants are not caused by differences in stomatal aperture or ABA-induced stomatal closure.
Fully expanded leaves of gtl1 plants had fewer stomata than did the wild type (Figure 4), which was determined on the same leaves used for gas exchange analyses (Figure 3). Leaf abaxial stomatal density was 24.1% ± 2.5% lower in gtl1 plants compared with the wild type (Figures 4A and 4B), which correlated with a 26.0% ± 1.9% lower transpiration rate at saturating light levels (Figure 3A). The larger leaf trichome phenotype associated with gtl1 plants (see Supplemental Figure 7 online; Breuer et al., 2009) is unlikely a principal cause of reduced transpiration because any effects on boundary layer conductance due to trichomes would be minimal for small leaves such as those of Arabidopsis plants (Jarvis and McNaughton, 1986). These results indicate that higher WUE of gtl1 plants is due to reduced transpiration that is attributable to a reduction in leaf abaxial stomatal density.
Figure 4.
gtl1 Plants Have Lower Stomatal Densities and Indices and Have Stomatal Precursor Cells.
(A) Representative images of leaf abaxial epidermal layers from 8-week-old wild-type (Col-0) and gtl1-4 plants (8-h diurnal photoperiod). Pavement cells and stomata are illustrated in white and black, respectively. Stomatal precursor cells (gray) are indicated with arrows. Bars = 50 μm.
(B) to (E) Stomatal density (the number of stomata per area), stomatal index (the number of stomata per total epidermal cells), the number of stomata precursor cells, and pavement cell density were analyzed in the leaf abaxial epidermal layers from wild-type and gtl1-4 plants. Data are the mean of seven individual plants (mean ± se, n = 7); * and ** indicate significant difference from the wild type at P < 0.05 and P < 0.01, respectively.
Lower stomatal indices (number of stomata per total number of epidermal cells) caused by gtl1 (Figure 4C) suggested altered stomatal development in these plants. Stomatal precursor cells, such as meristemoids or GMCs, were detected in leaves of gtl1 but not wild-type plants (Figures 4A, arrows, and 4D), indicating that stomatal development may be delayed in gtl1 plants (Bergmann and Sack, 2007; Casson and Hetherington, 2010). Pavement cells were larger in leaves of gtl1 plants (Figure 4A), which resulted in a lower pavement cell density (Figure 4E). Larger pavement cells in fully expanded leaves of gtl1 plants may be attributable to unrepressed endoreduplication (Breuer et al., 2009). These results implicate GTL1 as a regulator of stomatal and pavement cell development and stomatal density that affects transpiration.
GTL1 Is Expressed in the Abaxial Epidermis and Negatively Regulates SDD1 Expression
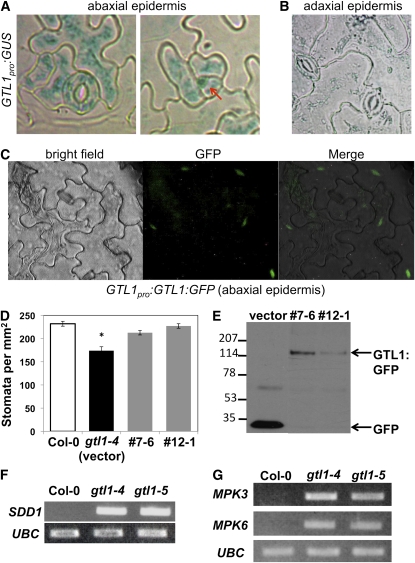
Stomatal development genes are expressed primarily in epidermal cells (Bergmann and Sack, 2007; Hunt and Gray, 2009). Analysis of plants transformed with a 2.9-kb GTL1 promoter-β-glucuronidase (GUS) reporter gene fusion (GTL1pro:GUS) indicated that the gene is expressed predominantly in guard cells, pavement cells, and meristemoids in the leaf abaxial epidermis (Figures 5A and 5B). GTL1 localization was determined by monitoring a C-terminal green fluorescence protein (GFP)–tagged fusion protein (GTL1pro:GTL1:GFP) (Figure 5C). GTL1-GFP expression, which was driven by the same GTL1 promoter that was used to drive GUS expression (Figures 5A and 5B), suppressed the reduced stomatal density phenotype of gtl1-4 (Figure 5D). The GTL1-GFP fusion protein was detected by protein gel blot analysis using anti-GFP (Figure 5E). GTL1 protein localized to nuclei in abaxial epidermal cells and was detected primarily in pavement cells (Figure 5C). These results indicate that GTL1 is expressed in guard cells throughout the abaxial epidermis, although GTL1 accumulates predominantly in pavement cell nuclei.
Figure 5.
GTL1 Is Expressed in the Abaxial Epidermis and Negatively Regulates SDD1 Expression.
(A) GUS activity in the abaxial epidermis of the first rosette leaf from 10-d-old seedlings harboring GTL1pro:GUS was analyzed by histochemical staining. Arrow indicates a triangle-shaped meristemoid.
(B) GUS activity was not detected in the adaxial epidermis of fully expanded rosette leaves from 4-week-old transgenic plants harboring GTL1pro:GUS.
(C) GTL1-GFP protein localization was analyzed in the leaf abaxial epidermis of 4-week-old transgenic plants harboring GTL1pro:GTL1:GFP (#12-1) by fluorescence microscopy.
(D) Stomatal density was measured in 6-week-old wild-type (Col-0), gtl1-4 (empty vector control line), and GTL1pro:GTL1:GFP complementation lines (#7-6 and #12-1) (mean ± se, n = 4; * indicates significant difference from the wild type at P < 0.05).
(E) GTL1-GFP protein expression in 6-week-old complementation lines and GFP protein expression in 6-week-old empty vector control plants were detected by immunoblot using anti-GFP antibody.
(F) and (G) RT-PCR analysis of SDD1, MPK3, MPK6, or UBC was performed on total RNA from fully expanded leaves of 8-week-old wild-type and gtl1 plants.
SDD1 activates the MAP kinase pathway that negatively regulates stomatal development (see Supplemental Figure 10 online; Bergmann and Sack, 2007; Casson and Hetherington, 2010). SDD1 transcript abundance in fully expanded leaves was much higher in gtl1 compared with wild-type plants (Figure 5F). Similarly, MPK3 and MPK6 expression was higher in leaves of gtl1 plants (Figure 5G). ER, TMM, and YDA expression was similar in gtl1 and wild-type plants (see Supplemental Figure 8 online). Thus, GTL1 appears to function as a stomatal development determinant by negatively regulating SDD1, MPK3, and MPK6 expression.
GTL1 Binds to the GT Element in the SDD1 Promoter
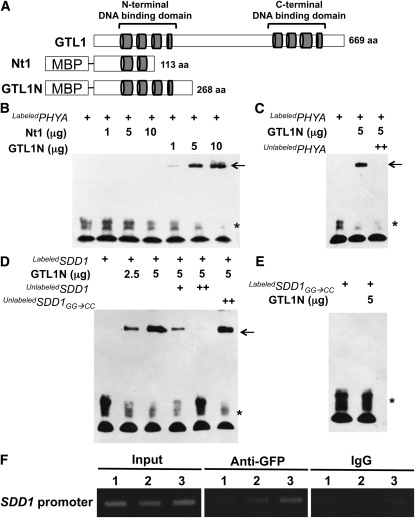
GT-2 transcription factor family proteins bind to GT elements (GT1 box, GGTTAA; GT2 box, GGTAAT; GT3 box, GGTAAA) in the promoters of target genes to activate or repress transcription (Kuhn et al., 1993; Ni et al., 1996; Zhou, 1999). Initially, GTL1 DNA binding activity was assessed using the rice PHYA promoter fragment that has been reported to interact with Arabidopsis and rice GT2 proteins (Kuhn et al., 1993; Ni et al., 1996). Two partial fragments of GTL1 (Nt1 and GTL1N polypeptides) were fused with maltose binding protein (MBP) to enhance solubility during purification (Figure 6A). An electrophoretic mobility shift assay (EMSA) determined that only GTL1N interacted with the rice PHYA promoter fragment (Figure 6B). The Nt1 polypeptide that was predicted to have an incomplete DNA binding domain did not interact with the PHYA promoter (Figures 6A and 6B). Unlabeled probe was an effective competitor (Figure 6C), indicating that GTL1N specifically binds to the PHYA promoter. Furthermore, the GTL1 N-terminal DNA binding domain effectively bound all three types of GT elements (see Supplemental Figure 9 online).
Figure 6.
N-Terminal DNA Binding Domain of GTL1 Binds to PHYA and SDD1 Promoters.
(A) Schematic diagram of the GTL1 protein, showing the four helical, N- and C-terminal DNA binding domains (represented as gray cylinders). Two protein fragments (Nt1 and GTL1N) were fused with MBP.
(B) Nt1 and GTL1N polypeptides and a biotin-labeled rice PHYA promoter fragment (400 ng) were used for EMSA. In (B) to (E), free probe and GTL1N-probe complexes are indicated by an asterisk and arrows, respectively.
(C) Unlabeled PHYA promoter (2 μg) was used as a competitor to determine the specificity of DNA binding activity for GTL1N.
(D) GTL1N polypeptide was used for EMSA with a biotin-labeled SDD1 promoter fragment (250 ng). Unlabeled probes (250 ng [+] and 1 μg [++]) were used as competitors. An unlabeled mutant version of the SDD1 promoter (GG → CC) was used as a noncompetitor.
(E) The mutant version of the SDD1 promoter (GG → CC) was labeled with biotin (250 ng) and used for EMSA with GTL1N polypeptides.
(F) ChIP analysis was conducted to determine the in vivo interaction between GTL1 with the SDD1 promoter. Input is chromatin before immunoprecipitation. Anti-GFP antibody was used to precipitate chromatin associated with GTL1-GFP. Mouse IgG was used as a negative control for the specificity of immunoprecipitation. The SDD1 promoter region associated with GTL1 was amplified by PCR using SDD1 promoter-specific primers for three different regions (1, 2, and 3). Region 1 (−1136 to −1400) is distal to the GT3 box. Region 2 (−538 to −824) is adjacent to, but does not contain, the GT3 box. Region 3 (−279 to −556) includes the GT3 box in the SDD1 promoter.
We hypothesized that GTL1 binds directly to the GT3 box (GGTAAA; −428 to −423) of the SDD1 promoter to repress SDD1 transcription. Migration of labeled SDD1 promoter was shifted by the addition of GTL1N polypeptide. However, this gel shift was abolished by adding unlabeled SDD1 promoter (Figure 6D), indicating that GTL1 specifically binds to the SDD1 promoter. Two guanine residues in the GT3 box (GGTAAA) are critical nucleotides in the interaction of GT elements with GT2 family proteins (Kuhn et al., 1993; Ni et al., 1996). We substituted CC for GG in the GT3 box (GGTAAA→ CCTAAA) in the SDD1 promoter fragment to determine specificity of the GT3 box for GTL1 DNA binding activity. The mutated SDD1 promoter was used as a noncompetitor (unlabeled mutated SDD1 promoter) and did not inhibit the interaction between GTL1 and the SDD1 promoter (Figure 6D). Furthermore, GTL1N did not bind to the labeled mutated SDD1 promoter (GG→CC) (Figure 6E). The results of an in vitro EMSA indicate that GTL1 specifically binds to the GT3 box in the SDD1 promoter.
In vivo interaction of GTL1 with the SDD1 promoter was tested by chromatin immunoprecipitation (ChIP) analysis using transgenic plants harboring the GTL1pro:GTL1:GFP construct that suppressed the reduced stomatal density phenotype of gtl1-4 plants (Figures 5D and 5E). Chromatin associated with GTL1-GFP was immunoprecipitated with anti-GFP antibody and subjected to PCR analysis using primers specific for different regions of the SDD1 promoter (see Supplemental Table 1 online). Region 3 (−279 to −556) included the GT3 box in the SDD1 promoter. Region 2 (−538 to −824) was adjacent to, but did not contain, the GT3 box. Region 1 (−1136 to −1400) was distal to the GT3 box. Region 3 primers resulted in the greatest amount of PCR product (Figure 6F). The amount of PCR amplification product was reduced with primers for region 2 and even more so with those for region 3 (Figure 6F). These results are consistent with the GTL1 fragment binding to the GT3 box in the SDD1 promoter determined by in vitro DNA binding (Figure 6D). Together, these results indicate that GTL1 binds to the SDD1 promoter to repress SDD1 expression, which regulates stomatal density.
DISCUSSION
The trihelix transcription factor GTL1, previously identified as a trichome development regulator (Breuer et al., 2009), is shown herein to regulate stomatal development. Furthermore, GTL1 loss-of-function mutations enhanced water deficit tolerance, which was associated with maintenance of leaf water status, even when plants were grown in media with low SWC (Figure 1). gtl1 mutations caused higher instantaneous and integrated plant WUE because of reduced transpiration that occurred without a difference in net CO2 assimilation (Figures 2 and 3). Lower transpiration, drought tolerance, and higher WUE were associated with reduced stomatal density in the leaf abaxial epidermis and higher SDD1 expression (Figures 4 and 5F), which suggests that gtl1 reduced stomatal density by increasing expression of SDD1, a negative regulator of stomatal development (Berger and Altmann, 2000; von Groll et al., 2002). GTL1 expression was downregulated by plant water deficit (see Supplemental Figure 2 online), and GTL1 localized to the nucleus in abaxial epidermal pavement cells (Figure 5C). EMSA indicated that GTL1 physically interacts with the GT3 box in the SDD1 promoter, and ChIP analysis confirmed that GTL1 was associated with the GT3 box of the SDD1 promoter (Figures 6D and 6F). Together, these results indicate that GTL1 regulates plant water use by modulating stomatal development through SDD1 transrepression (Figure 7).
Figure 7.
Model of GTL1 Function as a Determinant of WUE by Regulating Stomatal Density through Transrepression of SDD1.
GTL1 binds to the GT element (GT3 box: GGTAAA) in the SDD1 promoter to transrepress SDD1, which is a negative regulator of stomatal development. GTL1 negatively regulates WUE by positively regulating stomatal density and transpiration through transrepression of SDD1. GTL1 is expressed in water-sufficient conditions to facilitate stomatal development and is downregulated under water deficit to reduce transpiration.
GTL1 Affects Plant Water Use and CO2 Assimilation through Regulation of Stomatal Development
Transpiration and CO2 uptake occur primarily through stomatal pores, and conductance is dependent on stomatal density and pore aperture (Hetherington and Woodward, 2003; Nilson and Assmann, 2007). gtl1 caused a reduction in leaf abaxial surface stomatal density (Figure 4) and decreased transpiration (Figures 2 and 3), which resulted in enhanced water deficit tolerance (Figure 1D). Reduced stomatal density caused by gtl1 did not affect CO2 assimilation and biomass accumulation and resulted in higher integrated and instantaneous plant WUE (Figures 2 to 4). For most C3 plants, net CO2 assimilation rate saturates as stomatal conductance increases because of nonstomatal limitations, such as the regeneration of ribulose 1,5-bisphosphate (Farquhar and Sharkey, 1982). However, over a similar stomatal conductance range, transpiration will continue to increase linearly with stomatal conductance (Yoo et al., 2009). Therefore, a moderate decrease in stomatal density can reduce transpiration significantly without a concomitant effect on CO2 assimilation and result in higher WUE (Yoo et al., 2009). It is plausible that a more substantial reduction in stomatal density would reduce CO2 uptake substantially and lower both WUE and biomass accumulation. Consistent with this hypothesis, cauliflower mosaic virus 35S-driven SDD1 expression in C24 plants resulted in an ~60% reduction in stomatal density that was linked to a 20% reduction in net CO2 assimilation rate (Büssis et al., 2006).
Genetic variation for WUE is negatively correlated with transpiration in primary gene pools (Van den Boogaard et al., 1997; Impa et al., 2005; Yoo et al., 2009). ER, GPA1, CA1/4, and HDG11 are Arabidopsis genetic determinants known to regulate WUE through modulation of stomatal density (Masle et al., 2005; Yu et al., 2008; Hu et al., 2010; Nilson and Assmann, 2010). ER alleles were the cause of quantitative trait loci associated with natural variation in WUE between Col-4 and Landsberg erecta (Masle et al., 2005). ER mutations resulted in lower WUE, which was associated with a number of measurable phenotypes, including increased stomatal density and reduced photosynthetic capacity and mesophyll development (Masle et al., 2005). GPA1 (G protein α-subunit 1) loss of function resulted in reduced stomatal density and stomatal conductance but similar net CO2 assimilation leading to higher WUE (Nilson and Assmann, 2010). Improved WUE of gpa1 plants was due primarily to reduced stomatal density rather than to insensitivity to ABA-induced stomatal closure or to inhibition of stomatal opening (Fan et al., 2008; Nilson and Assmann, 2010). CA1 and CA4 (carbonic anhydrase 1 and 4) have also been implicated in the regulation of WUE (Hu et al., 2010). ca1 ca4 double mutations increased stomatal density, whereas guard cell–targeted overexpression of CA1 or CA4 in ca1 ca4 plants decreased stomatal density and improved WUE (Hu et al., 2010). The target determinants and molecular mechanisms by which GPA1, CA1, and CA4 regulate stomatal development and WUE remain to be elucidated.
A forward genetic screen of a T-DNA activation-tagged population for drought tolerance resulted in the identification of enhanced drought tolerance1 (edt1). The T-DNA in edt1 was inserted in the 5′-untranslated region of HOMEODOMAIN GLABROUS 11 (HDG11) and resulted in overexpression of HDG11, reduced stomatal density, and higher WUE. Drought tolerance and higher WUE were attributable to reduced stomatal density and transpiration and to enhanced root growth, photosynthesis, and ABA and Pro biosynthesis (Yu et al., 2008). ER expression was greater in edt1, and HDG11 transactivated the ER promoter when cotransformed into onion cells, implying that HDG11 directly activates ER expression and may regulate stomatal development through the negative regulatory pathway as does GTL1. The recent characterization of these genetic determinants that affect WUE indicates that plants have the capacity to optimize water use and CO2 assimilation by regulating stomatal development.
GTL1 Is a Positive Regulator of Stomatal Development as a Transrepressor of SDD1
SDD1 and MPK3/6 transcripts were misupregulated in gtl1 plants (Figures 5F and 5G), indicating that GTL1 functions upstream of these determinants in the negative regulatory pathway (see Supplemental Figure 10 online; Bergmann and Sack, 2007). GTL1 interacted with the GT3 box in an SDD1 promoter fragment in vitro (Figure 6D) and was associated with an SDD1 promoter fragment in chromatin that contained the GT3 box (Figure 6F). These data indicate that GTL1 is a transcriptional repressor of SDD1 and, through this function, positively regulates stomatal density (see Supplemental Figure 10 online; Bergmann and Sack, 2007; Casson and Hetherington, 2010).
The developmental program that determines stomatal density is sufficiently plastic to enable plants to regulate CO2 uptake and transpiration in response to changing environments (Casson and Gray, 2008). Climatic factors (e.g., light, CO2, relative humidity, and water deficit) and hormones alter stomatal density (Lake et al., 2001; Bergmann, 2004; Casson and Gray, 2008; Xu and Zhou, 2008; Ahmed et al., 2010), although the mechanism(s) by which these effectors modulate stomatal development has not been identified (Casson and Hetherington, 2010). GTL1 transcript was abundant when plants were growing with adequate water supply but expression was downregulated by water stress or dehydration (see Supplemental Figure 1 online). This drought-responsive downregulation of GTL1 expression may indicate that the transcription factor is an integrative node that links stomatal development to environmental regulation of density and gas exchange (Figure 7). Further investigation is necessary to determine the mechanisms by which other environmental factors regulate stomatal development and to determine the adaptive significance of these as plants cope with climatic fluctuations.
GTL1 was expressed in both pavement and guard cells of the leaf abaxial epidermis, but expression was not detected in the adaxial epidermis (Figures 5A and B), which correlates with GTL1 regulation of abaxial stomatal density (Figure 4A). GTL1 accumulated in the nuclei of pavement cells (Figure 4C), where SDD1 is not expressed (von Groll et al., 2002), which is consistent with the notion that GTL1 transrepresses this negative regulator of stomatal development. These GTL1 expression and protein localization data, together with high expression of SDD1 in gtl1 plants, indicate that SDD1 repression by GTL1 is necessary to allow formation of stomata during development and, in response to environmental changes, to optimize transpiration and CO2 assimilation. Further research is needed to determine whether tissue- and cell type–specific regulation of SDD1 is mediated by GTL1 and environmental changes.
GTL1 Fine-Tunes Stomatal Development to Regulate WUE
ER and TMM are known receptors of the signal cascade that negatively regulates the basal stomatal development pathway (see Supplemental Figure 10 online; Shpak et al., 2005; Bergmann and Sack, 2007). SDD1 encodes a subtilisin-like protease that activates ER and TMM to repress stomatal development (von Groll et al., 2002; Shpak et al., 2005). These receptors are negatively regulated by EPF1/2 ligands and positively regulated by STOMAGEN, independent of SDD1 (von Groll et al., 2002; Hara et al., 2007; Hunt and Gray, 2009; Sugano et al., 2010). It is plausible that convergent functions of EPF1/2, STOMAGEN, and SDD1 on ER and TMM receptors are necessary for precise regulation of stomatal number and plant water use in different ecosystems and in response to developmental and environmental cues (Casson and Hetherington, 2010). We posit that GTL1 is a determinant that integrates the effects of plant water-deficit stress on stomatal development to regulate carbon assimilation and transpiration. It is possible that other members of the GT-2 family that are closely related to GTL1 may have similar functions as transcriptional repressors of SDD1 expression. The existence of multiple regulatory determinants to optimize gas exchange and improve WUE in different environments is analogous to fine-tuning of signal transduction that is necessary for amplification and/or maintenance of a signal and for reducing noise fluctuations of signals (Thattai and van Oudenaarden, 2001; Kashiwagi et al., 2006; Saez et al., 2006; Alon, 2007).
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana genetic resources were wild type (Col-0 ecotype), gtl1-4 (SALK_005972), and gtl1-5 (SALK_044308). gtl1 mutant seeds were obtained from the ABRC at Ohio State University. Transgenic plants harboring and expressing GTL1pro:GUS and GTL1pro:GTL1:GFP were generated using the Agrobacterium tumefaciens–mediated floral dip method (Zhang et al., 2006). Single-copy and homozygous T3 plants were identified by genetic segregation on agar medium containing hygromycin. Seeds were stratified for 3 d at 4°C in the dark and then sown onto medium (1× Murashige and Skoog basal salt mixture, 2% sucrose, 2.5 mM MES, pH 5.7, and 0.8% agar) or in a 2:1 mixture of ProMix PGX soilless media (Premier Horticulture) and Turface calcined clay (Profile Products) in containers (200 mL, 1 liter, or 1.7 liter) or Falcon tubes (50 mL). Plants grown on agar were maintained in a growth room (temperature: 22°C [light]/18°C [dark]; photoperiod: 16 h [light]/8 h [dark]; light intensity: ~80 μmol quanta m−2 s−1; provided by fluorescent bulbs). Plants grown in soilless media were maintained in a controlled environment growth chamber (temperature: 22°C [light]/18°C [dark]; light intensity: ~125 μmol quanta m−2 s−1 provided by fluorescent bulbs; relative humidity: ~60%). Photoperiods for soilless media-grown plants are described in the corresponding figure legends.
Plasmid Construction
GTL1 promoter (GTL1pro):GUS was constructed in the pCAMBIA1303 vector (CAMBIA). The 2973-bp GTL1 promoter was amplified by designated primers (GTL1p-F-XmaI and GTL1p-R-NcoI) (see Supplemental Table 1 online). After the 35S promoter in pCAMBIA1303 was removed by XmaI and NcoI digestion, the GTL1 promoter was inserted to the XmaI/NcoI site. For GTL1pro:GTL1:GFP construction, the open reading frame of GTL1 was amplified by designated primers (GTL1-F-XmaI and GTL1-R-SpeI) from cDNA. The amplified GTL1 open reading frame was inserted into the XmaI and SpeI sites in the pCAMBIA1302 vector (CAMBIA) after the 35S promoter was removed and was fused to GFP in frame (pCAMBIA1302-GTL1:GFP). Then, the GTL1 promoter, amplified by designated primers (GTL1p-F-XmaI and GTL1p-R-XmaI), was inserted into the XmaI site in pCAMBIA1302-GTL1:GFP. MBP:GTL1 was constructed in the pMAL-C2 vector (Kim et al., 2007). Two GTL1 fragments (Nt1 and GTL1N) were amplified by designated primers (Nt1, Nt1-F-EcoRI and Nt1-R-PstI; GTL1N, Nt1-F-EcoRI and Nt2-R-PstI). The two amplified fragments were inserted into the EcoRI and PstI sites in the pMAL-C2 vector. Sequence information for all designated primers used for plasmid construction is provided in Supplemental Table 1 online.
Physiological Analyses
Water deficit stress was imposed by withholding water from containers (1 liter) with 288.7 ± 3.1 g (dry weight) of soilless media containing 20 plants (3 weeks old). Containers were irrigated with water to saturation and weighed at the start of water deficit stress treatment (initial weight) and then periodically throughout the treatment period. Relative SWC was calculated as (final fresh weight – dry weight)/(initial weight – dry weight)×100. After 14 d, plants were rewatered and survival was assessed 4 d after rewatering. In a separate experiment, leaf RWC of fully expanded leaves from 4-week-old plants grown in 200-mL containers at different relative SWC was assessed. Leaves were removed and immediately weighed to obtain leaf FW. Leaves were then placed into vials filled with distilled water for 24 h, blotted to remove excess water, and then weighed to obtained leaf turgid weight (TW). Leaves were then dried to a constant weight at 65°C and reweighed to obtain leaf dry weight (DW). Leaf RWC was calculated as (FW − DW)/(TW − DW)×100.
Whole-plant transpiration of 5- to 6-week-old plants was determined by a gravimetric method. Individual plants were grown in 200-mL containers. During the period that transpiration was measured, each container was covered with a polyethylene wrap to prevent evaporation from the soil surface. An individual plant was placed onto a balance, and the weight of each container was determined every 5 min. At the end of the experiment, total leaf area was determined from photographs of excised leaves using the ImageJ program (National Institutes of Health). Transpiration rate (mmol water m−2 s−1) was calculated based on gravimetric water loss rate and leaf area data. A curve was fit to the data using a 12-point moving average. A cubic polynomial was then fit to the smoothed data separately for the day period. The equation for the light period transpiration curve was then differentiated and solved for zero to give the time at which maximum transpiration occurs. This time was then used to calculate maximum light period transpiration rate.
Integrated WUE was calculated as final shoot dry weight divided by total water loss over a period of 6 weeks. Individual containers were covered with plastic wrap containing a central hole through which three to four seeds for each genotype were sown. Seven days after germination, containers were thinned to one seedling per container. Individual containers were weighed before and after irrigating with water every 5 to 6 d to determine moisture loss. Water loss from control containers (without plants) was subtracted from treatment containers. Shoot dry weight was determined at the end of the experiment.
Leaf gas exchange of fully expanded leaves, including transpiration, stomatal conductance, and net CO2 assimilation, was determined using a LI6400XT infrared gas analyzer (LI-COR Biosciences). Plants were grown for 60 d in 1.7-liter containers under short-day conditions (8 h light). These conditions resulted in plants with leaves large enough to fill the 6-cm2 LI-6400 chamber. Gas exchange was measured at PAR levels of 1500, 1000, 800, 500, 200, 150, 100, 80, 50, 25, 20, and 0 μmol m−2 s−1. A nonrectangular hyperbola curve was fit to the net CO2 assimilation data (Lambers et al., 1998). Quantum efficiency is the initial slope of the curve, dark respiration is the point at which the curve crosses the y axis at PAR = 0, and the light compensation point is the point at which the curve crosses the x axis (Lambers et al., 1998). VPD during measurement was ~1 kPa. Instantaneous WUE was calculated as net CO2 assimilation rates divided by transpiration rates.
Anatomical Analysis
Abaxial epidermal anatomy of fully expanded leaves from plants that were used for leaf gas exchange analyses was characterized. Transparent cellulose adhesive tape was attached to the middle portion of the adaxial lamina, and mesophyll layers of the leaf were removed by pulling the tape so that only the abaxial epidermis remained. The abaxial epidermis was placed on a slide and images were obtained under ×200 magnification using a Nikon-OptiPhot2 microscope. Stomatal density (stomatal number per area), stomatal index (ratio of stomata to total epidermal cells, including stomata, stomatal precursor cells, and pavement cells), the number of stomatal precursor cells, and pavement cell density (pavement cells per area) were obtained from a leaf area of 0.069 mm2. In all calculations, stomata were considered to be a pair of guard cells. Stomatal precursor cells were identified as cells at the meristemoid or GMC stage. These were not counted as stomata. The OminiGraffle Pro program was used to transform microscopy images to simplified black and white images.
Stomatal aperture was analyzed in abaxial epidermal peels from fully expanded leaves (Li et al., 2006). The epidermis was incubated in buffer solution (20 mM KCl, 5 mM MES-KOH, and 1 mM CaCl2, pH 6.15) under ~200 μmol m−2 s−1 for 4 h. Subsequently, the epidermis was placed onto a slide and images were photographed using a Nikon-OptiPhot2 microscope (×400 magnification). The aperture width of each stomatal pore was determined from the image.
Histochemical Assay for GUS Activity
Epidermal peels from T3 transgenic plants harboring GTL1pro:GUS were incubated in a fixation solution (0.3% formaldehyde, 10 mM MES, pH 5.6, and 0.3 M mannitol) for 1 h, washed with 50 mM sodium phosphate buffer, and then incubated further in staining solution (50 mM sodium phosphate buffer, 2.5 mM potassium ferricyanide, 2.5 mM potassium ferrocyanide, 0.3% Triton X-100, and 1.9 mM 5-bromo-4-chloro-3-indolyl β-d-glucuronide) for 12 h at 37°C. Tissues were then washed several times in 70% ethanol until chlorophyll was completely extracted.
Fluorescence Microscopy for GFP Imaging
Subcellular localization of GTL1-GFP fusion protein in the abaxial epidermis of 4-week-old transgenic plants harboring GTL1pro:GTL1:GFP was determined. A Nikon-OptiPhot2 microscope with epifluorescence was used to detect GFP expression using a B1E filter (excitation 470 to 490 nm; dichronic mirror 515 nm; barrier filter 520 to 560 nm). Images were photographed with a digital camera.
Purification of MBP-GTL1 Fusion Proteins
MBP-GTL1 fusion protein was expressed in BL21 Escherichia coli cells after induction with 0.1 mM of isopropyl β-d-thiogalactopyranoside and incubation for 2 h at 30°C. The culture was harvested by centrifugation for 5 min at 5000g. The pellet was resuspended in 50 mL of ice-cold MBP resuspension buffer (10 mM Tris-Cl, pH 7.5, 30 mM NaCl, 1 mM EDTA, 1 mM PMSF, and 1× Protease inhibitor cocktail), sonicated with a 550 Sonic Dismembrator (Fisher Scientific), and then centrifuged for 20 min at 14,000g. The supernatant was incubated with prewashed amylose resin (NEB) for 2 h at 4°C and washed five times with buffer (10 mM Tris-Cl, pH 7.5, and 1 M NaCl). MBP-GTL1 protein was eluted with buffer (10 mM Tris-Cl, pH 7.5, and 10 mM maltose). The eluate was collected with a centrifugal filter (Amicon Ultra 50K; Millipore) for buffer exchange according to the manufacturer’s protocol. The fusion protein was suspended in buffer (100 mM potassium phosphate, pH 7.8, 1 mM EDTA, 1% Triton X-100, and 10% glycerol).
EMSA
MBP-GTL1 fusion proteins purified on amylose resin were used to determine DNA binding by EMSA. Single-stranded complementary oligonucleotide fragments corresponding to regions of Arabidopsis SDD1 or rice (Oryza sativa) PHYA promoters (see Supplemental Table 1 online) that included the GT elements were synthesized (Macrogen) and biotinylated using the Biotin 3′-end DNA labeling kit (Thermo Fisher Scientific). Biotinylated complementary oligonucleotide pairs were annealed to make double-stranded and biotin-labeled probes (100 ng μL−1) by mixing in a buffer (10 mM Tris and 1 mM EDTA), boiling for 5 min, and cooling slowly overnight. Unlabeled complementary oligonucleotide pairs were also annealed to make double-stranded competitor probes (500 ng μL−1). EMSA reaction solutions were prepared by adding the following components in order according to the manufacturer’s protocol (LightShift Chemiluminescent EMSA kit; Thermo Fisher Scientific): 1× binding buffer, 50 ng poly (dI-dC), 2.5% glycerol, 0.06% Nonidet P-40, 5 mM MgCl2, 19 μg BSA, proteins, competitor, noncompetitor, and biotin-labeled probes. Reaction solutions were incubated for 20 min at room temperature. The protein-probes mixture was separated in a 6% polyacrylamide native gel and transferred to a Biodyne B Nylon membrane (Thermo Fisher Scientific). Migration of biotin-labeled probes was detected on x-ray film using streptavidin-horseradish peroxidase conjugates that bind to biotin and chemiluminescent substrate according to the manufacturer’s protocol.
ChIP Assay
Leaves of 3-week-old T3 transgenic plants harboring GTL1pro:GTP1:GFP were used for ChIP analysis. Anti-GFP antibody was used to pull down the chromatin, as described previously (Jin et al., 2008). Leaves were incubated in buffer (0.4 M sucrose, 10 mM Tris, pH 8.0, 1 mM EDTA, 1 mM PMSF, and 1% formaldehyde) under vacuum for 15 min to cross-link the chromatin. Then, 0.1 M Gly was added to the mixture, which was incubated for an additional 5 min to terminate the reaction. Leaves were ground in liquid nitrogen and resuspended in lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate, 0.1% SDS, 1 mM PMSF, 10 mM Na-butyrate, and 1× complete protease inhibitor [Roche]). Chromatin was sheared to ~200- to 1000-bp fragments by sonication and then centrifuged. Supernatants were precleared with protein G/salmon sperm DNA (protein G agarose beads) for 1 h at 4°C. After centrifugation, supernatant including chromatin (input material) was used for immunoprecipitation with anti-GFP antibody (Santa Cruz Biotechonology) or mouse IgG antibody (Abcam). Anti-GFP antibody bound to GTL-GFP-chromatin complexes was incubated with protein G agarose beads for 1 h at 4°C, washed several times, and eluted with elution buffer according to the manufacturer’s protocol (EZ ChIP chromatin immunoprecipitation kit; Millipore). Input and immunoprecipitated chromatin were uncross-linked for 5 to 6 h at 65°C with 12 μL of 5 M NaCl. Associated proteins were degraded by proteinase K. Chromatin were purified using spin columns and eluted in 50 μL of TE buffer. Input and immunoprecipitated chromatin were used for PCR analysis using various SDD1 promoter-specific primers that were designed to amplify SDD1 promoter fragments (see Supplemental Table 1 online).
Immunoblots
Total protein was extracted from leaves of 6-week-old plants grown in soilless media under the 12-h photoperiod conditions. Plant tissues were ground in liquid nitrogen and resuspended in extraction buffer (100 mM potassium phosphate buffer, pH 7.8, 1 mM EDTA, 1% Triton X-100, 10% glycerol, and 1 mM DTT). Ten micrograms of total protein from each plant was separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Thermo Fisher Scientific). The membrane was probed with anti-GFP antibody (Santa Cruz Biotechnology) and detected by the enhanced chemiluminescence Plus protein gel blotting detection system (GE Healthcare).
Genomic DNA Extraction, RNA Extraction, and RT-PCR Analysis
Genomic DNA from the leaves of 3-week-old plants was extracted by the cetyltrimethyl-ammonium bromide (CTAB) method (Richards et al., 2001) and used to determine homozygosity of T-DNA insertion in gtl1-4 and gtl1-5 by PCR analysis using GTL1-specific primers and a T-DNA–specific primer (see Supplemental Table 1 online). Total RNA was extracted from shoots of 4-week-old plants or fully expanded leaves of 8-week-old plants using TRIzol (Invitrogen) according to the manufacturer’s protocol. One microgram of total RNA was used to synthesize cDNA by the ThermoScript RT-PCR system for first-strand cDNA synthesis (Invitrogen). The same amount of cDNA was used for PCR analysis using primers for UBC (26 or 28 cycles) or ACT2 (28 cycles; internal control), GTL1 (35, 28, or 25 cycles for Figure 1C, Supplemental Figure 2A online, and Supplemental Figure 2B online, respectively), Cor15a (28 cycles), and stomatal development genes including SDD1 (40 cycles), MPK3 (28 cycles), and MPK6 (28 cycles) (see Supplemental Table 1 online).
Statistical Analysis
All bar graphs were analyzed by Student’s t test for pairwise comparison.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: GTL1 (At1g33240; NM_103052) and SDD1 (At1g04110; NM_100292).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. gtl1 Plants Were Less Wilted in Response to Water Deficit Stress.
Supplemental Figure 2. GTL1 Expression Is Downregulated by Water Stress and Dehydration.
Supplemental Figure 3. gtl1 Plants Have Reduced Light Period Transpiration Rates under Both Well-Watered and Water Deficit Stress Conditions.
Supplemental Figure 4. Leaf Morphology and Area of gtl1 Plants Are Similar to Those of Wild-Type Plants.
Supplemental Figure 5. Quantum Efficiency, Dark Respiration, and Light Compensation Point Were Similar Among Genotypes.
Supplemental Figure 6. gtl1 Plants Exhibited Similar Stomatal Aperture, and Transpiration, Germination, and Root Growth Were Similarly Affected by ABA in Wild-Type and gtl1 Plants.
Supplemental Figure 7. gtl1 Plants Have Increased Trichome Branch Length.
Supplemental Figure 8. GTL1 Does Not Regulate The Expression of TMM, ER, and YDA.
Supplemental Figure 9. The N-Terminal DNA Binding Domain of GTL1 Binds to Three Types of GT Elements.
Supplemental Figure 10. Stomatal Development Pathways Include a Basal Pathway and a Negative Regulatory Pathway for Fine-Tuning of Stomatal Development in Epidermal Cells.
Supplemental Table 1. Sequence Information Used for Plasmid Construction, RT-PCR, ChIP Analyses, and EMSA.
Acknowledgments
National Science Foundation Award MCB-0424850 and Binational Agricultural Research and Development Awards 103314 supported research in the laboratory of P.M.H. We thank Fernando Alemán for assistance with gene expression analysis and Hua Weng for reviewing this article.
References
- Ahmed F.E., Abusam S.M.A., Ahmed E.E.A. (2010). The bases of Blepharis sp. adaptation to water-limited environment. Asian J. Crop Sci. 2: 12–19 [Google Scholar]
- Alon U. (2007). Network motifs: Theory and experimental approaches. Nat. Rev. Genet. 8: 450–461 [DOI] [PubMed] [Google Scholar]
- Bacon M.A. (2004). Water Use Efficiency in Plant Biology. (Oxford, UK: Blackwell Publishing; ). [Google Scholar]
- Baker S.S., Wilhem K.S., Thomashow M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought-, and ABA-regulated gene expression. Plant Mol. Biol. 24: 701–713 [DOI] [PubMed] [Google Scholar]
- Berger D., Altmann T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14: 1119–1131 [PMC free article] [PubMed] [Google Scholar]
- Bergmann D.C. (2004). Integrating signals in stomatal development. Curr. Opin. Plant Biol. 7: 26–32 [DOI] [PubMed] [Google Scholar]
- Bergmann D.C., Lukowitz W., Somerville C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Bergmann D.C., Sack F.D. (2007). Stomatal development. Annu. Rev. Plant Biol. 58: 163–181 [DOI] [PubMed] [Google Scholar]
- Blum A. (1996). Crop responses to drought and the interpretation of adaptation. Plant Growth Regul. 20: 135–148 [Google Scholar]
- Breuer C., Kawamura A., Ichikawa T., Tominaga-Wada R., Wada T., Kondou Y., Muto S., Matsui M., Sugimoto K. (2009). The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell 21: 2307–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer P.B., Howles P.A., Dorian K., Griffith M.E., Ishida T., Kaplan-Levy R.N., Kilinc A., Smyth D.R. (2004). PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 131: 4035–4045 [DOI] [PubMed] [Google Scholar]
- Büssis D., von Groll U., Fisahn J., Altman T. (2006). Stomatal aperture can compensate altered stomatal density in Arabidopsis thaliana at growth light conditions. Funct. Plant Biol. 33: 1037–1043 [DOI] [PubMed] [Google Scholar]
- Casson S., Gray J.E. (2008). Influence of environmental factors on stomatal development. New Phytol. 178: 9–23 [DOI] [PubMed] [Google Scholar]
- Casson S.A., Hetherington A.M. (2010). Environmental regulation of stomatal development. Curr. Opin. Plant Biol. 13: 90–95 [DOI] [PubMed] [Google Scholar]
- Chaerle L., Saibo N., Van Der Straeten D. (2005). Tuning the pores: Towards engineering plants for improved water use efficiency. Trends Biotechnol. 23: 308–315 [DOI] [PubMed] [Google Scholar]
- Chaves M.M., Maroco J.P., Pereira J.S. (2003). Understanding plant responses to drought – From genes to the whole plant. Funct. Plant Biol. 30: 239–264 [DOI] [PubMed] [Google Scholar]
- Dehesh K., Bruce W.B., Quail P.H. (1990). A trans-acting factor that binds to a GT-motif in a phytochrome gene promoter. Science 250: 1397–1399 [DOI] [PubMed] [Google Scholar]
- Dong J., Bergmann D.C. (2010). Stomatal patterning and development. Curr. Top. Dev. Biol. 91: 267–297 [DOI] [PubMed] [Google Scholar]
- Dong J., MacAlister C.A., Bergmann D.C. (2009). BASL controls asymmetric cell division in Arabidopsis. Cell 137: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L.M., Zhang W., Chen J.G., Taylor J.P., Jones A.M., Assmann S.M. (2008). Abscisic acid regulation of guard-cell K+ and anion channels in Gbeta- and RGS-deficient Arabidopsis lines. Proc. Natl. Acad. Sci. USA 105: 8476–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar G.D., Sharkey T. (1982). Stomatal conductance and photosynthesis. Annu. Rev. Plant Physiol. 33: 317–345 [Google Scholar]
- Green P.J., Kay S.A., Chua N.H. (1987). Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J. 6: 2543–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Kajita R., Torii K.U., Bergmann D.C., Kakimoto T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington A.M., Woodward F.I. (2003). The role of stomata in sensing and driving environmental change. Nature 424: 901–908 [DOI] [PubMed] [Google Scholar]
- Hu H., Boisson-Dernier A., Israelsson-Nordström M., Böhmer M., Xue S., Ries A., Godoski J., Kuhn J.M., Schroeder J.I. (2010). Carbonic anhydrases are upstream regulators of CO2-controlled stomatal movements in guard cells. Nat. Cell Biol. 12: 87–93, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.Y., Chao D.Y., Gao J.P., Zhu M.Z., Shi M., Lin H.X. (2009). A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L., Bailey K.J., Gray J.E. (2010). The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol. 186: 609–614 [DOI] [PubMed] [Google Scholar]
- Hunt L., Gray J.E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19: 864–869 [DOI] [PubMed] [Google Scholar]
- Impa S.M., Nadaradjan S., Boominathan P., Shashidhar G., Bindumadhava H., Sheshshayee M.S. (2005). Carbon isotope discrimination accurately reflects variability in WUE measured at a whole plant level in rice. Crop Sci. 45: 2517–2522 [Google Scholar]
- Jarvis P.G., McNaughton K.G. (1986). Stomatal control of transpiration: Scaling up from leaf to region. Adv. Ecol. Res. 15: 1–49 [Google Scholar]
- Jin J.B., et al. (2008). The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 53: 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka M.M., Pillitteri L.J., Fujii H., Yoshida Y., Bogenschutz N.L., Takabayashi J., Zhu J.K., Torii K.U. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi A., Urabe I., Kaneko K., Yomo T. (2006). Adaptive response of a gene network to environmental changes by fitness-induced attractor selection. PLoS ONE 1: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.I., et al. (2007). yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 145: 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Böhmer M., Hu H., Nishimura N., Schroeder J.I. (2010). Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R.M., Caspar T., Dehesh K., Quail P.H. (1993). DNA binding factor GT-2 from Arabidopsis. Plant Mol. Biol. 23: 337–348 [DOI] [PubMed] [Google Scholar]
- Lake J.A., Quick W.P., Beerling D.J., Woodward F.I. (2001). Plant development. Signals from mature to new leaves. Nature 411: 154. [DOI] [PubMed] [Google Scholar]
- Lambers H., Chapin F.S., Pons T.L. (1998). Plant Physiological Ecology. (New York: Springer; ). [Google Scholar]
- Lampard G.R., Macalister C.A., Bergmann D.C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Li S., Assmann S.M., Albert R. (2006). Predicting essential components of signal transduction networks: A dynamic model of guard cell abscisic acid signaling. PLoS Biol. 4: e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister C.A., Ohashi-Ito K., Bergmann D.C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540 [DOI] [PubMed] [Google Scholar]
- Masle J., Gilmore S.R., Farquhar G.D. (2005). The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436: 866–870 [DOI] [PubMed] [Google Scholar]
- Morison J.I.L., Baker N.R., Mullineaux P.M., Davies W.J. (2008). Improving water use in crop production. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363: 639–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Dehesh K., Tepperman J.M., Quail P.H. (1996). GT-2: In vivo transcriptional activation activity and definition of novel twin DNA binding domains with reciprocal target sequence selectivity. Plant Cell 8: 1041–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson S.E., Assmann S.M. (2007). The control of transpiration. Insights from Arabidopsis. Plant Physiol. 143: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson S.E., Assmann S.M. (2010). The α-subunit of the Arabidopsis heterotrimeric G protein, GPA1, is a regulator of transpiration efficiency. Plant Physiol. 152: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobel P.S. (1999). Physicochemical & Environmental Plant Physiology. (San Diego, CA: Academic Press; ). [Google Scholar]
- Ohashi-Ito K., Bergmann D.C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera I.Y., Hung C.Y., Moore C.D., Stevenson-Paulik J., Boss W.F. (2008). Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 20: 2876–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505 [DOI] [PubMed] [Google Scholar]
- Richards E., Reichardt M., Rogers S. (2001). Preparation of genomic DNA from plant tissue. Current Protocols in Molecular Biology, Vol. 1, Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K., (New York: John Wiley and Sons; ), pp. 2.3.1–2.3.7 [DOI] [PubMed] [Google Scholar]
- Saez A., Robert N., Maktabi M.H., Schroeder J.I., Serrano R., Rodriguez P.L. (2006). Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 141: 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E.D., McAbee J.M., Pillitteri L.J., Torii K.U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Sinclair T.R., Tanner C.B., Bennett J.M. (1984). Water use efficiency in crop production. Bioscience 34: 36–40 [Google Scholar]
- Song X.J., Matsuoka M. (2009). Bar the windows: An optimized strategy to survive drought and salt adversities. Genes Dev. 23: 1709–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S.S., Shimada T., Imai Y., Okawa K., Tamai A., Mori M., Hara-Nishimura I. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463: 241–244 [DOI] [PubMed] [Google Scholar]
- Thattai M., van Oudenaarden A. (2001). Intrinsic noise in gene regulatory networks. Proc. Natl. Acad. Sci. USA 98: 8614–8619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayakumar M., Sheshshayee M.S., Nataraj K.N., Bindumadhava H., Devendra R., Aftab Hussain I.S., Prasad T.G. (1998). Why breeding for water use efficiency has not been successful? An analysis and alternate approach to exploit this trait for crop improvement. Curr. Sci. 74: 994–1000 [Google Scholar]
- Van den Boogaard B., Alewijnse D., Veneklaas E.J., Lambers H. (1997). Growth and water use efficiency of 10 Triticum aestivum cultivars at different water availability in relation to allocation of biomass. Plant Cell Environ. 20: 200–210 [Google Scholar]
- von Groll U., Berger D., Altmann T. (2002). The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14: 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z.M., Zou H.F., Lei G., Wei W., Zhou Q.Y., Niu C.F., Liao Y., Tian A.G., Ma B., Zhang W.K., Zhang J.S., Chen S.Y. (2009). Soybean Trihelix transcription factors GmGT-2A and GmGT-2B improve plant tolerance to abiotic stresses in transgenic Arabidopsis. PLoS ONE 4: e6898. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xu Z., Zhou G. (2008). Responses of leaf stomatal density to water status and its relationship with photosynthesis in a grass. J. Exp. Bot. 59: 3317–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C.Y., Pence H.E., Hasegawa P.M., Mickelbart M.V. (2009). Regulation of transpiration to improve crop water use. Crit. Rev. Plant Sci. 28: 410–431 [Google Scholar]
- Yu H., Chen X., Hong Y.Y., Wang Y., Xu P., Ke S.D., Liu H.Y., Zhu J.K., Oliver D.J., Xiang C.B. (2008). Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zhou D.X. (1999). Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci. 4: 210–214 [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]