Abstract
Hypotonia–cystinuria syndrome (HCS) and 2p21 deletion syndrome are two recessive contiguous gene deletion syndromes associated with cystinuria type I. In HCS patients, only SLC3A1 and PREPL are disrupted. In the 2p21 deletion syndrome, two additional genes (C2orf34 and PPM1B) are lost. Molecular analysis of the SLC3A1/PREPL locus was performed in the patients using quantitative polymerase chain reaction (PCR) methods. HCS in both siblings was confirmed with the deletion screen of the SLC3A1/PREPL locus. Fine mapping of the breakpoint revealed a deletion of 77.4 kb, including three genes: SLC3A1, PREPL and C2orf34. Features not present in classical HCS were a mild/moderate mental retardation and a respiratory chain complex IV deficiency. We report the first patients with a deletion of SLC3A1, PREPL and C2orf34. They present with a phenotype intermediate between HCS and 2p21 deletion syndrome.
BACKGROUND
Recently, two distinct recessive contiguous gene deletion syndromes associated with cystinuria type I have been described. The first, called hypotonia–cystinuria syndrome (HCS, MIM 606407 [OMIM] ), is characterised by neonatal and infantile hypotonia, poor feeding in neonates, growth retardation due to growth hormone deficiency, mild facial dysmorphism and cystinuria type I.1 The second, referred to as 2p21 deletion syndrome,2 shows in addition neonatal seizures, severe developmental delay, elevated serum lactate concentrations, and a reduced activity of the respiratory chain complexes I, III, IV and V.2,3 The difference in phenotype can be explained by the number of genes involved. HCS has been described in 14 families, which all have in common that both alleles of SLC3A1 and PREPL were deleted.1 The 2p21 deletion syndrome has been described in seven patients of a small Bedouin clan and is caused by a homozygous deletion of 179 kb. This deletion disrupts the coding region of at least four genes (SLC3A1, PREPL, PPM1B and C2orf34), explaining the more severe phenotype.2,4 A detailed comparison of both syndromes has been described.5
Cystinuria type I is due to the loss of SLC3A1, which encodes the heavy chain subunit of the cystine and dibasic amino acid transporter in the renal proximal tubule and small intestine.6 Furthermore, the smallest region of overlap of the different deletions causing HCS encompasses only 7.7 kb, and the flanking genes C2orf34 and PPM1B are normally expressed in Epstein–Barr virus (EBV) transformed lymphocytes of HCS patients.1,7 Therefore, it is assumed that loss of PREPL causes the remaining phenotype in HCS patients. The specific contributions for the loss of C2orf34 and PPM1B to the 2p21 deletion syndrome remains unclear.3,5
In this report, we describe two siblings with a homozygous deletion of SLC3A1, PREPL and C2orf34. The clinical presentation aids to the refinement of the genotype–phenotype correlation in both HCS and 2p21 deletion syndrome.
CASE PRESENTATION
This boy is the fifth child of healthy non-consanguineous parents, although they are from the same Moroccan village. Two older brothers died (one at birth and the other at 6 months) with severe unexplained hypotonia of an unknown cause. A sister is patient 2. Clinical observations are summarised in table 1.
Table 1. Clinical observations in the atypical hypotonia–cystinuria syndrome (HCS) patients.
| Patient 1 | Patient 2 | |
| Age | 17 years | 10.5 years |
| Sex | Male | Female |
| Poor sucking, nasogastric tube feeding as neonates | + | + |
| Sitting alone | 13 months | 10 months |
| Walking alone | 3.5 years | 17 months |
| Neonatal hypotonia | + | + |
| Facial dysmorphy | + | + |
| Psychomotor retardation | Moderate | Moderate |
| Growth retardation | + | + |
| GH stimulation (glucagon) | Not performed | Normal |
| Hyperphagia | − | − |
| Cystinuria type I | +1 calculus observed | +1 calculus observed |
| Lithiasis | + | + |
| Nerve conduction velocities | Not performed | Normal |
| Electromyography | Normal (at 5 months) | Normal (at 5 months) |
| Chronic denervation (17 years) | Chronic denervation (9 years) | |
| Muscle biopsy | Disproportionate fibre size | |
| Hypotrophy of type I fibres | ||
| Nuclear inclusions | Nuclear inclusions | |
| Hypotrophy of type II fibres | Hypotrophy of type II fibres | |
| Respiratory chain not studied | Complex IV deficiency | |
| Brain MRI | Normal | Specific abnormalities |
GH, growth hormone; MRI, magnetic resonance imaging.
He was born after a full term uneventful pregnancy with a birth weight of 3.2 kg and head circumference of 35.5 cm. The Apgar score was 10 at 1 min and 5 min. Within an hour after birth the child showed severe hypotonia and inability to suck. He had a Moro reflex but no grasping reflex. There was slight craniofacial dysmorphy including dolichocephaly, frontal bossing, mild ptosis of the eyelids, slight epicanthal folds, arched filtrum, and retrognathia. Brain computed tomography (CT) scan was normal. Electromyography (EMG) at 5 months was normal. A muscle biopsy at 6 months showed disproportionate fibre size, hypotrophy of type I fibres, and a few nuclear inclusions consistent with myotonic dystrophy. This diagnosis was ruled out by molecular studies. On histochemical staining a number of fibres were negative for cytochrome oxidase, but the respiratory chain was not studied. At the age of 13 months, he was able to sit, albeit with a persisting axial and peripheral hypotonia. He had no sucking reflex and was still fed by nasogastric tube. At the age of 24 months a urinary bladder stone was removed. He started walking alone by the age of 3 years and 6 months. Metabolic investigation was normal except for a pronounced increase in the urinary amino acids cystine, lysine, arginine and ornithine (with normal amino acid excretion in the parents).
He was lost from follow-up until at the age of 17 years when he showed moderate mental retardation with learning disabilities and episodes of weakness and fatigability. Height was 6 cm below the third centile, while weight and head circumference were at the third centile. Pubertal development was at Tanner stage IV. There was no hyperphagia. He was treated with citrate. Renal function was normal. Brain magnetic resonance imaging (MRI) showed no abnormalities. Nerve conduction velocity was normal while EMG showed a few myotonic activities, and features of chronic denervation. A repeated muscle biopsy showed mild lesions with hypotrophy of type I fibres, and immunohistochemistry showed a normal expression of dystrophin, caveolin 3 and dysferlin.
Patient 2 is 10.5 years of age and is the sister of patient 1. She was born after a full term uneventful pregnancy with a birth weight of 2.9 kg. The Apgar score was 10 at 1 min and 5 min. She was transferred to the neonatal intensive care unit for poor sucking, arthrogryposis, muscular hypotrophy and hypotonia, and absent deep tendon reflexes.
Urinary amino acid chromatography showed increased cystine, lysine, arginine and ornithine. Urine alkalinisation was started, and she showed weight gain after nasogastric tube feeding was initiated. Arthrogryposis started to regress. At the age of 5 months weight and height were at –3 SD. Hypotonia, poor sitting position, and brisk deep tendon reflexes persisted. EMG was normal and a musculus deltoideus biopsy showed nuclear inclusions, hypotrophy of type II fibres and cytochrome c oxidase (complex IV) deficiency by spectrometric analysis. The other respiratory chain complexes were normal. The child was able to sit at 10 months, and started walking at 17 months. At 8 years, she still showed muscular weakness, hypotonia, and obvious mental retardation. Brain MRI showed localised unspecific white matter subcortical signal anomalies. Renal ultrasonography showed a renal calculus (which was absent at 4 years of age). At 9 years, motor and sensory nerve conduction velocities were normal but EMG showed chronic denervation of the proximal end of the lower limbs. At 10.5 years, height was 118 cm (below the third centile of 127 cm) and weight was 22 kg (below the third centile of 23.8 kg). She showed no signs of puberty, and bone age was 7 years. A growth hormone stimulation test with glucagon showed normal growth hormone concentrations (maximal value 23.7 mIU/l).
INVESTIGATIONS
Quantitative polymerase chain reaction (PCR) was performed using qPCR Master Mix for SYBR Green I detection and fluorescein as internal standard (Eurogentec, Seraing, Belgium) on the MyIQ system (Bio-Rad, Nazareth-Eke, Belgium), in accordance with the manufacturer’s guidelines. PCR conditions were as follows: 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Primers were developed with Primer Express software (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). The relative number of alleles were calculated using the Livak (2−deltaCt) method.8 An amplicon within the p53 gene was used as reference. Primer sequences are listed in table 2.
Table 2. Primer sequences used for quantitative polymerase chain reaction (PCR).
| Forward | Reverse | |
| PREPL_exon2 | AACTTTTTCCAGGATCTTCAGCC | CACTTTTTCAAATGCATCCATGTT |
| PREPL_exon14 | GGAACTTGGACTTGACAGCACC | TCCCAGTTGAATGCAGTGTTTC |
| C2orf34_exon1 | GGCAGGTAAGGGAGAACCTG | GAGGAGTGACCAGAGGCAAA |
| c2orf34_intron1-1kb | AGCTGACCGACATAGGCTGT | GGACCACATGTGCATCATAGA |
| c2orf34_intron1-3kb | CATTGACAGTCTTTCCAGACCA | CCATTTGTAACAATAGCAAAACCA |
| C2orf34_exon2 | AGGCAAAGAAAGGGAAACTGA | TATATTGGACCCATGCACCA |
| SLC3A1_exon2 | ATCGTCCCTTAAAGATTTCAGATATGG | TCAACTTCCCGGAAATCTTCAA |
| SLC3A1_exon4 | TCAGGGAGGGCAATGATCTT | CAGTGACCTATAATGGTATTGCCCA |
| SLC3A1_intron4-9kb | CGTGATGTCATGGGAGTCAG | ATGTAGGTGCCGTGGACTCT |
| SLC3A1_exon9 | AAAGTCACCAATGCAGTGGGA | GCTTCAGAAAAACCAGCATTTGA |
| p53_reference | CCCAAGCAATGGATGATTTGA | GAGCTTCATCTGGACCTGGGT |
Genomic DNA (150 ng) was amplified using the Advantage 2 PCR kit (Clontech, Mountain View, California, USA). Junction fragments were purified with the Montage PCR Centrifugal Filter Device (Millipore, Billerica, Massachusetts, USA) before sequencing (Bigdye Terminator v3.1, Applied Biosystems) on the ABI3130 system (Applied Biosystems). Primers for deletion F are 5′-TCAGGGAGGGCAATGATCTT-3′ and 5′-GGACCACATGTGCATCATAGA-3′.
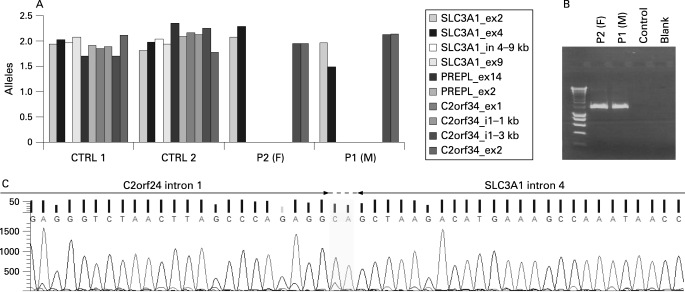
The diagnosis for HCS in both siblings was confirmed by the loss of amplicons for SLC3A1 exon 9 and PREPL exon 14. Fine mapping of the deletion using qPCR pinpointed the proximal breakpoint to a 9 kb interval in SLC3A1 intron 4 (fig 1A). The distal breakpoint was localised in a 3 kb interval of intron 1 of C2orf34, thus completely deleting PREPL (fig 1A). A junction fragment spanning the breakpoints was obtained with primers located in SLC3A1 exon 4 and C2orf34 intron 1 (fig 1B) and sequenced subsequently with the primer located in C2orf34 intron 1. The obtained sequence was compared with the reference sequence (genome assembly 36.2, NCBI) using BLAST (http://www.ncbi.nlm.nih.gov/blast) in order to determine the exact boundaries of the deletion. The breakpoints of the deletion, referred to as deletion F, are located in SLC3A1 intron 4 at position 23330027–28 (genome assembly 36.2, NCBI) and C2orf34 intron 1 at position 2340761–62 (fig 2C, HUGO nomenclature (http://www.hgvs.org/mutnomen/) with SLC3A1 as the first gene involved: c.891+799_C2orf34:138+2245del). Due to a lack of parental DNA, homozygosity for deletion F in both siblings was determined using microsatellite markers spanning a genomic region over 1 Mb (fig 2A). Homozygosity for the five microsatellites was observed (fig 2B), indicative for the presence of deletion F on both alleles.
Figure 1. Genetic analysis of the atypical hypotonia–cystinuria syndrome (HCS) patients.
(A) Quantitative polymerase chain reaction (PCR) analysis on genomic DNA from both siblings and control samples. Only relevant amplicons are shown. (B) Junction fragment PCR spanning the breakpoint. P1 (M), patient 1 (male); P2 (F), patient 2 (female). (C) The sequence at the joining of the deletion ends. The junction fragment PCR product derived from patient DNA was subjected to sequencing with the primer located in C2orf34 intron 1 used for the generation of the junction fragment. The two bases CA could be derived from either side, as they are present in both.
Figure 2. Genetic analysis of the atypical hypotonia–cystinuria syndrome (HCS) patients (continued).
(A) Schematic representation of the 2p21 locus (1.6 Mb interval). The microsatellite marker used for the haplotype analysis are indicated relative to the gene at the 2p21 locus. (B) Haplotype analysis with microsatellite markers flanking the breakpoint. (C) Schematic representation of deletion F (c.891+799_C2orf34:138+2245del) at the 2p21 locus. Sequences flanking the breakpoint are shown and are numbered in accordance with the numbering in the database (Homo sapiens, build 36.2, http://www.ncbi.clm.nih.gov/).
DIFFERENTIAL DIAGNOSIS
Differential diagnosis is that of hypotonia.
TREATMENT
Treatment is that of classical cystinuria. There is no treatment available for the remaining clinical problems apart from symptomatic treatment (e.g. growth hormone replacement therapy).
DISCUSSION
HCS has previously been described in 13 families, distributed across Europe, USA and Canada.1,7 Five different deletions have been described, four of which had in common that only PREPL and SLC3A1 have been deleted. Deletion E also showed a loss of the flanking gene C2orf34, but the loss of only one copy of this gene does not alter the HCS phenotype.1,7 A homozygous deletion of PREPL, SLC3A1, C2orf34 and PPM1B, on the other hand, causes the severe 2p21 deletion syndrome.2 The two siblings described here represent the first patients with an intermediate phenotype. Genetic analysis of the SLC3A1/PREPL locus showed a homozygous loss of both genes. In-depth analysis of the deletion revealed that the first exon of C2orf34 was also missing (which contains the ATG startcodon). The breakpoint in C2orf34 lies 2 kb distal to the breakpoint observed in the 2p21 deletion syndrome.2 It has previously been shown that the loss of exon 1 abolishes the expression of C2orf34 in a lymphoblast cell line of 2p21 deletion patients.4 However, as the 5′ primer was located in exon 1, it was not ruled out that an alternative, in-frame start was used to generate a partial C2orf34 protein. The first in-frame methionine is located in exon 5 of the long isoform, which would give rise to a truncated protein of 167 amino acids, whereas the full length protein contains 323 amino acids. In addition, this would disrupt the predicted methyltransferase domain at the aminoterminus.
As expected in a contiguous gene deletion syndrome, the loss of an additional gene (that is, C2orf34) adds to the severity and/or complexity of the phenotype. The present patients showed the classical symptoms of HCS, such as severe hypotonia at birth, poor feeding, slight facial dysmorphism, growth retardation and cystinuria type I. Surprisingly, only one calculi formation has been reported in both patients, despite the loss of both SLC3A1 alleles. In addition, both atypical HCS patients showed a mild to moderate mental retardation. Muscle biopsies on both patients showed abnormalities, but the respiratory chain has not been studied for patient 1. A cytochrome C oxidase deficiency (complex IV) was observed in patient 2. However, one classical HCS patient (family 13, homozygous for deletion B7) was reported to have a complete complex V deficiency.9 It was postulated that complex V deficiency was caused by additional modifier genes, either in cis or in trans.7 Respiratory chain deficiency was also observed in all patients with the 2p21 deletion syndrome. Only complex II, which is the only complex that does not contain any mitochondrial encoded subunit, had normal activity.2,3
Remarkably, the growth hormone test performed in patient 2 was normal, while in the HCS patients it is mostly abnormal. On the other hand, patients diagnosed with the 2p21 deletion syndrome also show normal growth hormone levels.2 Finally, the early death of two brothers in the present family remains unexplained. It should, however, be noted that no increased incidence of early death has been observed in the known HCS families.
The identification of patients with a loss of SLC3A1, PREPL and C2orf34 is important to gain insight into the contribution of each gene to the phenotype. Absence of a functional SLC3A1 allele causes cystinuria type I.10 Other features of HCS, like hypotonia, minor facial dysmorphism and growth retardation are likely to be caused by the loss of PREPL. This idea was supported by the fact that the flanking genes (C2orf34 and PPM1B) were normally expressed in lymphoblast cell lines of HCS patients.1 Although PREPL shows homology with members of the prolyl oligopeptidase family and harbours an active catalytic machinery, substrates have not been identified so far.1,5,11 This makes it impossible to explain the phenotype caused by the absence of PREPL. Since PREPL might be involved in the activation or degradation of neuropeptides and peptide hormones, it remains to be established if the phenotype is caused by the absence or accumulation of a bioactive peptide
No biochemical data are available for C2orf34 yet. It is predicted to have an S-adenosyl-L-methionine binding domain (COG3897),4 often found in S-adenosyl-L-methionine dependent methyltransferases.12 The lack of C2orf34 causes mild to moderate mental retardation, which could be explained if C2orf34 encodes a genuine methyltransferase. Various methyltransferases, like guanidine acetate methyltransferase (GAMT),13 euchromatin histone methyl transferase1 (Eu-HMTase1)14,15 and FTSJ116 have been associated with mental retardation. Interestingly, FTSJ1 encodes a protein belonging to the same superfamily of S-adenosyl-L-methionine dependent methyltransferases (SCOP ID 53335).16
A putative second phenotype that appears with the loss of C2orf34 is a deficiency in complex IV of the respiratory chain, which was only confirmed in patient 2. Complex IV is composed of 13 subunits, and the biogenesis of a functional complex IV is extremely complicated.17 Mutations in assembly factors cause various autosomal recessive complex IV deficiencies—for example, classical Leigh syndrome (SURF1)18 and French-Canadian Leigh syndrome (LRPPRC).19 Thus, C2orf34 may well have a function in the assembly of complex IV or in the regulation of factors involved.
Finally, PPM1B is absent only in the 2p21 deletion patients. It encodes for protein phosphatase 2Cβ (PP2Cβ), a member of the PPM family of eukaryotic protein serine/threonine phosphatases. The additional phenotypes due to the loss of PPM1B are neonatal convulsions and hypocalcaemia, a moderate to severe psychomotor retardation and a reduced activity of the respiratory chain complexes I, III, V.2,4 It is, however, not clear what the endogenous substrates for PP2Cβ are. In mouse, PP2Cβ has essential functions in early development, as a PPM1B knock-out mouse model shows an early pre-implantation lethality.20 Most likely, redundancy of family members of PPM1B rescues the early lethality in human. Nevertheless, a loss of PPM1B seems to contribute to the severity of the phenotype in the 2p21 deletion patients.
In conclusion, we have identified the first two patients with a homozygous deletion of three genes (SLC3A1, PREPL and C2orf34) at the HCS/2p21 deletion syndrome locus, which has enabled us to delineate further the contribution of each gene to the different phenotypes.
LEARNING POINTS
Atypical hypotonia–cystinuria syndrome (HCS) is caused by the homozygous deletion of SLC3A1, PREPL and C2orf34.
Atypical HCS patients present with a phenotype intermediate between HCS and 2p21 deletion syndrome.
Loss of C2orf34 may cause mental retardation.
Acknowledgments
This article has been adapted with permission from Chabrol B, Martens K, Meulemans S, Cano A, Jaeken J, Matthijs G, Creemers JWM. Deletion of C2orf34, PREPL and SLC3A1 causes atypical hypotonia–cystinuria syndrome. J Med Genet 2008;45:314–8.
Footnotes
Competing interests: None.
REFERENCES
- 1.Jaeken J, Martens K, Francois I, et al. Deletion of PREPL, a gene encoding a putative serine oligopeptidase, in patients with hypotonia-cystinuria syndrome. Am J Hum Genet 2006; 78: 38–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parvari R, Brodyansky I, Elpeleg O, et al. A recessive contiguous gene deletion of chromosome 2p16 associated with cystinuria and a mitochondrial disease. Am J Hum Genet 2001; 69: 869–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parvari R, Hershkovitz E. Chromosomal microdeletions and genes functions: a cluster of chromosomal microdeletions and the deleted genes functions. Eur J Hum Genet 2007; 15: 997–8 [DOI] [PubMed] [Google Scholar]
- 4.Parvari R, Gonen Y, Alshafee I, et al. The 2p21 deletion syndrome: characterization of the transcription content. Genomics 2005; 86: 195–211 [DOI] [PubMed] [Google Scholar]
- 5.Martens K, Derua R, Meulemans S, et al. PREPL: a putative novel oligopeptidase propelled into the limelight. Biol Chem 2006; 387: 879–83 [DOI] [PubMed] [Google Scholar]
- 6.Font-Llitjos M, Jimenez-Vidal M, Bisceglia L, et al. New insights into cystinuria: 40 new mutations, genotype-phenotype correlation, and digenic inheritance causing partial phenotype. J Med Genet 2005; 42: 58–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martens K, Heulens I, Meulemans S, et al. Global distribution of the most prevalent deletions causing hypotonia-cystinuria syndrome. Eur J Hum Genet 2007; 15: 1029–33 [DOI] [PubMed] [Google Scholar]
- 8.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25: 402–8 [DOI] [PubMed] [Google Scholar]
- 9.Zaffanello M, Beghini R, Zamboni G, et al. A sporadic case of cystinuria, respiratory chain and growth hormone deficiencies. Pediatr Nephrol 2003; 18: 846–7 [DOI] [PubMed] [Google Scholar]
- 10.Bisceglia L, Calonge MJ, Dello Strologo L, et al. Molecular analysis of the cystinuria disease gene: identification of four new mutations, one large deletion, and one polymorphism. Hum Genet 1996; 98: 447–51 [DOI] [PubMed] [Google Scholar]
- 11.Szeltner Z, Alshafee I, Juhasz T, et al. The PREPL A protein, a new member of the prolyl oligopeptidase family, lacking catalytic activity. Cell Mol Life Sci 2005; 62: 2376–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pintard L, Kressler D, Lapeyre B. Spb1p is a yeast nucleolar protein associated with Nop1p and Nop58p that is able to bind S-adenosyl-L-methionine in vitro. Mol Cell Biol 2000; 20: 1370–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stockler S, Holzbach U, Hanefeld F, et al. Creatine deficiency in the brain: a new, treatable inborn error of metabolism. Pediatr Res 1994; 36: 409–13 [DOI] [PubMed] [Google Scholar]
- 14.Kleefstra T, Brunner HG, Amiel J, et al. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet 2006; 79: 370–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleefstra T, Smidt M, Banning MJ, et al. Disruption of the gene euchromatin histone methyl transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J Med Genet 2005; 42: 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freude K, Hoffmann K, Jensen LR, et al. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am J Hum Genet 2004; 75: 305–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zee JM, Glerum DM. Defects in cytochrome oxidase assembly in humans: lessons from yeast. Biochem Cell Biol 2006; 84: 859–69 [DOI] [PubMed] [Google Scholar]
- 18.Zhu Z, Yao J, Johns T, Fu K, et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet 1998; 20: 337–43 [DOI] [PubMed] [Google Scholar]
- 19.Mootha VK, Lepage P, Miller K, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci U S A 2003; 100: 605–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki M, Ohnishi M, Tashiro F, et al. Disruption of the mouse protein Ser/Thr phosphatase 2Cbeta gene leads to early pre-implantation lethality. Mech Dev 2007; 124: 489–99 [DOI] [PubMed] [Google Scholar]