Abstract
We report on a fragile X mosaic male full mutation/normal allele detected by PCR and methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA). This combined analysis provides a diagnostic approach for fragile X syndrome (FXS). The method assesses the presence of expansion (full mutation), the CpG methylation status and could determine copy number changes (large deletions/duplications) along the FMR1 and FMR2 (fragile X mental retardation) genes. The method avoids detection of premutations, which makes it applicable for newborn screening. It can also be used in clarification of mosaic cases. The PCR results in our patient showed one normal allele; three repeats larger than his mother’s one. The MS-MLPA showed hypermethylated full mutation pattern in the proband. Both results are compatible with FXS mosaic case full mutation/normal allele. The patient demonstrates atypical mild clinical manifestation of the disease, which correlates to the presence of a normal size allele in the patient’s cells.
BACKGROUND
Fragile X syndrome (FXS, OMIM#300624), the most frequent cause of X-linked mental retardation is provoked mostly by expansion of unstable trinucleotide CGG repeat in the 5′UTR of the FMR1 (fragile X mental retardation 1) gene. As a result of this expansion, the promoter of the FMR1 gene is hypermethylated resulting in gene silencing. The polymorphic CGG repeat varies between 6 and 54±2 copies in normal individuals. The carriers of CGG repeats between 55±2 and 200, the so called “grey zone”, will not develop the disease, but the repeat may expand in the next generations.1,2 The individuals who possess >200 CGG copies are missing the gene product and the symptoms of FXS are present. It is well known that mosaicism for methylated full mutation and an unmethylated premutation account for more than 40% of affected males.3 To the best of our knowledge, mosaic cases of full mutation/normal allele or full mutation/deletion are rarely detected.3–7 The precise DNA diagnosis and assessment of the expansion is technically very complicated. It is mainly done by classical Southern blot analysis and radioactive or chemiluminescent detection. There are also some attempts described in the literature to use methylation-specific PCR-based assay after sodium bisulphate treatment of the genomic DNA to discriminate normal, premutated and full mutated alleles.8 Very promising is betaine-based-PCR in combination with “chimeric” CGG-targeted primer used by Tassone et al.9 The method could be applied for testing both males and females, it could distinguish normal homozygous females from hemizygous carriers of a pre/full mutation and it is suitable for a population screening method.9
Here we report on a FXS mosaic male full mutation/normal allele detected by a combination of PCR and methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA).10 This combined analysis provides a powerful, fast and easy to perform diagnostic approach for FXS, which assesses not only the presence of expansion but also the CpG methylation status and gives the possibility to detect mosaic cases. The MS-MLPA kit is designed to detect also large deletions/duplications along the FMR1 and FMR2 genes. Together with the FMR1 specific probes, FMR2 probes (FRAXE, OMIM#309548) are also included in the kit. Moreover, the MS-MLPA method provides crucial distinction between premutation and full mutation alleles, as premutation displays no promoter hypermethylation, whereas in full mutation the promoter is hypermethylated to varying degrees.11 This advantage makes the method extremely applicable in screening programmes, like newborn screening.
CASE PRESENTATION
The affected patient we report here is a 23-year-old male. He belongs to a Bulgarian family with two affected males (the proband we discuss here and his maternal first cousin). The cousin has been more severely affected (as reported by the proband’s mother) with severe problems in language management and profound mental retardation. Both boys have never been considered as having FXS.
Our patient has demonstrated intellectual impairment since childhood. No physical delay has been reported. The mental/behavioural problems have been noticed at about 3 years of age: partial avoidance of eye contact, attention deficit, from time to time hyperactivity and aggression has been present. The patient has no facial abnormalities and his mother does not show any dysmorphic features. No speech delay has been noticed by the parents, but the language skills have been impaired. From the very beginning, in school he has shown cognitive decline. He has graduated school for problematic children and at present is socially engaged as a construction worker. The karyotype analysis has been performed at an early age and the result has shown 46, XY normal male karyotype. Recently, the clinicians’ attention has been focused on this patient again regarding his sister’s pregnancy. For the first time, a kind of X-linked mental retardation has been suspected and the patient has been referred to DNA analysis for FXS.
INVESTIGATIONS
The PCR amplification was performed using the following primers and protocol: the sample DNA was subjected to chemical denaturation by 2 M NaOH and then precipitated by 2 M ammonium acetate and absolute ethanol. The amplification reaction was carried out in a 50 μl mixture, containing 0.2 μM of each primer9 (one of them was 6-FAM end labelled); 0.2 mM dNTPs; 2.2 M betaine; 2% DMSO; 1× supplied PCR reaction buffer (GENET BIO, Chungnam, Korea) and 2.5U Prime Taq (GENET BIO). The amplification conditions were denaturation at 97°C for 35 s; annealing at 64°C for 35 sec and synthesis at 68°C for 4 min (10 cycles); denaturation at 97°C for 35 sec; annealing at 64°C for 35 sec and synthesis at 68°C for 6 min (25 cycles).
The PCR fragments were analysed on ABI 310 genetic analyser (Applied Biosystem, California, USA) in the presence of ROX500 size standard (Applied Biosystems).
For confirmation of the PCR obtained results, the PCR fragments were separated in acrylamide gel followed by Southern blot and hybridisation with (CGG)5-DIG labelled probe (DIG-labelling and detection kit; Roche Applied Sciences, Indianoplis, USA).
The MS-MLPA was performed following the manufacturer’s instructions.10 The DNA samples were purified by standard phenol/chloroform extraction. Between 50 and 200 ng of DNA were diluted in TE buffer up to 4 μl total volume. The diluted samples were subjected to a short denaturation for 10 min in 1 μl denaturing buffer supplied within the MLPA kit. The hybridisation with the FMR1 specific probes as well as a number of control probes was performed at 60°C for overnight. Each sample was divided in two for two separate tests: the ligation reaction, which is the copy number test (CNT) for deletions/duplications detection, and the ligation-digestion reaction, which is the methylation test for methylation quantification at CpG islands.10 In both tests, the hybridised probes were ligated by a specific ligase mix provided by the manufacturer. In the methylation test, the methylation-sensitive restriction enzyme HhaI (Pharmacia Biotech, Sunnyvale, California USA) was added. The final step represents PCR amplification of the obtained fragments. The PCR buffer, PCR primers, the enzyme dilution buffer and the polymerase are provided in the kit.
The obtained PCR products were analysed on ABI 310 genetic analyser (Applied Biosystem) in the presence of ROX500 size standard (Applied Biosystems). Each patient sample is analysed simultaneously with at least two normal male samples and a full mutated male control.
The MLPA data interpretation for the CNT has to be performed by Coffalizer software10 or by Excel in order to assess copy number changes in comparison with the normal controls. This analysis is not presented here as we do not communicate on deletion/duplication pattern.
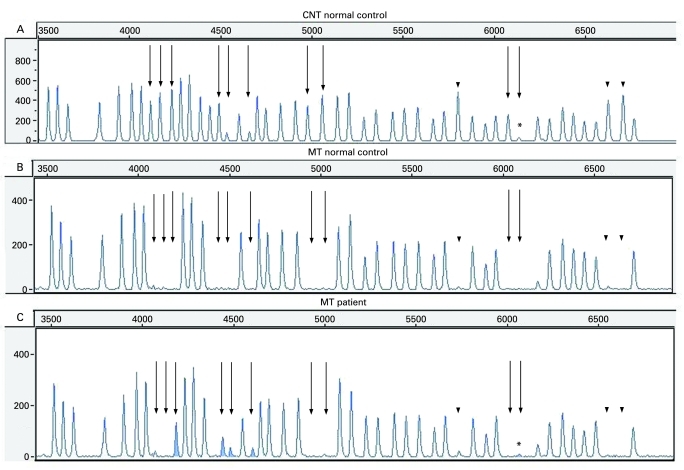
The methylation test results are easy to interpret as the five FMR1 exon 1 methylation-sensitive fragments (see fig 1A, the straight arrows) are absent in the normal controls after HhaI digestion (see fig 1B), but they appear as a result of hypermethylation in the full mutated patients (see fig 1C).
Figure 1.
The methylation-specific multiplex ligation-dependent probe amplification (MS-MLDPA) profiles. (A) Copy number test (CNT) in a normal male control. Straight arrows: methylation-specific FMR1 probes; dotted arrows: methylation-specific FMR2 probes; arrowheads: digestion controls. The fragment marked by asterisk is FMR1 exon 1 specific probe, which gives low signal and might be less reliable (manufacturer’s communication). (B) Methylation test in a normal control. All methylation-specific bands (pointed by different arrows) are missing as a result of HhaI digestion. (C) Methylation test (MT) in the presented patient. The five FMR1 exon 1 methylation-specific probes gave a signal (peaks are given in grey and pointed by straight arrows), as a result of hypermethylated full mutation, not digested by HhaI. The remaining methylation-specific fragments (FMR2 and digestion controls) are missing showing that the digestion was successful and there is no hypermethylation along the FMR2 gene.
DIFFERENTIAL DIAGNOSIS
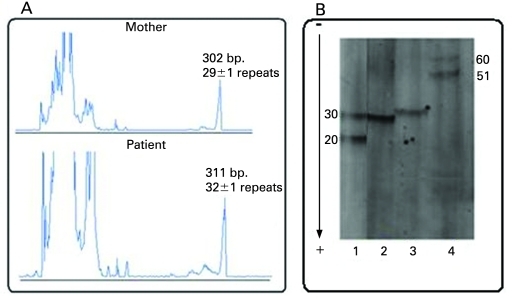
The PCR was performed on DNA from the mother, the patient, a normal control 20/30 CGG repeats and a premutated control about 51/60 CGG repeats. The PCR results showed one allele of about 29±1 CGG repeats in the mother and one allele in the affected son, but three repeats larger (about 32±1 CGG repeats) (fig 2A and B).
Figure 2.
(A) ABI electrophoretic profile of the PCR amplified CGG repeats in the mother and the patient. The fragment’s size in base pairs and the number of CGG repeats are provided above each peak. (B) PCR profile after acrylamide separation and Southern blot hybridisation with (CGG)5-DIG labelled probe. 1: control sample 20/30 CGG repeats; 2: mother 29±1 CGG repeats; 3: patient 32±1 CGG repeats; 4: control sample 51/60 CGG repeats.
The MS-MLPA was preliminary tested on 10 normal male controls and 6 full mutated FXS patients. The obtained methylation test results showed crucial discrimination between the control samples and the full mutated ones (data not shown). The patient presented here showed hypermethylated full mutation pattern compared with the normal control (fig 1). The panel A represents all specific fragments obtained after probes hybridisation10 (the CNT with no methylation specific enzyme added). The dotted arrows indicate methylation-specific FMR2 probes; the straight arrows point to methylation-specific FMR1 probes. The peaks used as digestion controls are marked by arrowheads. The panel B is the methylation test result in a normal control where HhaI methylation-specific enzyme digested the non-methylated FMR1 exon 1 specific fragments and they disappear. The same happens with the non-methylated FMR2 specific fragments included in the kit and the digestion control fragments. All methylation-sensitive fragments disappear in a normal control where they are not hypermethylated. The panel C represents the methylation test profile of the presented patient, where the five FMR1 exon 1 specific fragments are visible, as a result of a full mutation and a hypermethylation. The HhaI recognition site has been methylated and, hence, not digested by the enzyme. The remaining FMR2 methylation-specific fragments, as well as the digestion control fragments, are not present in the patient’s profile demonstrating that the HhaI digestion reaction was successful and there was no hypermethylation along the FMR2 gene-specific regions.
The obtained results by PCR/MS-MLPA were confirmed by the classical Southern blot, which showed normal allele of about 29±1 repeats and premutation of about 90 repeats in the mother and mosaic pattern of 33 and 500 CGG repeats in the patient (data not shown).
OUTCOME AND FOLLOW-UP
To the best of our knowledge, this is the first report on successful application of MS-MLPA for diagnostic purposes in FXS.
The atypical mild clinical manifestation of the disease in the present case correlates to the presence of a normal-size allele in the patient’s cells.
DISCUSSION
The combined application of PCR and MS-MLPA gives the possibility in few steps to detect normal FMR1 alleles, to prognose the expanded ones, to assess the CpG islands methylation, as well as to determine copy number changes like large deletions/duplications along the FMR1/FMR2 genes. Moreover, the MS-MLPA method detects only full mutation alleles, while premutation ones are omitted,11 which makes it suitable for screening programmes like newborn screening. As it can be seen from the present study, this combined analysis gives the possibility also successfully to detect mosaic cases.
In our opinion, the mosaic pattern of normal size/full mutation alleles was a result from inheritance of a maternal unstable premutated allele. The most logical mechanism for normal-size allele generation in our mosaic case is a deletion/regression within the frame of the full mutation restricted to the CGG repeat itself3,6,7,12 as the primers we used for PCR were designed in the repeat flanking regions. Only one case of maternal premutation reversion to a normal-size allele resulting in a non-mosaic male is reported so far,13 but in our opinion the frequency of such cases might be underestimated because of lack of clinical manifestation.
LEARNING POINTS
Fragile X mosaic male full mutation/normal allele was detected by PCR and methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA).
The patient demonstrates atypical mild clinical manifestation of the disease, which correlates to the presence of a normal size allele in the patient’s cells.
This is the first report on successful application of MS-MLPA method for diagnostic purposes in fragile X syndrome.
The MS-MLPA method assesses the presence of expansion (full mutation), the CpG methylation status and could determine copy number changes (large deletions/duplications) along the FMR1 and FMR2 genes.
The method avoids detection of premutations, which makes it applicable for newborn screening.
Acknowledgments
A fellowship from the Alexander von Humboldt Foundation to A Todorova is gratefully acknowledged.
Footnotes
Competing interests: none.
Patient consent: Patient/guardian consent was obtained for publication.
REFERENCES
- 1.Fu YH, Kuhl DP, Pizzuti A, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 1991; 67: 1047–58 [DOI] [PubMed] [Google Scholar]
- 2.Jin P, Warren ST. Understanding the molecular basis of fragile X syndrome. Hum Mol Genet 2000; 9: 901–8 [DOI] [PubMed] [Google Scholar]
- 3.Nolin SL, Glicksman A, Houck GE, et al. Mosaicism in fragile X affected males. Am J Med Genet 1994; 51: 509–12 [DOI] [PubMed] [Google Scholar]
- 4.van der Ouweland AMW, de Vries BBA, Bakker PLG, et al. DNA diagnosis of the fragile X syndrome in a series of 236 mentally retarded subjects and evidence for a reversal mutation in the FMR1 gene. Am J Med Genet 1994; 51: 482–5 [DOI] [PubMed] [Google Scholar]
- 5.Mingroni-Netto RC, Haddad LA, Vianna-Morgante AM. The number of CGG repeats of the FMR1 locus in premutated and fully mutated heterozygotes and their offspring: implications for the origin of mosaicism. Am J Med Genet 1996; 64: 270–3 [DOI] [PubMed] [Google Scholar]
- 6.Schmucker B, Seidel J. Mosaicism for a full mutation and a normal size allele in two fragile X males. Am J Med Genet 1999; 84: 221–5 [PubMed] [Google Scholar]
- 7.Grasso M, Faravelli F, Lo Nigro C, et al. Mosaicism for the full mutation and a microdeletion involving the CGG repeat and flanking sequences in the FMR1 gene in eight fragile X patients. Am J Med Genet 1999; 85: 311–16 [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, Law HY, Boehm CD, et al. Robust fragile X (CGG)n genotype classification using a methylation specific triple PCR assay. J Med Genet 2004; 41: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tassone F, Pan R, Amiri K, et al. A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J Mol Diagn 2008; 10(1): 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MRC-Holland. http://www.mlpa.com (accessed 7 March 2009) [Google Scholar]
- 11.Nygren A, Lens S, Carvalho R. Methylation-specific multiplex ligation-dependent probe amplification enables a rapid and reliable distinction between male FMR1 premutation and full mutation alleles. J Mol Diagn 2008; 10: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mila M, Castellvi-Bel S, Sanchez A, et al. Mosaicism for the fragile X syndrome full mutation and deletions within the CGG repeat of the FMR1 gene. J Med Genet 1996; 33: 338–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabolacci E, Pomponi MG, Pietrobono R, et al. A unique case of reversion to normal size of a maternal premutation FMR1 allele in a normal boy. Eur J Hum Genet 2007; 16: 209–14 [DOI] [PubMed] [Google Scholar]