Abstract
Vitellogenin, an egg-yolk protein precursor common to oviparous animals, is found abundantly in honeybee workers – a caste of helpers that do not usually lay eggs. Instead, honeybee vitellogenin (180 kDa) participates in processes other than reproduction: it influences hormone signaling, food-related behavior, immunity, stress resistance and longevity. The molecular basis of these functions is largely unknown. Here, we establish and compare the molecular properties of vitellogenin from honeybee hemolymph (blood) and abdominal fat body, two compartments that are linked to vitellogenin functions. Our results reveal a novel 40 kDa vitellogenin fragment in abdominal fat body tissue, the main site for vitellogenin synthesis and storage. Using MALDI-TOF combined with MS/MS mass-spectroscopy, we assign the 40 kDa fragment to the N terminus of vitellogenin, whereas a previously observed 150 kDa fragment corresponded to the remainder of the protein. We show that both protein units are N glycosylated and phosphorylated. Focusing on the novel 40 kDa fragment, we present a homology model based on the structure of lamprey lipovitellin that includes a conserved β-barrel-like shape, with a lipophilic cavity in the interior and two insect-specific loops that have not been described before. Our data indicate that the honeybee fat body vitellogenin experiences cleavage unlike hemolymph vitellogenin, a pattern that can suggest a tissue-specific role. Our experiments advance the molecular understanding of vitellogenin, of which the multiple physiological and behavioral effects in honeybees are well established.
Keywords: vitellogenin, hemolymph, fat body, functional protein fragments, homology modeling, glycosylation, phosphorylation, honeybee
INTRODUCTION
Vitellogenins are female-specific egg-yolk precursors (Spieth et al., 1991), relatives of the vertebrate lipoproteins (Baker, 1988; Mann et al., 1999). These precursors are synthesized by most oviparous animals for transfer into oocytes, where they serve as nourishment for embryos. In honeybees (Apis mellifera), however, vitellogenin is also expressed by workers, a caste of essentially sterile female helper bees (Engels and Fahrenhorst, 1974). Vitellogenin circulates in hemolymph (blood) and is stored at its site of production, the fat body – an insect organ functionally homologous to the vertebrate liver and white fat (Arrese and Soulages, 2009).
During the life course of worker honeybees, vitellogenin levels in the hemolymph and fat body drop, and these reduced concentrations influence several aspects of the life history of the bee (Munch and Amdam, 2010). RNA interference-mediated knockdown of the vitellogenin gene shows that, as vitellogenin protein levels decline, titers of the life-shortening juvenile hormone (JH) increase, and workers show immune senescence, susceptibility to oxidative stress and reduced survival, in addition to a higher probability of abandoning nest-tasks in favor of foraging for nectar (rather than pollen) from flowering plants (Amdam et al., 2003b; Amdam et al., 2004b; Guidugli et al., 2005; Nelson et al., 2007; Seehuus et al., 2006).
The specific, pleiotropic effects of vitellogenin on honeybee physiology, longevity and food-related behavior suggest that this protein can suppress insulin/insulin-like signaling (IIS) in workers (Corona et al., 2007; Hunt et al., 2007; Seehuus et al., 2006). IIS is eukaryotic pathway for nutrient sensing that integrates responses in metabolism, growth, feeding, reproduction, immunity, stress tolerance and survival (Kenyon, 2010). This possibility has implications beyond honeybee biology, because of the conserved nature of IIS; IIS is also a focus of human biomedical research (Munch and Amdam, 2010). However, it is largely unknown how the honeybee vitellogenin molecule exerts its many functions.
In terms of biochemistry, honeybee vitellogenin is grossly described as a 180 kDa monomeric phospholipoglycoprotein (Wheeler and Kawooya, 1990). All insect vitellogenins, excluding those of the honeybee suborder Apocrita, are cleaved in vivo, typically close to the polyserine track(s) at an RXXR consensus-sequence motif by subtilisin-like endoproteases (Barr, 1991; Rouille et al., 1995). The proteins diverge in their specific location of the polyserine tracts and cleavage sites but, overall, cleavage results in one smaller N-terminal fragment and a larger C-terminal fragment (Tufail and Takeda, 2008). Honeybee vitellogenin lacks the RXXR consensus sequence in the vicinity of the polyserine region, as well as the typical presence of cleavage products in hemolymph. The protein has, nonetheless, been observed in two forms: the mature 180 kDa protein in the hemolymph and fat body, and a lighter, 150 kDa fragment in the ovaries of queens (Seehuus et al., 2007) and in the hypopharyngeal head glands of workers, where vitellogenin constituents are processed into food secretions for larval feeding (Amdam et al., 2003a). The 150 kDa unit also appeared during purification of vitellogenin from queen hemolymph (Wheeler and Kawooya, 1990) and, therefore, was perceived as a degradation product (Wheeler and Kawooya, 1990).
Residue range 351-381 of honeybee vitellogenin comprises 13 serine residues, and comparative work from several taxa (Don-Wheeler and Engelmann, 1997; Tufail and Takeda, 2002) point to phosphorylation patterns in this region that can be important in protein-receptor interactions (Miller et al., 1982; Raikhel and Dhadialla, 1992). Glycosylation of vitellogenin has been connected to its secretion from the fat body of a cockroach (Leucophaea maderae) (Don-Wheeler and Engelmann, 1997), but this mechanism is not verified in bees. Overall, the placement and role of post-translational modifications on honeybee vitellogenin are unexplored topics in research.
Here, we first improve and develop purification protocols that allow us to compare vitellogenin derived from the hemolymph with that derived from the fat body of honeybee workers. We report a novel 40 kDa fragment that is abundant in sample purified from the fat body compartment. We assign the 40 kDa fragment to the vitellogenin N terminus using mass spectroscopy, and establish that the corresponding polypeptide is missing from the previously reported 150 kDa vitellogenin unit. Our interpretation is that the honeybee vitellogenin molecule is cleaved into a small N-terminal fragment (40 kDa) and a larger C-terminal fragment (150 kDa) and, thus, the reported 150 kDa unit does not correspond to a degradation product in fat body. We test the overall phosphorylation and glycosylation status of the 40 kDa and 150 kDa fragments, as well as that of the full-length 180 kDa protein. Finally, based on the known structure of lamprey (Ichthyomyzon unicuspis) lipovitellin (Raag et al., 1988; Thompson and Banaszak, 2002), we prepare a homology model for the 40 kDa unit, identifying a large positively charged groove that may be involved in ligand binding, as well as two insect-specific loops and a cavity with the potential to bind fatty acids. The functional significance of these structures is discussed. Our results represent the first major step forward in understanding the functional structure and composition of honeybee vitellogenin since the publication of its coding sequence eight years ago (Piulachs et al., 2003).
MATERIALS AND METHODS
Bees
To establish reliable sources of worker honeybees with ample levels of vitellogenin, four colonies were induced to develop diutinus bees (winter bees). This worker subcaste is naturally occurring in Northern Europe, and is characterized by a considerable accumulation of vitellogenin in the hemolymph and fat body (Fluri et al., 1977; Smedal et al., 2009). In brief, this development was achieved by caging the queen in each colony, which effectively blocks brood rearing. The technique was established and used before (Amdam et al., 2004a; Amdam et al., 2005; Maurizio, 1950).
Sample collection
Mature workers (minimum 2 weeks old) were collected from the nest into small cages for transfer to the laboratory. Bees were anesthetized in a refrigerator (4°C) until immobile. Next, hemolymph was collected through a small incision through the abdominal wall, as described previously (Amdam et al., 2004a). Hemolymph (10 μl) was dissolved in 100 μl ice-cold PBS (phosphate-buffered saline tablet in 200 ml ddH2O; Sigma-Aldrich, St Louis, MO, USA) containing 1 mmol l-1 disodium EDTA and protease inhibitor cocktail (Mini tablets; Roche, Indianapolis, IN, USA). To obtain abdominal fat body tissue, the last body segment, including the stinger apparatus and adhering rectum, gut and ovaries, was removed from the anesthetized bees by gripping the segment with tweezers. The remaining abdominal carcass (the body wall lined with fat body) was disconnected from the thoracic body-segment, placed in a cryotube and immediately frozen in liquid N2. All samples were stored at –80°C until use.
Immunofluorescence
Bee abdomens, dissected on ice, were fixed in formaldehyde and embedded in London Resin White (Electron Microscopy Science, Hatfield, USA), as described before (Seehuus et al., 2007), to provide resin blocks with embedded abdominal tissue ready for sectioning. Sections (1-2 μm) of resin-embedded material were cut with a diamond knife using a Reichert Jung ultra-microtome (Leica, Wetzlar, Germany). Fat body sections were dried onto SuperFrost/Plus slides (Thermo Scientific, Walldorf, Germany), rinsed and washed with PBS-T (PBS with 0.02% Triton X-100) and blocked with 2% BSA (Sigma-Aldrich) in PBS-T for 60 min at room temperature. After 3×15 min washes with PBS-T, sections were incubated overnight at 4°C with a polyclonal (rabbit) anti-vitellogenin antibody at 1:500 (raised against 180 kDa honeybee vitellogenin; Pacific Immunology, Ramona, CA, USA); specificity had been tested and confirmed previously (Seehuus et al., 2007). The negative control was incubated with PBS-T, as verified before (Seehuus et al., 2007). After washing in PBS-T, all samples were incubated with a polyclonal anti-rabbit antibody coupled to the fluorochrome Cy3 (AffiniPure Goat Anti-Rabbit IgG, Jackson ImmunoResearch Europe Ltd, Suffolk, UK) diluted 1:200, overnight at 4°C. Thus, for negative control staining, sections were incubated with secondary antibody only (Seehuus et al., 2007). Finally, samples were washed in PBS-T and subsequently mounted in 50% glycerol/PBS. Images were acquired with a confocal laser scanning microscope (TCS SP5, Leica).
Protein purification
Hemolymph
After quick thawing of the samples, debris was removed by centrifuging at 20,800 g at 4°C for 20 min, and thereafter filtering the supernatant with a syringe filter (Supor Membrane pore size 0.2 μm; Pall Life Sciences, Port Washington, NY, USA). Ion-exchange chromatography was performed as described previously for queen hemolymph using 50 mmol l-1 NaPO4 buffer and a 0-0.45 mol l-1 NaCl gradient (Wheeler and Kawooya, 1990) in order to remove apolipophorins and hexamerins, which are the major hemolymph proteins in insects. Sample collection was restricted to those fractions containing no apolipophorins or hexamerins; this procedure makes it possible to exclude the density ultracentrifugation step of the purification protocol described by Wheeler and Kawooya (Wheeler and Kawooya, 1990). Purification was completed after the Concanavalin A (ConA; Sigma-Aldrich) affinity chromatography step, as described before (Wheeler and Kawooya, 1990); the buffer was changed to ConA buffer (Wheeler and Kawooya, 1990) containing EDTA-free inhibitor cocktail (Mini pills; Roche) and a PD-10 desalting column (Amersham Biosciences, Piscataway, NJ, USA) was used. The purification steps were monitored by running samples on 4-20% gradient SDS-PAGE gels (Bio-Rad Laboratories, Hercules, CA, USA) with a Precision Plus Protein Standard molecular weight marker (Bio-Rad). The gels were stained with Coomassie Blue (Bio-Rad).
Fat body
Abdominal carcasses stored at –80°C were ground directly under liquid N2 without thawing and, thereafter, were transferred quickly to ice-cold 20 mmol l-1 Hepes buffer (pH 7) containing 0.2 M NaCl and protease inhibitor cocktail (Mini pills; Roche). Homogenates were prepared such that 1.5 ml buffer was added to materials from 7-10 abdominal carcasses. Next, samples were mixed by vortexing and sonicated (10 watts, 5 s, 10 times) with a Vibra Cell ultrasonic processor (Sonics & Materials Inc., Danbury, CT, USA) using a 2 mm probe on ice. Sonicated samples were centrifuged at 20,800 g, 4°C, 20 min, and the supernatant filtered as described above. Thereafter, samples were frozen (-80°C), thawed, and centrifuged again, before 1 ml of sample with a total protein concentration of approximately 5 mg ml-1 as defined by Bio-Rad protein assay (Bradford, 1976) was applied to a size-exclusion Superdex 200 10/300 GL column (GE Healthcare, Piscataway, NJ, USA). A total of 33 ml Hepes buffer (same as the homogenization buffer above) were used as a solvent. The size-exclusion column separates proteins by their size. The fractions containing largely vitellogenin were collected and the buffer changed to ion-exchange buffer in a PD-10 desalting column (Amersham Biosciences). Ion-exchange and thereafter ConA affinity purification was performed as with the hemolymph samples, and the purification steps were similarly monitored by SDS-PAGE gels.
To test whether the mechanical processing of fat bodies, per se, could influence our results, hemolymph sample (non-fragmented vitellogenin) was vortexed and sonicated like the fat body extract as a control.
To estimate the relative amount of vitellogenin fragments in fat body raw extract, five independent observers without prior expectations of band intensities were asked to assess 150 and 40 kDa vitellogenin bands relative to standard protein bands that covered a similar range of intensities. The observers were presented with four distinct 4-20% gradient gels (Bio-Rad) with a total of six gel lanes of fat body extract run together with a dilution series of a standard with proteins of known concentrations (Precision Plus Protein Standard, Bio-Rad). Each gel contained a separately prepared fat body extract to avoid pseudo-replication; i.e. each extraction used different diutinus abdomens (16 bees were used in total). From this material, the observers estimated the ratio of 40/150 kDa vitellogenin for every gel independently from each other.
Vitellogenin identification by western blot
Aliquots from each purification step were run on a 4-20% gradient SDS-PAGE gel (Bio-Rad). Separated proteins were transferred to nitrocellulose membrane and incubated with polyclonal antibody against vitellogenin (1:100,000; Pacific Immunology) as described before (Amdam et al., 2003a). Immun-Star horseradish peroxidase-conjugated secondary antibody kit (Bio-Rad) and Gel doc imager (Bio-Rad) were used for visualization.
Vitellogenin identification by mass spectroscopy
Proteolytic preparation of the samples
Purified protein from fat body was run on a SDS gel and stained as outlined above. The bands corresponding to 180, 150 and 40 kDa vitellogenin and sections of gel between 180 and 150 kDa and between 150 and 40 kDa that did not contain visible bands of separated proteins (i.e. ‘blank’ controls) were excised and prepared for digestion following existing protocols (Nasrin et al., 2009). Porcine trypsin (Promega, Madison, WI, USA), 180 ng per gel band in 10 mmol l-1 ammoniumbicarbonate (pH 8) was added to digest the samples overnight at 37°C. Digestion was stopped with 1% trifluoroacetic acid. Samples of tryptic peptides were purified on nC18 StageTip microcolumns, as described by Rappsilber et al. (Rappsilber et al., 2003). For tandem matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF/TOF) mass spectrometry, samples were eluted using 1 μl of matrix [6 mg ml-1 α-cyano-4-hydroxycinnamic acid in 60% acetonitrile (ACN)], 15% methanol, 0.1% trifluoroacetic acid (TFA), onto a ground steel MALDI target plate. For liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis, peptides were eluted twice with 10 μl each of 80% ACN, 0.1% formic acid. ACN was removed by vacuum concentrating.
MALDI-TOF/TOF
Experiments were conducted on an Ultra-flex MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) at an acceleration voltage of 20 kV. A peptide calibration standard kit II (Bruker Daltonics) covering a mass range from 757-3147 Da was used for calibration of the mass scale. The spectra were processed with FlexAnalysis (Bruker Daltonics) and the annotated peaks were targeted for peptide mass fingerprint analyses with Mascot (Matrix Sciences, London, UK; in-house Mascot server) using the NCBI database (Apis mellifera). The seven most-intense peaks of the 40 kDa vitellogenin sample were also analyzed in the TOF/TOF mode for fragmentation analysis. These were: 70-77, VWNGQYAR; 152-168, VNSVQVPTDDEPYASFK; 218-225, NFDNCDQR; 226-237, INYHFGMTDNSR; 265-276, HFTIQSSVTTSK; 287-294, QNGLVLSR; and 524-533, SMEAANIMSK.
Vitellogenin peptide identification by LC-MS/MS
Peptides (see preparation of samples above) were separated by an Ultimate 3000 nano-HPLC (Dionex Corporation, Sunnyvale, CA, USA), using a nC18 reverse-phase column coupled to a 4000 Q-Trap mass spectrometer (Applied Biosystems/MDS SCIEX, Concord, ON, Canada). The nC18 enrichment column was a C18 Pepmap 100, 5 μm particle diameter, 100 Å pore size, 5000×300 μm i.d. column (Dionex) and the nC18 analytical column was a 15,000×75 μm i.d. fused silica capillary column packed in-house with ReproSil-Pur 5 μm C18 resin (Dr Maisch Gmbh, Ammerbuch-Entringen, Germany). The peptides were eluted from the column at a constant flow rate of 300 nl min-1 using a mobile phase gradient [solvent A, aqueous 2% in 0.1% fluoroacetic acid (FA); solvent B, aqueous 90% ACN in 0.1% FA] as follows: solvent B was increased rapidly from 5-12% (0-2 min), followed by a slow increase from 12-30% (2-30 min) and from 30-50% (30-43 min). Elution of very hydrophobic peptides and conditioning of the column were performed during a 10 min isocratic elution with 95% solvent B and a 16 min isocratic elution with 5% B, respectively. The Q-Trap was operated in ‘Information Dependent Acquisition’ mode using automatic MS to MS/MS switching to acquire data. The instrument settings for ion-spray voltage was 2800 V and the collision energy was automatically adjusted based upon the ion charge state and mass of the compound. Data were processed using Analyst (AB SCIEX, Concord, ON, Canada) and analyzed with Mascot (Matrix Sciences).
Phosphostaining
Centrifuged hemolymph and fat body extracts, as well as vitellogenin from fat body (purified by size-exclusion, ion-exchange and ConA chromatography) were run along with PeppermintStick phosphoprotein standard/control (Invitrogen, Carlsbad, CA, USA) on a 4-20% gradient SDS-PAGE gel (Bio-Rad). The gel was treated according to the manufacturer's instructions, i.e. with the Pro-Q Diamond (Invitrogen) phosphostaining protocol for specific detection of phosphorylated proteins. The gel was visualized in trans-UV light with a Gel Doc imager (Bio-Rad).
Glycosylation
Fat body protein samples purified with size-exclusion and ion-exchange chromatography were deglycosylated in order to test the presence of glyco-groups in the 180, 150 and 40 kDa vitellogenin fractions. PNGaseF cleaves N-linked oligosaccharides from glycoproteins (Maley et al., 1989; Plummer and Tarentino, 1991). A control sample without the deglycosylation enzyme PNGaseF amidase (New England BioLabs, Inc., Beverly, MA, USA) was treated like the deglycosylated sample, as suggested by the manufacturer. The incubated (37°C, 1 h) samples were subjected to electrophoretic separation and vitellogenin bands were monitored for size shifts (conferring the presence vs absence of glycans) after Coomassie staining. For optimal protein separation, 4-20% SDS-PAGE gels were used for the 40 kDa band target, whereas 5% SDS-PAGE gels were used for the 180 and 150 kDa band targets.
Sequence alignment and modelling
The modeling software Bodil (Lehtonen et al., 2004) was used to align honeybee vitellogenin residues 22-323 with lamprey lipovitellin residues 20-281. N-terminal vitellogenin sequences from the following species were also aligned to guide the development of a consensus alignment: Bombus ignitus (Swiss-prot accession number B9VUV6), Pimpla nipponica (O17428), Pteromalus puparum (B2BD67), Leucophaea maderae (Q5TLA5), Acipenser transmontanus (Q90243) and Xenopus laevis (P18709). Also, a previously published multiple alignment of 20 vitellogenins (including honeybee vitellogenin) (Avarre et al., 2007) was used as a reference. The homology model was built with program HOMODGE in Bodil. Two putative loop sequences could not be properly modeled because lamprey does not contain these sequences; these residues were aligned with corresponding stretches of vitellogenin from 19 other insect species and nine vertebrate species using ClustalW (Thompson et al., 1994) and was manually adjusted to detect sequence conservation. An all-atom model was prepared for minimization and molecular dynamics (MD) simulation with Amber10 (Case et al., 2008) and the corresponding Amber03 (Duan et al., 2003; Lee and Duan, 2004) force field. The protein was solvated in a periodic truncated octahedron box with TIP3 water molecules (Jorgensen et al., 1983), providing 12 Å of water between the protein surface and the periodic box edge. The solute was minimized for 10,000 steps, followed by 10,000 steps of the system yielding an energetically refined homology model. The system was heated to 300 K in 300 ps, with restraints, followed by 2 ns of equilibration with constant pressure and temperature (NPT) in order to ensure correct water density. The production phase lasted for 10 ns and was performed with constant volume and energy (NVE) with a 1 fs time step, using SHAKE on hydrogen-heavy atom bonds. Analysis of the MD trajectories was performed with the Bio3d package in order to observe the relative stability of the secondary structure elements (Grant et al., 2006). The rotamer, geometry and Ramachandran plot location of single amino acid residues were adjusted with the validation tools of Coot (Emsley and Cowtan, 2004). An electrostatic map based on residue charges was created with PyMOL (Version 1.2r3pre, Schrödinger, LLC).
RESULTS
Vitellogenin immunolocalization, purification and western verification
Immunofluorescence staining confirmed that the hemolymph and fat body contained vitellogenin (Fig. 1A-C). Of the diverse tissues of the abdominal carcass (fat body, muscle, connective tissue, epithelium), vitellogenin immunoreactivity was exclusively detected in fat body trophocyte cells (Fig. 1A,B), which previously were identified as the sole source of the vitellogenin gene transcript in the abdominal carcass of honeybees (Corona et al., 2007).
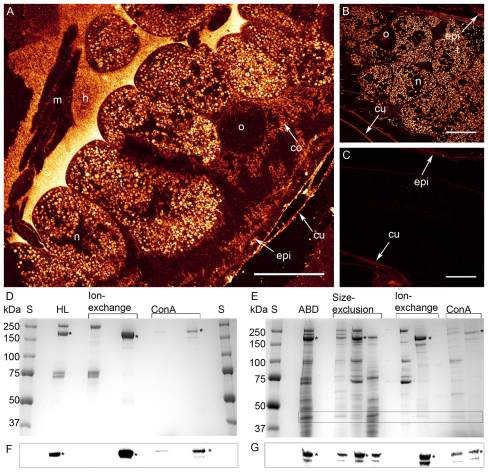
Fig. 1.
Immunolocalization of vitellogenin in the abdominal carcass of diutinus workers and purification of vitellogenin by SDS-PAGE, as verified by western blots. (A,B) Vitellogenin immunoreactivity (white/yellow) is detected in hemolymph (h) and the cytoplasm of trophocyte cells (t), but is not present in the cell nuclei, muscle cells (m), or the oenocyte cells of the fat body (o). Epidermis (epi, epithelial tissue), connective tissue (co) and cuticle (cu) are autofluorescent with confocal imaging, and thus visible also in the control (C). (D-G) Asterisks indicate full-length vitellogenin in the vitellogenin purification steps. S, molecular weight standard. (D) Purification from hemolymph. HL: raw hemolymph (3 μg protein). Ion-exchange: first lane shows flow-through (2 μg); second lane shows high salt elute (2 μg). ConA: first lane is Concanavalin A affinity chromatography column flow through (1 μg); second lane is eluted pure vitellogenin (0.8 μg). (E) Purification from abdomen. The 40 kDa vitellogenin is framed. ABD: sonicated abdominal protein extract (10 μg). Size-exclusion: chromatography fractions eluted by their size (4 μg each lane). Ion-exchange: chromatography flow through and subsequently high salt elute (2 μg each). ConA: affinity chromatography column flow through and on the last lane the pure vitellogenin eluate (1.4 μg each). (F,G) Western blots confirming the presence of vitellogenin in each purification step. The lanes are identical to the lanes for D and E above. (F) There is 2.5 μg protein on the HL lane, 2 μg on both ion-exchange lanes and 0.8 μg on the ConA lanes. (G) There is 1.5 μg abdominal protein on the gel, 1 μg on the size-exclusion lanes, 1.1 μg on the ion-exchange lanes and 1 μg on the ConA lanes.
From hemolymph, vitellogenin was purified in two steps: ion-exchange and ConA chromatography (Fig. 1D). From the abdominal fat body, successful purification required an additional size-exclusion chromatography step preceding the ion-exchange and ConA steps (Fig. 1E). The presence of full-length vitellogenin at 180 kDa in the hemolymph and abdominal fat body, as well as the previously detected 150 kDa fragment in the fat body, was verified by western blots with a vitellogenin-specific antibody (Fig. 1F,G). Partial fragmentation of the purified whole-length vitellogenin into 150 kDa was visible on the gels and the western blots (Fig. 1D,F).
A less intensely stained band of ∼40 kDa was observed in purified vitellogenin fractions from fat body (Fig. 1E). A band of the same size was observed to be abundantly present in the unprocessed fat body protein extract and throughout vitellogenin purification (Fig. 1E). Although the 40 kDa band was not recognized as vitellogenin by western blots (Fig. 1F,G), the closeness of 40 kDa to the size difference between 180 and 150 kDa vitellogenin made the small protein product interesting, and we followed it up for further identification.
We initially tested whether the mechanical stress or buffer conditions that were used during protein extraction from fat body, but not from hemolymph, could induce cleavage in some of the 180 kDa vitellogenin to produce the 150 and 40 kDa pieces observed during gel separation of fat body proteins. For example, a harshly treated protein might be denatured and, thus, become more vulnerable to enzymatic cleavage. To test this, we vortexed and sonicated hemolymph proteins (180 kDa vitellogenin only) under the same conditions and buffer with protease inhibitors as the abdominal fat body extract (see Materials and methods), but untreated and sonicated hemolymph vitellogenin remained equally intact at 180 kDa (supplementary material Fig. S1A). Semiquantitive estimation of the 150 and 40 kDa vitellogenin bands (using standard proteins for comparison) suggested an excess of the 40 kDa fragment (Materials and methods; see also supplementary material Fig. S1B). The average observed ratio was 8.9-fold more 40 than 150 kDa vitellogenin (standard deviation = 3.9).
In summary, our vitellogenin purification protocols for honeybee hemolymph and fat body yielded a 180 kDa protein product in hemolymph with some fragmentation into a 150 kDa product, and two immunoreactive products in fat body (180 and 150 kDa). In addition, a 40 kDa protein present in all fat body vitellogenin purification steps was identified, and could not be ascribed to a technical artifact.
Vitellogenin identification by mass spectrometry
We initially used MALDI-TOF analysis to test amino acid sequence correspondence between the bands detected at 180 and 150, as well as at 40, kDa in extracts of honeybee abdominal fat body (Fig. 2). The peptides identified from the 180 kDa band (Fig. 2A) were spread along the entire sequence of vitellogenin (Fig. 2D), confirming correspondence to the full-length protein. The 150 kDa vitellogenin mostly lacked hits at the N terminus (Fig. 2B,E). Conversely, in the case of the 40 kDa band, the peptide hits were heavily concentrated at the vitellogenin N terminus (residues 54-294) (Fig. 2C,F). This pattern of hits and misses is consistent with vitellogenin that has fragmented into a smaller N-terminal (40 kDa) and a larger C-terminal (150 kDa) piece. No hits to honeybee proteins were verified for the control samples (gel sections picked between the respective bands; see Materials and methods).
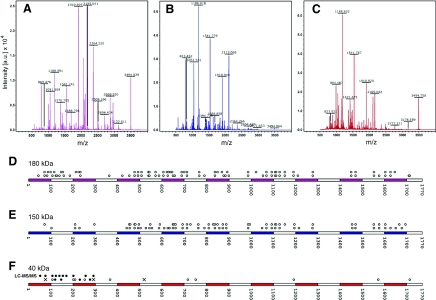
Fig. 2.
Mass-spectrometry identification of vitellogenin-derived peptides from the 180 kDa, 150 kDa and 40 kDa SDS-PAGE bands. Representative spectra of the 180 kDa (A), 150 kDa (B) and 40 kDa (C) vitellogenin bands. Selected peaks denoted by their mass/charge ratio are indicated in the figure panels. (D-F) Mapping of the peptides onto the vitellogenin primary sequence identifies which regions of vitellogenin can be assigned to each fragment. Each circle above a sequence denotes a peptide matching vitellogenin fragment that can be found in this position along the primary sequence. In D and E, the two rows of circles above sequences are independent repeats of a MALDI-TOF experiment on 180 and 150 kDa vitellogenin, respectively. (F) Vitellogenin peptide matches, 40 kDa gel band. LC-MS/MS peptide hits on the first lane indicated with black circles. MALDI-TOF peptide hits on the second lane are marked by seven peptides that were either confirmed (black circle) or not supported (crosses) by fragmentation analysis.
The 40 kDa sample MALDI-TOF peptide hits were subjected to further fragmentation analysis to establish whether they were supported: 70-77, VWNGQYAR (not supported); 152-168, VNSVQVPTDDEPYASFK (confirmed); 218-225, NFDNCDQR (confirmed); 226-237, INYHFGMTDNSR (confirmed); 265-276, HFTIQSSVTTSK (confirmed); 287-294, QNGLVLSR (not supported); and 524-533, SMEAANIMSK (not supported) (see Fig. 2F). Finally, we analyzed the 40 kDa sample with LC-MS/MS and obtained a profile of 10 peptides (Fig. 2F), which were all confirmed to match exclusively to the N-terminal vitellogenin sequence from residue 53 to 294.
Thus, we observed a correspondence between three vitellogenin products in honeybee abdominal fat body: the 180 kDa band corresponds to full-length protein, the 150 kDa band corresponds to vitellogenin lacking the N terminus and the 40 kDa band corresponds to the N terminus of the protein.
Phosphorylation and glycosylation status of vitellogenin and its fragments
We found that full-length vitellogenin (180 kDa), and the 150 and 40 kDa fragments, are phosphorylated. This result was obtained from raw hemolymph and fat body proteins, as well as from purified abdominal vitellogenin samples (Fig. 3).
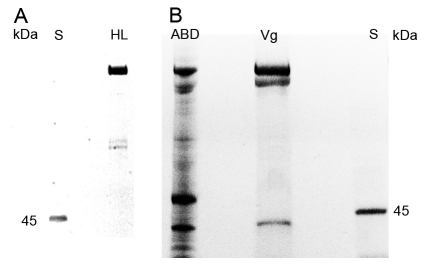
Fig. 3.
Phosphorylated proteins in hemolymph (A), abdominal protein extract and purified vitellogenin (B). ProQ phosphostaining with PeppermintStick (S) standard that provides both a positive and negative control (showing here only phosphorylated ovalbumin at 45 kDa). (A) HL: the diutinus worker hemolymph sample (2.5 μg protein) is dominated by full-length vitellogenin. (B) ABD: the abdominal protein extract (14 μg protein) shows 180, 150 and 40 kDa vitellogenin. Additionally, other abdominal proteins appear as putatively phosphorylated. Vg: the vitellogenin purified from diutinus abdomens (4.7 μg) shows 180, 150 and 40 kDa vitellogenin bands to be phosphorylated.
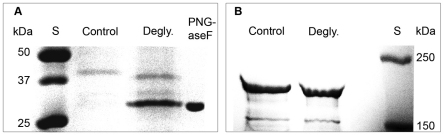
Full-length vitellogenin was previously found to be glycosylated, as the protein binds to ConA columns, in which lectins will crosslink to N-linked glyco groups (Ohyama et al., 1985; Wheeler and Kawooya, 1990). Likewise, we found that both the 150 and 40 kDa vitellogenin bind to the ConA column, as they were retained in the final step of the fat body purification protocol (Fig. 1E). Furthermore, in our deglycosylation test, the sizes of the 180, 150 and 40 kDa vitellogenin species were reduced (by ∼4.2, ∼2.4 and ∼2.5 kDa, respectively) as a result of the loss of this sugar group (Fig. 4). Thus, both the ConA binding and the size shift after deglycosylation support the presence of two glyco groups in 180 kDa vitellogenin that will segregate to one group each in the 40 and 150 kDa fragments.
Fig. 4.
Effects of deglycosylation by PNG-aseF on abdominal vitellogenin on SDS-PAGE. S, molecular weight standard. (A) Deglycosylation of the 40 kDa vitellogenin. The deglycosylated sample appears as a ∼2.5 kDa lighter molecular weight band. PNGaseF, the deglycosylation enzyme. (B) Deglycosylation of the 180 and 150 kDa vitellogenin. The full-length vitellogenin shifts ∼4.2 kDa and the 150 kDa vitellogenin shifts ∼2.4 kDa after the removal of N-linked glycans.
A positively charged patch, a lipophilic cavity and an indication of insect-specific coils in the honeybee N-terminal domain
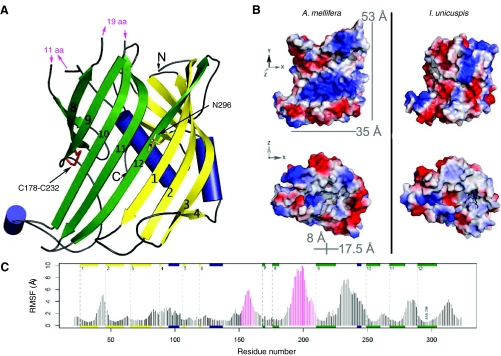
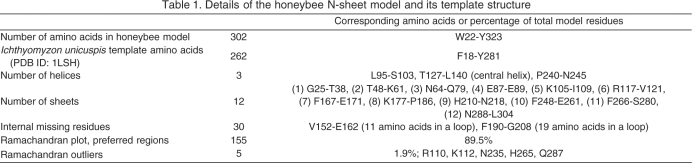
We succeeded in building a β-barrel-like homology model of the N-terminal honeybee vitellogenin region (N-sheet) (Anderson et al., 1998; Raag et al., 1988) (Fig. 5A; Table 1). There were no Ramachandran outliers in the β-sheets or α-helices indicating an excellent protein back-bone chemistry (Ramachandran et al., 1963). Five outliers were located in coils (Table 1). Loops have more evolutional plasticity than β-sheets or α-helices (Panchenko et al., 2005), which makes these regions more difficult to model accurately by homology, as their sequence differs from the template structure. Our electrostatic map of the model shows a patch of positively charged residues on the β-sheets (Fig. 5B). The hydrophobic inner cavity is predominately positively charged – contrary to lamprey lipovitellin, in which the cavity is partly negatively charged (Fig. 5B). Simulation of the model was performed in order to test the stability of the predicted secondary structures, and of the whole protein fragment. A relative mobility of the loops of the model compared with its more stationary α-helixes and β-sheets was observed, as is expected for a stable protein (Fig. 5C; supplementary material Fig. S2 for RMSD).
Fig. 5.
Structural characterization of the honeybee vitellogenin N-sheet. (A) A homology model based on the X-ray structure of lamprey lipovitellin. Helices are shown in blue, β-sheets are yellow and green (green for those corresponding to the homodimerization site of lamprey lipovitellin), the disulphide bridge (C178-C232) is shown as a red stick and the putatively glycosylated asparagine 296 residue is shown as an orange stick. The N and C termini are indicated and the β-sheets are numbered starting at the N terminus. The missing loop residues of 11 amino acids (from central helix to β-sheet 7) and 19 amino acid residues (from β-sheets 8 to 9) are indicated by pink arrows. (B) Electrostatic comparison between the honeybee (A. mellifera) vitellogenin N-sheet model and the lamprey (I. unicuspis) lipovitellin N-sheet. The insect-specific loops, with energetically minimized but otherwise random structure, are included. Red color represents the negatively charged Asp and Glu residues; blue color represents the positively charged His, Arg and Lys residues. Honeybee N-sheet accommodates a large positively charged patch (upper left; residues R33, R35, K53, R256, K276 and R294 on β-sheets 1-2 and 10-12), whereas the β-sheets of lamprey show a more even charge distribution (upper right). The cavity inside the N-sheet also differs in charge: the honeybee vitellogenin cavity (bottom left) contains a positive charge (K205, K 207, R251), whereas the lamprey cavity (bottom right) contains a negative charge due to two glutamic acid residues not present in honeybee. The black rod inside the lamprey cavity is a lipid found in the crystal structure. The dimensions of the honeybee N-sheet inner cavity are circa 8×17.5 Å, and the whole half-β-barrel is approximately 53×35 Å in size. (C) The positional fluctuations as determined by a 10 ns long MD simulation. Secondary structure elements are indicated schematically with helices in dark blue and strands in yellow and green, as in A. Pink bars depict the 11- and 19-residue long loops.
Table 1.
Details of the honeybee N-sheet model and its template structure
We found two insect-specific loops in the 40 kDa vitellogenin fragment (location indicated with pink arrows in Fig. 5A). These two regions could not be properly modelled, as the vertebrate template structure lacks the corresponding sequences. The regions were considered as loops based on PSIPRED (Jones, 1999) and Gor4 (Garnier et al., 1996) secondary structure predictions (predictions not shown) and because of the observation of the conserved coil-indicator residues glycine and proline (Krieger et al., 2005) in these regions. A multiple alignment of 29 species shows that these loops are conserved in many insect species but are absent in vertebrates (supplementary material Fig. S3).
DISCUSSION
Vitellogenin influences hormone signaling, food-related behavior, immunity, stress resistance and longevity in honeybee workers (Amdam et al., 2004a; Amdam et al., 2004b; Fluri et al., 1982; Nelson et al., 2007; Seehuus et al., 2006). The molecular properties of the protein, however, are not well known. Here, we identify a previously uncharacterized 40 kDa vitellogenin fragment in the worker fat body, and reject the hypothesis that a complimentary 150 kDa unit represents a breakdown product. We describe the post-translational modification of the 40 kDa and 150 kDa vitellogenin units, as well as of the full-length vitellogenin, in two body compartments: hemolymph and fat body. Finally, we present the first homology model for the 40 kDa unit of this vitellogenin.
Initially, we established compartment-specific purification protocols for the target protein: from hemolymph and fat body. Immunofluorescence staining confirmed vitellogenin to be abundantly and specifically present in these compartments. Improved yields of vitellogenin were facilitated by our choice of source bees: workers that were induced to accumulate vitellogenin based on a natural process in which vitellogenin-rich worker bees develop in the absence of brood rearing (Maurizio, 1950). Our protocol for hemolymph vitellogenin purification is based on an extraction protocol optimized for honeybee queens (Wheeler and Kawooya, 1990). Building from this method, we were able to omit a time-consuming (16 h) density ultra-centrifugation step by pooling ion-exchange chromatography fractions that do not contain apolipophorin or hexamerins. Our protocol for hemolymph may not only be time efficient, but also yield more intact vitellogenin as the protein has less time to degrade during handling – although we did experience limited vitellogenin fragmentation. Our procedure for fat body was built on the same original protocol (Wheeler and Kawooya, 1990), to which we added size-exclusion chromatography as the first step.
After purification, we found three vitellogenin products in fat body that separated on SDS-PAGE gels as bands of 180, 150 and 40 kDa in size. Western blots confirmed the 180 and 150 kDa bands to be vitellogenin, as seen previously (Amdam et al., 2003a). The antibody, however, did not immunostain the 40 kDa band. The lack of recognition of this fragment, which we identified as vitellogenin by other means (see below), is likely to be due to a smaller fraction of the polyclonal antibody population being specific to an epitope or epitopes on the N terminus of vitellogenin relative to the remainder of the protein, as seen in vinculin antibody-binding pattern (Kilic and Ball, 1991).
We used MALDI-TOF mass-spectrometry to verify that the 40 kDa protein band was a vitellogenin fragment. Further LC-MS/MS experiment showed peptide hits exclusively at the N terminus of vitellogenin (residues 54-294). The MALDI-TOF peptide hits of the 150 kDa vitellogenin, reciprocally, were centered between residues 427-1700, supporting our proposition that the 40 and 150 kDa vitellogenin fragments correspond to non-overlapping products of the full-length 180 kDa vitellogenin. Note that some regions in the full-length and 150 kDa vitellogenin products seem to systematically lack MALDI-TOF peptide hits (see Fig. 2A,B). We believe these misses are caused by post-translational modification. For instance, phospho groups at phosphorylated peptides, particularly at the so-called polyserine linker between the residues 351-381 of vitellogenin, tend to complicate detection (Steen et al., 2006).
The 180, 150 and 40 kDa vitellogenin units were shown to be phosphorylated by phosphoprotein staining. Strong candidates for phosphorylation are the 13 serine residues situated between residues 351 and 381 (Don-Wheeler and Engelmann, 1997; Tufail and Takeda, 2002). This region approximately lies at the intersection of the 40 kDa and the 150 kDa vitellogenin. In some cases, protein cleavage is known to be regulated by phosphorylation (Alexandru et al., 2001; Walter et al., 1999), and this cannot be excluded in the case of honeybee vitellogenin. Where is the exact cleavage site located? Honeybee vitellogenin sequence lacks the RXXR consensus cleavage site near the polyserine linker present in many other insect vitellogenins (Barr, 1991; Rouille et al., 1995). The LC-MS/MS peptide hits end at residue 294, but this does not prove it to be the terminal amino acid of 40 kDa vitellogenin. According to a theoretical molecular mass calculation, a molecular mass of 40 kDa corresponds to the residues 17-370 (1-16 is a cleaved signal sequence (Piulachs et al., 2003). Amino acid 370 is in the middle of the polyserine motif: nslssseekl kqdilnlrtd issssssisss (residue 370 bolded). Hypothetically, if vitellogenin were cut at residue 370, there would be multiple serine tracts in both the resulting fragments, 40 and 150 kDa, which is in accordance with the finding that both fragments are clearly phosphorylated. However, there are many possible phosphorylation sites along the whole vitellogenin sequence. Therefore, further studies are needed to trace the exact site, and to understand the regulation of the cleavage.
Another post-translational modification – glycosylation – of 180, 150 and 40 kDa vitellogenin is supported by binding to Concanavaline A that has affinity for N-glycosylated proteins (Ohyama et al., 1985). The deglycosylation experiment also showed size shifts of the fragments on a SDS-PAGE gel. The vitellogenin sequence contains three Asn-X-Ser/Thr sequences for possible N glycosylation (Marshall, 1974): one in the N terminus (Asn296; highlighted in Fig. 5A) and two towards the C terminus (Asn1067 and Asn1153). The sum of our size-shift estimates for 150 and 40 kDa vitellogenin (∼2.4 and ∼2.5 kDa, respectively) differs from the 180 kDa vitellogenin estimate (∼4.2 kDa) by 0.7 kDa, which could be due to a minor inaccuracy on our SDS-PAGE analysis. For example, decapod Cherax quadricarinatus vitellogenin contains a glycan of a similar size, 2067.7 Da Glc3Man9GlcNAc2, among other glycans (Khalaila et al., 2004).
Our homology model based on the lamprey lipovitellin crystal structure (Anderson et al., 1998; Mann et al., 1999; Raag et al., 1988; Thompson and Banaszak, 2002) of N-terminal vitellogenin, comprises residues 22-323 that are present in the 40 kDa fragment. Interestingly, the N-sheet accommodates two putative loop regions that are found in insects only, with the exception of the vitellogenin of the cockroaches Blattella germanica and Leucophaea maderae (supplementary material Fig. S3). These loops contain several conserved proline and glycine residues that usually hinder α-helix or β-sheet formation (Krieger et al., 2005), and there are also some conserved charged residues in the loops. The loops are located at the lamprey homodimerization region, and might thus be connected to possible dimerization differences between insect and vertebrate vitellogenins. The presence of many charged residues raises a question of a more catalytical role of these loops. There are many examples of loops with important biological functions – some loops even possess active site residues (Hedstrom et al., 1992; Joseph et al., 1990; Williams and McCammon, 2009; Williams and Essex, 2009; Xu et al., 2007). As they are clearly conserved, a structural or catalytic role of some kind could be hypothesized for these N-terminal loops in insects.
A comparison between the lamprey lipovitellin and the honeybee vitellogenin N-sheet reveals differences in their charge distribution. There are six Arg and His residues on honeybee N-sheet β-sheets that form a positively charged patch. In lamprey, the positive and negative charges are evenly distributed. A positively charged patch might attract negatively charged ligands (Cherstvy, 2009; Corsico et al., 2005; Grigg et al., 2010; Honig and Nicholls, 1995; Horii et al., 2004). The interior of the N-sheet is lined by a mainly hydrophobic cavity, in which a lipid molecule is present in the lamprey lipovitellin crystal structure (Thompson and Banaszak, 2002). It seems likely that the 40 kDa vitellogenin of honeybee also has the ability to bind a lipid, as the cavity is even more exposed to ligands when the rest of the protein is removed. The honeybee N-sheet cavity is positively charged at one side. This could offer an ideal site for a fatty acid with a negatively charged headgroup to bind, like in the case of intestinal fatty acid binding protein (Cistola et al., 1989; Jakoby et al., 1993; Sacchettini et al., 1989).
According to our study, the honeybee vitellogenin fragmentation pattern differs in fat body and hemolymph. Vitellogenin cleavage is abundant in abdominal (fat body containing) protein extract, whereas we only observed the intact 180 kDa protein in fresh worker hemolymph. However, the hemolymph vitellogenin also formed a 150 kDa unit after sampling and during purification (see Results) (see also Wheeler and Kawooya, 1990). In Oreochromis aureus, the N terminus of vitellogenin was identified as the vitellogenin receptor-binding site (Li et al., 2003). If this is also the case with honeybee vitellogenin, then 150 kDa vitellogenin formation in hemolymph would be undesirable, as, thereafter, the protein could not be taken up by tissues via receptor-mediated endocytosis. It is an open question as to how vitellogenin integrity remains intact in the hemolymph of a living bee.
Lack of 40 or 150 kDa units in fresh hemolymph might also suggest that cleaved vitellogenin is not secreted from the fat body. The intracellular role or roles of the 40 kDa unit are currently unknown, but it is tempting to speculate on a mechanism that links the putative lipid binding properties of 40 kDa vitellogenin to a biologically relevant context in which worker bees store increasing amounts of vitellogenin in their fat bodies. A fatty acid mixture from larvae, called brood pheromone (Le Conte et al., 1994), inhibits fat body vitellogenin accumulation. In accordance with this, larvae must be absent from the colony milieu before vitellogenin-rich bees develop (Smedal et al., 2009). In theory, the lipophilic cavity of the 40 kDa vitellogenin might bind such pheromone fatty acids. When larval brood is present, this interaction could stabilize full-length vitellogenin and perhaps facilitate vitellogenin secretion. Reciprocally, more vitellogenin would remain in fat body when brood and pheromone is absent. We also observed an excess of the 40 kDa vitellogenin in our abdominal extracts when compared with the 150 kDa vitellogenin (Fig. 1E; see also supplementary material Fig. S1B). The intensity of any one protein band separating on a one-dimentional gel can be cofounded by overlapping or partly overlapping bands. However, we did not observe other 40 kDa proteins during the various steps of vitellogenin purification, and mass-spectrometry analysis detected only 40 kDa vitellogenin in the same range. We therefore conclude that the 40 kDa band might represent an almost pure fraction of vitellogenin. A bias towards the 40 kDa fragment, relative to the 150 kDa fragment, could indicate that the 150 kDa fragment is consumed in the fat body, whereas the remaining 40 kDa vitellogenin is involved in an additional mechanism for preserving vitellogenin stores; for instance, by repressing exocytosis via membrane binding.
The findings presented here represent a step forward in the understanding of honeybee vitellogenin. Knowledge about this protein could, eventually, provide unique illustrations on how insect social organization can be achieved at the level of protein structure. Moreover, insights into the molecular properties of vitellogenins are of broader general interest, as alternative functions of this family of yolk precursor proteins are now surfacing in diverse taxa: including in the immune defense against viruses and bacteria in fish (Garcia et al., 2010; Li et al., 2008; Shi et al., 2006), and in aging in Caenorhabditis elegans (Nakamura et al., 1999).
Supplementary Material
Acknowledgments
The work was performed at the Department of Biochemistry at University of Bergen in the laboratory headed by professor Aurora Martinez. H.H., Ø.H. and G.V.A. conceived and designed the experiments. H.H., Ø.H. and B.S. collected the samples. H.H. performed the experiments with the exception of mass-spectroscopy and immunohistochemistry. Ø.H., H.H. and G.V.A. analyzed the mass-spectroscopy data. B.S. performed the immunohistochemistry. L.S. performed model minimization and simulation. H.H., Ø.H. and G.V.A. wrote the paper. L.S. wrote the model simulation and minimization section of the paper, and B.S. wrote the immunohistochemistry section. We thank Professor Martinez for kindly supporting and assisting the project, and for access to instruments and reagents. We also thank Randi Svebak for technical support. We thank C. Kreibich, M. Speth and A. H. Søyland for providing bees and assisting the sampling. The mass-spectroscopy experiments – from sample extraction from gel pieces until data collection and protein identification – and assistance with data interpretation and writing were performed by the PROBE Proteomic Unit at University of Bergen. We thank Dr Anne Døskeland for expert technical support.
LIST OF ABBREVIATIONS
- ABD
abdominal protein extract
- ACN
acetonitrile
- ConA
concanavalin A
- Degly.
deglycosylated
- HL
hemolymph
- IIS
insulin/insulin-like signaling
- JH
juvenile hormone
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- MALDI-TOF/TOF
tandem matrix-assisted laser desorption/ionization time-of-flight
- MD
molecular dynamics
- S
molecular weight standard
- TFA
trifluoroacetic acid
- Vg
vitellogenin
Supplementary material available online at http://jeb.biologists.org/cgi/content/full/214/4/582/DC1
The PROBE work was partly supported by the National Program for Research in Functional Genomics (FUGE) funded by the Norwegian Research Council. Ø.H. was supported by the Research council (#185306) and Norwegian Cancer Society (#58240001). G.V.A. was supported by the Research Council of Norway (#180504, 185306 and 191699), the National Institute on Aging (NIA P01 AG22500) and the PEW Charitable Trust.
REFERENCES
- Alexandru G., Uhlmann F., Mechtler K., Poupart M. A., Nasmyth K. (2001). Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105, 459-472 [DOI] [PubMed] [Google Scholar]
- Amdam G. V., Norberg K., Hagen A., Omholt S. W. (2003a). Social exploitation of vitellogenin. Proc. Natl. Acad. Sci. USA 100, 1799-1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam G. V., Simoes Z. L., Guidugli K. R., Norberg K., Omholt S. W. (2003b). Disruption of vitellogenin gene function in adult honeybees by intra-abdominal injection of double-stranded RNA. BMC Biotechnol. 3, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam G. V., Hartfelder K., Norberg K., Hagen A., Omholt S. W. (2004a). Altered physiology in worker honey bees (Hymenoptera: Apidae) infested with the mite Varroa destructor (Acari: Varroidae): a factor in colony loss during overwintering? J. Econ. Entomol. 97, 741-747 [DOI] [PubMed] [Google Scholar]
- Amdam G. V., Simoes Z. L. P., Hagen A., Norberg K., Schroder K., Mikkelsen O., Kirkwood T. B. L., Omholt S. W. (2004b). Hormonal control of the yolk precursor vitellogenin regulates immune function and longevity in honeybees. Exp. Gerontol. 39, 767-773 [DOI] [PubMed] [Google Scholar]
- Amdam G. V., Norberg K., Omholt S. W., Kryger P., Lourenco A. P., Bitondi M. M. G., Simoes Z. L. P. (2005). Higher vitellogenin concentrations in honey bee workers may be an adaptation to life in temperate climates. Insectes Soc. 52, 316-319 [Google Scholar]
- Anderson T. A., Levitt D. G., Banaszak L. J. (1998). The structural basis of lipid interactions in lipovitellin, a soluble lipoprotein. Structure 6, 895-909 [DOI] [PubMed] [Google Scholar]
- Arrese E. L., Soulages J. L. (2009). Insect fat body: energy, metabolism, and regulation. Annu. Rev. Entomol. 55, 207-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarre J. C., Lubzens E., Babin P. J. (2007). Apolipocrustacein, formerly vitellogenin, is the major egg yolk precursor protein in decapod crustaceans and is homologous to insect apolipophorin II/I and vertebrate apolipoprotein B. BMC Evol. Biol. 7, 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. E. (1988). Is Vitellogenin an ancestor of Apolipoprotein-B-100 of human low-density lipoprotein and human lipoprotein-lipase. Biochem. J. 255, 1057-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr P. J. (1991). Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell 66, 1-3 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248-254 [DOI] [PubMed] [Google Scholar]
- Case D. A. D. T. A., Cheatham T. E., III, Simmerling C. L., Wang J., Duke R. E., Luo R., Crowley M., Walker Ross C., Zhang W., Merz K. M., et al. (2008). AMBER 10. San Francisco, CA: University of California, San Francisco; [Google Scholar]
- Cherstvy A. G. (2009). Positively charged residues in DNA-binding domains of structural proteins follow sequence-specific positions of DNA phosphate groups. J. Phys. Chem. B 113, 4242-4247 [DOI] [PubMed] [Google Scholar]
- Cistola D. P., Sacchettini J. C., Banaszak L. J., Walsh M. T., Gordon J. I. (1989). Fatty acid interactions with rat intestinal and liver fatty acid-binding proteins expressed in Escherichia coli. A comparative 13C NMR study. J. Biol. Chem. 264, 2700-2710 [PubMed] [Google Scholar]
- Corona M., Velarde R. A., Remolina S., Moran-Lauter A., Wang Y., Hughes K. A., Robinson G. E. (2007). Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc. Natl. Acad. Sci. USA 104, 7128-7133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsico B., Franchini G. R., Hsu K. T., Storch J. (2005). Fatty acid transfer from intestinal fatty acid binding protein to membranes: electrostatic and hydrophobic interactions. J. Lipid Res. 46, 1765-1772 [DOI] [PubMed] [Google Scholar]
- Don-Wheeler G., Engelmann F. (1997). The biosynthesis and processing of vitellogenin in the fat bodies of females and males of the cockroach Leucophaea maderae. Insect Biochem. Mol. Biol. 27, 901-918 [DOI] [PubMed] [Google Scholar]
- Duan Y., Wu C., Chowdhury S., Lee M. C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T., et al. (2003). A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24, 1999-2012 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126-2132 [DOI] [PubMed] [Google Scholar]
- Engels W., Fahrenhorst H. (1974). Alters- und kastenspezifische Veränderungen der Haemolymph-protein-spectren bei Apis mellifera. Wilhelm Roux Arch. Entwickl. Mech. Org. 174, 285-296 [DOI] [PubMed] [Google Scholar]
- Fluri P., Wille H., Gerig L., Luscher M. (1977). Juvenile hormone, vitellogenin and hemocyte composition in winter worker honeybees (Apis-Mellifera). Experientia 33, 1240-1241 [Google Scholar]
- Fluri P., Luscher M., Wille H., Gerig L. (1982). Changes in weight of the pharyngeal gland and hemolymph titers of juvenile-hormone, protein and vitellogenin in worker honey bees. J. Insect Physiol. 28, 61-68 [Google Scholar]
- Garcia J., Munro E. S., Monte M. M., Fourrier M. C., Whitelaw J., Smail D. A., Ellis A. E. (2010). Atlantic salmon (Salmo salar L.) serum vitellogenin neutralises infectivity of infectious pancreatic necrosis virus (IPNV). Fish Shellfish Immunol. 29, 293-297 [DOI] [PubMed] [Google Scholar]
- Garnier J., Gibrat J. F., Robson B. (1996). GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266, 540-553 [DOI] [PubMed] [Google Scholar]
- Grant B. J., Rodrigues A. P., ElSawy K. M., McCammon J. A., Caves L. S. (2006). Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics 22, 2695-2696 [DOI] [PubMed] [Google Scholar]
- Grigg J. C., Cooper J. D., Cheung J., Heinrichs D. E., Murphy M. E. (2010). The Staphylococcus aureus siderophore receptor HtsA undergoes localized conformational changes to enclose staphyloferrin A in an arginine-rich binding pocket. J. Biol. Chem. 285, 11162-11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidugli K. R., Nascimento A. M., Amdam G. V., Barchuk A. R., Omholt S., Simoes Z. L., Hartfelder K. (2005). Vitellogenin regulates hormonal dynamics in the worker caste of a eusocial insect. FEBS Lett. 579, 4961-4965 [DOI] [PubMed] [Google Scholar]
- Hedstrom L., Szilagyi L., Rutter W. J. (1992). Converting trypsin to chymotrypsin: the role of surface loops. Science 255, 1249-1253 [DOI] [PubMed] [Google Scholar]
- Honig B., Nicholls A. (1995). Classical electrostatics in biology and chemistry. Science 268, 1144-1149 [DOI] [PubMed] [Google Scholar]
- Horii K., Okuda D., Morita T., Mizuno H. (2004). Crystal structure of EMS16 in complex with the integrin alpha2-I domain. J. Mol. Biol. 341, 519-527 [DOI] [PubMed] [Google Scholar]
- Hunt G. J., Amdam G. V., Schlipalius D., Emore C., Sardesai N., Williams C. E., Rueppell O., Guzman-Novoa E., Arechavaleta-Velasco M., Chandra S., et al. (2007). Behavioral genomics of honeybee foraging and nest defense. Naturwissenschaften 94, 247-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M. G., Miller K. R., Toner J. J., Bauman A., Cheng L., Li E., Cistola D. P. (1993). Ligand-protein electrostatic interactions govern the specificity of retinol- and fatty acid-binding proteins. Biochemistry 32, 872-878 [DOI] [PubMed] [Google Scholar]
- Jones D. T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195-202 [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983). Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926-935 [Google Scholar]
- Joseph D., Petsko G. A., Karplus M. (1990). Anatomy of a conformational change: hinged “lid” motion of the triosephosphate isomerase loop. Science 249, 1425-1428 [DOI] [PubMed] [Google Scholar]
- Kenyon C. J. (2010). The genetics of ageing. Nature 464, 504-512 [DOI] [PubMed] [Google Scholar]
- Khalaila I., Peter-Katalinic J., Tsang C., Radcliffe C. M., Aflalo E. D., Harvey D. J., Dwek R. A., Rudd P. M., Sagi A. (2004). Structural characterization of the N-glycan moiety and site of glycosylation in vitellogenin from the decapod crustacean Cherax quadricarinatus. Glycobiology 14, 767-774 [DOI] [PubMed] [Google Scholar]
- Kilic F., Ball E. H. (1991). Partial cleavage mapping of the cytoskeletal protein vinculin. Antibody and talin binding sites. J. Biol. Chem. 266, 8734-8740 [PubMed] [Google Scholar]
- Krieger F., Moglich A., Kiefhaber T. (2005). Effect of proline and glycine residues on dynamics and barriers of loop formation in polypeptide chains. J. Am. Chem. Soc. 127, 3346-3352 [DOI] [PubMed] [Google Scholar]
- Le Conte Y., Streng L., Trouiller J. (1994). The recognition of larvae by worker honeybees. Naturwissenschaften 81, 462-465 [Google Scholar]
- Lee M. C., Duan Y. (2004). Distinguish protein decoys by using a scoring function based on a new AMBER force field, short molecular dynamics simulations, and the generalized born solvent model. Proteins 55, 620-634 [DOI] [PubMed] [Google Scholar]
- Lehtonen J. V., Still D. J., Rantanen V. V., Ekholm J., Bjorklund D., Iftikhar Z., Huhtala M., Repo S., Jussila A., Jaakkola J., et al. (2004). BODIL: a molecular modeling environment for structure-function analysis and drug design. J. Comput. Aided. Mol. Des. 18, 401-419 [DOI] [PubMed] [Google Scholar]
- Li A., Sadasivam M., Ding J. L. (2003). Receptor-ligand interaction between vitellogenin receptor (VtgR) and vitellogenin (Vtg), implications on low density lipoprotein receptor and apolipoprotein B/E. The first three ligand-binding repeats of VtgR interact with the amino-terminal region of Vtg. J. Biol. Chem. 278, 2799-2806 [DOI] [PubMed] [Google Scholar]
- Li Z., Zhang S., Liu Q. (2008). Vitellogenin functions as a multivalent pattern recognition receptor with an opsonic activity. PloS One 3, e1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley F., Trimble R. B., Tarentino A. L., Plummer T. H., Jr (1989). Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180, 195-204 [DOI] [PubMed] [Google Scholar]
- Mann C. J., Anderson T. A., Read J., Chester S. A., Harrison G. B., Kochl S., Ritchie P. J., Bradbury P., Hussain F. S., Amey J., et al. (1999). The structure of vitellogenin provides a molecular model for the assembly and secretion of atherogenic lipoproteins. J. Mol. Biol. 285, 391-408 [DOI] [PubMed] [Google Scholar]
- Marshall R. D. (1974). The nature and metabolism of the carbohydrate-peptide linkages of glycoproteins. Biochem. Soc. Symp. 1974, 17-26 [PubMed] [Google Scholar]
- Maurizio A. (1950). The influence of pollen feeding and brood rearing on the length of life and physiological conditions of the honeybee. Bee World 31, 9-12 [Google Scholar]
- Miller M. S., Benore-Parsons M., White H. B., III (1982). Dephosphorylation of chicken riboflavin-binding protein and phosvitin decreases their uptake by oocytes. J. Biol. Chem. 257, 6818-6824 [PubMed] [Google Scholar]
- Munch D., Amdam G. V. (2010). The curious case of aging plasticity in honey bees. FEBS Lett. 584, 2496-2503 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Yasuda K., Adachi H., Sakurai Y., Ishii N., Goto S. (1999). Vitellogenin-6 is a major carbonylated protein in aged nematode, Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 264, 580-583 [DOI] [PubMed] [Google Scholar]
- Nasrin N., Kaushik V. K., Fortier E., Wall D., Pearson K. J., de Cabo R., Bordone L. (2009). JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PloS One 4, e8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. M., Ihle K. E., Fondrk M. K., Page R. E., Amdam G. V. (2007). The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 5, e62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama Y., Kasai K., Nomoto H., Inoue Y. (1985). Frontal affinity chromatography of ovalbumin glycoasparagines on a concanavalin A-sepharose column. A quantitative study of the binding specificity of the lectin. J. Biol. Chem. 260, 6882-6887 [PubMed] [Google Scholar]
- Panchenko A. R., Wolf Y. I., Panchenko L. A., Madej T. (2005). Evolutionary plasticity of protein families: coupling between sequence and structure variation. Proteins 61, 535-544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piulachs M. D., Guidugli K. R., Barchuk A. R., Cruz J., Simoes Z. L., Belles X. (2003). The vitellogenin of the honey bee, Apis mellifera: structural analysis of the cDNA and expression studies. Insect Biochem. Mol. Biol. 33, 459-465 [DOI] [PubMed] [Google Scholar]
- Plummer T. H., Jr., Tarentino A. L. (1991). Purification of the oligosaccharide-cleaving enzymes of Flavobacterium meningosepticum. Glycobiology 1, 257-263 [DOI] [PubMed] [Google Scholar]
- Raag R., Appelt K., Xuong N. H., Banaszak L. (1988). Structure of the lamprey yolk lipid-protein complex lipovitellin-phosvitin at 2.8 A resolution. J. Mol. Biol. 200, 553-569 [DOI] [PubMed] [Google Scholar]
- Raikhel A. S., Dhadialla T. S. (1992). Accumulation of yolk proteins in insect oocytes. Annu. Rev. Entomol. 37, 217-251 [DOI] [PubMed] [Google Scholar]
- Ramachandran G. N., Ramakrishnan C., Sasisekharan V. (1963). Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 7, 95-99 [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M. (2003). Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663-670 [DOI] [PubMed] [Google Scholar]
- Rouille Y., Duguay S. J., Lund K., Furuta M., Gong Q., Lipkind G., Oliva A. A., Jr, Chan S. J., Steiner D. F. (1995). Proteolytic processing mechanisms in the biosynthesis of neuroendocrine peptides: the subtilisin-like proprotein convertases. Front. Neuroendocrinol. 16, 322-361 [DOI] [PubMed] [Google Scholar]
- Sacchettini J. C., Gordon J. I., Banaszak L. J. (1989). Refined apoprotein structure of rat intestinal fatty acid binding protein produced in Escherichia coli. Proc. Natl. Acad. Sci. USA 86, 7736-7740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehuus S. C., Norberg K., Gimsa U., Krekling T., Amdam G. V. (2006). Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc. Natl. Acad. Sci. USA 103, 962-967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehuus S. C., Norberg K., Krekling T., Fondrk K., Amdam G. V. (2007). Immunogold localization of vitellogenin in the ovaries, hypopharyngeal glands and head fat bodies of honeybee workers, Apis mellifera. J. Insect Sci. 7, 1-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Zhang S., Pang Q. (2006). Vitellogenin is a novel player in defense reactions. Fish Shellfish Immunol. 20, 769-772 [DOI] [PubMed] [Google Scholar]
- Smedal B., Brynem M., Kreibich C. D., Amdam G. V. (2009). Brood pheromone suppresses physiology of extreme longevity in honeybees (Apis mellifera). J. Exp. Biol. 212, 3795-3801 [DOI] [PubMed] [Google Scholar]
- Spieth J., Nettleton M., Zucker-Aprison E., Lea K., Blumenthal T. (1991). Vitellogenin motifs conserved in nematodes and vertebrates. J. Mol. Evol. 32, 429-438 [DOI] [PubMed] [Google Scholar]
- Steen H., Jebanathirajah J. A., Rush J., Morrice N., Kirschner M. W. (2006). Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol. Cell. Proteomics 5, 172-181 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. R., Banaszak L. J. (2002). Lipid-protein interactions in lipovitellin. Biochemistry 41, 9398-9409 [DOI] [PubMed] [Google Scholar]
- Tufail M., Takeda M. (2002). Vitellogenin of the cockroach, Leucophaea maderae: nucleotide sequence, structure and analysis of processing in the fat body and oocytes. Insect Biochem. Mol. Biol. 32, 1469-1476 [DOI] [PubMed] [Google Scholar]
- Tufail M., Takeda M. (2008). Molecular characteristics of insect vitellogenins. J. Insect. Physiol. 54, 1447-1458 [DOI] [PubMed] [Google Scholar]
- Walter J., Schindzielorz A., Grunberg J., Haass C. (1999). Phosphorylation of presenilin-2 regulates its cleavage by caspases and retards progression of apoptosis. Proc. Natl. Acad. Sci. USA 96, 1391-1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D. E., Kawooya J. K. (1990). Purification and characterization of honey bee vitellogenin. Arch. Insect Biochem. Physiol. 14, 253-267 [DOI] [PubMed] [Google Scholar]
- Williams S. L., McCammon J. A. (2009). Conformational dynamics of the flexible catalytic loop in Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate reductoisomerase. Chem. Biol. Drug. Des. 73, 26-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. L., Essex J. W. (2009). Study of the conformational dynamics of the catalytic loop of WT and G140A/G149A HIV-1 integrase core domain using reversible digitally filtered molecular dynamics. J. Chem. Theory Comput. 5, 411-421 [DOI] [PubMed] [Google Scholar]
- Xu W., Yuan X., Beebe K., Xiang Z., Neckers L. (2007). Loss of Hsp90 association up-regulates Src-dependent ErbB2 activity. Mol. Cell. Biol. 27, 220-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.