Abstract
Deletion of a single nucleotide (7630delT) within MT-CO2, the gene of subunit II of cytochrome c oxidase (COX), was identified in a clinically typical MELAS case. The deletion-induced frameshift results in a stop codon close to the 5′ end of the reading frame. The lack of subunit II (COII) precludes the assembly of COX and leads to the degradation of unassembled subunits, even those not directly affected by the mutation. Despite mitochondrial proliferation and transcriptional upregulation of nuclear and mtDNA-encoded COX genes (including MT-CO2), a severe COX deficiency was found with all investigations of the muscle biopsy (histochemistry, biochemistry, immunoblotting). The 7630delT mutation in MT-CO2 leads to a lack of COII with subsequent misassembly and degradation of respiratory complex IV despite transcriptional upregulation of its subunits. The genetic and pathobiochemical heterogeneity of MELAS appears to be greater than previously appreciated.
BACKGROUND
MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes) is a common clinical presentation caused by mutations in mitochondrial DNA (mtDNA).1–5 This multisystem disorder is characterised by stroke-like episodes, seizures, recurrent headaches, mental retardation, muscle weakness and short stature. Diagnostic laboratory abnormalities are lactic acidosis and deficiencies in the respiratory chain of muscle mitochondria. Most patients first present between the ages of 5 and 15 years.
About 80% of MELAS cases are caused by transition 3243A→G in the mitochondrial tRNALeu(UUR) gene (MT-TL1).4 Other mutations in the same gene, a few other mitochondrial tRNA genes and some of the mitochondrial-encoded subunits of respiratory complex I (MT-ND1, MT-ND5, MT-ND6) account for the remaining cases.1,4,6,7
We report a patient with all the clinical symptoms and laboratory abnormalities typical of MELAS, but devoid of any mutation in a gene previously associated with this disease. Instead, the deletion of a single nucleotide (7630delT) within MT-CO2, the gene of cytochrome c oxidase (COX) subunit II (COII), was identified as the cause of his condition.
CASE PRESENTATION
This patient has previously been described in detail,8 and is thus only briefly summarised and updated here. At the age of 13 years, the patient experienced his first stroke-like episode with reversible aphasia, right hemiparesis and hemianopsia. Exercise intolerance, progressive mental impairment and short stature had previously been noted. Serum lactate was constantly raised. The patient developed secondarily generalised and myoclonic seizures, left ventricular hypertrophy, retinopathy and progressive nephropathy, and had recurrent headaches and vomiting.
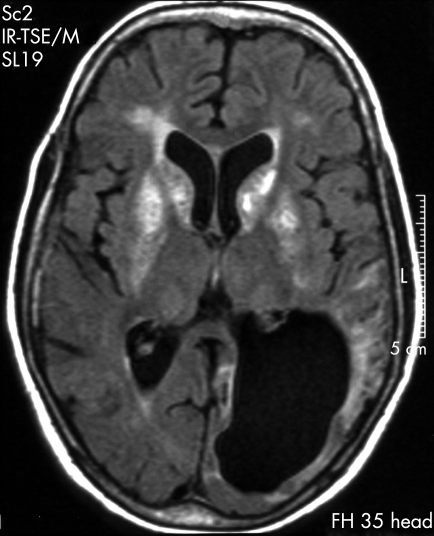
Repeated MRI of the brain over the years revealed an increasing number of focal lesions and progressive cortical atrophy throughout the cerebral cortex, beginning and most pronounced in the left parieto-occipital lobe (fig 1). Brain angiography showed no angiopathic alterations. Basal ganglia calcifications were noted from the first neuroradiological examination (fig 1), and the most last MRI scan taken showed moderate cerebellar lesions. The patient died at the age of 23 years; no postmortem examination was carried out. There is no family history of any kind of mitochondrial disease.
Figure 1. MRI of the patient’s brain at the age of 16 years, almost 4 years after his first stroke-like episode.
The image from an axial fluid-attenuated inversion recovery sequence shows characteristic hyperintensities in the basal ganglia (particularly in the caput nuclei caudati and putamina), a residual infarction-like cortical lesion in the severely atrophic left posterior lobe and the resultant widening of the posterior horn of the left lateral ventricle.
INVESTIGATIONS
A biopsy from the vastus lateralis muscle taken at 13 years of age was processed for routine histochemical staining.9 A small fibre bundle was fixed and processed for transmission electron microscopy. Activities of respiratory chain enzymes and citrate synthase (CS) were measured in the homogenate of the frozen muscle.10
DNA was isolated by standard procedures.11 Southern hybridisation was performed after linearisation of mtDNA with the restriction enzymes BamHI or PvuII.11 Direct sequencing of PCR-amplified segments of mtDNA by dye-terminator chemistry was used to screen the complete mitochondrial genome for mutations.11 Deviations from the revised Cambridge reference sequence (AC_000021)12 were confirmed by reamplification and sequencing. A primer-introduced PCR restriction fragment length polymorphism (PCR-RFLP) assay was used to screen for and quantify 7630delT (see supplementary material).10,13
Proteins were extracted from frozen muscle, separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis, blotted onto polyvinylidene fluoride membranes and detected by antibody staining and ECL Advance chemiluminescence (GE Healthcare Bio-Sciences, Uppsala, Sweden) according to standard protocols (see supplementary materials for antibody details).11
RNA was extracted from frozen muscle and cDNA synthesised by reverse transcription according to standard protocols using oligo d(T)18 primers and M-MuLV reverse transcriptase (Fermentas, St Leon-Rot, Germany).11 Diluted cDNAs were subjected to relative quantification by real-time RT-PCR using SYBR green I on an ABI Prism sequence detection system (Applied Biosystems, Foster City, California, USA) essentially as described previously (see supplementary materials for primer sequences)14,15 and analyses were carried out in triplicate.
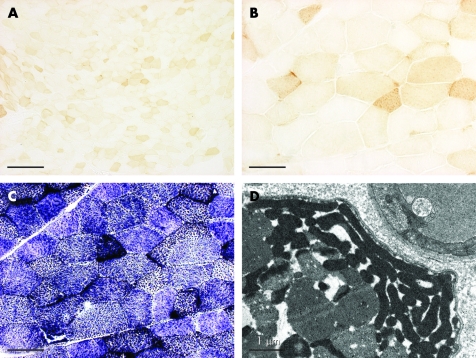
The muscle biopsy revealed pronounced, generalised COX hyporeactivity (>90% of muscle fibres) with numerous fibres completely non-reactive and only a few with near-normal reactivity (fig 2A,B). Succinate dehydrogenase (SDH) staining showed the complementary pattern of mitochondrial proliferation in muscle fibres (fig 2C). However, of the few arterioles identified in the muscle section, only one displayed pronounced SDH reactivity. The presence of ragged-red fibres (∼2%) indicated subsarcolemmal as well as sarcoplasmic accumulation of mitochondria, which was also seen by transmission electron microscopy (fig 2D). Like most muscle fibres of normal appearance, ragged-red fibres were hyporeactive or non-reactive for COX.
Figure 2. Histopathology of the muscle biopsy.
(A) COX staining of transverse section (overview); (B) detail from the centre of (A). (C) Succinate dehydrogenase staining of serial section of (B). (D) Transmission electron micrographic detail of a muscle fibre. Bars: (A) 200 µm; (B, C) 50 µm; (D) 1 µm.
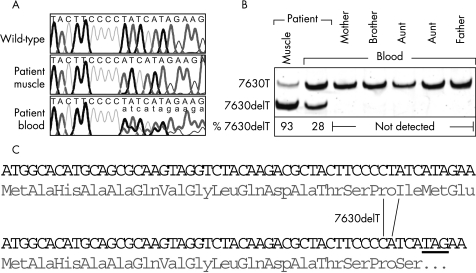
Mitochondrial proliferation was also reflected by the marked increase in CS activity in the muscle homogenate (table 1). Assessment of respiratory chain enzymes corroborated the complex IV deficiency already denoted by histology; COX activity was barely detectable. Other respiratory chain enzyme measurements independent from complex IV were either in the normal range or were also increased (table 1). Southern blotting excluded large-scale rearrangements of muscle mtDNA. As none of the previously reported MELAS mutations was found by the initial, partial sequence analysis,8 the entire muscle mtDNA was finally sequenced. In total, 15 discrepancies from the revised Cambridge reference sequence were found,12 14 of which were previously reported polymorphisms16,17 (see supplementary materials for a complete listing). The 15th, a deletion of T at position 7630 (7630delT; fig 3A), however, is obviously pathogenic. This mutation, 7630delT, gives rise to a frameshift in the reading frame of MT-CO2, resulting in a premature stop of translation (fig 3C). A peptide translated from MT-CO2 harbouring 7630delT will thus consist of only 16 amino acids instead of 227. This mutation was not found in the two major collections of mtDNA mutations.16,17
Table 1. Biochemical assessment of the patient’s muscle biopsy.
| Enzyme | Representing | Activity | Normal range |
| Citrate synthase | Mitochondrial matrix marker | 232 | 43 to 132 |
| NADH:CoQ oxidoreductase | Respiratory complex I | 54 | 17.3 to 45.1 |
| NADH:O2 oxidoreductase | Respiratory complex I/III/IV | 8.4 | 11.4 to 48 |
| Succinate:cytochrome c oxidoreductase | Respiratory complex II/III | 11.2 | 8.8 to 35.9 |
| Cytochrome c oxidase | Respiratory complex IV | 2.9 | 105 to 279 |
Figure 3. Mitochondrial DNA analysis.
(A) Identification of 7630delT. Traces from mtDNA sequencing of the patient’s muscle and blood and from an unaffected control subject; nucleotides 7621 to 7640 shown. Bold black, T; bold grey, A; fine line black, G; fine line grey, C; uppercase, predominant sequence; lowercase, inferior sequence (in case of heteroplasmy). (B) Quantification of 7630delT. PCR restriction fragment length polymorphism analysis of mtDNA from the patient’s muscle and blood and from blood of relatives; percentage of the mutant allele indicated. (C) Effects of 7630delT on the translation of MT-CO2. The first 18 codons/amino acids of MT-CO2 are shown; top, wild-type sequence; bottom, 7630delT sequence; underlined, stop codon resulting from the frameshift.
PCR-RFLP analysis showed that 7630delT was in fact heteroplasmic in the patient’s muscle also (fig 3B). The deletion was also found in the patient’s blood, although at a much lower frequency. None of the patient’s relatives contained the mutation in their blood mtDNA.
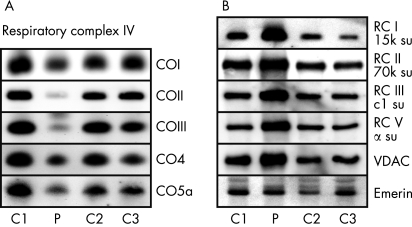
Given the overt pathogenic role of 7630delT, we refrained from doing a single-fibre PCR-RFLP correlation analysis, but instead used the scarce amount of biopsy material to analyse protein and mRNA steady-state levels of various subunits of the respiratory chain to throw light on the pathogenic mechanism of 7630delT. As predicted, the protein levels of COII were dramatically reduced (fig 4A). However, levels of other subunits of respiratory complex IV were also diminished, although not all to the same extent. In stark contrast, (selected) subunits of the other four complexes of the respiratory chain, as well as a mitochondrial marker protein, voltage-dependent anion channel (VDAC), were generally upregulated (fig 4B). Expression of a nuclear protein (emerin) was comparable with controls.
Figure 4. Amounts of protein of various subunits of the respiratory chain in the patient’s muscle.
Immunoblot analysis of (A) selected subunits of respiratory complex IV (COn) and (B) representative subunits (su) of respiratory complex (RC) I, II, III and V and of VDAC and emerin, from the patient (P) and three control subjects (C1–3). See supplementary materials for antibody details and abbreviations.
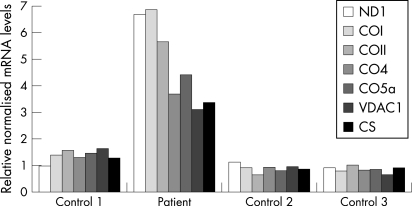
At the transcription level, the picture was quite different (fig 5). Relative quantification of mRNA levels by real-time reverse transcriptase PCR revealed that the transcripts of various subunits of COX, including COII, were all upregulated in the patient’s muscle relative to controls. Consistent with the results of the immunoblot and the biochemical assessment, the mRNA levels of subunit 1 of respiratory complex I (ND1) and the mitochondrial markers VDAC and CS were also increased. Mitochondrial gene expression was about six-fold higher than in controls, whereas expression of genes encoded by the nuclear genome was 3–4-fold increased.
Figure 5. mRNA levels of various subunits of the respiratory chain in the patient’s muscle.
Relative quantification of subunit 1 of respiratory complex I (ND1) mRNA, of selected mRNAs of subunits of respiratory complex IV (COn), of VDAC and of CS from the patient and three control subjects by quantitative real-time reverse transcriptase PCR. The mRNA levels normalised to glyceraldehyde-3-phosphate dehydrogenase mRNA are given relative to the mean of the three controls.
DISCUSSION
COX is the terminal enzyme of the mitochondrial respiratory chain.18 The complex embedded in the inner mitochondrial membrane is composed of 13 subunits. Its highly conserved active core is formed by the three subunits encoded by the mitochondrial genome (COI–III). The CuA centre of COII, chelating two copper ions, forms the entry point for electrons coming from respiratory complex III via cytochrome c. A lack of subunit COII is thus lethal for COX function. The 7630delT mutation excludes >90% of the MT-CO2 reading frame from translation, thereby immediately disclosing its pathogenic role and mechanism.
Besides the almost complete lack of COII, other subunits of COX, which were not directly affected by the mutation, were also reduced. However, the sequential nature of COX assembly offers an explanation for this observation.18–20 Notably, assembly begins with the association of COI and CO4, forming an intermediate complex. Next, after addition of COII, COIII and most of the remaining nuclear encoded components join. Apparently, lack of COII precludes assembly of COX beyond the first intermediate stage and leads to the degradation of unassembled subunits by the mitochondrial proteolytic system.21 Accordingly, COX subunits were generally reduced in the patient’s muscle despite their strong transcriptional upregulation; association of COI and CO4 in the first intermediate assembly complex could explain their less pronounced reduction. A very similar pattern of COX subunit destabilisation was previously reported for a patient with a heteroplasmic missense mutation in MT-CO2.20,22
Typically for an mtDNA mutation impairing respiratory chain function, the patient’s muscle had reacted with a pronounced mitochondrial proliferation; the increase in mitochondria and in mitochondrial gene expression was evident using several investigative methods. Interestingly, transcriptional upregulation of mtDNA-encoded genes was generally stronger than that of nuclear mitochondrial genes, thereby substantiating the idea that a greater abundance of mitochondrial transcripts compensates for the poor mitochondrial translation initiation efficiency, resulting in similar levels of respiratory chain subunits synthesised in different cellular compartments.23
Cytoplasmic mRNAs containing a premature stop codon are subjected to degradation by a quality-control mechanism termed “nonsense-mediated decay”.24 No such surveillance pathway appears to operate in mitochondria, as relative mRNA levels of mutant COII were increased to the same extent as other mitochondrial mRNAs.
The clinical symptoms and the neuroradiological findings of our patient were all most characteristic of MELAS, with all the cardinal symptoms and all the common (present in >50% of cases) features as defined and listed by Hirano and Pavlakis, except hearing loss, being present.3,4 Nephropathy was the only one of the rarer MELAS features recorded. The histopathology and mitochondrial biochemistry of the patient’s muscle biopsy, however, reflected the underlying molecular defect more specifically than exemplifying MELAS. Specifically, respiratory complex I, which appears to be the primary biochemical target in 3243A→G-MELAS,3–5,25 was not affected, but in fact increased as a result of the mitochondrial proliferation. Furthermore, all ragged-red fibres were COX-negative, just like most other muscle fibres, whereas ragged-red fibres in 3243A→G-MELAS are often COX-positive or even hyper-reactive.3–5,26 Obviously, massive mitochondrial proliferation in ragged-red fibres suffices to restore cellular COX activity to normal or greater in cases of protein biosynthesis deficiency caused by, for example, 3243A→G, but is insufficient to compensate for a genetic COX deficiency. Accordingly, intramuscular arteries, which are typically COX-positive in 3243A→G-MELAS as a result of mitochondrial proliferation in the smooth-muscle cells of the arterial walls,3–5,26,27 were COX-negative in the muscle biopsy taken from our patient. However, only a single small arteriole displayed the increased SDH reactivity characteristic of mitochondrial proliferation,3–5,26,27 and the biopsy did not contain any vessels as large as those originally reported as being strongly SDH-reactive vessels.27
MELAS caused by a deleterious mutation in an essential COX subunit gene with its associated morphological and biochemical peculiarities as discussed above, questions current concepts of MELAS pathophysiology. Specifically, a critical role for “elevated” COX in (strongly SDH-reactive) blood vessels through its ability to sequester nitric oxide was proposed.5,28 The predicted shortage in the concentration of circulating nitric oxide was suggested to be fatal by occasionally preventing vasodilatation in times or places needed, thereby triggering the stroke-like episodes characteristic of MELAS. The 7630delT mutation challenges this concept. Owing to the detrimental nature of this mutation, mitochondrial proliferation in skeletal muscle fibres or blood vessels could not restore COX activity. In fact, COX peptides and activity were barely detectable in the muscle homogenate, and histochemistry evidence indicated that it was restricted to a few muscle fibres only. Nonetheless, the patient suffered from the pathognomonic stroke-like episodes as well as all the other devastating clinical features of MELAS.
Acknowledgments
This article has been adapted with permission from Rossmanith W, Freilinger M, Roka J, Raffelsberger T, Moser-Their K, Prayer D, Bernert G, Bittner RE. Isolated cytochrome c oxidase deficiency as a cause of MELAS. J Med Genet 2007;45:117–21.
Footnotes
Competing interests: None.
REFERENCES
- 1.McFarland R, Taylor RW, Turnbull DM. Mitochondrial disease—its impact, etiology and pathology. Curr Top Dev Biol 2007; 77: 113–55 [DOI] [PubMed] [Google Scholar]
- 2.Pavlakis SG, Phillips PC, DiMauro S, et al. Mitochondrial myopathy, encephalopathy, lactic acidosis and strokelike episodes: a distinctive clinical syndrome. Ann Neurol 1984; 16: 481–8 [DOI] [PubMed] [Google Scholar]
- 3.Hirano M, Pavlakis SG. Mitochondrial myopathy, encephalopathy, lactic acidosis and strokelike episodes (MELAS): current concepts. J Child Neurol 1994; 9: 4–13 [DOI] [PubMed] [Google Scholar]
- 4.Thambisetty M, Newman NJ. Diagnosis and management of MELAS. Expert Rev Mol Diagn 2004; 4: 631–44 [DOI] [PubMed] [Google Scholar]
- 5.Scaglia F, Northrop JL. The mitochondrial myopathy encephalopathy, lactic acidosis with stroke-like episodes (MELAS) syndrome: a review of treatment options. CNS Drugs 2006; 20: 443–64 [DOI] [PubMed] [Google Scholar]
- 6.Kirby DM, McFarland R, Ohtake A, et al. Mutations of the mitochondrial ND1 gene as a cause of MELAS. J Med Genet 2004; 41: 784–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liolitsa D, Rahman S, Benton S, et al. Is the mitochondrial complex I ND5 gene a hot-spot for MELAS causing mutations? Ann Neurol 2003; 53: 128–32 [DOI] [PubMed] [Google Scholar]
- 8.Barisic N, Bernert G, Ipsiroglu O, et al. Effects of oral creatine supplementation in a patient with MELAS phenotype and associated nephropathy. Neuropediatrics 2002; 33: 157–61 [DOI] [PubMed] [Google Scholar]
- 9.Taylor RW, Schaefer AM, Barron MJ, et al. The diagnosis of mitochondrial muscle disease. Neuromuscul Disord 2004; 14: 237–45 [DOI] [PubMed] [Google Scholar]
- 10.Rossmanith W, Raffelsberger T, Roka J, et al. The expanding mutational spectrum of MERRF: substitution G8361A in the mitochondrial tRNALys gene. Ann Neurol 2003; 54: 820–3 [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J, Russell DW. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2001 [Google Scholar]
- 12.Andrews RM, Kubacka I, Chinnery PF, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 1999; 23: 147. [DOI] [PubMed] [Google Scholar]
- 13.Yoneda M, Tanno Y, Tsuji S, et al. Detection and quantification of point mutations in mitochondrial DNA by PCR. Methods Enzymol 1996; 264: 432–41 [DOI] [PubMed] [Google Scholar]
- 14.Vandesompele J, De Paepe A, Speleman F. Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT-PCR. Anal Biochem 2002; 303: 95–8 [DOI] [PubMed] [Google Scholar]
- 15.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingman M. mtDB– Human Mitochondrial Genome Database. http://www.genpat.uu.se/mtDB (accessed January 2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallace DC, Lott MT.MITOMAP: a Human Mitochondrial Genome Database. http://www.mitomap.org (accessed January 2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khalimonchuk O, Rödel G. Biogenesis of cytochrome c oxidase. Mitochondrion 2005; 5: 363–88 [DOI] [PubMed] [Google Scholar]
- 19.Shoubridge EA. Cytochrome c oxidase deficiency. Am J Med Genet 2001; 106: 46–52 [DOI] [PubMed] [Google Scholar]
- 20.Taanman JW, Williams SL. Assembly of cytochrome c oxidase: what can we learn from patients with cytochrome c oxidase deficiency? Biochem Soc Trans 2001; 29: 446–51 [DOI] [PubMed] [Google Scholar]
- 21.Koehler CM. Mitochondrial biogenesis. : Holt IJ, ed. Genetics of mitochondrial diseases. Oxford, NY: Oxford University Press, 2003: 47–68 [Google Scholar]
- 22.Rahman S, Taanman JW, Cooper JM, et al. A missense mutation of cytochrome oxidase subunit II causes defective assembly and myopathy. Am J Hum Genet 1999; 65: 1030–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taanman JW. Mitochondrial DNA expression. : Holt IJ, ed. Genetics of mitochondrial diseases. Oxford, NY: Oxford University Press, 2003: 27–46 [Google Scholar]
- 24.Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol 2005; 17: 316–25 [DOI] [PubMed] [Google Scholar]
- 25.Jacobs HT. Disorders of mitochondrial protein synthesis. Hum Mol Genet 2003; 12(Suppl 2): R293–301 [DOI] [PubMed] [Google Scholar]
- 26.Goto Y. Clinical features of MELAS and mitochondrial DNA mutations. Muscle Nerve 1995; (Suppl 3): S107–12 [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa H, Matsuoka T, Goto Y, et al. Strongly succinate dehydrogenase-reactive blood vessels in muscles from patients with mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes. Ann Neurol 1991; 29: 601–5 [DOI] [PubMed] [Google Scholar]
- 28.Naini A, Kaufmann P, Shanske S, et al. Hypocitrullinemia in patients with MELAS: an insight into the “MELAS paradox”. J Neurol Sci 2005; 229–230: 187–93 [DOI] [PubMed] [Google Scholar]