Abstract
Our approach to protein-protein docking includes three main steps. First we run PIPER, a rigid body docking program based on the Fast Fourier Transform (FFT) correlation approach, extended to use pairwise interactions potentials. Next, the 1000 best energy conformations are clustered, and the 30 largest clusters are retained for refinement. Third, the stability of the clusters is analyzed by short Monte Carlo simulations, and the structures are refined by the medium-range optimization method SDU. The first two steps of this approach are implemented in the ClusPro 2.0 protein-protein docking server. Despite being fully automated, the last step is computationally too expensive to be included in the server. Comparing the models obtained in CAPRI rounds 13–19 by ClusPro, by the refinement of the ClusPro predictions, and by all predictor groups, we arrived at three conclusions. First, for the first time in the CAPRI history, our automated ClusPro server was able to compete with the best human predictor groups. Second, selecting the top ranked models, our current protocol reliably generates high quality structures of protein-protein complexes from the structures of separately crystallized proteins, even in the absence of biological information, provided that there is limited backbone conformational change. Third, despite occasional successes, homology modeling requires further improvement to achieve reliable docking results.
INTRODUCTION
The goal of our research is a computationally effective docking protocol to generate high accuracy structures of protein-protein complexes from the structures of unbound component proteins, preferably without any human intervention. We use a multi-stage approach to docking, which first globally explores the energy landscape using a simplified energy model and limited flexibility to discover regions of interest, and then focuses on these sites using detailed scoring and sampling. Several steps of this algorithm have been substantially improved during the last few years. These include a novel Fast Fourier Transform (FFT) docking method with pairwise potentials implemented in PIPER,1 a novel method of generating reference states for molecular recognition potentials (Decoys as the Reference State or DARS),2 a clustering technique for the detection of near native conformations,3 a method of eliminating some of the non-native clusters by analyzing the stability of funnels in the free energy landscape,4 and a new medium-range optimization method using Semi-Definite programming based Underestimation (SDU).5 In parallel with these methodological innovations we have been improving and developing our protein-protein docking server ClusPro. Since its introduction as the first public protein docking server in 2004, ClusPro has run over 30,000 jobs for more than 3,000 users. Structures generated by the server have been reported in over 100 research papers.
Here we briefly describe our methodology and discuss both the performance of our group and that of the ClusPro server in CAPRI rounds 13–19. We argue that we arrived at an important point: the top 10 models generated by the server included acceptable or better predictions for almost all targets that involved the docking of separately crystallized proteins (i.e., without the need for homology modeling) if the binding did not lead to substantial conformational change. We note that in CAPRI servers can use only the atomic coordinates of the component proteins, and no literature or other data that are available to human predictors. Nevertheless, ClusPro outperformed not only the other 9 participating servers, but also 53 of the 63 participating predictor groups. Using stability analysis and the SDU method we were able to refine the ClusPro models to achieve medium or high accuracy. Learning from our previous mistakes, for most targets we avoided the use of any biological information. Arguably the six targets we have correctly predicted were relatively easy, but we think that the results nevertheless show substantial progress relative to our performance in the previous rounds. Our predictions were much less satisfying for targets that involved the docking of homology models, indicating that substantial improvement of both modeling and docking is required to achieve similar levels of reliability and accuracy in this more difficult case.
METHODS
Our multi-stage approach to protein docking
The first step of our approach is a global soft rigid body search using PIPER,1 a Fast Fourier Transform (FFT) based docking program. The major advantage of PIPER is the inclusion of arbitrary pairwise interaction potentials. Such structure-based (or knowledge-based) potentials are commonly used for ranking docked conformations,6–12 but their use directly for docking is novel. A pairwise interaction potential is usually represented using a KxK matrix where K is number of different atom or residue types. In order to evaluate the function effectively, we perform eigenvector decomposition of this matrix. Each of the eigenvectors can be evaluated as a single correlation, and we can limit ourselves to eigenvectors corresponding to significant eigenvalues. We demonstrated2 that the majority of existing potentials have only 4 significant eigenvalues, thus making PIPER’s approach extremely efficient.1 The top 1000 structures are retained from PIPER for further analysis. We note that PIPER is available free for academic users upon request.
The second step of the algorithm is clustering the retained structures using pairwise Root Mean Square Deviation (RMSD) as the distance measure.3 The biophysical meaning of clustering is isolating highly populated low energy basins of the energy landscape. Many studies, including ours, have demonstrated that clustering algorithms generally perform better for isolating near native structures as compared to selecting low energy conformations, if used in conjunction with exhaustive energy-based sampling such as in PIPER.
For refinement we use two different methods which enhance each other. The first is a medium-range optimization method called Semi-Definite programming based Underestimation (SDU). Taking advantage of the funnel-like behavior of the free energy, SDU constructs a convex quadratic underestimator based on a set of local energy minima. The tightest underestimator is obtained by solving a semi-definite programming problem, which is used to bias further sampling. The process is iterated with the set of local minima being updated, and the search region being reduced until convergence conditions are satisfied.5 Second, for each retained local minimum we investigate whether it is surrounded by a broad region of attraction. The analysis is similar to determining the stability of fixed points in dynamical systems. A fixed point is stable and it is an attractor if all trajectories started in a neighborhood of the fixed point remain in the neighborhood. In agreement with this analogy, we examine the “stability” of each local minimum by starting Monte Carlo simulations from random points around the cluster center in order to determine whether the trajectories converge to some small region within the region defined by the cluster. The cluster is considered stable if a strong attractor exists, it is close to the original cluster center, and contains several low energy structures.4 This results in many non-native clusters being discarded.
The ClusPro server
In parallel with our method development, we were the first group to provide the community with a protein-docking server. Since 2004, ClusPro has run more than 30,000 jobs for thousands of academic users. Due to computational limitations, ClusPro implements only the first two steps of our protocol, as refinement is still computationally expensive. ClusPro participated in CAPRI and was consistently one of the best servers, but its performance had been inferior to that of human predictors. At the end of the previous CAPRI assessment, we overhauled the server and released the beta version of ClusPro 2.0, which included some of the recent advances in our methodology starting with PIPER. In CAPRI rounds 13–19 we have used both versions of ClusPro, and based on the spectacular performance of ClusPro 2.0 we have moved it beyond the beta version phase. The new ClusPro 2.0 server is hosted at http://cluspro.bu.edu/. Currently the server has three different scoring schemes, the first for enzyme-inhibitor complexes, the second for antigen-antibody pairs, and the third is for other, primarily signal transduction, complexes. Although in the current version of ClusPro the scoring scheme for a particular complex is selected by the user, this step can be automated and we plan to do so in the future.
According to the usage pattern of ClusPro, one of the frequent applications is predicting the structure of oligomeric assemblies from the structures of single subunits. Indeed, the structures of these assemblies may be difficult to resolve experimentally. We have implemented an algorithm within the PIPER docking program to predict molecular assemblies of homo-oligomers. Given the number of subunits and the 3D structure of one monomer, the method samples all the possible N-mer symmetries. Based on a scoring function that clusters the low free energy structures at each binding interface, the algorithm predicts the complex structure as well as the symmetry of the protein assembly. The method is general and does not involve any free parameters. Using this application, we validated predictions for trimers, tetramers (discriminating between dimer of dimers and 4-fold symmetry structures), pentamers and hexamers (discriminating between trimer of dimers, dimer of trimers, and 6-fold symmetry structures). For complexes that involve more than one binding interface, the cluster size at each surface provides a strong indication as to which interface forms first. The method was fully implemented in the ClusPro server.
Ranking of the predictions
The ClusPro predictions were obtained within the first 24–48 hours after releasing a target. The solutions were clustered and ranked on the basis of cluster size. The centers of the 10 largest clusters were submitted as predictions without any further processing. For the manual predictions we have refined the clusters generated by ClusPro using stability analysis and optimization by SDU. We have discarded all clusters that were unstable or had high energy after the optimization. The remaining clusters were ranked based on their level of stability, SDU energy, cluster size, and in some cases the available biological information. As in the previous rounds, our attempts to incorporate additional information were not useful. In particular, for Target 39 a misleading publication prevented us from ranking the correct model highly, although it was the center of the largest cluster from ClusPro and was very stable. As will be discussed, apart from target T39 and from target T42 with some open questions, the scoring scheme enables us to find our best quality models as model M01, with models M02 frequently representing another good model.
Homology modeling
A number of CAPRI targets required the construction of homology models for docking (Table II). We use multiple approaches for homology modeling that have been developed by other groups. For targets with existing structures with high sequence similarity (¿40%), we use these as templates for Modeller13 to generate a number of models, which are then docked using our methodology. For targets without high sequence similarity structures, structure prediction services such as I-TASSER14 are used to generate structures.
Table II.
Performance on Complexes Involving Homology Modeling
| Target | Name | Type | Our group | Cluspro | Community | ||
|---|---|---|---|---|---|---|---|
| Score | Model | Score | Model | ||||
| T29 | Trm8/Trm82 | Homology/bound | - | - | - | - | 9**+8* |
| T33 | Methyltrans./RNA | Homology/unbound | - | - | - | - | - |
| T34 | Methyltrans./RNA | Homology/bound | 2** + 1* | M01+M02 | 1* | M05 | 25**+40* |
| T35 | Linked Modules | Homology/homology | - | - | - | - | 1* |
| T36 | Linked Modules | Homology/bound | - | - | - | - | 1* |
| T37 | Arf6/LZ2 | Unbound/homology | - | - | - | - | 1***+7**+13* |
| T38 | Centaurin-α1/FHA | Unbound/homology | - | - | - | - | - |
Acceptable (*), medium (**) and high (***) accuracy predictions.
RESULTS AND DISCUSSION
In order to separate docking performance from the potential difficulties due to low accuracy in homology modeling, we focus on targets that did not require homology modeling. As shown in Table I our methods work very well for these classical docking targets. In contrast, our performance is far less satisfactory for the targets that required the construction and docking of homology models (Table II). Since our approach needs substantial further improvement to obtain reliable predictions in these combined modeling and docking problems, the targets in Table II will not be discussed in detail. The only exception is target T34, which includes an RNA molecule as one of the components of the complex, and hence required the integration of RNA parameters into the ClusPro server.
Table I.
Performance on Unbound/Unbound and Unbound/Bound Complexes
| Target | Name | Type | Our group | Cluspro | Community | ||
|---|---|---|---|---|---|---|---|
| Score | Model | Score | Model | ||||
| T30 | Rnd1/Plexin B1 RBD | Unbound/unbound | almost | M03 | - | - | 2* |
| T32 | Savinase/BASI | Unbound/unbound | 2** + 1* | M01+M02 | almost | - | 15***+13**+7* |
| T39 | Centaurin-α1/FHA | Unbound/bound | 1*** | M07 | 1** | M10 | 1***+2** |
| T40 | Trypsin/api-a | Bound/unbound | |||||
| T40.CA | 2***+3** | M01 | 1** | M06 | 47***+33**+17* | ||
| T40.CB | 2***+1**+1* | M02 | 1* | M02 | 31***+21**+14* | ||
| T41 | Colicin E9/Im2 | Unbound/unbound | 2** | M01 | 1** | M01 | 24***+57**+64* |
| T42 | Designed TPR Dimer | Unbound/unbound | |||||
| AB result | 1*** | M09 | 1*** | M08 | 7***+4**+4* | ||
| BC result | - | - | - | - | 2***+1**+2* | ||
Acceptable (*), medium (**) and high (***) accuracy predictions.
Target T30
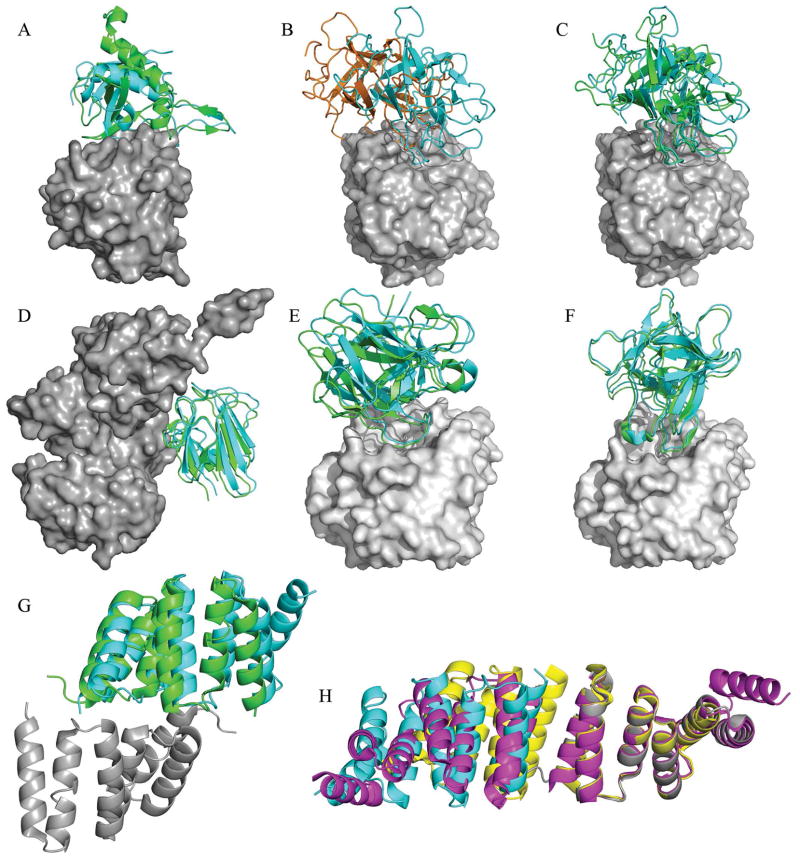
Target T30 is a complex between a Rho subfamily G-protein Rnd1, and the Rho-binding domain (RBD) of plexin B1. The binding induces substantial conformational changes in the interface such that the superimposition of unbound structures on the bound complex is discarded due to clashes. Neither ClusPro nor our group generated acceptable models, but we came close, and some of the models were in the correct region. In particular, model M08 by ClusPro had 14 Å ligand RMSD. The model was refined to 8.9 Å ligand RMSD and 5.4 Å backbone interface RMSD by SDU, and was submitted as model M03. This model would be acceptable if it met the condition for the native contacts, demonstrating the need for backbone refinement. As shown in Table I, the entire CAPRI community generated only 2 acceptable predictions for this clearly difficult target. Our model M03, together with the crystal structure, is shown in Figure 1A.
Figure 1.
(A) Rnd1/RBD of plexin B1 (target T30). Refined prediction of the Plexin-B1 RBD domain (cyan), superimposed on the bound structure of RBD (green). The bound receptor, Rnd1, is shown in gray (PDB code 2REX). (B) Savinase - BASI complex (target T32). Comparison of the ClusPro starting model M05 (brown) and the top refined model M01 (cyan). The refinement results in a shift of the interacting loop of the ligand along the receptor cavity (gray). (C) Savinase - BASI complex (target T32). The high quality prediction M01 (cyan) is very close to the position of the bound ligand (green) in the x-ray structure of the complex (PDB code 3BX1). The receptor surface is shown in gray. (D) Centaurin-α1/KF13B FHA complex (target T39). Refined prediction of FHA (cyan), superimposed on the bound structure of FHA (green). The receptor, Centaurin-α1, is shown in gray (PDB code 3FM8). (E) Bovine trypsin - double arrowhead protease inhibitor complex (target T40), interaction mode CA. We compare our high quality prediction, model M01 (cyan), to the experimental ligand position shown in green (pdb code 3E8L). The Lys145 loop of the ligand is involved in the interface. Trypsin is shown as gray surface. (F) Bovine trypsin - double arrowhead protease inhibitor complex (target T40), interaction mode CB. We compare our high quality prediction, model M02 (cyan), to the experimental ligand position shown in green (pdb code 3E8L). The Leu87 loop of the ligand is involved in the interface. Trypsin is shown as gray surface. (G) Designed TPR trimer (target T42). ClusPro model M08 of one subunit (cyan), superimposed on the bound dimer subunits shown in green and gray (PDB code 2WQH). (H) Designed TPR trimer (Target T42). ClusPro model M01 (cyan), and symmetry mates in 1NA0 (magenta) and 1NA3 (cyan) relative to bound receptor (gray).
Target T32
Target T32 is a complex between the protease savinase (pdb code 1SVN) and the bifunctional barley alpha-amylase/subtilisin inhibitor BASI (pdb code 1AVA, chain C). ClusPro produced an almost acceptable model as M05 with the backbone interface RMSD of less than 5 Å, providing a good starting point for refinement. ¿From this model refinement by stability analysis generated two high quality models (M01 and M02), and refinement by SDU provided one medium prediction (M03). The difference between the ClusPro model M05 and a high quality refined model M01 is mostly due to a translational shift as illustrated in Figure 1B. Our best refined model (M01) has a ligand RMSD of 2.1 Å and a backbone interface RMSD of 0.5 Å Its closeness to an X-ray complex structure (pdb code 3BX1) is shown in Figure 1C. As indicated in Table I, target T32 was relatively easy, with many good predictions by the CAPRI community. Nevertheless, it is remarkable that the stability analysis and SDU methods were able to refine a near-acceptable model to high and medium quality, respectively, and then to correctly rank the best models.
Target T34: Methyltransferase/RNA complex
Target 34 involved a homology model of a methyltransferase from , and hence results are shown in Table II. The interacting partner was a triple hairpin structure of rRNA given in bound form. To model RNA we added all-hydrogen nucleic acids topology and CHARMM 27 parameters to the ClusPro 2.0 parameter set. Although sequence alignment was given, alternative alignments were possible. We relied on the provided alignment and used the Modeller program to generate solutions for the ClusPro server prediction stage. The server was able to generate an acceptable model (model M05). In the manual refinement stage we produced another acceptable model (model M08) using the same alignment. We also explored an alternative homology model, which together with SDU refinement gave a pair of medium predictions (models M01 and M02). Notice that we again correctly ranked the best models. As shown in Table II, for this target there were many medium quality solutions, but no high accuracy ones.
Target T39
In target T39 unbound Centaurin-α1 (pdb 3FEH) was docked to a bound FHA domain. In this case our group provided two of the three correct predictions submitted by the entire CAPRI community, with our manual submission providing the only high quality model. ClusPro was able to provide a medium quality model as the 10th ranked result for this protein. Stability analysis showed this cluster to be extremely stable, and it was refined to the high quality model shown in Figure 1D. However, since this structure was not compatible with experimental information indicating that the binding occurred in the GAP domain between residues 1–126, the model was submitted as M07, ranked worse than some other models that agreed better with the reported experiments. This clearly illustrates the danger of relying on experimental information. In fact, without misleading data, our high accuracy model would have been submitted as model M01 on the basis of cluster size and stability.
Target T40: Trypsin/api-a
This target focuses on the interaction between bovine trypsin (pdb 1BTY) and a double arrowhead protease inhibitor (bound). The ClusPro server identified the canonical trypsin electrostatic interface CA around Lys145 of the ligand as a medium quality model (model M06), and also the second interface CB around Leu87 of the ligand) as an acceptable model (model M02). Subsequent refinement at the manual stage yielded two high quality predictions, one from SDU and another from stability analysis (models M01 and M03) and three medium quality predictions (models M05, M07, and M09) for the interface CA, and two high quality (models M02 and M04), one medium (model M08), and one acceptable (model M06) predictions for the interface CB. Figure 1E shows the proximity of the refined model M01 to the experimental CA interface (pdb code 3E8L). Figure 1F illustrates the closeness of the high quality model M02 to the X-ray structure of the trypsin-double arrowhead complex (pdb code 3E8L). Notice that out of the 10 submitted models from refinement, only one (model M10) was incorrect, and that models M01 and M02 again were our best submissions.
Target T41: Colicin E9/Im2
This target seemed to be easy due to previously solved structures of colicin E9 with Im9, a protein with high homology and sequence identity to the Im2 component of the complex. We used the crystal structure of colicin E9 (PDB code 1BXI) as the receptor. It was a challenge to choose a ligand structure as the NMR structure file of the unbound Im2 (PDB code 2NO8) contained 60 separate conformers, and our rigid body docking in PIPER can only use one conformer for docking. Two methods were used for choosing conformers, first by the RMSD to the bound Im9, and the second by the “openness” of the binding site, a criterion based on previous observations made when docking NMR models. Using the Im2 conformer with the most open pocket, as the best prediction ClusPro returned a model which retained the interface observed in the E9/Im9 complex, indicating that we are very likely in the right region of the conformational space. Therefore our ClusPro submissions were based on this conformer, resulting in the top ranked model M01 being medium quality with 2.6 Å ligand RMSD and 1.6 Å backbone interface RMSD to the crystal structure (pdb 2WPT15). Refinement using stability analysis improved the results to 2.3 Å ligand RMSD and 1.2 Å backbone interface RMSD, which was again submitted as model M01. A second medium quality model M09 was obtained by SDU.
Target T42: Designed Dimer/Trimer
The stated problem was to find a dimer interface for this artificial protein containing three tetratricopeptide repeats. The repeat was highly similar to the one used in previous constructs (PDB codes 1NA0 and 1NA3). We used the 1NA0 structure for the docking, with an Asp residue mutated to Tyr in each repeat. The docking involved the multimer option of ClusPro as described in the methods. The crystal structure of the dimer (pdb 2WQH16) showed two different interfaces, referred to as dimer A and dimer B, and it was reported that in solution the protein forms large oligomers. ClusPro directly provided a high quality result for dimer A as model M08, which was also submitted unmodified as model M09 by our predictor group. This model, shown in Figure 1G, has 2.6 Å ligand RMSD and 0.9 Å backbone interface RMSD. Of far more interest to our group was a different interface we have predicted and submitted as our model M01, shown in Figure 1H. This structure was the top model obtained by ClusPro not only for target T42, but also for both 1NA0 and 1NA3, and it showed high level of stability in our analysis. In addition, this interface is actually seen in the dimer unit cell for both 1NA0 and 1NA3. Thus, we speculate that the protein would use these interactions to form the oligomers seen in solution. However, since this interface was not seen in the X-ray structure, at this point there is no evidence supporting the validity of our top prediction.
DISCUSSION
As described by the report of the evaluators in this issue, our group correctly predicted the structures for six targets in rounds 13–19 of CAPRI. We note, however, that seven other groups also made six successful predictions, and that we distinguished ourselves only by the slightly higher number of high or medium accuracy models. However, we believe that together with the performance of the ClusPro automated protein-protein docking server our results represent a certain breakthrough. By definition, servers could use only the atomic coordinates of two component proteins, and no literature or other data that were available to human predictors. Nevertheless, with five correct predictions, four of which were at least medium accuracy, ClusPro outperformed not only the other 9 servers, but also 53 of the 63 participating groups. This was for the first time in the history of CAPRI that an automated server was able to compete with human predictors.
The second improvement is in reliability. As shown in Table I, ClusPro provided at least acceptable prediction for almost all unbound/unbound or unbound/bound targets for which correct predictions were submitted by several groups. The only exception was target T32 for which the ClusPro prediction was not acceptable, but was good enough to be refined to high accuracy. Achieving this level of reliability appears to be an important change: in previous rounds we have always missed some of the targets that were not particularly difficult for other groups. More generally, it was largely assumed that each method works best on certain types of problems and may be less optimal for others. Here ClusPro correctly predicted all targets that were also predicted by other groups. The best solutions provided by ClusPro were further refined using SDU and stability analysis. As with ClusPro, we generally avoided the use of additional information, or at least kept the best models that our methods predicted. Due to this strategy ClusPro was one of the two correct predictors for target T39 on which incorrect experimental data was published, misleading most of the groups. The model was then refined to high accuracy, which was the only such model for this target.
The third important improvement is in the scoring scheme. As described in the methods, ClusPro clusters the low energy structures based on pairwise RMSD, and returns the centers of the largest clusters as top predictions. The clusters are then refined by SDU and subjected to stability analysis. For submission we selected the large clusters that also showed high level of stability (i.e., a large fraction of Monte Carlo trajectories stayed within the cluster), and after refinement included a number of relatively low energy structures. As shown in Table I, this strategy selected the best models as M01 for four (or five if we consider the two interfaces in T40) of the six targets we have correctly predicted. We followed the same selection strategy also for target T39, but due to the misleading information we have submitted the model, considered the best, as M07. For target T42 our best prediction was submitted as M09, but we believe that our model M01 is relevant for forming the oligomers reported in solution. The best predictions were also models M01 and M02 for Target T34 (Table II). Thus, we believe that combining the three decision criteria we made substantial progress toward reliable ranking of the predictions, at least for the relatively easy targets.
Our automated docking method predicted correct structures for almost all unbound/unbound and unbound/bound targets that had only moderate conformational change upon binding. For over 60% of the targets the ClusPro models were refined to high quality prediction, and in almost all other cases we obtained medium quality structures. Moreover, this level of accuracy was largely achieved without the use of biological information. Although the present web-based version of ClusPro does not perform the computationally very demanding refinement steps, it will be included when appropriate computational resources will be available. Thus, our results indicate the feasibility of high accuracy automated docking for a substantial fraction of proteins.
As shown in Table II, however, the server, our group, and even the entire CAPRI community failed to obtain similarly good predictions for the majority of targets that required homology modeling. It is clear that extending reliable docking to homology models requires substantial method development. Traditional homology modeling focuses on the backbone conformation and possibly on the packing of side chains in the core, but not much emphasis is given to correctly predicting surface side chains such as lysines and arginines that usually are not even well defined in the x-ray structure. However, the position of these side chains is frequently crucial for docking. Thus, we need both homology modeling tools that focus more on surface loops and side chains, and adaptive docking tools in which the search strategy is optimized depending on the accuracy of the protein structures being docked.
Acknowledgments
This work has been supported by grants GM61867 and GM93147 from the National Institute of Health. For the CPU time used for this paper we are grateful to grant MRI DBI-0116574 and to the Boston University Scientific Computing and Visualization Center for the opportunity of running the PIPER program on the Blue Gene/L supercomputer.
References
- 1.Kozakov D, Brenke R, Comeau SR, Vajda S. Piper: An fft-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 2.Chuang G-Y, Kozakov D, Brenke R, Comeau SR, Vajda S. Dars (decoys as the reference state) potentials for protein-protein docking. Biophys J. 2008 Nov;95(9):4217–27. doi: 10.1529/biophysj.108.135814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozakov D, Clodfelter K, Vajda S, Camacho C. Optimal clustering for detecting near-native conformations in protein docking. Biophys J. 2005;89:867–875. doi: 10.1529/biophysj.104.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozakov D, Schueler-Furman O, Vajda S. Discrimination of near-native structures in protein-protein docking by testing the stability of local minima. Proteins. 2008 Aug;72(3):993–1004. doi: 10.1002/prot.21997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen Y, Paschalidis IC, Vakili P, Vajda S. Protein docking by the underestimation of free energy funnels in the space of encounter complexes. PLoS Comput Biol. 2008 Oct;4(10):e1000191. doi: 10.1371/journal.pcbi.1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camacho C, Gatchell D, Kimura S, Vajda S. Scoring docked conformations generated by rigid-body protein-protein docking. Proteins. 2000;40:525–537. doi: 10.1002/1097-0134(20000815)40:3<525::aid-prot190>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 7.Li L, Cheng R, Weng Z. RDOCK: Refinement of rigid-body protein docking predictions. Proteins. 2003;53:693–707. doi: 10.1002/prot.10460. [DOI] [PubMed] [Google Scholar]
- 8.Comeau S, Gatchell D, Vajda S, Camacho C. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 9.Moont G, Gabb H, Sternberg M. Use of pair potentials across protein interfaces in screening predicted docked complexes. Proteins. 1999;35:364–373. [PubMed] [Google Scholar]
- 10.Murphy J, Gatchell D, Prasad J, Vajda S. Combination of scoring functions improves discrimination in protein-protein docking. Proteins. 2003;53:840–854. doi: 10.1002/prot.10473. [DOI] [PubMed] [Google Scholar]
- 11.Lu H, Lu L, Skolnick J. Development of unified statistical potentials describing protein-protein interactions. Biophys J. 2003;84:1895–1901. doi: 10.1016/S0006-3495(03)74997-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, Zhang C, Zhou H, Zhou Y. A physical reference state unifies the structure-derived potential of mean force for protein folding and binding. Proteins. 2004;56:93–101. doi: 10.1002/prot.20019. [DOI] [PubMed] [Google Scholar]
- 13.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen M-Y, Pieper U, Sali A. Curr Protoc Bioinformatics. Unit 5.6. Chapter 5. Oct, 2006. Comparative protein structure modeling using modeller. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y. I-tasser server for protein 3d structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meenan NAG, Sharma A, Fleishman SJ, Macdonald CJ, Morel B, Boetzel R, Moore GR, Baker D, Kleanthous C. The structural and energetic basis for high selectivity in a high-affinity protein-protein interaction. Proc Natl Acad Sci U S A. 2010 May; doi: 10.1073/pnas.0910756107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krachler AM, Sharma A, Kleanthous C. Self-association of tpr domains: Lessons learned from a designed, consensus-based tpr oligomer. Proteins. 2010 Jul;78(9):2131–43. doi: 10.1002/prot.22726. [DOI] [PubMed] [Google Scholar]