Interactive proteomics technology was used to identify Arabidopsis nucleoporins, which are components of the nuclear pore complex (NPC). Nucleoporin domain organization is similar in plants, vertebrates, and yeast. This finding suggests that most NPC structures are conserved throughout eukaryotes.
Abstract
The nuclear pore complex (NPC) facilitates nucleocytoplasmic transport, a crucial process for various cellular activities. The NPC comprises ~30 nucleoporins and is well characterized in vertebrates and yeast. However, only eight plant nucleoporins have been identified, and little information is available about the complete molecular structure of plant NPCs. In this study, an interactive proteomic approach was used to identify Arabidopsis thaliana nucleoporins. A series of five cycles of interactive proteomic analysis was performed using green fluorescent protein (GFP)-tagged nucleoporins. The identified nucleoporins were then cloned and subcellular localization analyses were performed. We found that the plant NPC contains at least 30 nucleoporins, 22 of which had not been previously annotated. Surprisingly, plant nucleoporins shared a similar domain organization to their vertebrate (human) and yeast (Saccharomyces cerevisiae) counterparts. Moreover, the plant nucleoporins exhibited higher sequence homology to vertebrate nucleoporins than to yeast nucleoporins. Plant NPCs lacked seven components (NUCLEOPORIN358 [Nup358], Nup188, Nup153, Nup45, Nup37, NUCLEAR DIVISION CYCLE1, and PORE MEMBRANE PROTEIN OF 121 kD) that were present in vertebrate NPCs. However, plants possessed a nucleoporin, Nup136/Nup1, that contained Phe-Gly repeats, and sequence analysis failed to identify a vertebrate homolog for this protein. Interestingly, Nup136-GFP showed greater mobility on the nuclear envelope than did other nucleoporins, and a Nup136/Nup1 deficiency caused various defects in plant development. These findings provide valuable new information about plant NPC structure and function.
INTRODUCTION
The nucleus is the most important organelle in the eukaryotic cell. It contains the cell’s genetic material and directs cellular activity by regulating gene expression. The nuclear pore complex (NPC) is found on the nuclear envelope. This structure represents the sole gateway to the nucleus, mediating the traffic of proteins and RNAs between the cytoplasm and nucleoplasm (Xu and Meier, 2008; Meier and Brkljacic, 2009a). The nucleocytoplasmic transport of macromolecular cargo is crucial for cellular function. This process depends on both the recognition of the NPC and interaction with the NPC. The NPC is the largest multiprotein complex in the cell and comprises multiple copies of ~30 different proteins called nucleoporins (Rout et al., 2000; Cronshaw et al., 2002). The molecular masses of mammal and yeast NPCs are estimated to be 125 and 50 MD, respectively. Ultrastructural analysis of the NPC reveals that its basic architecture is conserved among vertebrates (Goldberg and Allen, 1996), yeast (Allen and Douglas, 1989), and plants (Roberts and Northcote, 1970; Fiserova et al., 2009). Vertebrate NPC subunits form a doughnut-shaped channel with an eightfold radial symmetry that can be divided into three parts: a nuclear basket, a central pore, and cytoplasmic filaments. In addition, the NPC has been shown to undergo conformational changes during the development of tobacco (Nicotiana tabacum) BY-2 cells (Fiserova et al., 2009).
The NPC is well characterized in vertebrates and yeast, and their nucleoporins share structural motifs and specific sequences. Transmembrane domains are found in a small number of nucleoporins. In vertebrate cells, GLYCOPROTEIN OF 210 kD (gp210), PORE MEMBRANE PROTEIN OF 121 kD (Pom121), and NUCLEAR DIVISION CYCLE1 (NDC1) have been identified as integral pore membrane proteins (Cronshaw et al., 2002). They appear to function as a scaffold for the NPC and, following mitosis, are involved in reconstructing the nuclear pore in the newly formed nuclear envelope (Güttinger et al., 2009). At least a third of the total NPC mass is comprised of nucleoporins that contain a functionally significant Phe-Gly (FG) repeat domain. This domain binds directly to transport receptors and thereby mediates active transport through the NPC (Ryan and Wente, 2000). FG repeats also play a role in NPC permeability, preventing nonkaryophilic proteins larger than 40 kD from entering (or exiting) the nucleus by simple diffusion (Patel et al., 2007). Some nucleoporins are members of the Trp-Asp (WD) repeat family. These repeats form a β-propeller structure that is thought to be important for the assembly of large multiprotein complexes (Smith et al., 1999). In the NPC, WD repeats appear to mediate the assembly of NPC scaffold subdomains (Rabut et al., 2004) and facilitate interactions between transport complexes and the NPC (Cronshaw et al., 2002).
NPCs are stable throughout interphase (Daigle et al., 2001) but disassemble into subcomplexes during mitosis. At the end of mitosis, each subcomplex is sequentially recruited to the nuclear periphery, in association with nuclear envelope reformation (Bodoor et al., 1999). In vertebrate NPCs, three subcomplexes have been described (reviewed in Tran and Wente, 2006): the Nup62 subcomplex (Nup62, Nup58, Nup54, and Nup45); the Nup107-160 subcomplex (Nup160, Nup133, Nup107, Nup96, Nup75, Nup43, Nup37, Seh1, Sec13, and ALADIN); and the Nup93 subcomplex (Nup205, Nup188, Nup155, Nup93, and Nup35). Each subcomplex performs specific functions.
Currently, detailed information about NPC molecular structure is only available for a limited number of vertebrates and yeasts that belong to the Opisthokonta group. The NPCs of plants, which belong to the Archaeplastida, have been largely neglected; however, a deeper understanding of plant NPC structure is important for developing a broader perspective of NPC origin and evolution. Sequence similarity searches have failed to identify plant homologs for about half of the nucleoporins identified in metazoan NPCs (Xu and Meier, 2008). To date, only eight nucleoporins (Nup160, Nup133, Nup96, Nup88, Nup75, Tpr/NUA, Nup1, and RAE1) have been identified and characterized experimentally in plants, including Arabidopsis thaliana, Lotus japonicus, and tobacco (Nicotiana tabacum) (Meier and Brkljacic, 2009a, 2009b). These nucleoporins have been shown to be involved in different events in the plant life cycle, including flowering, pathogen interaction, nodulation, cold response, and hormone signaling (Meier and Brkljacic, 2009a, 2009b). Among the eight plant nucleoporins identified, four (i.e., Nup160, Nup133, Nup96, and Nup75) correspond to components of the evolutionarily conserved Nup107-160 subcomplex, which functions as a structural scaffold in vertebrate NPCs (Harel et al., 2003).
To determine NPC composition, fractionation procedures have been designed to enrich nucleoporins from the nuclei of yeast (Rout et al., 2000) and vertebrates (Cronshaw et al., 2002). Intact NPCs have been purified biochemically via sequential solubilization of nuclear substructures. Individual nucleoporins were then separated by combining high-performance liquid chromatography and SDS-PAGE, followed by identification with mass spectrometry. Although this is a comprehensive strategy, it requires a large investment of time and effort to isolate sufficient quantities of high-quality NPCs.
In this study, a green fluorescent protein (GFP) tag was used to visualize nucleoporins in living cells and immunoaffinity purification enabled a detailed investigation of nucleoporin–nucleoporin interactions. Since this approach allows rapid and efficient isolation of complexes, it substantially reduces the time required for analysis and represents a proven cost-saving strategy compared with the classical complex purification procedures described for yeast (Rout et al., 2000) and vertebrates (Cronshaw et al., 2002). Here, we describe the use of mass spectrometry–based interactive proteomics for the comprehensive identification of Arabidopsis NPC components. The plant-specific nucleoporin Nup136 was also characterized. Our results suggest that the basic NPC structure is conserved in plants, yeast, and vertebrates, although each organism uses specialized nucleoporins with functions specific to their own NPCs. These findings contribute to the broader understanding of nuclear development and evolution in eukaryotes.
RESULTS
Identification and Characterization of 30 Arabidopsis Nucleoporins Using Interactive Proteomics
The nucleoporin RAE1 (RNA export factor 1) is encoded by a wide range of plant genomes and is known to function in mRNA export and spindle assembly in tobacco (Lee et al., 2009) and vertebrates (Blower et al., 2005). Transgenic Arabidopsis plants expressing GFP-tagged Arabidopsis RAE1 were generated to analyze the NPC in living cells. Imaging indicated that RAE1-GFP localized predominantly to the nuclear envelope in root tip cells (Figure 1A). The nuclear surface exhibited dot-like fluorescent signals (Figure 1B), which were similar to the NPC fluorescent signals obtained from other eukaryotes (Rabut et al., 2004). These results suggest that RAE1-GFP localizes to the NPC.
Figure 1.
Arabidopsis Nucleoporins That Coimmunoprecipitate with RAE1-GFP in Transgenic Arabidopsis Plants.
(A) Root tip cells from transgenic Arabidopsis plants stably expressing RAE1-GFP. The images were obtained by confocal laser scanning microscopy. Bar = 20 μm.
(B) The nuclear surface of a root cell from transgenic Arabidopsis plants stably expressing RAE1-GFP. The image was obtained by confocal laser scanning microscopy. Bar = 10 μm.
(C) and (D) Silver staining (C) and immunoblot analysis (D) of lysates from wild-type Arabidopsis (wt) and transgenic (RAE1-GFP) plants immunoprecipitated with anti-GFP antibody. Immunoprecipitates were separated by SDS-PAGE followed by silver staining (C) or immunoblot analysis with an anti-GFP antibody (D). Asterisks indicate nonspecific signals.
(E) Identification of Arabidopsis nucleoporins by mass spectrometry. Extracts from transgenic Arabidopsis plants expressing RAE1-GFP were immunoprecipitated with anti-GFP antibody and then subjected to LTQ-Orbitrap mass spectrometry. Arabidopsis Genome Initiative (AGI) codes and annotations were obtained from the TAIR database (http://www.Arabidopsis.org). Scores were calculated by Mascot (Matrix Science). We assigned a name to each nucleoporin candidate identified in this study.
Anti-GFP antibody was used to immunoprecipitate detergent-solubilized fractions from wild-type and transgenic RAE1-GFP plants. Immunoprecipitates were separated by SDS-PAGE, followed by either silver staining (Figure 1C) or immunoblot analysis with anti-GFP antibody (Figure 1D). A single band on the immunoblot confirmed the presence of intact RAE1-GFP in the immunoprecipitate (Figure 1D). Silver staining indicated that the RAE1-GFP sample contained numerous bands that were not present in the wild-type sample, suggesting that the immunoprecipitate contained many components of the NPC.
To identify the proteins present in the immunoprecipitate, mass spectrometry analysis was performed using an LTQ-Orbitrap for in-gel and in-solution digestions. A total of 200 proteins were identified in the RAE1-GFP sample, each containing one or more of the peptides detected in the mass spectrometry analysis. However, none of these proteins were present in the wild-type immunoprecipitate (see Supplemental Data Set 1 online). The 200 Arabidopsis proteins were compared with databases containing metazoan nucleoporin sequences, and 25 proteins were identified as candidate nucleoporins based on sequence similarity. These were named according to their size and similarity to human nucleoporins (Figure 1E).
To determine whether the 24 nucleoporin candidates localize to the nuclear envelope, cDNAs and/or genes encoding these proteins were cloned. The gene encoding Nup214 was incorrectly registered as two independent genes (At1g55540 and At1g55545) in the Arabidopsis genome database (http://www.Arabidopsis.org/). Constructs encoding GFP-nucleoporin fusion proteins were prepared for 17 of the 24 nucleoporins, and these expression constructs were transiently expressed in protoplasts derived from cultured Arabidopsis cells (see Supplemental Figure 1 online). In addition, constructs encoding 13 GFP-tagged nucleoporins were generated and then expressed stably in transgenic Arabidopsis lines (Figure 2). With the exception of Sec13-GFP, all of the stably expressed nucleoporins localized specifically to the nuclear envelope (Figure 2). Sec13-GFP was found on the nuclear envelope and in dot-like structures within the cytoplasm (Figure 2). In addition to nuclear envelope localization, most transiently expressed nucleoporins were distributed within the cytoplasm and/or nucleoplasm (see Supplemental Figure 1 online). This distribution pattern is consistent with that of endogenous nucleoporins in cultured animal cells (Bodoor et al., 1999; Guan et al., 2000; Griffis et al., 2002). The localization of transiently expressed Nup133-GFP differed from that of the other nucleoporins, as it was not detected on the nuclear envelope, but rather in the nucleoplasm (see Supplemental Figure 1 online). However, this unexpected localization pattern might represent an artifact caused by the GFP tag. The anti-GFP antibody coimmunoprecipitated endogenous Nup133 and GFP-tagged nucleoporins (RAE1-GFP, Nup93a-GFP, and Nup43-GFP) in the detergent-solubilized fraction (Figure 1E; see below and Supplemental Figure 2C and Supplemental Data Sets 1 to 3 online). In Arabidopsis Nup133, amino acids 651 to 1087 show 39% similarity to the corresponding region of human Nup133, which suggests that Nup133 is an NPC component on the nuclear envelope.
Figure 2.
Subcellular Localization of Stably Expressed GFP-Tagged Nucleoporins in Arabidopsis.
GFP fusion constructs were generated for 13 nucleoporins identified after anti-GFP antibody immunoprecipitation of a detergent-solubilized fraction from RAE1-GFP plants (Figure 1D). The constructs were then stably expressed in Arabidopsis plants. Images of epidermal root tip cells were all obtained at the same magnification using confocal laser scanning microscopy. Bar = 20 μm.
In mass spectrometry analysis, Nup93a exhibited the highest Mascot score, which indicates that, among the identified candidates, Nup93a is most likely to interact with RAE1 (Figure 1E). Extracts prepared from a transgenic plant stably expressing Nup93a-GFP were immunoprecipitated and then subjected to a second mass spectrometric analysis (see Supplemental Data Set 2 and Supplemental Table 1 online). Nup93a-GFP coimmunoprecipitated with 11 nucleoporins (see Supplemental Table 1 online), 10 of which were identified in the immunoprecipitate of plants expressing RAE1-GFP (Figure 1E). The remaining nucleoporin was identified as At1g52380, for which amino acids 231 to 378 show 46% similarity to the corresponding region of human Nup50. Arabidopsis was found to produce another homolog, At3g15970, which exhibits 59% sequence similarity to At1g52380. A RanBP1 domain is found in both of these Arabidopsis proteins (see Figure 4) and in human Nup50 (Lindsay et al., 2002). Therefore, At1g52380 and At3g15970 were designated as Nup50a and Nup50b, respectively.
Figure 4.
Schematic Representation of the Predicted Domain Features of Arabidopsis Nucleoporins.
The 30 Arabidopsis nucleoporins separate into four groups: (1) FG-bearing nucleoporins (FG-Nups); (2) Nup107-160 subcomplex proteins, corresponding to the outer ring structure of the NPC; (3) Nup93 subcomplex proteins, corresponding to the inner ring structure of the NPC; and (4) proteins with other functions. The scale at the top indicates the number of amino acids. A single orange vertical line in FG-Nups indicates the position of an FG repeat. The Max scores and E-values indicate sequence homologies with human (Homo sapiens) and yeast (Saccharomyces cerevisiae) counterparts. Different names for S. cerevisiae homologs are shown in parentheses. No homologs of Nup43, ALADIN, gp210, and Elys/HOS1 were identified in S. cerevisiae. Procedures outlining domain prediction are described in Methods, and details of the domains are shown in Supplemental Table 2 online. AGI, Arabidopsis Genome Initiative.
To determine the localization of Nup50a and Nup50b, GFP fusion constructs of these proteins were stably and transiently expressed in Arabidopsis. Nup50a-GFP and Nup50b-GFP localized to the nucleoplasm, rather than to the nuclear envelope (Figures 3A and 3B), a finding consistent with Nup50 localization in other eukaryotes (Lindsay et al., 2002). Following immunoprecipitation of extract from transgenic plants stably expressing Nup50a-GFP, we performed a third mass spectrometric analysis. Nup50a-GFP coimmunoprecipitated with one nucleoporin (Nup50b) and seven nucleocytoplasmic transport factors (six importins and one RAS-RELATED NUCLEAR [Ran] protein) (Figure 3C; see Supplemental Data Set 4 online). This result suggests that Nup50a is involved in nucleocytoplasmic transport via its interactions with importins and Ran, rather than by forming part of the NPC scaffolding.
Figure 3.
Identification of Arabidopsis Nup50a and Nup50b.
(A) and (B) Fluorescence images of root tip cells from transgenic plants stably expressing Nup50a-GFP (left panel) or Nup50b-GFP (right panel) (A). Fluorescence images of protoplasts transiently expressing Nup50a-GFP (left panel) or Nup50b-GFP (right panel) (B). Images were obtained by confocal laser scanning microscopy (GFP) and differential interference contrast (DIC) microscopy. Fluorescence–differential interference contrast overlays are shown. Bars = 20 μm.
(C) Mass spectrometry identification of Arabidopsis nucleoporins and nucleocytoplasmic transport factors that interact with Nup50a-GFP. Immunoprecipitates from transgenic Arabidopsis plants expressing Nup50a-GFP were subjected to LTQ-Orbitrap mass spectrometry. Arabidopsis Genome Initiative (AGI) codes and annotations were obtained from the TAIR database (http://www.Arabidopsis.org). Scores were calculated by Mascot. Assigned names represent nucleoporins identified and named in this study.
The Arabidopsis genome database was queried with human nucleoporin sequences and a specific motif for the nucleoporins identified as Nup43 and Nup35, which were not detected in the coimmunoprecipitation analysis of GFP-tagged nucleoporins. Nup43-GFP localized to the nuclear envelope in both transgenic plants (see Supplemental Figure 2A online) and protoplasts (see Supplemental Figure 2B online). A fourth mass spectrometric analysis showed that Nup43-GFP coimmunoprecipitated with 13 nucleoporins (see Supplemental Figure 2C and Supplemental Data Set 3 online), indicating that Nup43 is a component of the plant NPC. Arabidopsis Nup35 has a molecular mass of 35.4 kD and contains a mitotic phosphoprotein N' end domain (Table 1), which is also found in human Nup35. Neither Nup35-GFP nor GFP-Nup35 was expressed successfully in Arabidopsis cells, suggesting that the GFP-tagged Nup35 may be unstable in vivo.
Table 1.
Summary of Arabidopsis Nucleoporins Identified
| Name | AGI Code | Reference | Annotation | Bait |
| CGI | At1g75340 | This study | Zinc finger (CCCH-type) family protein | RAE1 |
| Nup214 | AAD10642*1 | This study | RAE1 | |
| Nup98a | At1g10390 | This study | Nucleoporin family protein | RAE1, Nup93a |
| Nup98b | At1g59660 | This study | Nucleoporin family protein | RAE1 |
| Nup62 | At2g45000 | This study | Expressed protein | RAE1, Nup93a, Nup43 |
| Nup58 | At4g37130 | This study | Hyp-rich glycoprotein family protein | RAE1 |
| Nup54 | At1g24310 | This study | Expressed protein | RAE1 |
| Nup136 | At3g10650 | (*2) | Expressed protein | RAE1 |
| Nup50a | At1g52380 | This study | Ran binding protein 1 domain-containing protein | Nup93a |
| Nup50b | At3g15970 | This study | Ran binding protein 1 domain-containing protein | |
| Nup160 | At1g33410 | (*3, *4) | Expressed protein | RAE1, Nup43 |
| Nup133 | At2g05120 | (*5) | Expressed protein | RAE1, Nup93a, Nup43 |
| Nup107 | At3g14120 | This study | Expressed protein | RAE1 |
| Nup96 | At1g80680 | (*4) | Nucleoporin family protein | |
| Nup75 | At4g32910 | (*6) | Expressed protein | RAE1, Nup43 |
| Nup43 | At4g30840 | This study | WD-40 repeat protein family | Nup43 |
| Seh1 | At1g64350 | This study | Transducin family protein/WD-40 repeat family protein (SEH1H) | RAE1, Nup93a, Nup43 |
| Sec13 | At2g30050 | This study | Transducin family protein/WD-40 repeat family protein | RAE1, Nup43 |
| Nup205 | At5g51200 | This study | Expressed protein | RAE1, Nup43 |
| Nup155 | At1g14850 | This study | Nonrepetitive/WGA-negative nucleoporin family protein (NUP155) | RAE1, Nup93a, Nup43 |
| Nup93a | At2g41620 | This study | Nucleoporin-interacting component family protein | RAE1, Nup93a, Nup43 |
| Nup93b | At3g57350 | This study | Nucleoporin-interacting component-related | RAE1, Nup93a |
| Nup35 | At3g16310 | This study | Mitotic phosphoprotein N' end (MPPN) family protein | |
| Nup88 | At5g05680 | (*7) | NPC protein-related | RAE1 |
| ALADIN | At3g56900 | This study | Aladin-related/adracalin-related | RAE1 |
| RAE1 | At1g80670 | (*8) | Transducin family protein/WD-40 repeat family protein | RAE1, Nup93a |
| gp210 | At5g40480 | This study | Expressed protein | RAE1, Nup93a, Nup43 |
| Eyls/HOS1 | At2g39810 | (*9) | HOS1 | RAE1, Nup43 |
| GLE1 | At1g13120 | This study | Expressed protein | RAE1 |
| Tpr/NUA | At1g79280 | (*10, *11) | Expressed protein | RAE1, Nup43 |
Arabidopsis Genome Initiative (AGI) codes and annotations are from the TAIR database (http://www.Arabidopsis.org), except for Nup214 (*1, accession number for GenBank). Bait indicates the GFP-tagged nucleoporins used in the immunoprecipitation experiments. References are as follows: *2, Lu et al. (2010); *3, Dong et al. (2006b); *4, Parry et al. (2006); *5, Kanamori et al. (2006); *6, Saito et al. (2007); *7, Cheng et al. (2009); *8, Lee et al. (2009); *9, Lee et al. (2001); *10, Jacob et al. (2007); *11, Xu et al. (2007b).
HOS1 (for HIGH EXPRESSION OF OSMOTICALLY RESPONSE GENE1) was immunoprecipitated with either RAE1-GFP (Figure 1E; see Supplemental Data Set 1 online) or Nup43-GFP (see Supplemental Data Set 3 online). HOS1, which functions as a negative regulator of cold signaling (Dong et al., 2006a), is distributed throughout the nucleus and cytosol of Arabidopsis (Lee et al., 2001). We found that HOS1 contains a region with homology to vertebrate nucleoporin EMBRYONIC LARGE MOLECULE DERIVED FROM YOLK SAC (Elys) (see Supplemental Figures 3A and 3B online), which is critical for recruiting the Nup107-160 complex to chromatin (Gillespie et al., 2007; Doucet et al., 2010). HOS1 homologs are found in the genome databases of other higher plants. However, it is interesting to note that these homologs are almost half the molecular size of vertebrate Elys proteins. Taken together, the results suggest that HOS1 may perform similar functions to Elys, acting as a component of plant NPCs.
Lamins are a major component of the nuclear laminae on the interior of the nuclear envelope in vertebrates. Although no lamin gene homologs have been detected in plant genomes (Meier, 2001), a filamentous lattice similar to vertebrate lamina was identified by ultrastructural analysis of the nuclear envelope of tobacco (Fiserova et al., 2009). Fiserova et al. (2009) reported that this filamentous structure was attached to the inner nuclear membrane and linked to the NPC (Fiserova et al., 2009), which indicates a physical interaction between these structures. Recently, LITTLE NUCLEI (LINC) proteins have been identified as potential lamin-like proteins in Arabidopsis (Dittmer et al., 2007). LINC proteins are predicted to form a large coiled-coil region similar to that observed in vertebrate lamin proteins, and they are involved in the maintenance of nuclear morphology (Dittmer et al., 2007). However, we were unable to identify LINC or filamentous proteins with a long coiled-coil region from our interactive proteomics studies using GFP-tagged nucleoporins (see Supplemental Data Sets 1 to 5 online). Our failure to detect these components could be due to technical limitations of the interactive proteomics method used.
In this study, eight known plant nucleoporins (Nup160, Nup133, Nup96, Nup88, Nup75, Tpr/NUA, Nup1, and RAE1) and 22 novel nucleoporins were isolated from Arabidopsis and characterized (Table 1; Meier and Brkljacic, 2009a). These Arabidopsis nucleoporins show homology to human nucleoporins. However, seven human NPCs (Nup358, Nup188, Nup153, Nup45, Nup37, NDC1, and Pom121) do not appear to have homologs in the genome databases of higher plants, including Arabidopsis, poplar (Populus trichocarpa), grape (Vitis vinifera), and rice (Oryza sativa). The absence of these nucleoporins might account for differences in size between plant and vertebrate NPCs (discussed later).
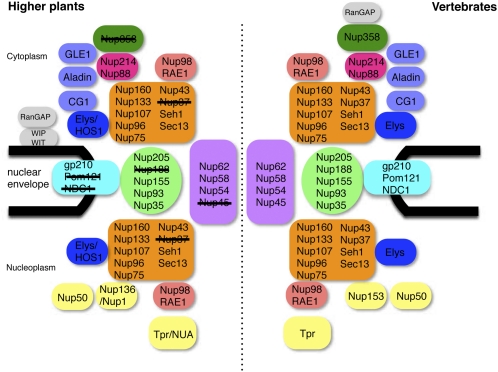
Higher sequence similarity scores and lower E-values were obtained for sequence homologies between Arabidopsis and human nucleoporins than between Arabidopsis and yeast nucleoporins (Figure 4). This finding suggests that the NPC structure of Arabidopsis is more similar to the human NPC structure than to the yeast structure. On the other hand, the domain organization of each nucleoporin was similar in Arabidopsis, human, and yeast (Figure 4; see Supplemental Table 2 online), which indicates a similar NPC architecture among these organisms (Figure 8).
Figure 8.
Comparison between the NPCs of Higher Plants and Vertebrates.
Subcomplexes are shown as single units. Plant nucleoporins that were not identified in this study are crossed out. WIT, WIP, and RanGAP were identified previously (Xu et al., 2007a; Zhao et al., 2008).
Nup136/Nup1 Shows Dynamic Behavior on the Nuclear Envelope and Is Required for Plant Development
The RAE1-GFP immunoprecipitation technique allowed identification of Nup136, a 136-kD nucleoporin with a sequence unique to higher plants (Figure 1E). In a recent study, yeast two-hybrid screening revealed that Nup136 interacts with the mRNA export complex and, in that study, Nup136 was named Arabidopsis Nup1 (Lu et al., 2010). Nup136/Nup1 homologs are present in various plants, but not in yeast or vertebrates, which suggests that Nup136/Nup1 is unique to higher plant species. Nup136/Nup1 has FG repeats in the C-terminal region (Figure 4, orange lines), but contains no other known functional motifs. The FG repeats mediate the translocation of transport factor-cargo complexes through the NPC by interacting with transport factors (Weis, 2003). The presence of such repeats in Nup136/Nup1 suggests that this protein may be required for active transport through the NPC. In transgenic Arabidopsis plants, Nup136-GFP localized to the nuclear envelope (Figure 2; see Supplemental Figure 1 online) and coimmunoprecipitated with three nucleoporins and four importin proteins (Figure 5A; see Supplemental Data Set 5 online). These results suggest that Nup136-GFP is incorporated into the NPC and that it interacts with importin proteins.
Figure 5.
Nup136/Nup1 Is a Nucleoporin with Dynamic Behavior on the Nuclear Envelope.
(A) Mass spectrometry identification of Arabidopsis nucleoporins and nucleocytoplasmic transport factors that interact with Nup136-GFP. Immunoprecipitates from transgenic Arabidopsis plants expressing Nup136-GFP were subjected to LTQ-Orbitrap mass spectrometry. Arabidopsis Genome Initiative (AGI) codes and annotations were obtained from the TAIR database (http://www.Arabidopsis.org). Scores were calculated by Mascot. Assigned names represent nucleoporins identified and named in this study.
(B) FRAP analysis of nucleoporin behavior on the nuclear envelope. Constructs encoding GFP fusions with gp210, Nup93a, Nup136, Nup54, Nup107, and Nup88 were stably expressed in tobacco BY-2 cells. An arrow indicates the photobleaching time point. Vertical bars represent sd (n ≥ 6).
(C) Cultured tobacco cells were cotransformed with constructs encoding Nup136-GFP and Histone2B-tdTomato to visualize the nucleoporin (green, top panels) and chromatin DNA (magenta, bottom panels), respectively. Living tobacco cultured cells were observed during cell division. The confocal time-lapse images (single planes) are shown from left to right, and the time is shown in seconds after breakdown of the nuclear envelope. Bar = 10 μm.
To determine if Nup136-GFP is mobile on the nuclear envelope, fluorescence recovery after photobleaching (FRAP) analysis was performed in transgenic tobacco BY-2 cells (Figure 5B). GFP-tagged nucleoporins from each subcomplex of the NPC were also selected and expressed as controls. After the nuclear envelopes of interphase cells were subjected to photobleaching, Nup136-GFP fluorescence recovered, whereas the fluorescence of other nucleoporins did not (Figure 5B). This finding suggests that, unlike other nucleoporins, Nup136 interacts dynamically with the NPC on the nuclear envelope.
The subcellular dynamics of Nup136-GFP were investigated in tobacco BY-2 cells to determine how Nup136 disassembles and reassembles during mitosis. Figure 5C shows confocal time-lapse microscopy of cells stably expressing Nup136-GFP together with Histone2B-tdTomato, a vital marker for chromatin DNA (Boisnard-Lorig et al., 2001). At interphase (−180 s), specific nuclear envelope labeling of Nup136-GFP was present. At metaphase (0 to ~460 s), Nup136-GFP fluorescence was dispersed throughout the cytosol, and no membranous structures were observed. At early anaphase (~780 s), Nup136-GFP–labeled particle-like structures were detected in the cytosol. Subsequently (~880 s), Nup136-GFP fluorescence was observed in membranous structures around each chromosome in daughter cells. By the end of telophase (~1060 s), Nup136-GFP fluorescence labeled the nuclear envelopes of each daughter cell, and cytoplasmic pools were almost entirely depleted. These results suggest that Nup136/Nup1 disassembles and disperses into the cytosolic pool during nuclear envelope breakdown and then reassembles as the nuclear envelope reforms in daughter cells.
To determine the physiological role of Arabidopsis Nup136, two T-DNA insertion mutant lines, nup136-1 and nup136-2, were isolated (Figure 6A). RT-PCR analysis revealed that no full-length NUP136 transcripts were present in the nup136-1 and nup136-2 mutants (Figure 6B). The nup136 mutants showed developmental defects. Although the plants grew normally before bolting, the nup136 mutants exhibited an early flowering phenotype (Figure 6C). Under long-day conditions, the mutants produced fewer rosette leaves (5.7 ± 0.8 for nup136-1 and 7.0 ± 1.1 for nup136-2) than wild-type plants (12.8 ± 0.6) at bolting (Figure 6D). The early flowering phenotype has been observed previously in Arabidopsis nucleoporin mutants, including nup96 (Parry et al., 2006), nup160 (Dong et al., 2006b; Parry et al., 2006), and tpr/nua (Jacob et al., 2007; Xu et al., 2007b). The tpr/nua mutant contains significantly reduced levels of several microRNAs (miR159, miR165, and miR393) that are important for regulating flowering time (Jacob et al., 2007). Taken together, these results suggest that nuclear pores are involved in the transport of unknown factors that might determine flowering time in Arabidopsis.
Figure 6.
Two Knockout Mutants, nup136-1 and nup136-2, Exhibit Various Defects in Plant Development.
(A) A schematic representation of the Nup136/Nup1 gene, which contains nine exons. The positions of T-DNA insertions in nup136-1 and nup136-2 are shown. Closed boxes represent exons and solid lines represent introns.
(B) RT-PCR analysis of NUP136/NUP1 and Actin2 (ACT2) transcripts in wild-type (WT), nup136-1, and nup136-2 cells. Full-length NUP136 transcripts were amplified by PCR. Amplification of NUP136/NUP1 and ACT2 required 30 and 27 PCR cycles, respectively. RT-PCR products were visualized with ethidium bromide. These data are representative of two biological replicates and two technical replicates.
(C) Four-week-old wild-type (wt), nup136-1, and nup136-2 plants.
(D) Flowering time was evaluated by the number of primary rosette leaves formed prior to flowering under long-day conditions. Data represent means ± sd (n ≥ 14).
(E) The two nup136/nup1 knockout mutants exhibited a defect in fruit maturation. Inset provides a magnified view of representative siliques of the same age.
(F) to (H) Scanning electron micrographs of pollen grains of wild-type (F), nup136-1 (G), and nup136-2 (H) plants. Arrows indicate the shrunken pollen grains of the mutants. Bar = 50 μm.
The nup136 mutants produced stunted fruits (Figure 6E, inset) with substantially fewer mature seed grains (5.5 ± 3.3 grains for nup136-1 and 6.9 ± 3.6 grains for nup136-2) than the wild type (32.1 ± 3.6 grains). Some nup136 pollen grains were shrunken (Figures 6F to 6H, arrows) and an in vitro pollen germination assay showed significantly lower germination rates for nup136 pollen (51.3% ± 10.0% for nup136-1 and 43.6% ± 16.7% for nup136-2) than the wild type (80.3% ± 6.3% for the wild type) (see Supplemental Figure 4 online). These results suggest that Nup136 is important for proper pollen development, which is required for seed production.
Transport receptors export mRNA from the nucleus, and these receptors interact specifically with nucleoporins in the NPC (Erkmann and Kutay, 2004). Arabidopsis nucleoporins, including Nup160 (Dong et al., 2006b) and Tpr/NUA (Xu et al., 2007b), are involved in mRNA export from the nucleus. To determine the functional role played by Nup136 in mRNA export, in situ hybridization analysis was performed with oligo(dT) probes to localize poly(A)+ RNA in wild-type Arabidopsis and nup136 mutants. A higher nuclear poly(A)+ RNA signal was detected in the leaf cells of nup136 mutants than in those of the wild type (see Supplemental Figure 5 online), which suggests that, in Arabidopsis, Nup136 is required for mRNA export through the NPC. This result is consistent with a previous study (Lu et al., 2010).
Arabidopsis Nup136 was also found to regulate nuclear morphology (Figure 7). In rosette leaf epidermal cells from the nup136-1 and nup136-2 mutants, the nuclei were more spherical and uniform than those in wild-type plants (Figures 7A), exhibiting a shorter major axis and a higher circularity index (Figures 7B and 7C). These results suggest that Nup136 is required for wild-type nuclear morphology.
Figure 7.
Effects of a NUP136/NUP1 Knockout on Nuclear Morphology.
(A) Nuclear morphology of rosette leaf epidermal cells stained with Hoechst 33342. Images were obtained by confocal laser scanning microscopy. Insets show magnified images of nuclei. Bars = 20 μm. Bars in insets = 10 μm. wt, wild type.
(B) and (C) The major axis length (B) and circularity index (C) of nuclei were measured in wild-type, nup136-1, and nup136-2 cells. Data represent means ± sd (n ≥ 63).
DISCUSSION
Structure of the NPC in Arabidopsis
A total of 30 Arabidopsis nucleoporins were identified in this study (Figure 8). Although almost all of these shared sequence similarity with vertebrate nucleoporins, a similar level of homology was not observed with yeast (Figure 4). This finding suggests that, in overall structure, the plant NPC is more similar to the vertebrate NPC than to the yeast NPC. In-lens field emission scanning microscopy (feSEM) analysis showed that the plant NPC (tobacco NPC = ~105 nm in diameter) (Fiserova et al., 2009) and the vertebrate NPC (Xenopus laevis NPC = 110 to ~120 nm in diameter) (Goldberg and Allen, 1996) are larger than the yeast NPC (~95 nm) (Kiseleva et al., 2004). No Arabidopsis homologs were found for seven of the vertebrate nucleoporins (Nup358, Nup188, Nup153, Nup45, Nup37, NDC1, and Pom121) (Table 1), and three of these proteins (Nup358, Nup45, and Pom121) are also absent in yeast. Thus, it is possible that Nup358, Nup45, and Pom121 represent vertebrate-specific nucleoporins and that they may be involved in determining species-specific differences in NPC function and complexity.
The vertebrate nucleoporin Nup358 is a component of NPC cytoplasmic filaments (Walther et al., 2002) and is involved in anchoring RanGAP (Ran GTPase-activating protein) to the nuclear envelope. RanGAP activates Ran GTPase and plays a crucial role in nucleocytoplasmic transport through the NPC (Clarke and Zhang, 2008). Our results and previous studies (reviewed in Xu and Meier, 2008) indicate that no ortholog of Nup358 is present in the genomes of higher plants (Table 1, Figure 8). Arabidopsis uses WIP (WPP-domain interacting protein) and WIT (WPP domain-interacting tail-anchored protein) to anchor RanGAP to the nuclear envelope, and immunolocalization analyses indicate that these proteins localize to the NPC (Xu et al., 2007a; Zhao et al., 2008) (Figure 8). Although our interactive proteomics technique did not detect WIP, WIT, or RanGAP (see Supplemental Data Set 1 to 5 online), it is possible that these proteins interact weakly with the NPC and that this weak interaction might contribute to the dynamic regulation of nucleocytoplasmic transport.
Alternatively, it is possible that WIP and WIT were not solubilized sufficiently by the detergent used in this study, since these proteins contain a single C-terminal membrane-spanning domain (Xu et al., 2007a; Zhao et al., 2008). Such technical limitations may have resulted in a failure to identify some low-solubility and/or low-affinity nucleoporins and interacting proteins.
Nup188 is a large protein in the Nup93 subcomplex, which is found in the inner ring of the NPC (Figures 4 and 8). Previous bioinformatics studies identified a putative Nup188 in Arabidopsis (At4g38760; Neumann et al., 2006); however, with a molecular mass of 74 kD, this potential homolog is significantly smaller than Nup188 in vertebrates (196 kD in human) or yeast (188 kD in Saccharomyces cerevisiae). Our interactive proteomics study was unable to identify At4g38760 or any other candidate homologs for Nup188.
Since three of the Arabidopsis nucleoporins (gp210, ALADIN, and Nup43) identified in this study have orthologs in the vertebrate NPC (Table 1) and no orthologs in the yeast NPC, it is possible that these nucleoporins perform functions specific to higher eukaryotes. Gp210 is an integral membrane protein that anchors the NPC to the nuclear envelope (Galy et al., 2008). Nup43 and ALADIN belong to the WD repeat protein family, and these repeats mediate the assembly of large protein complexes (Smith et al., 1999). WD repeats may be involved in the assembly of NPC subdomains or facilitate interactions between the NPC and transport complexes. In this study, three other WD repeat–containing nucleoporins (RAE1, Seh1, and Sec13) were identified in the Arabidopsis NPC (Figures 4 and 8). Although the function of ALADIN remains unknown, a human disease called triple-A syndrome is associated with several deletions and point mutations in this protein (Tullio-Pelet et al., 2000). However, since an ALADIN knockout produced a different phenotype in mice (Huebner et al., 2006), it is possible that the function of ALADIN varies with species. Arabidopsis GFP-tagged ALADIN localized to the nuclear envelope (see Supplemental Figure 1 online) and interacted with other nucleoporins (Figure 1E; see Supplemental Table 1 online). Further characterization of Arabidopsis ALADIN could provide valuable insight into the physiological role played by this protein in plants.
This analysis of candidate NPC-associated proteins (see Supplemental Data Sets 1 to 5 online) will contribute to a better understanding of the mechanisms underlying nucleocytoplasmic transport, which is a vital process in the plant life cycle.
Is Nup136 a Functional Homolog of Vertebrate Nup153?
Nup136/Nup1 is unique to higher plants, and it has three characteristics in common with vertebrate Nup153, for which no homolog has been identified in higher plants. Nup136/Nup1 and Nup153 contain FG repeats in their C-terminal regions (Figure 4) (Enarson et al., 1998; Partridge and Schwartz, 2009), and both proteins exhibit dynamic behavior on the nuclear envelope (Figure 5B) (Griffis et al., 2004; Rabut et al., 2004). Moreover, Nup136/Nup1 binds to importin proteins (importin-α and importin-β) (Figure 5A; see Supplemental Data Set 5 online), and vertebrate Nup153 has the ability to bind to importin-α in nuclei (Kodiha et al., 2008). Given these similarities, we propose that Arabidopsis Nup136 represents a functional homolog of vertebrate Nup153. However, plant Nup136/Nup1 does not contain two of the conserved functional domains in Nup153 (Nup153 domain and zinc-finger motif) (Enarson et al., 1998; Partridge and Schwartz, 2009); thus, it is possible that these domains perform a unique role in vertebrates.
In this study, we investigated how Nup136/Nup1 functions in the plant NPC. Nup136/Nup1 deficiency led to the alteration of nuclear morphology in the epidermal cells of rosette leaves (Figure 7). Nuclear morphology is rigidly maintained and regulated by several factors, including the cytoskeleton, nuclear skeleton, and nuclear lamina (Webster et al., 2009). Our results suggest that Nup136/Nup1 is involved in the structural maintenance of the nucleus via an association with these factors. Alternatively, it is possible that the abnormal nuclear morphology is caused by defects in nucleocytoplasmic transport of mRNAs and/or nuclear components, such as LINCs, since nup136 mutants exhibited defective mRNA export (see Supplemental Figure 5 online).
FRAP analysis revealed a dynamic association between Nup136 and the NPC, which was not observed between the NPC and any of the other nucleoporins (Figure 5B). In mammalian cells, peripheral NPC components exhibit more dynamic behavior than central components (Rabut et al., 2004). Since Nup136-GFP showed high levels of mobility on the nuclear envelope, it is likely that Nup136/Nup1 functions as an adaptor and/or regulator molecule in the periphery of the NPC rather than as part of a central structural scaffold.
Knockout of Arabidopsis NUP136/NUP1 led to an abnormal accumulation of poly(A)+ RNA in the nuclei (see Supplemental Figure 5 online), which suggests that Nup136/Nup1 may mediate nucleocytoplasmic transport in a similar manner to Nup153 in vertebrates (Kodiha et al., 2008). With respect to its physiological role in a multicellular organism, Nup136/Nup1 was also shown to be important for the regulation of flowering and fertilization (Figure 6; see Supplemental Figure 4 online). Based on these results, Nup136/Nup1 appears to regulate mRNA export and/or turnover in the NPC, and these functions are necessary for plant development. In the future, it will be important to clarify the molecular connections between mRNA export and the developmental defects observed in these nup136 mutants.
Following nuclear envelope breakdown during mitosis, Nup136-GFP localized to the cytosolic pool, and it did not associate with any membranous structures (Figure 5C). This result contrasts with a previous study, in which GFP-tagged human lamin B-receptor, a marker for the nuclear envelope, localized to the endoplasmic reticulum following nuclear envelope breakdown (Irons et al., 2003). This discrepancy could be due to the fact that the lamin B-receptor is an integral membrane protein, while Nup136 is a soluble protein. Our findings suggest that Nup136/Nup1 is released from the membrane in metaphase cells and then recruited to the newly forming nuclear envelope in daughter cells.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (ecotype Columbia) was used as the wild type. Protoplasts were prepared from cultured Arabidopsis cells that had been subcultured and were incubated in medium containing Murashige and Skoog salts and 0.4 M mannitol, as described previously (Tamura et al., 2003). Tobacco (Nicotiana tabacum) BY-2 cells were cultured as described previously (Mitsuhashi et al., 2000). T-DNA insertion mutants (SALK_104728 [nup136-1] and SAIL_796_H02 [nup136-2]) were obtained from the ABRC at Ohio State University.
Bioinformatics of the Arabidopsis Nucleoporins
The data sets of the Arabidopsis nucleoporins were queried against The Arabidopsis Information Resource (TAIR) database (http://www.Arabidopsis.org), ATTED-II (http://atted.jp/), or GenomeNet (http://www.genome.jp/) to obtain annotations, functional assignments, structural information, and sequence relationships. Domain architectures were predicted using a domain analysis tool on the EMBL-EBI website (InterProScan; http://www.ebi.ac.uk/Tools/InterProScan/), except that the transmembrane region of gp210 was predicted with the tool in the TMHMM server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/); SMC_prok_A and SMC_prok_B of Tpr/NUA were listed in the TIGR Rice Genome Annotation Resource, and RING-type zinc finger of Elys/HOS1 was listed in UniProt.
Transient and Stable Expression of GFP Fusions in Arabidopsis
Genomic DNAs and cDNAs of all 30 Arabidopsis nucleoporins identified were cloned into either pENTR/D-TOPO or pENTR1A (Invitrogen) using specific primers (see Supplemental Table 3 online). To generate the DNA constructs for GFP-tagged nucleoporins, each of the cloned nucleoporin DNAs was transferred from the entry clone to the destination vector pGWB405 (Nakagawa et al., 2007a, 2007b) by an LR reaction. Histone 2B (At5g22880) cDNA was amplified by PCR using gene-specific primers (see Supplemental Table 3 online) and was then cloned into the entry vector pENTR/D-TOPO (Invitrogen). To generate the DNA construct for Histone 2B-tdTomato, the cloned cDNA was transferred from the entry vector to destination vector pGWtd by an LR reaction.
Protoplasts derived from cultured Arabidopsis cells were transiently transformed with constructs encoding GFP-tagged nucleoporins using the polyethylene glycol method (Tamura et al., 2003). Arabidopsis plants and tobacco BY-2 cells were transformed by infection with Agrobacterium tumefaciens. The cells were inspected by confocal laser scanning microscopy and differential interference contrast microscopy (model LSM510 META; Carl Zeiss).
RT-PCR Analysis
Total RNA was isolated from 10-d-old seedlings using a RNeasy Plant Mini kit (Qiagen). Reverse transcription was performed using a kit (Ready-To-Go RT-PCR beads; GE Healthcare) with an oligo(dT)12-18 primer. Gene-specific primers are given in Supplemental Table 3 online. ACT2 was amplified in 27 PCR cycles and NUP136 in 30. PCR products were visualized with ethidium bromide.
In Vitro Pollen Germination Assay
An in vitro pollen germination assay was performed as described previously (Boavida and McCormick, 2007). Pollen grains were inspected by light microscopy 20 h after germination.
Confocal Laser Scanning Microscopy
Fluorescence confocal images were obtained using a laser scanning microscope (Zeiss LSM510 META; Carl Zeiss) equipped with the 405-nm line of a blue diode laser, 488-nm line of a 40-mW Ar/Kr laser, or the 544-nm line of a 1-mW He/Ne laser and a ×100 1.45–numerical aperture (NA) oil immersion objective (alpha Plan-Fluar, 000000-1084-514; Carl Zeiss), ×63 1.2-NA water immersion objective (C-Apochromat, 441777-9970-000; Carl Zeiss), or ×40 0.95-NA dry objective (Plan-Apochromat, 440654-9902-000; Carl Zeiss). Image analysis was performed using LSM image examiner software (Carl Zeiss). The data were exported as 8-bit TIFF files and processed using either Adobe Photoshop Elements 4.0 (Adobe Systems) or ImageJ 1.40g (National Institutes of Health).
FRAP Experiment
FRAP experiments were performed with a confocal laser scanning microscope (LSM510; Carl Zeiss) using a ×40 0.95-NA dry objective (Plan-Apochromat, 440654-9902-000; Carl Zeiss) and a completely open pinhole. Fluorescence on the particular region (22 × 22 pixels) of nuclear envelope was bleached with full laser power. After photobleaching, images were acquired for 790 s at 30-s intervals.
SDS-PAGE and Immunoblot Analysis
Protein extracts from seedlings and isolated nuclei were subjected to SDS-PAGE followed by either Coomassie Brilliant Blue staining or immunoblot analysis. Immunoreactive signals were detected with the ECL detection system (GE Healthcare) using anti-GFP antibody (JL-8; Clontech) (1:3000). Antibodies were diluted with Solution 1 of an immunostaining kit (Can Get Signal Immunoreaction Enhancer Solution; Toyobo). Immunoreactive signals were detected with the ECL detection system (GE Healthcare).
Immunoprecipitation
Immunoprecipitation was performed with μMACS Epitope Tag Protein Isolation Kits (Miltenyi Biotec). Whole seedlings of each transgenic Arabidopsis plant (~0.5 g) expressing GFP-tagged nucleoporins were homogenized on ice in 2 mL of buffer containing 50 mM HEPES-KOH, pH 7.5, 0.15 M NaCl, 0.5% (v/v) Triton X-100, and 0.1% (v/v) Tween 20. Homogenates were centrifuged at 10,000g for 15 min at 4°C to remove cellular debris. The supernatants were mixed with magnetic beads conjugated to an anti-GFP antibody (Miltenyi Biotec) and then incubated on ice for 10 min. The mixtures were applied to μ Columns (Miltenyi Biotec) in a magnetic field to capture the magnetic antigen-antibody complex. After extensive washing with the buffer, immunoaffinity complexes were eluted with 50 μL of either 0.1 M Na2CO3 solution, pH 11.0, or 2× SDS sample buffer containing 100 mM Tris-HCl, pH 6.8, 4% (w/v) SDS, 20% (w/v) glycerol, and 5% (v/v) 2-mercaptoethanol. Fractions eluted with 0.1 M Na2CO3 were subsequently neutralized with 5 μL of 1 M MES.
Peptide Preparation for Tandem Mass Spectrometry Analysis
The immunoprecipitates were reduced-alkylated and then treated with 0.01 mg/mL trypsin (sequence grade; Promega) in 50 mM ammonium bicarbonate and incubated at 37°C for 16 h. The digested peptides were recovered twice with 20 μL of 5% (v/v) formic acid in 50% (v/v) acetonitrile. The extracted peptides were combined and then evaporated to 10 μL in a vacuum concentrator.
For in-gel digestion, the protein components of the immunoprecipitates were separated on a 3-cm-long SDS gel. The gel slice isolated from each lane was cut into three fractions according to molecular mass: a <50-kD fraction, a 50- to 100-kD fraction, and a >100-kD fraction. Each excised gel fraction was treated twice with 25 mM ammonium bicarbonate in 30% (v/v) acetonitrile for 10 min followed by 100% (v/v) acetonitrile for 15 min, and then dried in a vacuum concentrator. The dried gel was treated with 0.01 mg/mL trypsin in 50 mM ammonium bicarbonate and incubated at 37°C for 16 h. The digested peptides were recovered twice with 20 μL of 5% (v/v) formic acid in 50% (v/v) acetonitrile. The extracted peptides were combined and then evaporated to 10 μL in a vacuum concentrator.
Mass Spectrometric Analysis and Database Search
Liquid chromatography–tandem mass spectrometry (MS/MS) analyses were performed using the LTQ-Orbitrap XL-HTC-PAL system. Trypsin digests were loaded on the column (75-μm internal diameter, 15-cm length; L-Column, CERI) using the Paradigm MS4 HPLC pump (Michrom BioResources) and HTC-PAL Autosampler (CTC Analytics), and were eluted by a gradient of 5 to 45% (v/v) acetonitrile in 0.1% (v/v) formic acid for 70 min. The eluted peptides were introduced directly into an LTQ-Orbitrap XL mass spectrometer (Thermo) with a flow rate of 300 nL/min and a spray voltage of 2.0 kV. The range of MS scan was m/z 450 to 1500. The top three peaks were subjected to MS/MS analysis. MS/MS spectra were analyzed by the Mascot server (version 2.2) in house (Perkins et al., 1999) (http://www.matrixscience.com/) and compared against proteins registered in TAIR8. The Mascot search parameters were set as follows: threshold of the ion score cutoff, 0.05; peptide tolerance, 10 ppm; MS/MS tolerance, ±0.8 D; and peptide charge, 2+ or 3+. The search was also set to allow one missed cleavage by trypsin, a carboxymethylation modification of Cys residues, and a variable oxidation modification of Met residues.
In Situ Hybridization
The poly (A)+ RNA in situ hybridization was performed as previously described (Gong et al., 2005). The 45-mer oligo(dT) labeled with one fluorescein molecule at the 5′-end (Hokkaido system science, Hokkaido, Japan) was used as a probe. The stained cells were inspected with a confocal laser scanning microscope (Carl Zeiss).
Hoechst Staining
The nuclei in rosette leaves from 2-week-old plants were stained for 30 min with 1 μg/mL Hoechst 33342 solution, 3.7% (w/v) paraformaldehyde, 10% (v/v) DMSO, 3% (v/v) Nonidet P-40, 50 mM PIPES-KOH, pH 7.0, 1 mM MgSO4, and 5 mM EGTA. The stained nuclei were inspected with a confocal laser scanning microscope by exciting with a 405-nm diode laser (Carl Zeiss).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the accession numbers shown in Table 1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Fluorescence Images of Protoplasts Derived from Cultured Cells Transiently Expressing GFP-Tagged Nucleoporins.
Supplemental Figure 2. Identification of Arabidopsis Nup43.
Supplemental Figure 3. Identification of Arabidopsis Elys/HOS1.
Supplemental Figure 4. The Pollen in nup136/nup1 Mutants Exhibits a Low Germination Rate.
Supplemental Figure 5. Abnormal Accumulation of Poly(A)+ RNA in the Nuclei of nup136 Mutants.
Supplemental Table 1. Eleven Nucleoporins Identified by Mass Spectrometry of Immunoprecipitates from Transgenic Arabidopsis Plants Expressing Nup93a-GFP.
Supplemental Table 2. Information for Domain Positions Used in Figure 4.
Supplemental Table 3. Primers Used in This Study.
Supplemental Data Set 1. Proteins Identified by Mass Spectrometry of Immunoprecipitates from RAE1-GFP Transgenic Plants.
Supplemental Data Set 2. Proteins Identified by Mass Spectrometry of Nup93a-GFP Immunoprecipitates.
Supplemental Data Set 3. Proteins Identified by Mass Spectrometry of Nup43-GFP Immunoprecipitates.
Supplemental Data Set 4. Proteins Identified by Mass Spectrometry of Nup50a-GFP Immunoprecipitates.
Supplemental Data Set 5. Proteins Identified by Mass Spectrometry of Nup136-GFP Immunoprecipitates.
Acknowledgments
We thank Iris Meier (Ohio State University) for a critical reading of the manuscript. This work was supported by a Kyoto University Startup Grant from Kyoto University to K.T., by Grants-in-Aid for Scientific Research (20570036, 21370094, 21770224, and 22000014), and by the Global Center of Excellence Program “Formation of a Strategic Base for Biodiversity and Evolutionary Research: from Genome to Ecosystem” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- Allen J.L., Douglas M.G. (1989). Organization of the nuclear pore complex in Saccharomyces cerevisiae. J. Ultrastruct. Mol. Struct. Res. 102: 95–108 [DOI] [PubMed] [Google Scholar]
- Blower M.D., Nachury M., Heald R., Weis K. (2005). A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 121: 223–234 [DOI] [PubMed] [Google Scholar]
- Boavida L.C., McCormick S. (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52: 570–582 [DOI] [PubMed] [Google Scholar]
- Bodoor K., Shaikh S., Salina D., Raharjo W.H., Bastos R., Lohka M., Burke B. (1999). Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112: 2253–2264 [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig C., Colon-Carmona A., Bauch M., Hodge S., Doerner P., Bancharel E., Dumas C., Haseloff J., Berger F. (2001). Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13: 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.T., Germain H., Wiermer M., Bi D., Xu F., García A.V., Wirthmueller L., Després C., Parker J.E., Zhang Y., Li X. (2009). Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21: 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P.R., Zhang C. (2008). Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 9: 464–477 [DOI] [PubMed] [Google Scholar]
- Cronshaw J.M., Krutchinsky A.N., Zhang W., Chait B.T., Matunis M.J. (2002). Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N., Beaudouin J., Hartnell L., Imreh G., Hallberg E., Lippincott-Schwartz J., Ellenberg J. (2001). Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer T.A., Stacey N.J., Sugimoto-Shirasu K., Richards E.J. (2007). LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell 19: 2793–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.-H., Agarwal M., Zhang Y., Xie Q., Zhu J.-K. (2006a). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.H., Hu X., Tang W., Zheng X., Kim Y.S., Lee B.H., Zhu J.K. (2006b). A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol. 26: 9533–9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet C.M., Talamas J.A., Hetzer M.W. (2010). Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell 141: 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enarson P., Enarson M., Bastos R., Burke B. (1998). Amino-terminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma 107: 228–236 [DOI] [PubMed] [Google Scholar]
- Erkmann J.A., Kutay U. (2004). Nuclear export of mRNA: From the site of transcription to the cytoplasm. Exp. Cell Res. 296: 12–20 [DOI] [PubMed] [Google Scholar]
- Fiserova J., Kiseleva E., Goldberg M.W. (2009). Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 59: 243–255 [DOI] [PubMed] [Google Scholar]
- Galy V., Antonin W., Jaedicke A., Sachse M., Santarella R., Haselmann U., Mattaj I. (2008). A role for gp210 in mitotic nuclear-envelope breakdown. J. Cell Sci. 121: 317–328 [DOI] [PubMed] [Google Scholar]
- Gillespie P.J., Khoudoli G.A., Stewart G., Swedlow J.R., Blow J.J. (2007). ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr. Biol. 17: 1657–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M.W., Allen T.D. (1996). The nuclear pore complex and lamina: Three-dimensional structures and interactions determined by field emission in-lens scanning electron microscopy. J. Mol. Biol. 257: 848–865 [DOI] [PubMed] [Google Scholar]
- Gong Z., Dong C.H., Lee H., Zhu J., Xiong L., Gong D., Stevenson B., Zhu J.K. (2005). A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E.R., Altan N., Lippincott-Schwartz J., Powers M.A. (2002). Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell 13: 1282–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E.R., Craige B., Dimaano C., Ullman K.S., Powers M.A. (2004). Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol. Biol. Cell 15: 1991–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T., Kehlenbach R.H., Schirmer E.C., Kehlenbach A., Fan F., Clurman B.E., Arnheim N., Gerace L. (2000). Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell. Biol. 20: 5619–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güttinger S., Laurell E., Kutay U. (2009). Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10: 178–191 [DOI] [PubMed] [Google Scholar]
- Harel A., Orjalo A.V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D.J. (2003). Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11: 853–864 [DOI] [PubMed] [Google Scholar]
- Huebner A., Mann P., Rohde E., Kaindl A.M., Witt M., Verkade P., Jakubiczka S., Menschikowski M., Stoltenburg-Didinger G., Koehler K. (2006). Mice lacking the nuclear pore complex protein ALADIN show female infertility but fail to develop a phenotype resembling human triple A syndrome. Mol. Cell. Biol. 26: 1879–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons S.L., Evans D.E., Brandizzi F. (2003). The first 238 amino acids of the human lamin B receptor are targeted to the nuclear envelope in plants. J. Exp. Bot. 54: 943–950 [DOI] [PubMed] [Google Scholar]
- Jacob Y., Mongkolsiriwatana C., Veley K.M., Kim S.Y., Michaels S.D. (2007). The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol. 144: 1383–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E., Allen T.D., Rutherford S., Bucci M., Wente S.R., Goldberg M.W. (2004). Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J. Struct. Biol. 145: 272–288 [DOI] [PubMed] [Google Scholar]
- Kodiha M., Tran D., Qian C., Morogan A., Presley J.F., Brown C.M., Stochaj U. (2008). Oxidative stress mislocalizes and retains transport factor importin-alpha and nucleoporins Nup153 and Nup88 in nuclei where they generate high molecular mass complexes. Biochim. Biophys. Acta 1783: 405–418 [DOI] [PubMed] [Google Scholar]
- Lee H., Xiong L., Gong Z., Ishitani M., Stevenson B., Zhu J.K. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 15: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Lee H.S., Wi S.J., Park K.Y., Schmit A.C., Pai H.S. (2009). Dual functions of Nicotiana benthamiana Rae1 in interphase and mitosis. Plant J. 59: 278–291 [DOI] [PubMed] [Google Scholar]
- Lindsay M.E., Plafker K., Smith A.E., Clurman B.E., Macara I.G. (2002). Npap60/Nup50 is a tri-stable switch that stimulates importin-alpha:beta-mediated nuclear protein import. Cell 110: 349–360 [DOI] [PubMed] [Google Scholar]
- Lu Q., Tang X., Tian G., Wang F., Liu K., Nguyen V., Kohalmi S.E., Keller W.A., Tsang E.W.T., Harada J.J., Rothstein S.J., Cui Y. (2010). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61: 259–270 [DOI] [PubMed] [Google Scholar]
- Meier I. (2001). The plant nuclear envelope. Cell. Mol. Life Sci. 58: 1774–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I., Brkljacic J. (2009a). The nuclear pore and plant development. Curr. Opin. Plant Biol. 12: 87–95 [DOI] [PubMed] [Google Scholar]
- Meier I., Brkljacic J. (2009b). Adding pieces to the puzzling plant nuclear envelope. Curr. Opin. Plant Biol. 12: 752–759 [DOI] [PubMed] [Google Scholar]
- Mitsuhashi N., Shimada T., Mano S., Nishimura M., Hara-Nishimura I. (2000). Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY-2 cells. Plant Cell Physiol. 41: 993–1001 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007a). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007b). Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Neumann N., Jeffares D.C., Poole A.M. (2006). Outsourcing the nucleus: Nuclear pore complex genes are no longer encoded in nucleomorph genomes. Evol. Bioinform. Online 2: 23–34 [PMC free article] [PubMed] [Google Scholar]
- Parry G., Ward S., Cernac A., Dharmasiri S., Estelle M. (2006). The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18: 1590–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge J.R., Schwartz T.U. (2009). Crystallographic and biochemical analysis of the Ran-binding zinc finger domain. J. Mol. Biol. 391: 375–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.S., Belmont B.J., Sante J.M., Rexach M.F. (2007). Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129: 83–96 [DOI] [PubMed] [Google Scholar]
- Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Rabut G., Doye V., Ellenberg J. (2004). Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6: 1114–1121 [DOI] [PubMed] [Google Scholar]
- Roberts K., Northcote D.H. (1970). Structure of the nuclear pore in higher plants. Nature 228: 385–386 [DOI] [PubMed] [Google Scholar]
- Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., Chait B.T. (2000). The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 148: 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.J., Wente S.R. (2000). The nuclear pore complex: A protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12: 361–371 [DOI] [PubMed] [Google Scholar]
- Saito K., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19: 610–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes C., Saxena K., Neer E.J. (1999). The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 24: 181–185 [DOI] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S.I., Watanabe M., Nishimura M., Hara-Nishimura I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35: 545–555 [DOI] [PubMed] [Google Scholar]
- Tran E.J., Wente S.R. (2006). Dynamic nuclear pore complexes: Life on the edge. Cell 125: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Tullio-Pelet A., et al. (2000). Mutant WD-repeat protein in triple-A syndrome. Nat. Genet. 26: 332–335 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Pickersgill H.S., Cordes V.C., Goldberg M.W., Allen T.D., Mattaj I.W., Fornerod M. (2002). The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 158: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M., Witkin K.L., Cohen-Fix O. (2009). Sizing up the nucleus: Nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 122: 1477–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. (2003). Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112: 441–451 [DOI] [PubMed] [Google Scholar]
- Xu X.M., Meier I. (2008). The nuclear pore comes to the fore. Trends Plant Sci. 13: 20–27 [DOI] [PubMed] [Google Scholar]
- Xu X.M., Meulia T., Meier I. (2007a). Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr. Biol. 17: 1157–1163 [DOI] [PubMed] [Google Scholar]
- Xu X.M., Rose A., Muthuswamy S., Jeong S.Y., Venkatakrishnan S., Zhao Q., Meier I. (2007b). NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19: 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Brkljacic J., Meier I. (2008). Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 20: 1639–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]