Abstract
Nucleocytoplasmic export and biogenesis of mRNPs are closely coupled. At the gene, concomitant with synthesis of the pre-mRNA, the transcription machinery, hnRNP proteins, processing, quality control and export machineries cooperate to release processed and export competent mRNPs. After diffusion through the interchromatin space, the mRNPs are translocated through the nuclear pore complex and released into the cytoplasm. At the nuclear pore complex, defined compositional and conformational changes are triggered, but specific cotranscriptionally added components are retained in the mRNP and subsequently influence the cytoplasmic fate of the mRNP. Processes taking place at the gene locus and at the nuclear pore complex are crucial for integrating export as an essential part of gene expression. Spatial, temporal and structural aspects of these events have been highlighted in analyses of the Balbiani ring genes.
Introduction
Gene expression in eukaryotic cells requires the coordinated action of many molecular processes and the machines that carry them out. Pre-mRNAs, synthesised by transcription of protein-coding genes, are assembled into pre-mRNA–protein (pre-mRNP) complexes, processed, transformed into export competent mRNA–protein (mRNP) complexes and exported to the cytoplasm. There is no sharp distinction between components present in pre-mRNPs and components present in mRNPs. Some components interact transiently either with pre-mRNPs or mRNPs, while other components are present in both pre-mRNPs and mRNPs. In addition, an mRNP does not consist of the same components when it enters the nuclear pore complex (NPC) as when it emerges into the cytoplasm.
Many of the pre-mRNP/mRNP components are abundant in the nucleus, such as small nuclear ribonucleoproteins (snRNPs) and several of the heterogeneous nuclear ribonucleoproteins (hnRNPs), and they recognise binding determinants in the transcripts, such as splice sites and poly(A) signals and more or less specific binding motifs. As a result, different pre-mRNPs contain largely the same components. However, gene-specific sequences and unique exon–intron organisation, in combination with the set of available binding components, result in pre-mRNPs that have gene-specific combinatorial compositions. In addition, the composition of a pre-mRNP along a transcribing gene changes as components bind, processing occurs and as components in some cases leave the pre-mRNP.
The spatial separation of making translatable mRNPs from the translation process in eukaryotic cells has made possible additional levels of regulation of gene expression. It has become more and more evident that the mechanisms for export of mRNPs are integrated in the biogenesis of the mRNP. The molecular machines that are responsible for biogenesis have evolved to interact with each other to achieve coordination, efficiency, regulation and quality control. The transcription machinery and the transcription process are at the centre of coordinating many of the processes and the molecular machines. It is moreover evident that the NPC provides a molecular environment that triggers rebuilding of the mRNP, essential for the future cytoplasmic functions.
Our knowledge of the chain of molecular events, from synthesis of pre-mRNPs at the gene to entrance of mRNPs into the cytoplasm, has been gained mainly by combining results from in vitro analyses of individual processing events, studies of gene constructs introduced into cells and yeast genetics. More knowledge about the expression of specific, endogenous genes in vivo is needed to understand the temporal, spatial and structural coordination of the different steps and of the different molecular machines that are involved. Analyses of the expression of the Balbiani ring (BR) genes in the intact polytene nuclei of the dipteran Chironomus tentans have contributed essential and unique information (reviewed in Daneholt 2001a). For the BR genes, it is possible to study most nuclear events at the gene, in the interchromatin space and at the NPCs. Here, we will use the BR genes and their pre-mRNPs/mRNPs as reference to summarise the knowledge about the steps of pre-mRNP/mRNP formation and export in eukaryotic cells. The overall intranuclear steps in gene expression as seen for BR1/BR2 genes are shown in Fig. 1. At the gene, the pre-mRNA is synthesised and assembled with hnRNP proteins, processing, export and quality control components. The released, export competent BR mRNP moves randomly by diffusion through the interchromatin space. At the NPC, the BR mRNP binds, moves into the central channel, goes through conformational and compositional changes and, at the cytoplasmic side, translation is rapidly initiated.
Fig. 1.
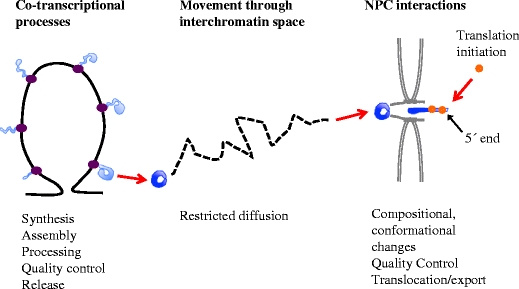
Overview of the intranuclear steps of expression of a BR1/BR2 gene. Processes taking place at the gene locus, in the interchromatin space and at the NPC are listed. BR gene chromatin in black, RNA polymerase II elongation complex in purple, pre-mRNPs in light blue, mRNPs in dark blue and ribosomes in orange. The NPC and nuclear membrane are in grey
The cotranscriptional formation of export competent mRNPs
According to structural determination of the elongating RNA polymerase II in yeast (Kornberg 2007), about eight bases of the pre-mRNA are base paired to the DNA. It is suggested that the nascent pre-mRNA chain appears on the surface of the RNA polymerase II after reaching approximately double this length. At this exit site, it is likely that the pre-mRNA rapidly associates with various proteins and snRNPs that have access to sequence motifs in the pre-mRNA on a competitive basis. Analyses of transcribing BR genes by electron microscopy (EM) show that pre-mRNP assembly is initiated very early and that the transcription elongation complex and the pre-mRNP form a closely connected multi-molecular structure (Skoglund et al. 1983; Wetterberg et al. 2001) (Fig. 2). The observed gene-specific combination of SR proteins bound to pre-mRNAs in a single nucleus support that binding affinities determines the combinations of proteins in each pre-mRNP (Björk et al. 2009). The sequential exposure of newly synthesised pre-mRNA sequences during transcription presumably contributes to order during assembly and processing. This is exemplified in the BR3 gene where introns are excised in an overall 5′ to 3′ order (Wetterberg et al. 1996).
Fig. 2.
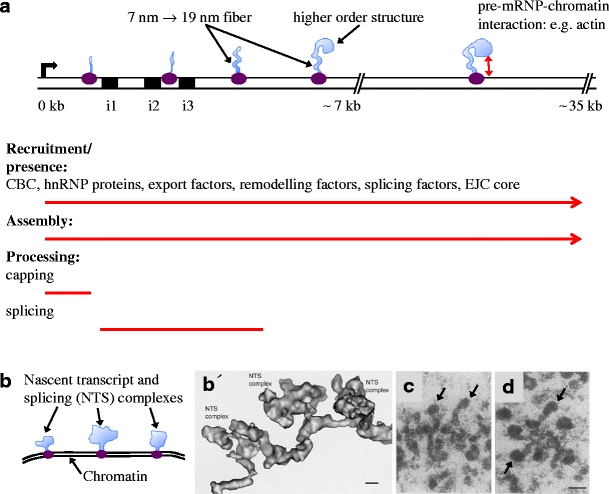
Cotranscriptional synthesis, assembly and processing of BR1/BR2 and BR3 pre-mRNPs. a The proximal (0–7 kb) and middle (7–35 kb) regions of the BR1/BR2 gene is shown schematically. The bent arrow indicates the transcription start site. The RNA polymerase II elongation complex in purple and the pre-mRNPs in light blue. The proximal region contains three introns (i1–i3, black boxes) and exons 1–3, followed by the long (approximately 35 kb) exon 4. The continuous recruitment of components to the pre-mRNPs (listed components have been demonstrated) and the continuous assembly of the pre-mRNPs are indicated by red arrows. The processing events of the pre-mRNPs are indicated (red lines). Assembly of the exon sequences results in a 7-nm pre-mRNP fibre that is further folded into a 19-nm fibre, followed by organisation into higher order structures. Interaction between the pre-mRNP and chromatin may involve pre-mRNP-bound actin. b Schematic representation of part of the BR3 gene. The BR3 gene contains multiple short introns and exons. Three nascent transcript and splicing (NTS) complexes are shown. Each complex consists of a pre-mRNP (light blue), including splicing factors and an RNA polymerase II elongation complex (purple). b′ EM 3D reconstruction of a corresponding BR3 gene segment with three NTS complexes (adopted from Wetterberg et al. 2001). The repeated assembly and structural dynamics of the spliceosome along the multi-intron gene dominate the structure of the pre-mRNPs. The scale bar represents 10 nm. c, d EM images of nascent BR1/BR2 pre-mRNPs in the middle region of the gene (adopted from Skoglund et al. 1983). The 19-nm fibre and the higher order granular structure are seen in several pre-mRNPs. The granular structure (arrows) gets bigger and denser as more RNA protein is added (compare c and d). The scale bar represents 50 nm
It is not understood how efficient access for all pre-mRNA interacting components is orchestrated close to the surface of the RNA polymerase II exit channel. It is possible that the components get access to the pre-mRNP because they continuously and randomly move within the interchromatin space. It is also possible that the local concentration of components is increased at active genes by intermolecular interactions. The RNA polymerase II C-terminal domain (CTD) is located in convenient proximity to the pre-mRNA exit channel, and it is implicated in loading components onto the pre-mRNA. Apparently, the CTD has evolved as a regulatable interaction surface to facilitate coordinated delivery of components to the pre-mRNP and to the chromatin (Yoh et al. 2008). In this way, the local concentration of specific components can be increased at the appropriate time. The CTD is long enough to simultaneously bind several proteins. The CTD is structurally versatile and an induced fit is important when it interacts with different components (reviewed in Meinhart et al. 2005). Furthermore, binding can be regulated because interacting proteins recognise specific serine phosphorylation patterns in the CTD (reviewed in Buratkowski 2009).
The coupling between pre-mRNP assembly, the transcription process and chromatin dynamics is not fully understood. One aspect is that assembly of the pre-mRNP may prevent formation of RNA–DNA hybrids and transcription-associated recombination (Huerta and Aguilera 2003). Another aspect might be that pre-mRNP assembly influences the chromatin structure to facilitate transcription elongation. Recent data imply that actin binds to certain hnRNP proteins in BR pre-mRNPs and recruits HATs (Fig. 2a), thereby directing histone acetylation (reviewed in Visa and Percipalle 2010). Also, the chromatin remodelling complex SWI/SNF associates with BR pre-mRNAs (Tyagi et al. 2009). It is suggested that this complex, as part of the pre-mRNPs, could in addition influence splicing and 3′ end processing.
As a result of evolving specific interactions between components of the different molecular machines, coupling between transcription and pre-mRNP assembly, processing and export has been established. In such a system, quality control can be reached by connecting different machines through regulating subunit interactions. Such a case could be the apparent coordination between assembly of the 3′ end machinery and delivery of export adaptors (Johnson et al. 2009). It is further likely that the many cotranscriptional interactions and rearrangements that include multi-component assemblies involve kinetic proofreading (Hopfield 1974) that increases fidelity and efficiency. Specific ATP-dependent steps in splicing have been suggested to be such examples (Burgess and Guthrie 1993; Xu and Query 2007).
In addition to the cotranscriptional environment, it is known that the localisation of genes inside the nucleus is important for gene expression. The presence of specific transcription factories in the nucleus has been suggested (reviewed in Sutherland and Bickmore 2009), but the molecular basis for such factories is not clear. In the polytene nucleus, multiple copies of each BR gene are closely packed, providing a local environment of high concentration of various factors, but there is no evidence for arranging different genes in close approximation. In yeast, a sub-compartment at the NPC facilitates gene expression. Direct interactions between several different components involved in transcription activation, transcription elongation, 3′ end processing, export as well as quality control and components of the NPC have been demonstrated in yeast for highly expressed and inducible genes (reviewed in Dieppois and Stutz 2010). The multitude of interactions suggest a direct coupling between the cotranscriptional processes and processes at the NPC. Such a direct interaction is apparently not the case for most active genes since they are not located at the NPCs. However, Nup153 and Tpr are dynamic NPC components, and in addition to being part of NPCs, they are associated with active chromatin within the nucleus (Vaquerizas et al. 2010). Interactions in the interior of the nucleus between chromatin and nucleoporins are also important for gene expression during cell differentiation (Capelson et al. 2010; Kalverda et al. 2010). Indirect coupling mechanisms between mRNP synthesis and NPC transport may furthermore exist, as suggested from studies of BR mRNP export (Kylberg et al. 2008).
Using immuno EM, it has been demonstrated that many components are recruited to the multi-molecular pre-mRNA–RNA polymerase II elongation complex on the active BR genes. There, they cooperate throughout transcription to facilitate and coordinate transcription, pre-mRNP assembly and processing. They also, in many cases, couple the cotranscriptional processes with export and cytoplasmic processes. These pre-mRNP/mRNP components and their functions will be briefly described below.
Pre-mRNP/mRNP components and their function
hnRNP proteins
More than 20 different hnRNP proteins, named A1 to U, are abundant components of pre-mRNPs/mRNPs and constitute a structurally and functionally diverse group of RNA binding proteins (reviewed in Dreyfuss et al. 2002). Different hnRNP proteins are present in widely different amounts in cell nuclei. Many of the hnRNP proteins bind to pre-mRNPs cotranscriptionally as demonstrated in lampbrush chromosomes (Wu et al. 1991) and for BR1 and BR2 pre-mRNPs (Visa et al. 1996b). In BR1 and BR2 pre-mRNPs, an hnRNP A1-like protein is present in multiple copies along the pre-mRNP (Kiseleva et al. 1997). The hnRNP proteins thus seem to be present throughout the length of pre-mRNPs and on individual transcripts they are likely to be present in specific combinations (reviewed in Singh and Valcárcel 2005), presumably determined by preferential sequence binding properties, post-translational modifications and cellular localisation. As they bind to the pre-mRNP, the hnRNP proteins can affect the structure of the pre-mRNP and the binding of other components, thereby influencing many aspects of synthesis, processing and function of pre-mRNPs/mRNPs, for example, polyadenylation, mRNA stability, export and translation. Several hnRNP proteins modulate splice site choices (reviewed in Martinez-Contreras et al. 2007), as exemplified by hnRNP A1 and hnRNP H that can both repress and stimulate splicing (Fisette et al. 2010). Hrp36, an hnRNP A1-like protein, and the Y-box p50 protein (Soop et al. 2003) both bind to BR1 and BR2 pre-mRNPs cotranscriptionally and remain associated with the BR mRNPs in polysomes, suggesting multiple roles for these proteins.
Nuclear cap binding proteins
All pre-mRNAs are capped and associated with cap binding proteins (CBPs) to form the cap binding complex (CBC). This universal 5′ end modification of the pre-mRNAs occurs soon after the start of elongation (presumably on 25- to 40-base-long pre-mRNAs) by the combined action of several enzymatic activities that also interact with the Ser5 phosphorylated CTD of RNA polymerase II (reviewed in Gu and Lima 2005). The cap binding proteins CBP20 and probably CBP80 are added early on BR1 and BR2 pre-mRNPs (Visa et al. 1996a), and thus the CBC is present in the pre-mRNP throughout most of the transcription elongation. This is in line with the observed interaction between the CBC and other cotranscriptional processing events (reviewed in Lewis and Izaurralde 1997), including the export machinery (Cheng et al. 2006).
The splicing machinery
SnRNPs and other splicing factors are efficiently recruited to intron-containing nascent transcripts (Kiseleva et al. 1994; Baurén et al. 1996; Lacadie and Rosbash 2005; Listerman et al. 2006) and are part of a nascent pre-mRNP/transcription complex (Wetterberg et al. 2001). Cotranscriptional splicing was originally observed in Drosophila (Beyer and Osheim 1988) and has been directly demonstrated for the BR1 (Baurén and Wieslander 1994) and BR3 (Wetterberg et al. 1996) pre-mRNAs. Observations in mammalian cells support that splicing is largely cotranscriptional (reviewed by Neugebauer 2002; Bentley 2005; Perales and Bentley 2009). EM visualisation of the BR3 pre-mRNPs in situ (Fig. 2b and b′) (Wetterberg et al. 2001) indicates that spliceosome components are repeatedly recruited to the multi-intron transcript during transcription elongation and that spliceosome components and introns leave the pre-mRNP after completion of splicing. It should be pointed out that spliceosome assembly is initiated cotranscriptionally, but splicing need not be completed until after 3′ end processing (Tardiff et al. 2006), depending on for example the position of the introns (Baurén and Wieslander 1994). Based on observations on the BR1 gene, splicing can be stimulated by 3′ end processing and may be essentially completed in the pre-mRNP while it is being polyadenylated, still retained at the gene locus (Baurén et al. 1998).
The SR proteins
The family of SR proteins fulfils multiple roles in the life of a pre-mRNP/mRNP (reviewed in Sanford et al. 2005; Long and Cáceres 2009). SR proteins are recruited to nascent pre-mRNPs (Baurén et al. 1996; Misteli et al. 1997; Mabon and Misteli 2005). During constitutive splicing more than one SR protein is required. SR proteins directly contact the pre-mRNA at several specific sites during the splicing reaction (Shen and Green 2004) and form important protein–protein bridges. They also influence alternative splice site choices, probably by contributing to specific combinations of splicing factors that associate with pre-mRNPs. Pre-mRNPs from different genes, including BR genes, bind several different members of the SR protein family in gene-specific combinations (Björk et al. 2009). At least four different types of SR proteins remain with BR mRNPs during export to the cytoplasm. Experimental interference with one of these SR proteins, SRSF2 (SC35), suggests that it is important for cotranscriptional events as well as for nucleocytoplasmic export for a single BR pre-mRNP/mRNP. One of the SR proteins, SRSF1 (ASF/SF2), even stays with the mRNP in polysomes, in agreement with its known role in translation initiation (Michlewski et al. 2008). These data underline the numerous findings that SR proteins are part of pre-mRNPs/mRNPs from the gene to the cytoplasm and influence many important processes such as synthesis, processing, export, stability and translation. In addition, an SR-like protein, RSF1/hrp23, is recruited to the pre-mRNP cotranscriptionally and inhibits spliceosomal early assembly at incorrect sites within exons (Björk et al. 2006).
The exon junction complex
A specific complex of proteins associates with the pre-mRNP/mRNP in a splicing-dependent but non-sequence-specific way, at a region about 20 nucleotides upstream of exon–exon junctions (reviewed in Le Hir and Andersen 2008). Assembly of this exon junction complex (EJC) represents a splicing-dependent change in pre-mRNP composition. It is assumed that an EJC is deposited upstream exon–exon junctions in a multi-intron pre-mRNA. The core of the EJC consists of eIF4AIII, a member of the DEAD-box family of RNA helicases that binds to the pre-mRNA, the Mago-Y14 heterodimer and Barentsz/MLN51. The order and timing of assembly of the EJC is not precisely known. The eIF4AIII is associated with both unspliced and spliced pre-mRNAs and may interact with spliceosomal proteins (Ideue et al. 2007). Detailed structural analyses indicate that eIF4AIII undergoes conformational changes and that Mago-Y14 is needed for the tight binding of eIF4AIII to the RNA. This may take place during spliceosomal C-complex formation (Herold et al. 2009). The four EJC core components associate with the BR1 and BR2 pre-mRNPs during cotranscriptional splicing (Björk et al. manuscript in preparation). The EJC core is stably bound to the mRNA and serves as an anchoring platform for many different proteins in the nucleus and in the cytoplasm. Among these proteins are splicing-associated proteins, export proteins and mRNA quality control proteins (Le Hir et al. 2001). In the cytoplasm, the EJC is involved in localisation of mRNPs, nonsense-mediated decay (NMD) and translational control (Isken et al. 2008; Ma et al. 2008). The EJC is present on CBC bound mRNPs and upon the first round of translation, the EJC is removed from the mRNP and eIF4E replaces the CBC and steady-state translation initiation can take place (reviewed in Isken and Maquat 2007).
The 3′ end processing machinery
The conserved 3′ end processing machinery (reviewed in Mandel et al. 2008; Millevoi and Vagner 2010) binds to a combination of sequence signals in the pre-mRNA and, in a coupled two-step process, cleaves the pre-mRNA and polyadenylates the free 3′ end. Even if it has been reported that 3′ end processing can occur after release from the gene (West et al. 2008), more commonly it takes place cotranscriptionally (reviewed in Perales and Bentley 2009). The recruitment of 3′ end processing factors appears to involve multiple mechanisms, pointing to interplay with the transcription process. Some 3′ end processing factors are recruited already at the 5′ end of the gene, possibly at the promoter, involving for example TFIID (Dantonel et al. 1997). Some factors, such as the cleavage polyadenylation specificity factor (CPSF), bind to RNA polymerase II (Nag et al. 2007). A maximal amount of 3′ end processing factors is found at the 3′ end of the gene, downstream the poly(A) site, probably recruited by the signals in the pre-mRNA (Glover-Cutter et al. 2008). At this position, Ser2 phosphorylation in the RNA polymerase II CTD is maximal and the cleavage stimulation factor (CstF) binds to the Ser2 phosphorylated CTD. In the BR1 gene, the RNA polymerase continues transcription approximately 600 bp downstream the position of the poly(A) site (Fig. 3). At this point, termination of transcription, 3′ end cleavage and the initial polyadenylation as well as stimulation of excision of the last intron take place (Baurén et al. 1998). As a result, the composition of the pre-mRNP changes; the functional 3′ end processing machinery temporarily associates and the poly(A)-binding protein (PABP) becomes part of the pre-mRNP. Ultrastructural analysis of the BR1/BR2 pre-mRNPs at the end of the genes has not been performed so far. Although initiated at the gene, polyadenylation is completed after release of the pre-mRNP from the gene since the length of the poly(A) tail gets longer in the interchromatin located BR pre-mRNPs/mRNPs.
Fig. 3.
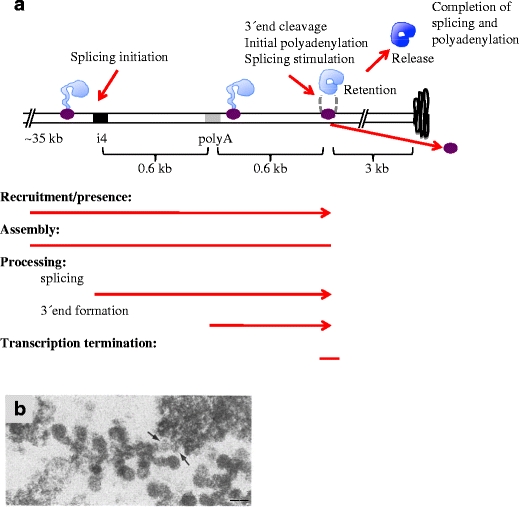
Cotranscriptional processes at the 3′ end of the BR1/BR2 gene. a Schematic representation. Colour code as in Fig. 2. Excision of intron 4 (i4, black box) is initiated rapidly. About 0.6 kb downstream the poly(A) signal (grey box), transcription termination, 3′ end cleavage and polyadenylation are coordinated. The pre-mRNP is retained at the gene during the initial polyadenylation, and splicing is stimulated at this stage. Approximately 3 kb of DNA, as extended chromatin, is present downstream the site of release of the pre-mRNP, before being folded into compact chromatin (black curved line). Released mRNP in dark blue. b EM image of the 3′ end of the gene (adopted from Ericsson et al. 1989). The arrows indicate the chromatin fibre, extending from the final pre-mRNP into the compact chromatin. The scale bar represents 50 nm
The transcription-export complex
The transcription-export complex (TREX) is associated with the elongating RNA polymerase II-nascent pre-mRNP and plays several roles. TREX consists of the multi-subunit THO complex and two export proteins, the RNA helicase Sub2 (in yeast)/UAP56 (in mammals) and Yra1 (in yeast)/Aly (in mammals). In yeast, the THO complex appears to be associated with the transcription elongation complex and is believed to facilitate loading of proteins onto the pre-mRNA (Jensen et al. 2004). In mammals, the THO complex contains additional subunits and one of these, Thoc5, has been reported to participate in binding the export receptor NXF1 (Katahira et al. 2009). Recruitment of Sub2/UAP56 and Yra/Aly to BR pre-mRNPs is cotranscriptional (Kiesler et al. 2002). Importantly, Sub2/UAP56 is yet an example of a protein that has multiple functions. Apart from its presence during transcription elongation, it is involved in splicing, where it plays a role in U2 snRNP recruitment (Kistler and Guthrie 2001). Moreover, as part of the TREX complex, it is involved in export.
The recruitment of Sub2/UAP56 to the pre-mRNA can apparently take place by several mechanisms, connecting export to transcription and processing. Hpr1, a subunit of the yeast THO complex, has been shown to be required for recruitment of Sub2 and Yra1 to the pre-mRNA (Zenklusen et al. 2002). Aly can bind to nascent transcripts dependent on the transcription elongation factor Spt6. Spt6 itself binds to the Ser2 phosphorylated RNA polymerase II CTD and recruits Aly via its interacting protein Iws1 (Yoh et al. 2007). Recruitment of the export proteins is moreover connected to 3′ end processing. Direct interaction between the subunit Pcf11 of the yeast 3′ end cleavage factor IA (CFIA) and Yra1 may couple delivery of Yra1 to Sub2 and assembly of a functional CFIA (Johnson et al. 2009). In mammals, it has been reported that UAP56 and Aly interact with the EJC (for example Le Hir et al. 2001). However, it has also been demonstrated that in humans, CBP80 recruits the TREX complex to the 5′ end of the pre-mRNP in a splicing-dependent manner via interaction with Aly (Cheng et al. 2006).
mRNA export adaptors and receptors
Export of mRNPs requires that specific proteins in the mRNPs serve as adaptors for binding to export receptors. The export receptors in turn mediate contact with the NPC and are essential for translocation of the mRNP through the NPC. The vast majority of mRNPs use a specific heterodimer export receptor called NXF1:NXT1 in mammals and Mex67:Mtr2 in yeast. NXF1 belongs to a family of proteins (Herold et al. 2000), and several NXF family members exhibit mRNA export function. Some members are expressed in specific tissues and may serve specialised functions in mRNA metabolism (Tan et al. 2005). In addition, a second receptor, the karyopherin Crm1, is involved in export of some mRNPs (Gallouzi and Steitz 2001; Cullen 2003; Carmody and Wente 2009).
A number of different adaptors to the NXF1:NXT1 export receptor have been identified. The recruitment of these adaptors is directly or indirectly connected to pre-mRNA processing steps. Yra1/Aly is a well-known adaptor, described above as part of the TREX complex. Besides Yra1/Aly, some SR proteins, SRSF3 (SRp20), SRSF7 (9G8) and SRSF1 (ASF/SF2), remain associated with the processed mRNP. In a hypophosphorylated form, they serve as adaptors for NXF1:NXT1 (reviewed in Huang and Steitz 2005). A BR mRNP contains multiple export adaptors. Aly (Kiesler et al. 2002) and several types of SR proteins (Björk et al. 2009) have been shown to be part of BR1 and BR2 mRNPs, suggesting that multiple copies of the export receptor NXF1:NXT1 are associated with the BR mRNP. In addition, Crm1 is part of the BR1 and BR2 mRNPs (Zhao et al. 2004).
In summary, multiple export adaptor proteins and mechanisms of loading them onto the pre-mRNPs/mRNPs are utilised. These mechanisms involve both transcription and different pre-mRNA processing steps and thus connect these processes to the formation of export competent mRNPs. It seems likely that individual mRNPs associate with more than one adaptor, loaded by different mechanisms, possibly to increase export efficiency. It is also likely that some mRNAs require specific arrangements. The HSP70 mRNA, for example, requires a co-adaptor, Thoc5, which is part of the TREX complex, for binding the export receptor NXF1 (Katahira et al. 2009). Furthermore, it is likely that biological variation accentuates one variant or the other, such that different conditions dominate in different tissues or species and that mRNPs from certain genes preferentially use specific export factors.
Gene-specific components
Although most components that bind to pre-mRNPs/mRNPs are not gene specific, there are exceptions. For example, some, but far from all, regulators of alternative splicing are expressed tissue specifically (reviewed in Nilsen and Graveley 2010). A second example is the transcriptional repressor CA150/hrp130 that is highly enriched in the BR3 gene in an RNA-dependent manner. This conserved protein is known to repress transcription elongation in mammals. It has therefore been suggested that CA150/hrp130 may adjust the transcription rate to the frequent cotranscriptional excision of the many introns in the BR3 pre-mRNAs (Sun et al. 2004). A third example is ADAR II that edits within double-stranded parts of specific pre-mRNAs cotranscriptionally (Laurencikiene et al. 2006), implying that this enzyme can be part of the pre-mRNP in the appropriate cases.
Multiple roles for individual components
An important finding from studies of individual components of pre-mRNPs/mRNPs is that the same protein can interact with different molecular assemblies and be involved in more than one process. Four examples, SR proteins, the CBPs, the core EJC and the Sub2/UAP56 described above, emphasise this principle of multi-functional proteins. Evolution has clearly shaped multiple contact possibilities between subunits involved in different processes. The functional diversity of these proteins and their affinities are likely to be influenced by for example mutually exclusive interactions as suggested between Yra1 and Sub2 and Pcf11, by the phosphorylation status as observed for SR protein recruitment in splicing versus interaction with export receptors and by the varying interaction partners depending on cellular localisation as suggested for the core EJC.
These examples show that proteins having multiple functions during transcription, processing, export and translation are essential for the coordination of gene expression and thus are important for reaching accuracy as well as efficiency.
The structure of pre-mRNPs and mRNPs
To understand mRNP export in its molecular context in vivo, it is important to analyse the dynamic structure and composition of pre-mRNPs and the corresponding mRNPs. The structure and composition of a pre-mRNP changes during transcription, reflecting the gradual synthesis of the pre-mRNA, the continuous assembly of the pre-mRNP and the transient interactions with processing machines (Figs. 2 and 3). Information about the structure of pre-mRNPs is generally lacking. In EM studies of mammalian cell nuclei, perichromatin fibrils and granules presumably represent pre-mRNPs, but detailed structural analyses of such gene-specific pre-mRNPs have not been performed (reviewed in Fakan and van Driel 2007). The most detailed structural information available is for BR pre-mRNPs (Skoglund et al. 1983; Wetterberg et al. 2001). During transcription elongation, the pre-mRNP structure changes as more RNA is made and incorporated together with additional proteins. In the BR1 and BR2 pre-mRNPs, distinct structural changes are seen (Figs. 2 and 3). In the very beginning of the 5′ proximal part of the about 35-kb-long genes, the pre-mRNP structure is not yet precisely analysed. Thin fibres and occasional granules are present. In this region, three introns are incorporated into the pre-mRNP and subsequently rapidly excised (Fig. 2a). The structure of a pre-mRNP can be highly influenced by the presence of processing machines, such as the spliceosome. The 11-kb-long BR3 gene contains 38 introns and the pre-mRNP repeatedly take several distinct structures during recruitment, assembly and function of the spliceosome (Fig. 2b and b′) (Wetterberg et al. 2001).
Further into the proximal region of the BR1/BR2 gene, excision of the three introns is complete. The pre-mRNP then appears as a 19-nm-thick irregular fibre (Fig. 2a). Based on Miller spreads and on analyses of released BR mRNPs, the 19-nm fibre is in fact built from a folded, basic 7-nm fibre. At the end of the proximal region, a second structural change appears in the pre-mRNP. The 19-nm-thick fibre reaches a length that from then on is constant throughout transcription of the middle region (a single approximately 30-kb exon) of the BR1 and BR2 genes (Fig. 2a, c and d). At the tip of the 19-nm fibre, the 5′ part of the pre-mRNP folds into a more compact granular higher order structure. In this higher order structure, the 7-nm fibre is still the basic unit, although arranged in a different way. The molecular mechanisms for the structural transitions (7 nm to 19 nm to compact granular structure) are not known. They could for example be a consequence of processing of introns 1 to 3 or they could reflect assembly properties of a long pre-mRNP fibre. In summary, the packing of the BR1 and BR2 exon sequences takes place in distinct and reproducible steps; a 7-nm fibre is folded into an irregular 19-nm fibre and subsequently folded into a more compact, eventually spherical structure. At the 3′ end of the BR1 and BR2 genes, the overall pre-mRNP structure is still dominated by the 19-nm fibre and the growing compact spherical structure (Fig. 3).
We predict from these results on the BR genes that, in general, pre-mRNPs are structurally diverse. The structure will depend on the size of the transcription unit, the exon–intron organisation, the presence of processing machineries and the dynamic rearrangements during the processing events.
Also for mRNPs in the interchromatin space, structural information is needed. The structure of the BR1 and BR2 mRNPs has been analysed by EM tomography (Skoglund et al. 1986). The mRNP consists of a 7-nm fibre (Lönnroth et al. 1992) that is folded into higher order structures, apparently folded in a back-and-forth manner (Daneholt 2001b). Overall, a spherical mRNP is formed, having a diameter of about 50 nm and a central hole (Fig. 4a and b). This structure largely reflects the build-up of the compact spherical structure seen late during transcription (Fig. 3b). All BR mRNPs from an individual BR1 or BR2 gene have the same structure, and pre-mRNPs from the related BR1 and BR2 genes are very similar. This suggests that the mRNP structure is the result of specific interactions. In yeast, a large population of presumably nuclear mRNPs have been isolated and characterised by EM (Batisse et al. 2009). Many different mRNPs shared a common architecture consisting of a constant 5–7-nm-thick elongated ribbon-like structure with a length that increased with the size of the mRNA. From these examples, it is difficult to know if there is a common architecture for all nuclear mRNPs. It is possible that mRNPs have a more unified structure than pre-mRNPs because processing factors are not present. It could then be that a 5–7-nm fibre-like structural element is present and that for longer mRNAs, such a fibre can be arranged in a higher order of folding. It is also possible that the CBC and the poly(A) tail with its PABPs adopt structures that are different from the body of the mRNP. As indicated from studies of BR mRNPs (Mehlin et al. 1992, 1995) and dystrophin mRNPs (Mor et al. 2010) during export through the NPCs, it is evident that the mRNP structure is flexible and can be rearranged (see ‘mRNP interaction with the NPC and release into the cytoplasm’ section).
Fig. 4.
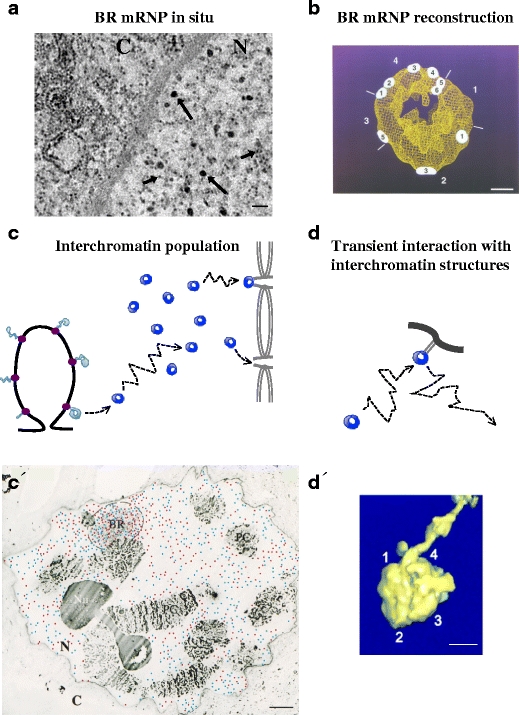
The BR1/BR2 mRNPs in the interchromatin space. a EM image of BR mRNPs in situ. The BR mRNPs (50 nm in diameter) in the interchromatin are indicated by long arrows. For comparison, short arrows indicate ribosomal subunits. N nucleus, C cytoplasm. The scale bar represents 100 nm. b The 3D structure of the BR1/BR2 mRNP (50 nm in diameter) (adopted from Mehlin et al. 1995, © The Rockefeller University Press. The Journal of Cell Biology, 1995, 129:1205–1216, Fig. 7). The numbers (1–6) on the mRNP indicate multiple contact points with the inner ring of the NPC during export. The numbers outside the mRNP (1–4) indicate described domains of the mRNP. The scale bar represents 10 nm. c The released BR1/BR2 mRNPs move randomly inside the interchromatin space and become part of a population of mRNPs, from which individual mRNPs (stochastically) bind to the basket of the NPCs. Colour code as in Fig. 1. c′ EM image of a section through a polytene nucleus (adopted from Singh et al. 1999). Newly synthesised (red dots) and old (blue dots) BR1/BR2 mRNPs are randomly distributed. The highest concentration of mRNPs is at the gene locus (BR). PC polytene chromosomes, Nu nucleolus, N nucleus, C cytoplasm. The scale bar represents 5 μm. d Within the interchromatin space, the random movement of the mRNP is occasionally interrupted by transient interactions (light grey, thin lines) with interchromatin structures (dark grey, thick line). d′ EM 3D reconstruction of a BR1/BR2 mRNP interacting with an interchromatin fibre structure (adopted from Miralles et al. 2000, © The Rockefeller University Press. The Journal of Cell Biology, 2000, 148:271–282, Fig. 2d, part IV). Numbers 1–4 refer to the described domains of the mRNP. The scale bar represents 20 nm
Quality control in the nucleus
Improperly processed or assembled pre-mRNPs are recognised and degraded in the nucleus, as has been most extensively analysed in yeast (reviewed in Schmid and Jensen 2008; Fasken and Corbett 2009; Dieppois and Stutz 2010). Although decapping and 5′–3′ degradation has been observed for poorly spliced pre-mRNAs, the nuclear exosome, having 3′–5′ exonucleolytic activity and its activator complex TRAMP, is the most well-characterised degradation complex. The exosome is associated with the elongating RNA polymerase II in active genes (Andrulis et al. 2002). In the BR genes, the exosome component Rrp4 (and presumably the exosome) is in addition present in pre-mRNPs throughout transcription, an interaction that is mediated by the hrp59/hnRNP M (Hessle et al. 2009). Pre-mRNPs with defects in assembly, splicing or 3′ end processing can be retained at the gene locus in an exosome-dependent manner (Hilleren et al. 2001). The molecular mechanism of this retention is not known, at least partly because the mechanisms for normal release of pre-mRNPs/mRNPs from the gene is not known. It has been observed that an appropriate poly(A) tail, presumably coupled to PABPs, is needed for release from the gene locus. In the BR1 gene (Fig. 3), correctly 3′ end cleaved transcripts with short, approximately 20-A long, poly(A) tails were present at the gene locus and extension of the poly(A) tails occurred as the mRNPs were released or shortly thereafter (Baurén et al. 1998). The initial polyadenylation thus occurs at the gene locus and unless efficiently performed leads to retention and exosome degradation (reviewed in Schmid and Jensen 2008). Since the different cotranscriptional events are coupled, defects in RNA polymerase II, pre-mRNP assembly and splicing could possibly all result in retention at the gene locus, mediated through a 3′ end processing pathway.
Unspliced and improperly assembled mRNPs are also retained and presumably degraded at the NPC. The basket-associated Mlp1 and Mlp2 (Tpr in vertebrates) and Pml39 proteins are involved in sorting proper and improper mRNPs for translocation and retention, respectively (reviewed in Fasken and Corbett 2009). Additional NPC-associated proteins are involved, probably indirectly because they are important for NPC assembly (Esc1) and Mlp1 anchoring (Nup60). The mRNPs may directly interact with Mlp1 via the poly(A)-binding protein Nab2. The SUMO protease Ulp1 is localised to the NPC and is involved in retaining unspliced mRNPs, suggesting that desumoylation is part of this process (Lewis et al. 2007).
mRNPs move inside the interchromatin space by diffusion
Different types of data support the view that active chromatin is unfolded and form loops of various lengths (reviewed in Cremer et al. 2004; Sutherland and Bickmore 2009). In mammalian cell nuclei, such actively transcribing chromatin loops may be found inside chromosome territories or at the surface of these territories. EM observations suggest that in both cases, active transcription occurs at the interface between chromatin and interchromatin (reviewed in Fakan and van Driel 2007). The nascent pre-mRNPs are accessible from the interchromatin space, and upon termination of transcription/processing, mRNPs are directly delivered into the interchromatin space. This situation is most evident in polytene nuclei. EM analyses show that the active BR genes loop out into the interchromatin space, that the nascent BR pre-mRNPs are in direct contact with the interchromatin space and that the pre-mRNPs/mRNPs are directly delivered into the interchromatin space (Fig. 4c and c′).
Biochemical analysis of the nuclear BR1/BR2 mRNPs suggests that each of these long mRNAs is associated with approximately 500 average-sized proteins (Wurtz et al. 1990). A number of different proteins have been identified by immuno EM (CBC, hnRNP proteins, export factors, PABP, EJC core complex, helicases, SR proteins and a splicing repressor), but the complete composition is not yet known. It is evident that each BR mRNP contains proteins that have specific functions during export, for example at the NPC and subsequently in the cytoplasm. In general, proteins present in mRNPs have been identified (Dreyfuss et al. 2002; Singh and Valcárcel 2005) and also characterised for mRNP populations (Batisse et al. 2009). More information about the protein composition in gene-specific mRNPs is however needed.
Before the movement and distribution of mRNPs inside the cell nucleus could be studied, it was believed that efficiency demanded structural mRNP transport systems. The current model proposes that mRNPs move in a non-directional manner away from the gene by diffusion. This diffusion, however, is restricted by the chromatin organisation and structures inside the interchromatin (Zachar et al. 1993). In mammalian nuclei, the interchromatin space forms a three-dimensional labyrinth and diffusion of the mRNPs occurs within this restricted space. It is possible that narrow channels in between volumes of chromatin increase the efficiency of directionality for mRNP movement. Direct measurements of distribution and rate of movement are compatible with a restricted diffusion (Politz and Pederson 2000; Shav-Tal et al. 2004; Ben-Ari et al. 2010; Mor et al. 2010). It has been observed that within the interchromatin space, mRNPs move unrestricted in and out of interchromatin granule clusters (speckles at the light microscope level), suggesting that, in general, mRNPs do not accumulate at specific regions for specific processing/modification steps, at least not for splicing (Ritland Politz et al. 2006).
Compared to mRNPs in mammalian nuclei, the movement of BR1/BR2 mRNPs is less restricted within the interchromatin space of the large polytene nucleus (Singh et al. 1999), and the diffusion coefficient is higher than reported for mRNPs in mammalian nuclei (Siebrasse et al. 2008). This is probably because chromatin restrains movement in mammalian nuclei. Tracking of individual BR mRNPs demonstrated that within the interchromatin space, the mRNPs are slowed down occasionally. This may reflect transient interactions with non-chromatin components in the interchromatin space (Fig. 4d and d′). Such interactions have been demonstrated (Miralles et al. 2000).
It has been estimated that movement through the nucleus of mammalian cells, from the gene to the NPC, occurs within a time frame of 5–40 min, i.e. not a fixed time (Mor et al. 2010). In the case of BR1/BR2 mRNPs, newly made mRNPs become part of a pool of nuclear mRNPs from which individual BR mRNPs are exported, presumably after randomly docking at NPCs (Fig. 4c and c′). We propose that this reflects a situation common to all mRNPs and that the nuclear residence time for individual mRNPs varies. It is possible that diffusion through narrow interchromatin channels increases the directionality for mRNP movement in mammalian nuclei.
mRNP interaction with the NPC and release into the cytoplasm
A decisive step in nucleocytoplasmic export is the recognition between the mRNP and the NPC. The NPC is a huge protein complex, comprising around 125 MDa in mammals and 60 MDa in yeast. The structure and function of the NPC has been reviewed in several recent reviews (for example Cook et al. 2007; D’Angelo and Hetzer 2008; Lim et al. 2008; Terry and Wente 2009; Strambio-De-Castillia et al. 2010). Briefly, the NPC is built from about 30 different proteins, called nucleoporins, divided into structural and FG nucleoporins. FG nucleoporins contain flexible domains rich in phenylalanine and glycine residue repeats, separated by characteristic spacer sequences. In total, it is estimated to be 500–1,000 nucleoporins/NPC. The NPC has a characteristic 8-fold symmetry and consists of a core embedded in the nuclear envelope and fibrillar structures on both the nuclear and cytoplasmic sides. The core consists of a central channel positioned in between a nuclear and a cytoplasmic ring. The central channel contains a meshwork of unfolded, flexible FG nucleoporins. The fibrillar structure on the nuclear side, called the basket, consists of eight fibrils attached to the nuclear ring of the core. The ends of the basket fibrils can interconnect with each other. On the cytoplasmic side, there are also eight fibrils but they are shorter than those on the nuclear side.
The binding of BR1 and BR2 mRNPs to the basket of the NPC and their translocation through the NPC has been studied in some detail (Fig. 5). Initially, the BR mRNP interacts with the tip of the basket fibrils and there are structural rearrangements of the basket (Fig. 5a and a′) (Kiseleva et al. 1996). The fibrils open up and a ring-like structure is formed around the centrally placed BR mRNP. It is possible that, before binding the mRNP, the basket fibrils are in fact quite flexible and that they constantly move and thereby contribute to the efficiency of recruitment of mRNPs to the basket (Kylberg et al. 2010). After binding, the BR mRNP enters the basket and the ring structure regresses (Fig. 5b and b′). Tpr and the FG nucleoporin Nup153 are present in the basket fibrils. Export receptors interact with FG repeats and several FG nucleoporins in addition to Nup153 are present on surface-accessible NPC positions including the basket (reviewed in Terry and Wente 2009). Export receptors bind directly to the FG repeats with domains that are different from the mRNP-binding domains (Stewart 2007), and different export receptors may require different subsets of FG nucleoporins. BR mRNPs are retained on top of the basket when Nup153 is experimentally interfered with, suggesting that Nup153 is involved in the transfer of the BR mRNP into the basket (Soop et al. 2005). Wheat germ agglutinin blocks the BR mRNPs at the basket, suggesting that GlcNAc-bearing nucleoporins, but presumably not Nup153, are involved in processes preceding the translocation through the central channel (Kylberg et al. 2010).
Fig. 5.
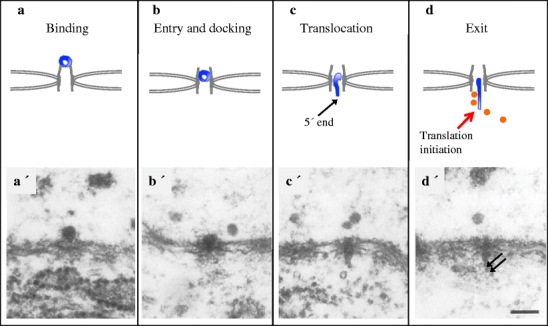
The behaviour of BR1/BR2 mRNPs at the NPC. Schematic representations and EM images (adopted from Mehlin et al. 1992) of the corresponding steps. a, a′ The mRNP (in dark blue) binds to the tip of the basket (in grey). b, b′ It then enters into the basket and docks at the entrance of the central channel. c, c′ Subsequently, the mRNP changes conformation as it is translocated through the central channel with the 5′ end first. d, d′ At the cytoplasmic side, initiation of translation can occur rapidly at the exit of the central channel. Orange dots in (d) and arrows in (d′) indicate ribosomes. The scale bar represents 100 nm
As previously pointed out, a BR mRNP contains multiple export adaptors for NXF1:NXT1 and it also contains Crm1. It has been observed that during translocation, the BR mRNP rotates and that it has multiple contact points with the NPC nuclear ring (Fig. 4b) (Mehlin et al. 1995). This may reflect the multiple export receptors and/or the mechanics of entering the central channel. At least for large mRNPs, such as the BR mRNPs, efficient NPC passage may require multiple export receptors. In agreement with such an interpretation, interference with SRSF2 (SC35) (Björk et al. 2009) or inhibition of formation of LMB–RanGTP complex (Zhao et al. 2004) only partially reduces export.
After docking at the entrance of the central channel, the BR mRNP strikingly changes structure as it continues to move into the channel and adopts a more extended shape, approximately 25 nm in diameter (Fig. 5c and c′). The NPC channel allows almost free passage of molecules smaller than 5 nm in diameter (Feldherr and Akin 1997; Keminar and Peters 1999) and appears to have an upper limit of transport of complexes with a diameter of 39 nm (Wente and Rout 2010). The structural rearrangement of the BR mRNP may be a consequence of this physical size limit and the translocation process. Compositional changes could facilitate these structural rearrangements. The BR mRNPs move through the NPC channel with its 5′ end first (Fig. 5c and c′) (Mehlin et al. 1992, 1995; Visa et al. 1996a). This may be a general property of mRNPs since it has also been observed for dystrophin mRNPs (Mor et al. 2010). The molecular details are not known. It is possible, though, that a number of proteins leave the mRNP at this point. In the case of the BR mRNP, immuno EM studies suggest that the TREX components UAP56 and Aly (Kiesler et al. 2002), the splicing regulator hrp23/RSF1 (Sun et al. 1998) and hrp59/hnRNP M (Kiesler et al. 2005) all leave the BR mRNP during docking or translocation through the NPC. It is also possible that proteins important for export, such as RAE1, interact transiently and specifically with the BR mRNP at this stage (Sabri and Visa 2000).
The translocation through the NPC channel is rapid. Real-time visualisation of individual mRNPs during export showed that the export time was about 0.5 s (Mor et al. 2010 and references herein). The movement of the mRNP through the NPC was 15-fold faster than simple diffusion, suggesting that the translocation is facilitated. Several mechanisms for translocation have been put forward (reviewed in Peters 2009).
At the cytoplasmic side of the NPC, BR mRNPs do not regain their spherical interchromatin structure. Instead, they form extended mRNP fibrils. In general, it has been suggested that a reorganisation of mRNPs takes place at the cytoplasmic side of the NPC. The DEAD-box helicase Dbp5 is enriched at this location. Dbp5 is important for the export process (Alcazar-Roman et al. 2006; Weirich et al. 2006), presumably by remodelling the mRNP and simultaneously releasing proteins, such as NXF1:NXT1, thereby creating a directionality of the export (Lund and Guthrie 2005). Dbp5 binds to RNA and to several protein partners that are present at the cytoplasmic side of the NPC (Moeller et al. 2009). Nup214 (Nup159 in yeast) binds Dbp5, but only in the RNA-free form. It is therefore possible that Dbp5 could be delivered from Nup214 to the mRNP. Immuno EM studies of translocating BR1 and BR2 mRNPs suggest that Dbp5 is added to the mRNP at this stage (Zhao et al. 2002). Gle1 and its cofactor InsP6, which are associated with another nucleoporin and positioned close to Nup214, stimulate the Dbp5 ATPase activity. This results in a local activation of Dbp5, leading to the mRNP remodelling and release of proteins. One such protein is the nuclear poly(A)-binding protein that is lost from BR mRNP at NPC translocation or shortly thereafter (Bear et al. 2003). This would be in line with that Dbp5 has been shown in vitro to remodel mRNP and displace the yeast Nab2 protein (Tran et al. 2007).
Dbp5 has been shown to also bind to pre-mRNPs cotranscriptionally in yeast (Estruch and Cole 2003) and to BR pre-mRNPs (Zhao et al. 2002). Such ‘pre-bound’ Dbp5 could function at the NPC. It is also possible that Dbp5 is involved in remodelling of the pre-mRNP in the nucleus, for example during cotranscriptional assembly.
The routes taken by mRNPs in the cytoplasm can be different. For efficiently translated mRNPs, such as BR mRNPs, initiation of translation can occur immediately after entrance into the cytoplasm (Fig. 5d and d′). CBP20, and presumably the entire CBC, is lost from the BR mRNP during translocation (Visa et al. 1996a). Furthermore, the translation initiation factor eIF4H (Björk et al. 2003) becomes associated with the BR mRNP at the cytoplasmic side of the NPC. Morphologically, ribosomes can be seen associated with the extended BR mRNP fibril (Mehlin et al. 1992). This implies that a pioneer round of translation and establishment of polysomes can occur already in the perinuclear region.
The NPC translocation process is an important step in transforming the intranuclear mRNP into a functional cytoplasmic mRNP. Understanding the molecular events at the NPC that deciphers the information built into the nuclear mRNP composition and structure will be important to understand gene expression. The intranuclear mRNP enters the NPC via the basket as an export competent mRNP, and during translocation, conformational and compositional changes are triggered. Changes occur at both the 5′ and 3′ ends of the mRNP. Export receptors and other proteins are displaced. The cotranscriptional assembly is, however, not entirely replaced. The EJC, hnRNP proteins and SR proteins are examples of cotranscriptionally added components that will influence localisation, translation and quality control in the cytoplasm.
Conclusions and future perspectives
The making of an export and translation competent mRNP requires the coordinated action of many multi-component protein and protein–RNA complexes. At the active gene locus and at the NPC, important events integrate the formation, processing and modification of the mRNP before delivery into the cytoplasm. Interactions between components in the different molecular machines and multiple roles for individual components at several steps result in a streamlined synthesis, processing and export pathway. In addition, the interplay between subcomponents improves efficiency and makes quality control possible at several steps.
Identical molecular components are present in different organisms, but variations in the pathway of mRNP biogenesis clearly exist that presumably tend to emphasise specific processes depending on the cell type or experimental system. We therefore need to analyse principles and mechanisms in favourable systems and learn how these mechanisms operate in specific cases and in general. We furthermore need experimental tools to better analyse the formation, possibly pre-formation, and reuse of the multi-molecular machines in intact cell nuclei. It is still largely unknown how integrated, efficient and regulated action of the machines and processes is brought about in the cell nucleus.
Acknowledgements
We thank N. Visa and B. Daneholt for critical reading of the manuscript. We apologise to our colleagues for not being able to cite all relevant publications due to space limitations. Many critical publications are included in cited reviews. This work was supported by funding from the Swedish Research Council.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Batisse J, Batisse C, Budd A, Böttcher B, Hurt E. Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J Biol Chem. 2009;284:34911–34917. doi: 10.1074/jbc.M109.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurén G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- Baurén G, Jiang W-Q, Bernholm K, Gu F, Wieslander L. Demonstration of a dynamic, transcription-dependent organization of pre-mRNA splicing factors in polytene nuclei. J Cell Biol. 1996;133:929–941. doi: 10.1083/jcb.133.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurén G, Belikov S, Wieslander L. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev. 1998;12:2759–2769. doi: 10.1101/gad.12.17.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear DG, Fomproix N, Soop T, Björkroth B, Masich S, Daneholt B. Nuclear poly(A)-binding protein PABPN1 is associated with RNA polymerase II during transcription and accompanies the released transcript to the nuclear pore. Exp Cell Res. 2003;286:332–344. doi: 10.1016/S0014-4827(03)00123-X. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y, Brody Y, Kinor N, Mor A, Tsukamoto T, Spector DL, Singer RH, Shav-Tal Y. The life of an mRNA in space and time. J Cell Sci. 2010;123:1761–1774. doi: 10.1242/jcs.062638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Rules of engagement: cotranscriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Björk P, Baurén G, Gelius B, Wrange Ö, Wieslander L. The Chironomus tentans translation initiation factor eIF4H is present in the nucleus but does not bind to mRNA until the mRNA reaches the cytoplasmic perinuclear region. J Cell Sci. 2003;116:4521–4532. doi: 10.1242/jcs.00766. [DOI] [PubMed] [Google Scholar]
- Björk P, Wetterberg-Strandh I, Baurén G, Wieslander L. Chironomus tentans–repressor splicing factor represses SR protein function locally on pre-mRNA exons and is displaced at correct splice sites. Mol Biol Cell. 2006;17:32–42. doi: 10.1091/mbc.E05-04-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk P, Jin SB, Singh OP, Persson J-O, Hellman U, Wieslander L. Specific combinations of SR proteins associate with single pre-messenger RNAs in vivo and contribute different functions. J Cell Biol. 2009;184:555–568. doi: 10.1083/jcb.200806156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratkowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem Sci. 1993;18:381–384. doi: 10.1016/0968-0004(93)90094-4. [DOI] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci. 2009;122:1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of the mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Cremer T, Küpper K, Dietzel S, Fakan S. Higher order chromatin architecture in the cell nucleus: on the way from structure to function. Biol Cell. 2004;96:555–567. doi: 10.1016/j.biolcel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Nuclear mRNA export: insights from virology. Trends Biochem Sci. 2003;28:419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt B. Assembly and transport of a premessenger RNP particle. Proc Natl Acad Sci U S A. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt B. Packing and delivery of a genetic message. Chromosoma. 2001;110:173–185. doi: 10.1007/s004120000127. [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Murphy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- Dieppois G, Stutz S. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. J Cell Sci. 2010;123:1989–1999. doi: 10.1242/jcs.053694. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Ericsson C, Mehlin H, Björkroth B, Lamb MM, Daneholt B. The ultrastructure of upstream and downstream regions of an active Balbiani ring gene. Cell. 1989;56:631–639. doi: 10.1016/0092-8674(89)90585-0. [DOI] [PubMed] [Google Scholar]
- Estruch F, Cole CN. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol Biol Cell. 2003;14:1664–1676. doi: 10.1091/mbc.E02-09-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S, van Driel R. The perichromatin region: a functional compartment in the nucleus that determines large-scale chromatin folding. Semin Cell Dev Biol. 2007;18:676–681. doi: 10.1016/j.semcdb.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Fasken MB, Corbett AH. Mechanisms of nuclear mRNA quality control. RNA Biol. 2009;6:237–241. doi: 10.4161/rna.6.3.8330. [DOI] [PubMed] [Google Scholar]
- Feldherr CM, Akin D. The location of the transport gate in the nuclear pore complex. J Cell Sci. 1997;110:3065–3070. doi: 10.1242/jcs.110.24.3065. [DOI] [PubMed] [Google Scholar]
- Fisette JF, Toutant J, Dugré-Brisson S, Desgroseillers L, Chabot B. hnRNP A1 and hnRNP H can collaborate to modulate 5′ splice selection. RNA. 2010;16:228–238. doi: 10.1261/rna.1890310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Steitz JA. Delineation of mRNA nuclear export pathways by the use of cell-permeable peptides. Science. 2001;294:1895–1901. doi: 10.1126/science.1064693. [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Lima CD. Processing the message: structural insights into capping and decapping mRNA. Curr Opin Struct Biol. 2005;15:99–106. doi: 10.1016/j.sbi.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Herold A, Suyama M, Rodrigues JP, Braun IC, Kutay U, Carmo-Fonseca M, Bork P, Izaurralde E. Tap (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol. 2000;20:8996–9008. doi: 10.1128/MCB.20.23.8996-9008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Lührmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol. 2009;29:281–301. doi: 10.1128/MCB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessle V, Björk P, Sokolowski M, Gonsález de Valdivia E, Silverstein R, Artemenko K, Tyagi A, Maddalo G, Ilag L, Helbig R, Zubarev RA, Visa N. The exosome associates cotranscriptionally with the nascent pre-mRNP through interactions with heterogeneous nuclear ribonucleoproteins. Mol Biol Cell. 2009;20:3459–3470. doi: 10.1091/mbc.E09-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. SRprises along a messenger's journey. Mol Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Huerta P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Ideue T, Sasaki YT, Hagiwara M, Hirose T. Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes Dev. 2007;21:1993–1998. doi: 10.1101/gad.1557907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Boulay J, Olesen JR, Colin J, Weyler M, Libri D. Modulation of transcription affects mRNP quality. Mol Cell. 2004;16:235–244. doi: 10.1016/j.molcel.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Johnson SA, Cubberley G, Bentley DL. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell. 2009;33:215–226. doi: 10.1016/j.molcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerud M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Katahira J, Inoue H, Hurt E, Yoneda Y. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J. 2009;28:556–567. doi: 10.1038/emboj.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keminar O, Peters R. Permeability of single nuclear pores. Biophys J. 1999;77:217–228. doi: 10.1016/S0006-3495(99)76883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesler E, Miralles F, Visa N. HEL/UAP56 binds cotranscriptionally to the Balbiani ring pre-mRNA in an intron independent manner and accompanies the BR mRNP to the nuclear pore. Curr Biol. 2002;12:859–862. doi: 10.1016/S0960-9822(02)00840-0. [DOI] [PubMed] [Google Scholar]
- Kiesler E, Hase ME, Brodin D, Visa N. Hrp59, an hnRNP M protein in Chironomus and Drosophila, binds to exonic splicing enhancers and is required for expression of a subset of mRNAs. J Cell Biol. 2005;168:1013–1025. doi: 10.1083/jcb.200407173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E, Wurtz T, Visa N, Daneholt B. Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fibre. EMBO J. 1994;13:6052–6061. doi: 10.1002/j.1460-2075.1994.tb06952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E, Goldberg MW, Daneholt B, Allen TD. RNP export is mediated by structural reorganization of the nuclear pore basket. J Mol Biol. 1996;260:304–311. doi: 10.1006/jmbi.1996.0401. [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Visa N, Wurtz T, Daneholt B. Immunocytochemical evidence for a stepwise assembly of Balbiani ring premessenger ribonucleoprotein particles. Eur J Cell Biol. 1997;74:407–416. [PubMed] [Google Scholar]
- Kistler AL, Guthrie C. Deletion of Mud2, the yeast homolog of U2AF65, can bypass the requirement for Sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. The molecular basis for eukaryotic transcription. Proc Natl Acad Sci U S A. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kylberg K, Björkroth B, Ivarsson B, Fomproix N, Daneholt B. Close coupling between transcription and exit of mRNP from the cell nucleus. Exp Cell Res. 2008;314:1708–1720. doi: 10.1016/j.yexcr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Kylberg K, Björk P, Fomproix N, Ivarsson B, Wieslander L, Daneholt B. Exclusion of mRNPs and ribosomal particles from a thin zone beneath the nuclear envelope revealed upon inhibition of transport. Exp Cell Res. 2010;316:1028–1038. doi: 10.1016/j.yexcr.2009.10.016. [DOI] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Laurencikiene J, Källman AM, Fong N, Bentley DL, Öhman M. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep. 2006;7:303–307. doi: 10.1038/sj.embor.7400621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Andersen GR. Structural insights into the exon junction complex. Curr Opin Struct Biol. 2008;18:112–119. doi: 10.1016/j.sbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore M. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur J Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- Lewis A, Felberbaum R, Hochstrasser M. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J Cell Biol. 2007;178:813–827. doi: 10.1083/jcb.200702154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RYH, Aebi U, Fahrenkrog B. Towards reconciling structure and function in the nuclear pore complex. Histochem Cell Biol. 2008;129:105–116. doi: 10.1007/s00418-007-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- Long JC, Cáceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Lönnroth A, Alexciev K, Mehlin H, Wurtz T, Skoglund U, Daneholt B. Demonstration of a 7 nm RNP fibre as the basic structural element in a premessenger RNP particle. Exp Cell Res. 1992;199:292–296. doi: 10.1016/0014-4827(92)90437-D. [DOI] [PubMed] [Google Scholar]
- Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Jülich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Mabon SA, Misteli T. Differential recruitment of pre-mRNA splicing factors to alternatively spliced transcripts in vivo. PLoS Biol. 2005;3:e374. doi: 10.1371/journal.pbio.0030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- Mehlin H, Daneholt B, Skoglund U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 1992;69:605–613. doi: 10.1016/0092-8674(92)90224-Z. [DOI] [PubMed] [Google Scholar]
- Mehlin H, Daneholt B, Skoglund U. Structural interaction between the nuclear pore complex and a specific translocating RNP particle. J Cell Biol. 1995;129:1205–1216. doi: 10.1083/jcb.129.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Cáceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucl Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Öfverstedt LG, Sabri N, Aissouni Y, Hellman U, Skoglund U, Visa N. Electron tomography reveals posttranscriptional binding of pre-mRNPs to specific fibres in the nucleoplasm. J Cell Biol. 2000;148:271–282. doi: 10.1083/jcb.148.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Cáceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- Nag A, Narsinh K, Martinson HG. The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat Struct Mol Biol. 2007;14:662–669. doi: 10.1038/nsmb1253. [DOI] [PubMed] [Google Scholar]
- Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley DL. Cotranscriptionality: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R. Translocation through the nuclear pore: Kaps pave the way. BioEssays. 2009;31:466–477. doi: 10.1002/bies.200800159. [DOI] [PubMed] [Google Scholar]
- Politz JC, Pederson T. Movement of mRNA from transcription site to nuclear pores. J Struct Biol. 2000;129:252–257. doi: 10.1006/jsbi.2000.4227. [DOI] [PubMed] [Google Scholar]
- Ritland Politz JC, Tuft RA, Prasanth KV, Baudendistel N, Fogarty KE, Lifshitz LM, Langowski J, Spector DL, Pederson T. Rapid diffusional shuttling of poly(A) RNA between nuclear speckles and the nucleoplasm. Mol Biol Cell. 2006;17:1239–1249. doi: 10.1091/mbc.E05-10-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri N, Visa N. The Ct-RAE1 protein interacts with Balbiani ring RNP particles at the nuclear pore. RNA. 2000;6:1597–1609. doi: 10.1017/S1355838200001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Ellis J, Cáceres JF. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem Soc Trans. 2005;33:443–446. doi: 10.1042/BST0330443. [DOI] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. Quality control of mRNP in the nucleus. Chromosoma. 2008;117:419–429. doi: 10.1007/s00412-008-0166-4. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Green MR. A pathway of sequential arginine–serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Siebrasse JP, Veith R, Dobay A, Leonardt H, Daneholt B, Kubitscheck U. Discontinuous movement of mRNP particles in nucleoplasmic regions devoid of chromatin. Proc Natl Acad Sci U S A. 2008;105:20291–20296. doi: 10.1073/pnas.0810692105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Valcárcel J. Building specificity with nonspecific RNA-binding proteins. Nat Struct Mol Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- Singh OP, Björkroth B, Masich S, Wieslander L, Daneholt B. The intranuclear movement of Balbiani ring premessenger ribonucleoprotein particles. Exp Cell Res. 1999;251:135–146. doi: 10.1006/excr.1999.4490. [DOI] [PubMed] [Google Scholar]
- Skoglund U, Andersson K, Björkroth B, Lamb MM, Daneholt B. Visualization of the formation and transport of a specific hnRNP particle. Cell. 1983;34:847–855. doi: 10.1016/0092-8674(83)90542-1. [DOI] [PubMed] [Google Scholar]
- Skoglund U, Andersson K, Strandberg B, Daneholt B. Three-dimensional structure of a specific pre-messenger RNP particle established by electron microscope tomography. Nature. 1986;319:560–564. doi: 10.1038/319560a0. [DOI] [PubMed] [Google Scholar]
- Soop T, Nashchekin D, Zhao J, Sun X, Alzhanova-Ericsson AT, Björkroth B, Ovchinikov L, Daneholt B. A p50-like Y-box protein with a putative translational role becomes associated with pre-mRNA concomitant with transcription. J Cell Sci. 2003;116:1493–1503. doi: 10.1242/jcs.00353. [DOI] [PubMed] [Google Scholar]
- Soop T, Ivarsson B, Björkroth B, Fomproix N, Masich S, Cordes VC, Daneholt B. Nup153 affects entry of messenger and ribosomal ribonucleoproteins into the nuclear basket during export. Mol Biol Cell. 2005;16:5610–5620. doi: 10.1091/mbc.E05-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- Sun X, Alzhanova-Ericsson AT, Visa N, Aissouni Y, Zhao J, Daneholt B. The hrp23 protein in the Balbiani ring pre-mRNP particles is released just before or at the binding of the particles to the nuclear pore complex. J Cell Biol. 1998;142:1181–1193. doi: 10.1083/jcb.142.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zhao J, Kylberg K, Soop T, Palka K, Sonnhammer E, Visa N, Alzhanova-Ericsson AT, Daneholt B. Conspicuous accumulation of transcription elongation repressor hrp130/CA150 on the intron-rich Balbiani ring 3 gene. Chromosoma. 2004;113:244–257. doi: 10.1007/s00412-004-0314-4. [DOI] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- Tan W, Zolotukhin AS, Tretyakova I, Bear J, Lindtner S, Smulevitch SV, Felber BK. Identification and characterization of the mouse nuclear export factor (Nxf) family members. Nucl Acids Res. 2005;33:3855–3865. doi: 10.1093/nar/gki706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell. 2009;8:1814–1827. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Tyagi A, Ryme J, Brodin D, Östlund-Farrants AC, Visa N. SWI/SNF associates with nascent pre-mRNPs and regulates alternative pre-mRNA processing. PLoS Genet. 2009;5:e1000470. doi: 10.1371/journal.pgen.1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins Nup153 and Megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harb Perspect Biol. 2010;2:a000620. doi: 10.1101/cshperspect.a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Alzhanova-Ericsson AT, Sun X, Kiseleva E, Björkroth B, Wurtz T, Daneholt B. A pre-mRNA protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/S0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Flick IS, Berger J, Thorner M, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2(10):a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ, Dye MJ. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell. 2008;29:600–610. doi: 10.1016/j.molcel.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterberg I, Baurén G, Wieslander L. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA. 1996;2:641–651. [PMC free article] [PubMed] [Google Scholar]
- Wetterberg I, Zhao J, Masich S, Wieslander L, Skoglund U. In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J. 2001;20:2564–2574. doi: 10.1093/emboj/20.10.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZA, Murphy C, Callan HG, Gall JG. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres and snurposomes. J Cell Biol. 1991;113:465–483. doi: 10.1083/jcb.113.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz T, Lönnroth A, Ovchinnikov L, Skoglund U, Daneholt B. Isolation and initial characterization of a specific premessenger ribonucleoprotein particle. Proc Natl Acad Sci U S A. 1990;87:831–835. doi: 10.1073/pnas.87.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-Z, Query CC. Competition between the ATPase Prp5 and branch region–U2 snRNA pairing modulates the fidelity of spliceosomal assembly. Mol Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar Z, Kramer J, Mims IP, Bingham PM. Evidence for channelled diffusion of pre-mRNAs during nuclear RNA transport in metazoans. J Cell Biol. 1993;121:729–742. doi: 10.1083/jcb.121.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Jin S, Björkroth B, Wieslander L, Daneholt B. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from the gene to cytoplasm. EMBO J. 2002;21:1177–1187. doi: 10.1093/emboj/21.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Jin SB, Wieslander L. CRM1 and Ran are present but a NES–CRM1–RanGTP complex is not required in Balbiani ring mRNP particles from the gene to the cytoplasm. J Cell Sci. 2004;117:1553–1566. doi: 10.1242/jcs.00992. [DOI] [PubMed] [Google Scholar]


