The discovery of the cell-adhesive properties of the Arg-Gly-Asp (RGD) sequence located in the 10th type III domain of fibronectin triggered widespread use of RGD-functionalized materials for directing cell behavior.[1–2] In studies of cell adhesion and migration, however, cell responses on RGD surfaces are never identical to those observed on fibronectin.[3] For example, we recently examined the attachment and patterning of Rat-1 fibroblasts on elastin-based artificial extracellular matrix (aECM) proteins bearing a fibronectin-derived RGD sequence, and found the average projected area of such cells to be approximately 60% of those spread on fibronectin.[4] We wondered whether it might be possible to elicit more nearly authentic cell responses by engineering aECM proteins that preserve the domain structure of fibronectin. Fibronectin type III domains 9 and 10 are ideal for such studies; the domains are relatively small (ca. 90 amino acids), and the PHSRN sequence derived from domain 9 has been reported to increase cell adhesion to RGD peptides derived from domain 10.[5] Garcia and coworkers have reported enhanced cell adhesion strength, signaling and proliferation on surfaces bearing tethered fibronectin fragments comprising domains 7–10,[6] and Ratner and Jiang have observed striking differences in the orientation and functionality of such fragments adsorbed on positively and negatively charged monolayer surfaces.[7] Fusion proteins containing fibronectin fragments have also been prepared. Mardon and Grant showed in 1994 that domains 9 and 10 support cell adhesion when fused to glutathione S-transferase.[8] More recently, Mrksich and coworkers completed a thorough study of cell adhesion to domains 9 and 10 immobilized as cutinase fusions on self-assembled alkanethiol monolayers.[9]
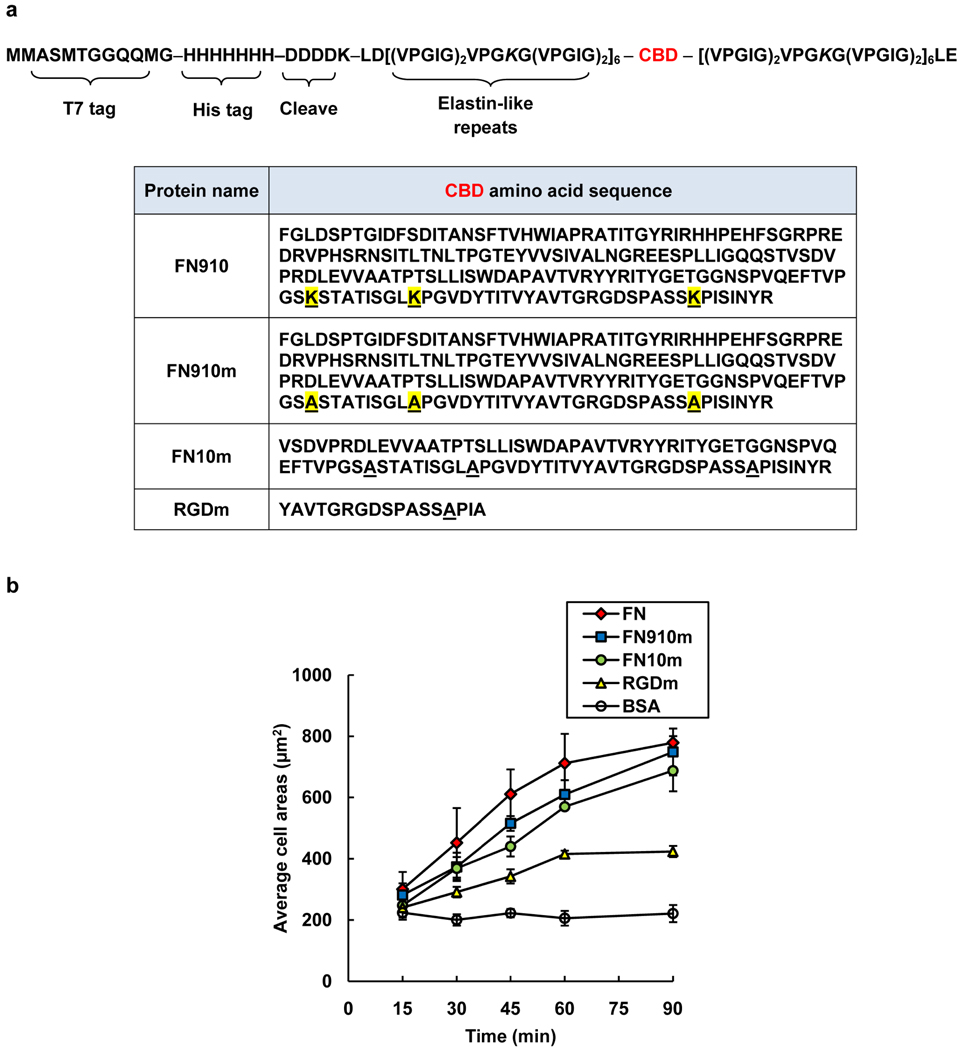
Here we report a genetic strategy to prepare elastin-based aECM proteins bearing full-length fibronectin domains. Each aECM protein carries a central cell-binding domain (CBD) flanked by relatively long (125-amino acid) elastin-like domains (Figure 1a) that contain lysine residues to facilitate crosslinking and fabrication of viscoelastic materials with tunable moduli.[10–12] For simplicity, each aECM protein is identified by its CBD.
Figure 1.
Artificial extracellular matrix (aECM) proteins containing full-length fibronectin domains. a) The general amino acid sequence of the aECM proteins. Each protein contains an N-terminal T7 tag, a hepahistidine tag, and an enterokinase cleavage site followed by six elastin-like repeats, a cell-binding domain (CBD, see table) and six elastin-like repeats. The differences between FN910 and FN910m are highlighted in yellow. The letter “m” denotes cell-binding domains containing lysine-to-alanine mutations. b) The average projected cell areas for the adsorbed protein surfaces at each time point. Data represent means ± SEM from three independent experiments.
We mutated all of the lysine residues in each CBD to alanine, to confine crosslinking to the elastin domains and to reduce the probability that crosslinking would compromise cell-binding behavior (Figure 1a). Mutated CBDs are identified by the suffix “m;” e.g., FN910m contains fibronectin domains 9 and 10 lacking all lysine residues. Bacterial expression typically yielded 500 mg to 2 g of purified aECM protein per 10 L of cell culture. The purity and molecular weight of each protein were verified by polyacrylamide gel electrophoresis (SDS-PAGE) (Figure S1).
Rat-1 fibroblasts were allowed to spread on both adsorbed and crosslinked aECM films. Cells spread faster on adsorbed FN910m and FN10m than on RGDm surfaces (Figure 1b). After 1.5 h, cell areas on FN910m and FN10m were comparable to those on the positive control fibronectin (FN). In contrast, the average cell area on RGDm was two-fold lower (Figure 1b). A similar trend was observed on crosslinked aECM films (Figure S2a). There were no differences in cell spreading behavior on crosslinked FN910 and FN910m, confirming that the lysine-to-alanine mutations did not affect cell binding (Figure S2b).
Binding of integrin α5β1 has been shown to promote rapid spreading of fibroblasts on biomaterials surfaces.[13] To determine whether aECM proteins containing full-length CBDs promoted increased integrin binding as compared to those containing the shorter RGDm domain, we used a modified solid-phase (ELISA) integrin-binding assay. We found the α5β1 binding affinity for FN910m to be enhanced approximately six-fold (KD = 17.2 nM) compared to FN10m (KD = 109 nM) and RGDm (KD = 100 nM). Under the same conditions, KD for FN was determined to be 6.3 nM (Figure S3), slightly higher than previously reported (2 – 4 nM),[14–16] perhaps reflecting differences in integrin presentation (i.e., immobilized integrins in refs 14–16 vs. soluble integrins in this work). The increase in α5β1 integrin binding affinity for FN910m is consistent with the reported synergistic effect of PHSRN, and suggests that full-length fibronectin domains remain functional when presented in the context of elastin-based aECM proteins. We did not see significant differences in α5β1 binding affinity for FN10m and RGDm (Figure S3), despite the fact that FN10m promoted faster cell spreading (Figure 1b and Figure S2a). It is likely that cell spreading behavior on FN10m and RGDm is mediated, at least in part, by integrin αvβ3.[6,17]
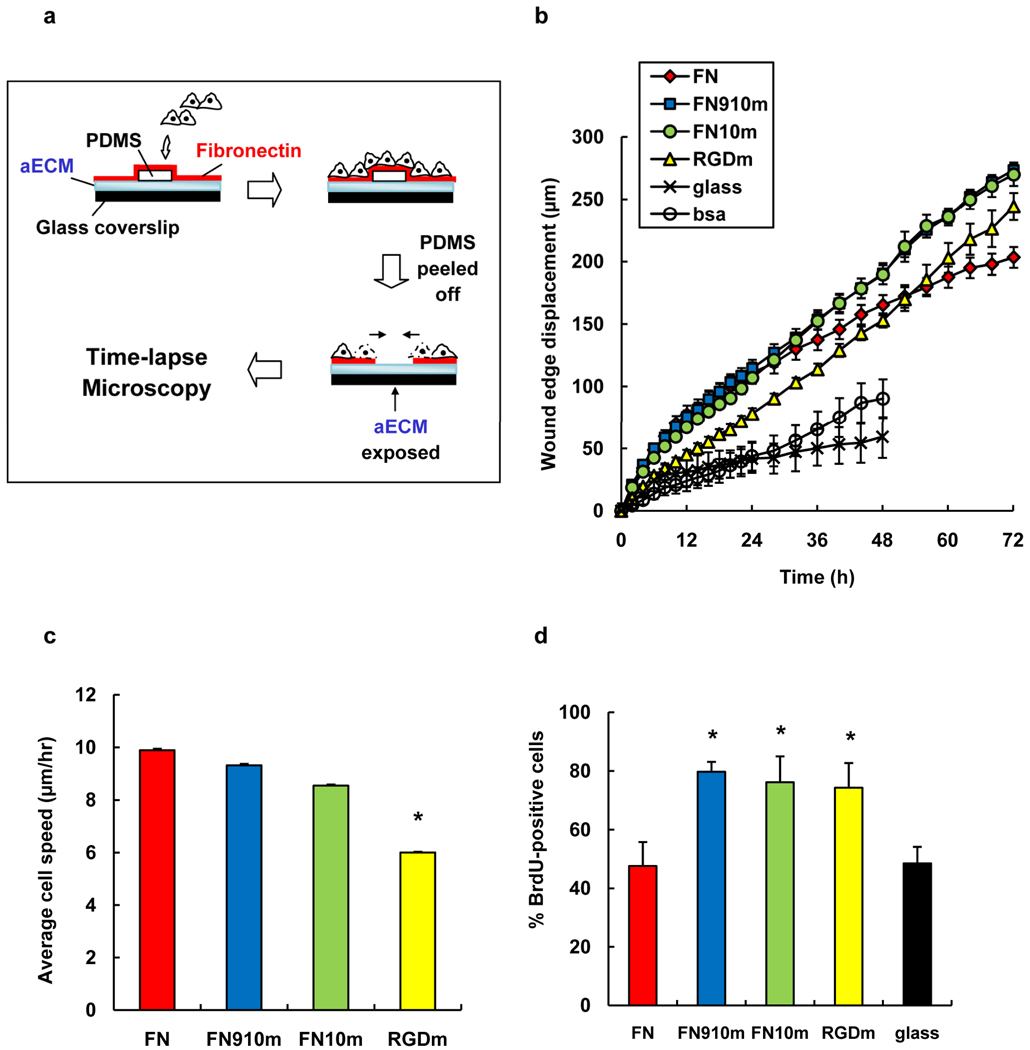
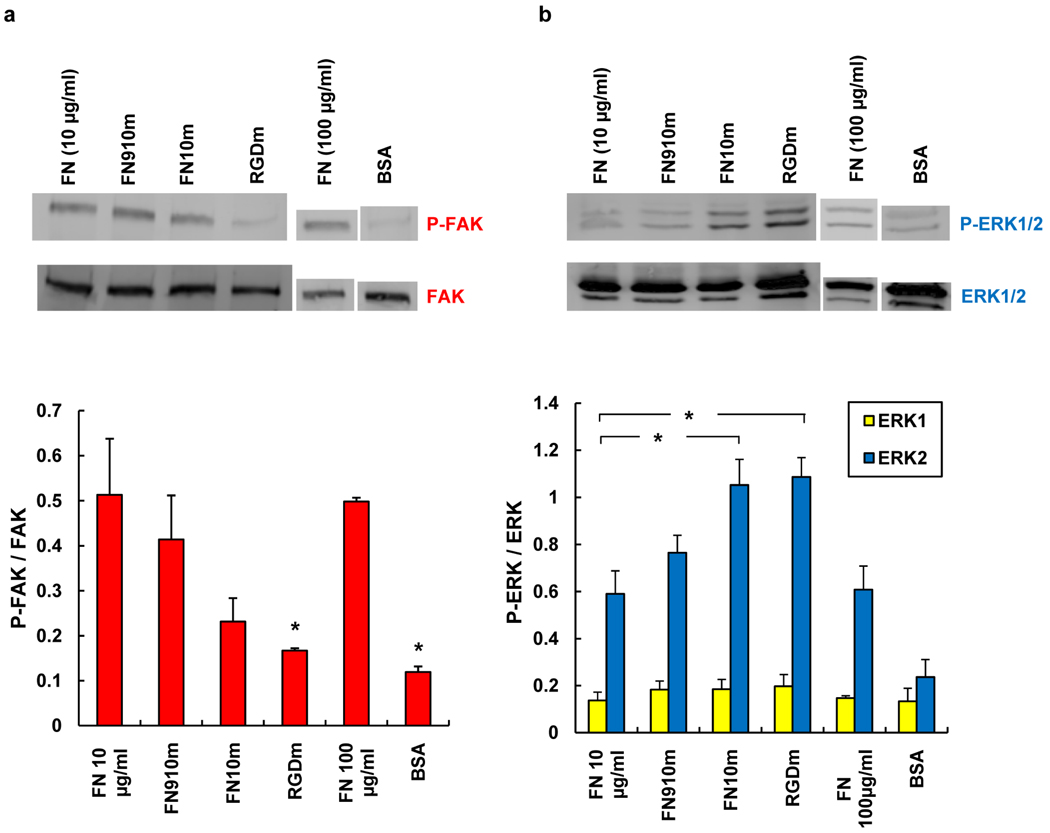
We next examined collective cell migration on aECM proteins, using the wound-healing assay shown in Figure 2a. Migration of fibroblast sheets was observed on all adsorbed protein surfaces except the BSA negative control (see supplemental movies S1 – S6). Figure 2b shows the displacement of the edge of the cell sheet on various surfaces over a period of 72 h. In the first 24 h of the experiment, cell sheets advanced at similar rates on FN, FN910m and FN10m. We observed no significant differences between FN910m and FN10m; both surfaces supported faster displacement of the wound edge than RGDm. Individual cells at the wound edge were tracked for 10 h from the start of wounding. We focused on cells at the wound edge to minimize the effects of substrate remodeling through deposition of secreted matrix proteins; cells at the leading edge constantly encounter virgin surface as the sheet advances. Average cell speeds are shown in Figure 2c. Cell speeds on FN910m (9.2 ± 0.8 µm h−1) and FN10m (8.3 ± 0.6 µm h−1) were comparable to that on FN (9.4 ± 0.8 µm h−1). In contrast, cells migrated significantly more slowly on RGDm (5.9 ± 0.4 µm h−1; P < 0.05), accounting for the slower wound closure on RGDm. The increase in cell speeds on proteins bearing full-length fibronectin domains was associated with increased phosphorylation of focal adhesion kinase (FAK), a protein required for integrin-mediated cell migration.[18] Increased levels of phospho-FAK (P-FAK) were observed in cells migrating on FN, FN910m and FN10m as compared to RGDm (Figure 3a), consistent with the lower cell speeds measured on the latter surface.
Figure 2.
Wound-healing behavior of Rat-1 fibroblasts on adsorbed protein surfaces. a) Schematic of wound-healing assay. b) Displacement of the wound edge as a function of time. c) Average speeds of cells from t = 0 to 10 h. Individual cells in the first row of the wound edge were tracked for 10 h post wounding. Cell speeds are slopes from linear fit of average distance traveled vs. time. Error bars are standard errors from fit. d) The percentage of BrdU-positive cells for the period t = 24 to 48 h (the cell sheet was wounded at t = 0). Data are means ± SEM from five independent experiments for each surface. *, significant difference from FN surface (P < 0.05).
Figure 3.
Determination of FAK and ERK phosphorylation in Rat-1 fibroblasts on adsorbed protein surfaces. Cell lysates were analyzed by Western blotting with a) anti-FAK, anti-phosphoFAK (pY397) and b) anti-phosphoERK1/2(p42/p44) and anti-total ERK1/2, antibodies. Band intensities were normalized to total-FAK or total-ERK bands. Reported data are means ± s.d. for three independent experiments. *, significantly different from FN surface (P < 0.05).
During the later stages of the wound-healing experiment (i.e., after 24 h), we were surprised to find that wound healing on aECM proteins with full-length fibronectin domains proceeded more rapidly than on FN (Figure 2b). The increased rate of wound closure appears to be a consequence of increased cell proliferation, as shown by a nearly two-fold increase in BrdU-labeling of cells on FN10m and FN910m as compared to FN (Figure 2d). This result is also consistent with higher levels of phosphorylated extracellular signal-regulated kinase (ERK) on the aECM protein surfaces (Figure 3b). To eliminate the possibility that the lower proliferation rate observed on FN was due to depletion of FN from the surface, we repeated these experiments with ten-fold higher FN concentrations. As expected, the extent of phosphorylation of both FAK and ERK was unchanged. We can offer no persuasive explanation for the reduced rate of proliferation on FN as compared to the aECM protein surfaces. We note, however, that Garcia and coworkers have observed reduced proliferation of C2C12 myoblasts on FN substrates that support increased levels of binding of α5 and β1 integrin subunits.[19]
In summary, we have developed aECM proteins that incorporate full-length cell-binding domains derived from fibronectin. In comparison with proteins that present shorter RGD sequences, the new aECM proteins promote more rapid cell spreading, tighter integrin binding, and faster wound healing in vitro.
Experimental
Cells, antibodies, and reagents
Dulbecco’s Modified Eagle Medium (DMEM), fetal bovine serum (FBS), penicillin/streptomycin, 0.05% Trypsin/0.25% EDTA, and PBS were obtained from Invitrogen (Carlsbad, CA). Rat-1 fibroblasts were generous gifts of the Asthagiri laboratory at Caltech. Cells were maintained in growth medium containing phenol-red, 10% fetal bovine serum (FBS) and 0.1% penicillin/streptomycin through passages 9 – 25. All experiments were performed in phenol-red-free and serum-free DMEM with 0.1% penicillin/streptomycin (SFM).
Restriction enzymes were obtained from New England Biolabs, Ipswich, MA. All ligations were performed using T4 DNA ligase (Roche Applied Science, 2.5 h, 25 °C). DNA was isolated using QIAprep Spin Miniprep Kits (Qiagen). DNA segments encoding various cell-binding domains were purchased from Genscript (Piscataway, NJ) or Integrated DNA Technologies (San Diego, CA). Cloning was performed in E. coli strain XL10-Gold (Stratagene). E. coli strain BL21 (DE3) pLysS (Novagen, Madison, WI) was used for protein expression.
Anti-phospho-FAK and anti-ERK 1/2 were purchased from Invitrogen. All other antibodies, human α5β1 integrin, human plasma fibronectin (FN), Hoechst 33342 dye, and TMB/E substrate were obtained from Chemicon (Temecula, CA). Bovine serum albumin (BSA) was obtained from Sigma. Bis[sulfosuccinimidyl]suberate (BS3) used for crosslinking aECM proteins was obtained from Pierce, Rockford, IL. The 5-bromo-2’-deoxyuridine (BrdU) labeling kit was purchased from Roche Applied Science (Indianapolis, IN). Round coverslips (12 mm diameter, No. 1) were from Deckgläser, Germany. All Western blotting reagents were obtained from GE Healthcare (Piscataway, NJ).
Expression and purification of aECM proteins
DNA fragments encoding various cell-binding domains were ligated into pET28aRW, which encodes an N-terminal T7 tag, a hexahistidine tag, and an enterokinase cleavage tag [20–22]. All products were verified by restriction digestion and DNA sequencing (Laragen, Los Angeles, CA). Large-scale expression and purification of proteins by thermal cycling were performed as previously described [11,21].
Cell spreading
Standard 24-well tissue culture plates were coated with aECM protein solutions (1 mg mL−1) or FN (10 µg mL−1) overnight at 4°C. Coverslips bearing crosslinked aECM films were also mounted separately into the wells using sterile grease. Wells containing adsorbed proteins were rinsed with PBS and subsequently blocked with 0.2 wt% heat-inactivated BSA solution (500 µL) at room temperature for 30 min. In each well, 4 × 104 cells mL−1 were added to 1 mL of SFM and incubated at 37 °C under 5% CO2/95% air. Images of five randomly-selected positions in each well were acquired every 15 min for 1.5 h. The projected cell areas for 200 cells were recorded using ImageJ for each surface at each time point.
α5β1 integrin binding assay (ELISA)
In ELISA binding assays, adsorbed aECM proteins were used instead of adsorbed integrins to eliminate high levels of non-specific adsorption of aECM to polystyrene during binding. Integrin binding conditions used were reported by Altroff et al. [23]. Briefly, FN, BSA, and aECM proteins were dissolved in 25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm MnCl2, 0.1 mm MgCl2, and 0.1 mm CaCl2 (EB) to obtain a final concentration of 0.1 µM. Clear, flat-bottom 96-well plates (Greiner, VWR) were incubated overnight with protein solutions (100 µL) at 4°C. The wells were washed once with EB containing 1% BSA and 0.1% Tween-20 (wash buffer), and blocked with 320 µl of EB with 5% BSA at 37°C for 30 min. The wells were washed twice with wash buffer (200 µL) and incubated with various concentrations of integrins (diluted in EB with 1% BSA) for 2 h at 37°C. The wells were washed five times with wash buffer and incubated with mouse anti-α5β1 (50 µL; clone JBS5, 1:200 in EB with 1% BSA) at room temperature for 1 h. Wells were rinsed with wash buffer five times before adding goat anti-mouse-HRP (50 µL; AP124P, 1:5000 in EB with 1% BSA) at room temperature for 30 min. Finally, wells were again washed five times with wash buffer and developed with 100 µL of TMB/E substrate (ES001) for 10 min at room temperature. The reaction was stopped by addition of 1 N H2SO4 (100 µL), and absorbance at 450 nm was read immediately using a Safire plate reader (Tecan, San Jose, CA). Assays were performed in triplicate and non-specific binding in the BSA wells was subtracted from the total binding values for each integrin concentration. The data were fitted to a sigmoidal curve using Origin v.8 (OriginLab, Northampton, MA). The apparent KD’s for various aECM proteins and FN were calculated using the molecular weights of fibronectin (250 kDa) and α5β1 integrin (265 kDa).
Wound healing
Proteins were adsorbed on cleaned glass coverslips at 4°C for up to 1 week and air-dried before use. A thin block of PDMS was placed in the center of the coverslip, which was mounted into a 24-well plate using silicone glue. To aid cell adhesion, FN solution (500 µL, 10 µg mL−1 in PBS) was added to each well and incubated overnight at 4 °C. Upon reaching confluence, the cell sheet was serum-starved for 24 h to arrest growth at the G0/G1 phase through contact inhibition [24–25]. Subsequently, the PDMS block was removed and cells were washed twice with SFM to remove cell debris. Images of several spots on each wound edge were acquired every 15 min for 72 h. Images were analyzed using ImageJ v1.37. The wound area was traced manually at various time points, and the displacement of the wound edge was calculated as the change in the wound area divided by the length of the wound.
Individual cell tracking
Cells at the edge of the wound sheet were tracked for 10 h from the start of wounding. Cell tracking was performed manually by tracking the centroid of each cell, using ImageJ with plug-in MTrackJ (http://www.bigr.nl/). The average distance traveled vs. time for all tracked cells was fit to a straight line, and the slope was reported as the average speed. An average of 100 cells was tracked for each protein surface.
BrdU-labeling
Wound-healing experiments were performed as previously described for 24 h, and the medium was replaced with SFM containing 10 µM BrdU. Cells were incubated for another 24 h, washed with pre-warmed PBS, and fixed in 70% ethanol/30% glycine (pH 2) at −20 °C for 20 min. After two washes with PBS, cells were incubated with 200 µL of anti-BrdU solution (100 µL anti-BrdU antibody stock diluted with 900 µL incubation buffer) for 30 min at 37 °C. After two PBS washes, cells were incubated with 1 µL Hoechst 33342 and 200 µL anti-mouse-FITC in PBS (1: 200) for 30 min at 37 °C. Cells within 350 µm of the wound edge were counted. The percent BrdU-positive cells was taken to be the number of BrdU-labeled cells as a percentage of the total Hoechst-labeled cells.
Immunoblotting
Standard 10-cm tissue culture Petri dishes were coated with 3 mL of FN (10 µg mL−1 or 100 µg mL−1 in PBS), BSA (2 mg mL−1 in PBS), or aECM proteins (1 mg mL−1 in PBS) for 2 – 3 days at 4°C. Dishes were rinsed with PBS and blocked with heat-inactivated BSA (2 mL; 2 mg mL−1 in PBS) for 30 min at room temperature. Confluent Rat-1 fibroblasts were serum-starved for 24 h to arrest cell growth [24,25], trypsinized and held in suspension in SFM for 45 min at 37°C to reduce contact-mediated signaling. Subsequently, 3.8 × 104 cells cm−2 were added to various protein surfaces for 1 h at 37°C under 5% CO2/95% air. Cells were placed on ice, washed twice with ice-cold PBS, and lysed in Laemmli buffer (300 µL; 62.5 mm Tris-Cl, pH 6.8, 20% glycerol, 10% 2-mercaptoethanol and 4% SDS). Cell lysates were centrifuged at 18000g for 15 min at room temperature. Equal amounts of proteins were boiled for 5 min and separated on 7% or 10% SDS-PAGE gels, transferred to nitrocellulose membranes, and blocked with TBST (20 mm Tris-Cl, pH 7.6, 0.9% NaCl, 0.1% Tween-20) with 5% BSA for 1 h at room temperature. Membranes were incubated with antibodies against FAK, phosphorylated FAK (pY397; 1:1000), ERK 1/2, and phosphorylated ERK 1/2 (Thr202/Tyr204, Thr185/Tyr187; 1:1000) in TBST with 3% BSA overnight at 4°C. After washing with TBST, secondary antibodies (horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG; 1:5000 in TBST) were incubated for 1 h at room temperature. Blots were washed three times for 15 min with TBST and developed according to the manufacturer’s instructions using the ECL Plus™ kit (GE Healthcare) and subsequently visualized on a Typhoon™ Trio molecular imager (GE Healthcare). Protein band intensities were measured using ImageQuant TL v7.0 and phosphorylation levels normalized to total FAK or total ERK1/2.
Statistical analysis
The statistical significance of differences was estimated by analysis of variance followed by the Tukey test. Differences were taken to be significant at P ≤ 0.05.
Supplementary Material
Acknowledgements
The authors thank Drs. Stacey Maskarinec and Shelly Tzlil for discussions. We acknowledge Dr. Anand Asthagiri for Rat-1 fibroblasts and generous access to the fluorescence microscope. E. F. is supported in part by the Nanyang Overseas Scholarship, Singapore. This work is supported by NIH EB001971, by the NSF Center for Science and Engineering of Materials, and by the Armed Forces Institute for Regenerative Medicine Wake Forest – Pittsburgh Consortium.
Contributor Information
Dr. Eileen Fong, Department of Bioengineering California Institute of Technology, Pasadena, California 91125, USA.
David A. Tirrell, Division of Chemistry and Chemical Engineering, Joseph J. Jacobs Institute for Molecular Engineering for Medicine, California Institute of Technology, Pasadena, California 91125, USA, tirrell@caltech.edu
References
- 1.Ruoslahti E, Pierschbacher MD. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 2.Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 3.Streeter HB, Rees DA. J. Cell Biol. 1987;105:507–515. doi: 10.1083/jcb.105.1.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrico IS, Maskarinec SA, Heilshorn SC, Mock ML, Liu JC, Nowatzki PJ, Franck C, Ravichandran G, Tirrell DA. J. Am. Chem. Soc. 2007;129:4874–4875. doi: 10.1021/ja070200b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aota S, Nomizu M, Yamada KM. J. Biol. Chem. 1994;269:24756–24761. [PubMed] [Google Scholar]
- 6.Petrie TA, Capadona JR, Reyes CD, Garcia AJ. Biomaterials. 2006;27:5459–5470. doi: 10.1016/j.biomaterials.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, He Y, Ratner BD, Jiang SY. J. Biomed. Mater. Res A. 2006;77A:672–678. doi: 10.1002/jbm.a.30586. [DOI] [PubMed] [Google Scholar]
- 8.Mardon HJ, Grant KE. FEBS Lett. 1994;340:197–201. doi: 10.1016/0014-5793(94)80137-1. [DOI] [PubMed] [Google Scholar]
- 9.Eisenberg JL, Piper JL, Mrksich M. Langmuir. 2009;25:13942–13951. doi: 10.1021/la901528c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMillan RA, Conticello VP. Macromolecules. 2000;33:4809–4821. [Google Scholar]
- 11.Nowatzki PJ, Tirrell DA. Biomaterials. 2003;25:1261–1267. doi: 10.1016/s0142-9612(03)00635-5. [DOI] [PubMed] [Google Scholar]
- 12.Trabbic-Carlson K, Setton LA, Chilkothi A. Biomacromolecules. 2003;4:572–580. doi: 10.1021/bm025671z. [DOI] [PubMed] [Google Scholar]
- 13.Massia SP, Stark J. J. Biomed. Mater. Res. 2001;56:390–399. doi: 10.1002/1097-4636(20010905)56:3<390::aid-jbm1108>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 14.Grant RP, Spitzfaden C, Altroff H, Campbell ID, Mardon HJ. J. Biol. Chem. 1997;272:6159–6166. doi: 10.1074/jbc.272.10.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altroff H, Choulier L, Mardon HJ. J. Biol. Chem. 2003;278:491–497. doi: 10.1074/jbc.M209992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mould AP, Askari JA, Aota S, Yamada KM, Irie A, Takada Y, Mardon HJ, Humphries MJ. J. Biol. Chem. 1997;272:17283–17292. doi: 10.1074/jbc.272.28.17283. [DOI] [PubMed] [Google Scholar]
- 17.Mao Y, Schwarzbauer JE. Cell Comm. Adhesion. 2006;13:267–277. doi: 10.1080/15419060601072215. [DOI] [PubMed] [Google Scholar]
- 18.Sieg DJ, Hauck CR, Ilic D, Klingbei CK, Schaefer E, Damsky CH, Schlaepfer DD. Nat. Cell Biol. 2000;2:249–256. doi: 10.1038/35010517. [DOI] [PubMed] [Google Scholar]
- 19.Garcia AJ, Vega MD, Boettiger D. Mol. Biol. Cell. 1999;10:785–798. doi: 10.1091/mbc.10.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heilshorn SC, Di Zio KA, Welsh ER, Tirrell DA. Biomaterials. 2003;24:4245–4252. doi: 10.1016/s0142-9612(03)00294-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu JC, Heilshorn SC, Tirrell DA. Biomacromolecules. 2003;5:497–504. doi: 10.1021/bm034340z. [DOI] [PubMed] [Google Scholar]
- 22.Liu JC, Tirrell DA. Biomacromolecules. 2008;9:2984–2988. doi: 10.1021/bm800469j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altroff H, van der Walle CF, Asselin J, Fairless R, Campbell ID, Mardon HJ. J. Biol. Chem. 2001;276:38885–38892. doi: 10.1074/jbc.M105868200. [DOI] [PubMed] [Google Scholar]
- 24.Schorl C, Sedivy JM. Methods. 2007;41:143–150. doi: 10.1016/j.ymeth.2006.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kues WA, Anger M, Carnwath JW, Paul D, Motlik J, Niemann H. Biol. Reprod. 2000;62:412–419. doi: 10.1095/biolreprod62.2.412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.