Abstract
OBJECTIVE
Obesity and associated pathologies are major global health problems. Transforming growth factor-β/Smad3 signaling has been implicated in various metabolic processes, including adipogenesis, insulin expression, and pancreatic β-cell function. However, the systemic effects of Smad3 deficiency on adiposity and insulin resistance in vivo remain elusive. This study investigated the effects of Smad3 deficiency on whole-body glucose and lipid homeostasis and its contribution to the development of obesity and type 2 diabetes.
RESEARCH DESIGN AND METHODS
We compared various metabolic profiles of Smad3-knockout and wild-type mice. We also determined the mechanism by which Smad3 deficiency affects the expression of genes involved in adipogenesis and metabolism. Mice were then challenged with a high-fat diet to study the impact of Smad3 deficiency on the development of obesity and insulin resistance.
RESULTS
Smad3-knockout mice exhibited diminished adiposity with improved glucose tolerance and insulin sensitivity. Chromatin immunoprecipitation assay revealed that Smad3 deficiency increased CCAAT/enhancer-binding protein β-C/EBP homologous protein 10 interaction and exerted a differential regulation on proliferator-activated receptor β/δ and proliferator-activated receptor γ expression in adipocytes. Focused gene expression profiling revealed an altered expression of genes involved in adipogenesis, lipid accumulation, and fatty acid β-oxidation, indicative of altered adipose physiology. Despite reduced physical activity with no modification in food intake, these mutant mice were resistant to obesity and insulin resistance induced by a high-fat diet.
CONCLUSIONS
Smad3 is a multifaceted regulator in adipose physiology and the pathogenesis of obesity and type 2 diabetes, suggesting that Smad3 may be a potential target for the treatment of obesity and its associated disorders.
Obesity is a global medical issue by virtue of its association with an array of metabolic abnormalities, including insulin resistance, hypertension, and hyperlipidemia, collectively termed “metabolic syndrome” (1). Thus, the need is urgent for elucidation of the molecular events underlying the development of metabolic syndrome and for identification of novel targets for disease prevention and therapy.
Obesity is primarily characterized by increased fat mass or white adipose tissue (WAT). WAT consists of adipocytes specialized in the storage of fat (2). In addition to its primary function as an energy reservoir, WAT is an endocrine organ that secretes adipocytokines (e.g., leptin and resistin) that have been shown to regulate glucose and lipid metabolism (3). In obesity, adipose secretion of adipocytokines is disturbed. The mechanisms that underlie obesity-associated pathologies, such as insulin resistance, are likely to involve communication among different organs, such as insulin-responsive skeletal muscle and WAT (4,5). It has been proposed that adipose lipid storage functions to prevent peripheral lipotoxicity (5). The excessive lipid accumulation in skeletal muscle and liver leads to insulin resistance resulting from the adverse effects of chronic lipotoxicity on these tissues (5,6). Indeed, free fatty acids (FFAs) can inhibit insulin activation of insulin receptor substrate-1–associated phosphatidylinositol-3-kinase activity in skeletal muscle (7).
Studies have shown that the three peroxisome proliferator–activated receptor isotypes (PPARα, β/δ, and γ) play central roles in this process (8–10). PPARα activation decreases dyslipidemia and regulates obesity in rodents by both increasing hepatic FFA oxidation and decreasing levels of circulating triglycerides responsible for adipocyte hypertrophy and hyperplasia (11,12). The transcriptional upregulation of PPARγ during adipogenesis is well studied. Adipogenic hormones, such as glucocorticoids, cyclic AMP, and insulin, induce a transient increase in the expression of the transcription factors CCAAT/enhancer-binding protein (C/EBP) β and δ early in adipocyte differentiation. Together they induce PPARγ expression in preadipocytes, subsequently triggering full-blown adipocyte differentiation (13). PPARβ/δ plays important functions in adipose tissue metabolism, weight control, and regulation of insulin sensitivity (14). PPARβ/δ protects against weight gain, hypertriglyceridemia, and insulin resistance in mice fed a high-fat diet (HFD) and in animals that are genetically predisposed to obesity (15). Thus, available information suggests that obesity and other facets of metabolic syndrome involve deregulation of signaling pathways mediated by PPAR.
Transforming growth factor (TGF)-β1 signals through a complex of two membrane-bound receptor serine/threonine kinases that recruit and phosphorylate Smad2 and Smad3. Once phosphorylated, Smad2 and Smad3 oligomerize with Smad4 and translocate to the nucleus to participate in transcriptional regulation (16). TGF-β1 has been reported to inhibit adipogenesis, although these findings were derived from in vitro preadipocyte models (17,18). Smad3 was shown to bind C/EBP and repress its transactivation potential, thus abolishing the expression of PPARγ2 (17). However, the in vivo effect of Smad3 on adiposity remains unclear. Elevated expression and plasma levels of TGF-β1 have been reported in WAT from obese mice and in diabetic patients, respectively (19,20). TGFβ1/Smad3 was also shown to regulate insulin gene expression and pancreatic β-cell function (21). Conceivably, this will have an impact on energy homeostasis. However, the effects of Smad3 on communications among insulin-responsive organs toward the development of diet-induced obesity and type 2 diabetes have not been examined in vivo.
We show that Smad3-knockout (KO) mice have reduced adiposity associated with impaired lipid accumulation and adipogenesis. We revealed that this distinct phenotype might be associated with altered expression of PPARγ2 and β/δ in the adipose tissue. Despite their reduced physical activity arising from muscle atrophy, these KO mice are resistant to HFD-induced obesity. The deficiency of Smad3 also confers improved glucose tolerance and insulin sensitivity.
RESEARCH DESIGN AND METHODS
Animals.
Smad3 heterozygous mice on a C57BL/6 J background were intercrossed to produce Smad3-KO and wild-type (WT) offspring (22). Male mice of the same age were used in all experiments.
Hyperinsulinemic-euglycemic clamp studies.
Clamp studies were performed as previously described (23).
In vitro insulin-induced glucose uptake assay.
After fasting (15 h), tissues were harvested, followed by incubation in a Krebs-Ringer bicarbonate buffer. The solution was then replaced with a Krebs-Ringer bicarbonate buffer solution containing [3H]2-deoxyglucose (2.25 mCi/mL) and [14C] mannitol (0.3 mCi/mL) in the presence or absence of insulin (12–14 nmol/L). The tissues were quickly frozen until use. The tissues were lysed with 1 mol/L NaOH and centrifuged. The supernatant was neutralized with 1 mol/L HCl, followed by measuring the radioactivity for [3H]2-deoxyglucose and [14C] mannitol by a scintillation counter. Net uptake of [3H]2-deoxyglucose uptake rates were corrected using [14C] mannitol.
Real-time quantitative PCR.
Quantitative PCR (qPCR) was performed on three paired sets of WT and KO WAT. The sequences of the primer pairs are summarized in Supplementary Table 1. qPCR was performed as previously described (24).
Fatty acid uptake.
WT and KO adipocytes were incubated in media containing 1 μCi/mL of [14C]palmitate complexed to BSA at a 6.6:1 molar ratio. The uptake and accumulation of [14C]palmitate were measured using a scintillation proximity assay (25).
Incorporation of [14C]palmitate into triglycerides.
WT and KO adipocytes were loaded with [14C]palmitate as described above. Lipids were extracted, including a recovery standard in each sample (0.02 μCi of [3H]cholesterol), and analyzed by thin-layer chromatography using hexane:ethyl ether:acetic acid (80:20:1) as previously described (26).
Fatty acid oxidation in adipocytes.
Substrate oxidation was monitored by incubating cells with [14C]palmitate, with subsequent capture of liberated 14CO2 as previously described (25).
Lipolysis in adipocytes.
Lipolysis was determined from freshly isolated adipocytes by measurement of glycerol released into the medium as previously described (27).
Lipid extraction from peripheral organs.
Lipid was extracted from tissues by a Folch extraction as previously described (28,29).
Statistical analysis.
Statistical analysis was performed using two-tailed Mann–Whitney tests. Values were expressed as mean ± SEM. P < 0.05 was considered statistically significant. Detailed description of various methods can be obtained from the corresponding author.
RESULTS
Smad3-KO mice exhibit reduced adiposity, hypoglycemia, and hyperinsulinemia.
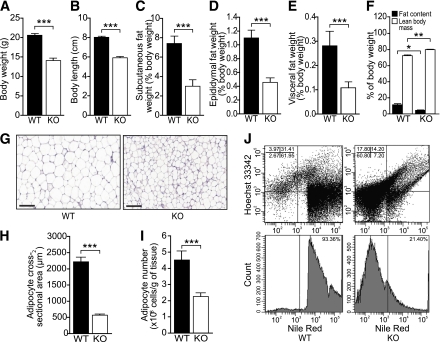
Eight-week-old Smad3-KO mice were characterized by reduced weight and short body length when compared with WT mice (Fig. 1A and B). To identify the cause of these anomalies in KO mice, we measured the weight of several organs, expressed as percent body weight. The weights of the liver, muscle, and brown adipose tissue were similar in WT and KO mice (Supplementary Fig. 1A–C). WAT weight from various fat pad depots was reduced by ∼63% in KO mice (Fig. 1C–E). Total body composition analysis by quantitative nuclear magnetic resonance further confirmed that fat content in KO mice was reduced by ∼63% (WT = 11.36 ± 1.51% vs. KO = 4.18 ± 0.29%; P < 0.05), whereas as expected from this observation, the proportion of the lean mass was consistently higher in KO mice (WT = 72.29 ± 1.24% vs. KO = 80.01 ± 0.49%; P < 0.01) (Fig. 1F). Smad3-KO epididymal WAT contained smaller adipocytes than their WT counterparts (WT = 2,206.03 ± 153.08 μm2 vs. KO = 567.49 ± 34.11 μm2; P < 0.001) (Fig. 1G and H). Fluorescence-activated cell sorting analysis comparing KO and WT WAT using Nile Red staining revealed a smaller and reduced number of adipocytes in KO mice (Fig. 1I and J). No significant difference in cell proliferation in WAT and liver histology was observed for both genotypes (Supplementary Fig. 1D and E).
FIG. 1.
Reduced adiposity in Smad3-KO mice. A–E: Mean body weight (A), overall mouth-to-anus body length (B), and relative weight of subcutaneous (C), epididymal (D), and visceral (E) white adipose fat pads of 8-week-old WT and Smad3-KO mice. Values represent percentage of body weight; n= 8/group. F: Body fat content and lean mass composition analysis of WT and KO mice; n= 8/group. G: Representative hematoxylin–eosin-stained paraffin-embedded section of WT and KO epididymal WAT. Scale bars, 100 μm. H: Mean cross-sectional area of WT and KO adipocytes (n= 2,000/group). I: Mean adipocyte number in WT and KO epididymal WAT. J: Flow cytometry analysis of adipocytes (Nile Red positive) and stromal/vascular cells (Hoechst positive) in WT and KO epididymal WAT. WAT was subjected to collagenase digestion. The adipocyte layer was gently recovered and stained for 5 min with 10 μL of Nile Red staining solution (Molecular Probes, Eugene, OR). Cells positive for Nile Red were counted using LSR II Flow Cytometer System (Becton Dickinson, Franklin Lakes, NJ). Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. (A high-quality color representation of this figure is available in the online issue.)
Smad3-KO mice show increased peripheral insulin sensitivity.
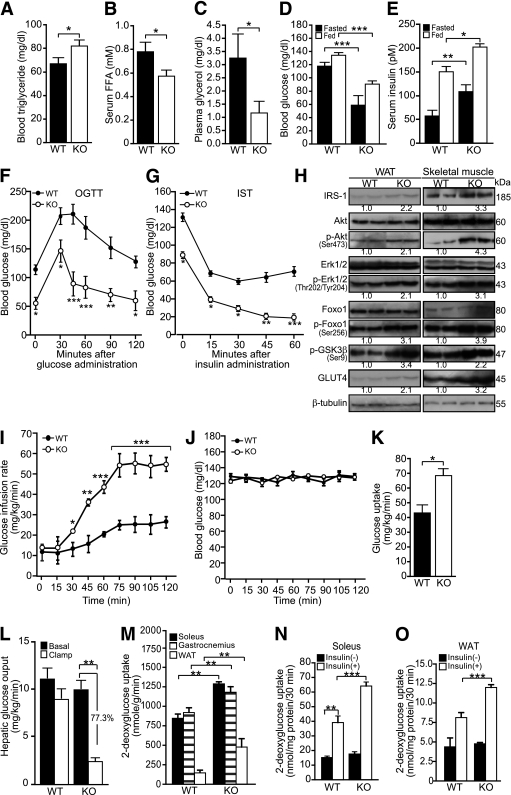
To uncover the systemic manifestations of reduced adiposity, we measured plasma lipid and glucose parameters in WT and KO mice. Although the plasma triglyceride level was elevated in KO mice, the FFA and glycerol levels were reduced, suggesting decreased lipolysis (Fig. 2A–C). Plasma cholesterol levels remained unchanged (Supplementary Fig. 1F). KO mice displayed hyperinsulinemia and were hypoglycemic regardless of feeding status (Fig. 2D and E). KO mice exhibited significantly faster reduction in blood glucose concentration after glucose administration (Fig. 2F), and they were more sensitive to insulin, as indicated by the prolongation of blood glucose–lowering effects of insulin in these animals (Fig. 2G).
FIG. 2.
Smad3-KO mice develop hypoglycemia and insulin hypersensitivity. A–E: Mean plasma triglyceride (A), FFA (B), glycerol (C), blood glucose (D), and blood insulin (E) in WT and KO mice. Blood glucose and insulin levels were measured at 6-h fasted and fed states. Blood glucose levels were measured using an Accu-Chek Advantage glucometer with glucose measurement strips (Roche Diagnostics, Indianapolis, IN). Serum insulin was measured by ELISA (Millipore, Billerica, MA). Serum total cholesterol, triglyceride, glycerol, and FFA concentrations were determined by quantitative enzymatic assays using l-type CHO-H (Wako Pure Chemical Industries, Osaka, Japan), serum triglyceride determination kit (Sigma-Aldrich, St. Louis, MO), free glycerol determination kit (Sigma-Aldrich), and serum/plasma FA and glycerol kit (Zen-Bio, Research Triangle Park, NC), respectively. F: Changes in blood glucose concentration at the indicated time points after oral glucose tolerance test (OGTT). G: Changes in blood glucose concentration at the indicated time points after IST. For the OGTT, mice were gavage fed with a 2 mg glucose/g body wt glucose load. For the IST, mice fasted for 6 h were intraperitoneally injected with 0.75 mU insulin/g body wt using an insulin syringe. OGTT and IST were performed on mice fasted for 6 h. Data are represented as mean ± SEM, n= 8/group. H: Immunoblot analysis of indicated proteins in WT and KO skeletal muscle and WAT. β-Tubulin was used as loading and transfer control. Representative blots are shown. I and J: Hyperinsulinemic-euglycemic clamp studies of Smad3-KO and WT mice. Glucose infusion rate required to maintained euglycemia in KO and WT mice (I). Blood glucose levels during the clamp study (J). K: Whole-body glucose uptake (peripheral insulin sensitivity). L: insulin-mediated suppression of hepatic glucose production rates (hepatic insulin sensitivity). Number represents percentage of suppression compared with basal hepatic glucose output. M: 2-DG uptake by soleus, gastrocnemius muscles, and WAT of KO and WT mice during clamp studies. Data are represented as mean ± SEM, n= 6/group. N and O: In vitro insulin-stimulated glucose uptake in skeletal muscles and WAT. 2-DG uptake into soleus muscle (N) and WAT (O) was measured for 30 min. The tissues from WT and KO were incubated in the absence [insulin(−)] or presence of insulin [insulin(+), 14 nmol/L], followed by measurement of 2-DG uptake. Net uptake of glucose was determined by subtracting the amount of [14C]mannitol from that of 2-DG. Data represent mean ± SEM, n= 10/group. *P < 0.05, **P < 0.01, ***P < 0.001 vs. WT controls.
These findings suggested that Smad3 deficiency improved glucose tolerance and insulin responsiveness. To understand the mechanisms of improved insulin sensitivity in KO mice, the protein expression levels of key mediators of insulin-signaling cascades were examined. Immunoblot analysis of WAT and skeletal muscle revealed increased GLUT4, insulin receptor substrate-1, phosphorylated FoxO1, GSK-3β, Akt, and ERK expression in Smad3-KO mice when compared with WT mice (Fig. 2H). Next, we measured in vivo insulin-stimulated glucose uptake by using hyperinsulinemic-euglycemic clamp studies. During constant hyperinsulinemia, a higher glucose infusion rate was required to maintain normal glucose levels in KO mice compared with WT mice (Fig. 2I and J). To determine the insulin-stimulated glucose uptake of skeletal muscle and WAT, we assessed 2-deoxy-D-[1,2-3H]glucose (2-DG) uptake during clamp studies. Whole-body glucose utilization (peripheral insulin sensitivity) and percentage of insulin-mediated suppression of hepatic glucose production (hepatic insulin sensitivity) were significantly increased in the KO mice (Fig. 2K and L). We also found a significant increase of 2-DG uptake into the soleus, gastrocnemius muscles, and WAT of KO mice (Fig. 2M). Data from in vitro 2-DG uptake of isolated soleus muscle and WAT were congruent with the in vivo findings that KO mice are more insulin-sensitive than WT (Fig. 2N and O).
Smad3-KO mice display decreased physical activity.
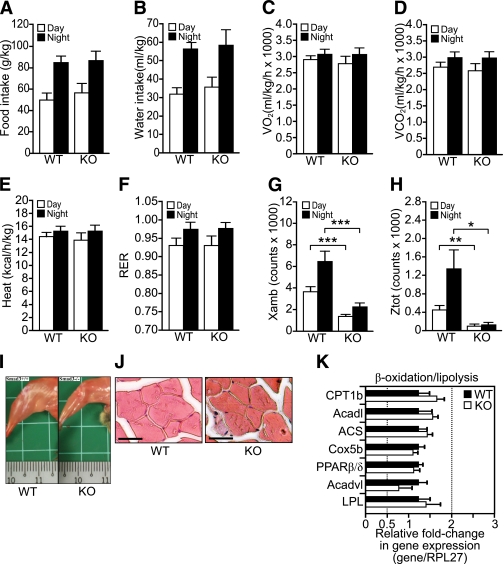
Next, we explored the impact of Smad3 deletion on whole-body metabolic activity in the absence of a dietary challenge. To this end, mice were fed a standard chow diet, and various metabolic parameters were monitored using metabolic cages. We observed no significant differences in food and water consumption (Fig. 3A and B), O2 consumption (Fig. 3C), CO2 production (Fig. 3D), and metabolic rate (heat production) (Fig. 3E) between WT and KO mice (Supplementary Fig. 2A–D). Both genotypes consumed carbohydrates as the main energy source given that their respiratory exchange ratio (RER), which equals Vo2/Vco2, was >0.92 (Fig. 3F). The RER indicates whether carbohydrates (RER = 1.0) or lipids (RER = 0.7) are being used to produce energy.
FIG. 3.
Smad3-KO mice exhibit reduced physical activity. A–D: The mean total food (A), water (B), O2 (C) consumptions, and CO2 emission (D) during a 24-h light and dark cycle in WT and KO mice. Values in C and D were measured by indirect calorimetry and expressed as average Vo2 and Vco2 per kg body wt per h during a 24-h monitoring session of a light-dark cycle. E: Heat production of WT and KO mice was calculated from the O2 production and the RER and expressed as average kcal/h/kg body wt. F: Fuel consumption was determined from the ratio of CO2 emitted to the amount of O2 consumed. Fuel type preference for carbohydrate has an RER of 1.0 versus fat of 0.7. G and H: Physical activity was assessed by the number of horizontal and vertical beam breaks, which represent the total horizontal (G) and rearing (H) movements during the 24-h monitoring period. I: Representative photograph showing the hind leg muscles of WT and KO mice. Small division on the ruler scale = 1 mm. J: Representative hematoxylin–eosin-stained histologic section of the WT and KO quadriceps muscles. Scale bars, 100 μm. K: Relative fold change in mRNA level of the indicated genes in WT and KO muscle was analyzed by real-time qPCR. Values were normalized to ribosomal protein L27. Results are represented as fold induction compared with WT. Primer sequences are summarized in Supplementary Table 1. Values represent mean ± SEM, n= 8/group. *P < 0.05, **P < 0.01, ***P < 0.001. (A high-quality color representation of this figure is available in the online issue.)
KO mice exhibited a decrease of ∼74% in both horizontal and rearing movements (Fig. 3G and H, Supplementary Fig. 2E and F). Macroscopic comparison between WT and KO showed a ∼10% reduction in muscle fiber size (Fig. 3I and J). Real-time qPCR analysis comparing WT and KO muscle revealed no difference in the expression of genes involved in β-oxidation or lipolysis (Fig. 3K).
The liver may contribute to the reduced adiposity in KO mice via increased fatty acid (FA) β-oxidation. qPCR analysis revealed no difference in mRNA expression level of some genes involved in β-oxidation between WT and KO liver (Supplementary Fig. 3A). In vitro total β-oxidation assay also showed no significant difference in the oxidation of [14C]palmitate in KO and WT hepatocytes (Supplementary Fig. 3B). These results indicate that the reduced adiposity in KO mice did not stem from increased energy expenditure arising from physical activity or increased hepatic β-oxidation.
Smad3-KO mice show increased β-oxidation and impaired lipolysis in WAT.
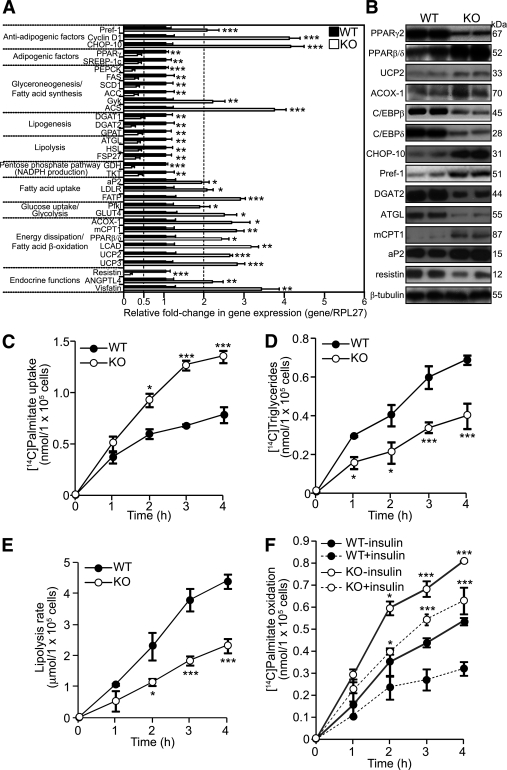
Two factors contribute to the expansion of WAT mass: increased size of existing adipocytes because of fat accumulation and the formation of new adipocytes through adipogenesis. Fat mass reflects the ratio between lipogenesis and lipolysis/β-oxidation. To examine the role of Smad3 in adipocyte physiology, we performed focused qPCR array on WT and KO WAT. The expression of several antiadipogenic factors, such as preadipocyte factor 1 (Pref-1), cyclin D1, and C/EBP homologous protein 10 (CHOP-10), was elevated by twofold or more in KO adipocytes, whereas there was a reduced PPARγ2 expression, suggesting that Smad3 deficiency inhibits adipogenesis (Fig. 4A). PPARγ2-regulated genes, such as PEPCK and fatty acid synthase (FAS), were also downregulated (Fig. 4A). The expression of the SREBP-1c, which stimulates many genes involved in FA metabolism and potentiates the transcriptional activity of PPARγ2, was decreased in KO mice. Furthermore, the expression of diacylglycerol acyltransferase-1 and 2 was reduced by approximately two- to threefold in KO WAT.
FIG. 4.
Smad3-KO adipose tissues have increased FA uptake and β-oxidation. A: Relative fold change in mRNA level of the indicated genes in WT and KO adipose tissue as determined by qPCR. Values are normalized to ribosomal protein L27. Normalized values from WT mice were arbitrarily assigned a value of 1. Only genes with twofold or greater change in expression are presented. Values represent mean ± SEM, n= 3/group. Primer sequences are summarized in Supplementary Table 1. B: Immunoblot analysis of indicated proteins in WT and KO adipose tissues. β-Tubulin was used as loading and transfer control. Representative blots from two mice for each genotype are shown. C–F: The rate of [14C]palmitate uptake (C), [14C]palmitate incorporation into triglycerides (D), lipolysis (E), and [14C]palmitate oxidation (F) at the indicated time points of adipocytes freshly isolated from the epididymal fat pads of WT and KO mice. Data are expressed as mean ± SEM, n= 3/group. *P < 0.05, **P < 0.01, ***P < 0.001 vs. WT controls.
In contrast with lipogenesis, mobilization of fat stores occurs predominantly through the hydrolysis of triglycerides into glycerol and FAs. The expression of adipose triglyceride lipase and hormone-sensitive lipase, the rate-limiting enzymes of this process, were decreased by approximately twofold in the KO WAT. Notably and in contrast with the reduced expression of PPARγ2, the expression of PPARβ/δ in the KO WAT was significantly increased. The mRNA levels of PPARβ/δ-regulated genes, such as uncoupling protein (UCP) 2, UCP3, and acyl-CoA oxidase 1 (14), were elevated in the KO WAT, suggesting an increase FA β-oxidation and thermogenesis (Fig. 4A). The protein levels of selected genes were similarly altered (Fig. 4B).
To strengthen the observations, we determined the effect of Smad3 deficiency on triglyceride metabolism. We measured [14C]palmitate uptake and its conversion into triglycerides in WT and KO adipocytes. We observed a 1.7-fold increase in [14C]palmitate uptake in KO adipocytes compared with WT (102.5 ± 9.4 nmol/105 cells vs. 61.0 ± 8.8 nmol/105 cells, respectively; Fig. 4C). Notably, the increased FA uptake was not accompanied by a concomitant increased conversion into triglycerides, which were reduced in KO adipocytes when compared with WT (0.26 ± 0.11 nmol/105 cells vs. 0.49 ± 0.17 nmol/105 cells, respectively; Fig. 4D). We also assayed the levels of glycerol released into the culture medium as an indicator of lipolysis. Smad3 deficiency led to a twofold decrease in glycerol release (WT = 2.9 ± 0.74 nmol/105 cells vs. KO = 1.45 ± 0.28 nmol/105 cells; Fig. 4E). We also found that the oxidation of [14C]palmitate in KO adipocytes was twofold higher than that of WT (0.61 ± 0.09 nmol/105 cells vs. 0.33 ± 0.08 nmol/105 cells, respectively; Fig. 4F). The incubation of adipocytes with insulin reduced palmitate oxidation in both WT and KO adipocytes; however, the oxidation level in KO adipocytes remained 1.8-fold higher than in WT (0.44 ± 0.09 nmol/105 cells vs. 0.24 ± 0.05 nmol/105 cells, respectively; Fig. 4F). Altogether, these observations show that Smad3 deficiency impairs adipogenesis and in parallel moderates fat accumulation by directing increased FA uptake to β-oxidation.
Repression of PPARβ/δ by C/EBPβ is relieved on upregulation of CHOP-10 resulting from Smad3 deficiency.
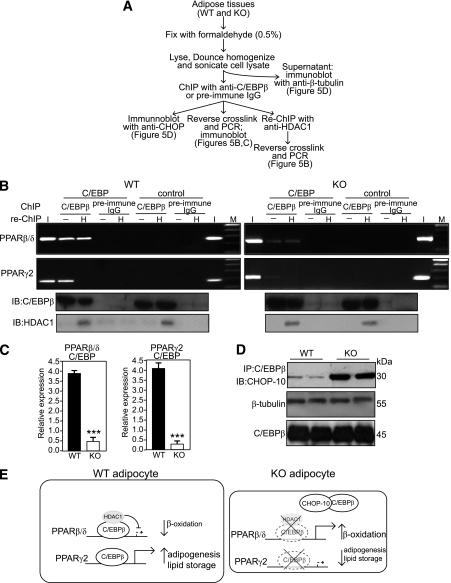
C/EBPβ represses the expression of PPARβ/δ in keratinocytes (30). To gain insight into the regulation of PPARβ/δ in adipocytes, we performed in vivo coimmunoprecipitation and chromatin immunoprecipitation (ChIP) using anti-C/EBPβ antibodies in WT and KO mice (Fig. 5A). ChIP and re-ChIP assays showed that in the WT WAT, C/EBPβ was associated with a specific C/EBP binding site at 494–485 bp upstream from the PPARβ/δ promoter. The data further showed that the association results in recruitment to this element of a transcriptional repressor complex containing histone deacetylase 1 (HDAC1) (Fig. 5B), indicating a C/EBPβ-mediated repression of PPARβ/δ. Notably, the association of C/EBPβ-HDAC1 complexes with this binding site was reduced in KO WAT (Fig. 5B). This association was further confirmed by qPCR normalized to chromatin before immunoprecipitation (i.e., input) (Fig. 5C). No amplification was observed using preimmune IgG or a control DNA sequence (−1179 to −894).
FIG. 5.
Increased C/EBPβ-CHOP-10 interaction in Smad3-KO adipocytes. A: Schematic outline of procedure for in vivo coimmunoprecipitation, ChIP, and re-ChIP. B: DNA-protein complexes from WT or KO adipocytes were immunoprecipitated (ChIP) with anti-C/EBPβ or preimmune IgG. Enrichment of the DNA fragment containing the C/EBP binding site on the endogenous PPARβ/δ, PPARγ2 promoter, or a control sequence upstream was evaluated by PCR. A second immunoprecipitation step (re-ChIP) was also performed with an anti-HDAC1-specific antibody (H). I = input chromatin before immunoprecipitation, − = negative control (without anti-HDAC1 antibody), M = DNA marker, IB = immunoblot. C: Real-time qPCR was performed on immunoprecipitates of C/EBPβ antibodies and normalized to input. Data are expressed as mean ± SEM, n= 5/group. ***P < 0.001. D: Western blot analyses were also performed on proteins after immunoprecipitation with anti-C/EBPβ antibody. Proteins were separated by SDS-PAGE, blotted, and probed with anti-CHOP-10 primary antibody as indicated. Bands were detected by chemiluminescence. Western blot control with β-tubulin antibody was performed using cell lysate before immunoprecipitation. E: Schematic diagram showing the repression of PPARβ/δ by C/EBPβ is relieved on upregulation of CHOP-10 resulting from depletion of Smad3.
Smad3 deficiency resulted in elevated expression of CHOP-10 (Fig. 4B). We examined possible involvement of CHOP-10 in the ability of C/EBPβ to regulate PPARβ/δ transcription. In vivo co-immunoprecipitation with anti-C/EBPβ followed by immunoblot with anti-CHOP-10 (Fig. 5A) revealed that the interactions between C/EBPβ and CHOP-10 are markedly enhanced in KO WAT (Fig. 5D). Thus, increased C/EBPβ-CHOP-10 interactions removed the inhibitory effect of C/EBPβ on PPARβ/δ promoter, resulting in increased PPARβ/δ expression in KO mice (Fig. 5E).
C/EBPβ is closely involved in the regulation of PPARγ in adipocytes (31). As expected, ChIP assay involving the PPARγ2 promoter showed that C/EBPβ was bound to the C/EBP binding site. In contrast with the PPARβ/δ promoter results, no amplification was detected after re-ChIP with anti-HDAC1 (Fig. 5B). In KO WAT, only weak amplification resulted with anti-C/EBPβ (Fig. 5B). The increased C/EBPβ-CHOP-10 interactions in the KO when compared with WT adipocytes (Fig. 5D) also diminished PPARγ2 promoter activity (Fig. 5B). These observations show that Smad3 deficiency alters adipose physiology, causing a change associated with diminished PPARγ2 and increased PPARβ/δ expression (Fig. 5E).
Smad3-KO mice are resistant to HFD-induced obesity and insulin resistance.
A chronic HFD treatment induces obesity and obesity-associated insulin resistance in mice (32). To examine the effect of Smad3 deficiency on HFD-induced obesity and insulin resistance, we placed KO and WT mice on a low-fat diet (LFD) or HFD for 18 weeks. On an HFD, WT mice became obese and were significantly heavier than those on an LFD (Fig. 6A and B). In contrast, no significant difference in body weight gain was observed in KO mice on either diet, indicative of their resistance to HFD-induced obesity (Fig. 6A and B). There was no difference in the mean daily food intake per mouse, regardless of diet, throughout the study (Supplementary Fig. 4A and B).
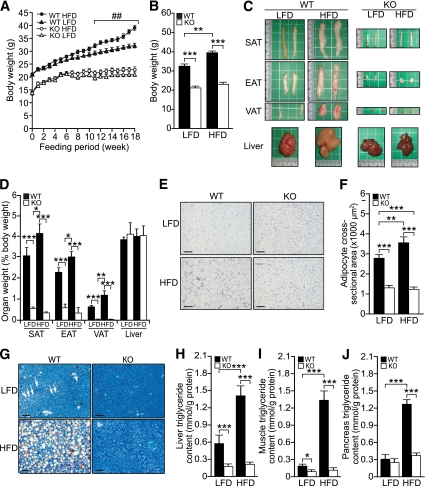
FIG. 6.
Smad3-KO mice are resistant to HFD-induced obesity and hepatic steatosis. A and B: Graph shows mean body weight of WT and KO mice on HFD or LFD over 18 weeks (A) and at the end of the 18-week diet regimen (B). C: Representative images of subcutaneous, epididymal, and visceral adipose tissues, and the liver of WT and KO mice from the two diet regimens. Small division on the ruler scale = 1 mm. D: Mean weights of the liver and the indicated adipose tissues of WT and KO mice after 18 weeks on HFD or LFD. Values are expressed as percentage of body weight. E: Representative hematoxylin–eosin-stained histologic section of the WT and KO EAT after 18 weeks on HFD or LFD treatment. Scale bars, 100 μm. F: Mean cross-sectional area of adipocytes (n= 400) from the WT and KO mice on the indicated diet regimen. G: Representative histologic section (oil red O and methylene blue staining) of the liver tissues. Scale bars, 100 μm. White arrows indicate oil droplets. H–J: Quantification of lipid accumulation in peripheral organs. Total triglyceride extracted from fresh-frozen liver (H), skeletal muscle (I), and pancreas (J) of WT and KO mice fed on HFD or LFD diet was expressed as amount in milimole per 1 g of protein. Data represent mean ± SEM, n = 10/group. ##P < 0.01 vs. WT LFD control; *P < 0.05, **P < 0.01, ***P < 0.001. EAT, epididymal adipose tissue; SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue. (A high-quality digital representation of this figure is available in the online issue.)
As expected, HFD induced an increase in adipose mass in all fat depots in WT mice (Fig. 6C and D). This increase of 33% in WAT was associated with hypertrophy of adipocytes (Fig. 6E and F) when compared with LFD controls. No significant difference in adipocyte size was observed between KO mice on either diet (Fig. 6E and F). Histologic analysis of the liver from WT mice on an HFD showed macrovesicular hepatic steatosis as evidenced by a fatty liver populated with abundant large vacuolar lipid droplets (Fig. 6G). We also observed small lipid droplets in the liver of WT mice on an LFD. Notably, the liver of KO mice had few or no lipid droplets and did not develop hepatic steatosis, regardless of treatment (Fig. 6G). HFD induced an increase in triglyceride level in liver, skeletal muscle, and pancreas of WT mice when compared with LFD controls. No significant difference in tissue triglyceride level was observed between KO mice on either diet (Fig. 6H–J, Supplementary Fig. 4C and D).
KO mice exhibited a higher plasma triglyceride level compared with WT mice on an LFD (Fig. 7A). As expected, the plasma triglyceride level was increased in WT and KO mice on an HFD, with the former reaching a level comparable to that of the KO mice under an LFD (Fig. 7A). The levels of glycerol and FFA were significantly elevated in WT and KO mice on an HFD (Fig. 7B and C), but remained lower in KO mice on either diet. Similarly, regardless of the diet and feeding status, KO mice consistently showed lower blood glucose levels (Fig. 7D). No significant difference in plasma cholesterol level was detected (Supplementary Fig. 4E). In agreement with these results, KO mice exhibited higher insulin levels under an LFD, but in contrast with WT mice, HFD treatment did not significantly increase these levels (Fig. 7E).
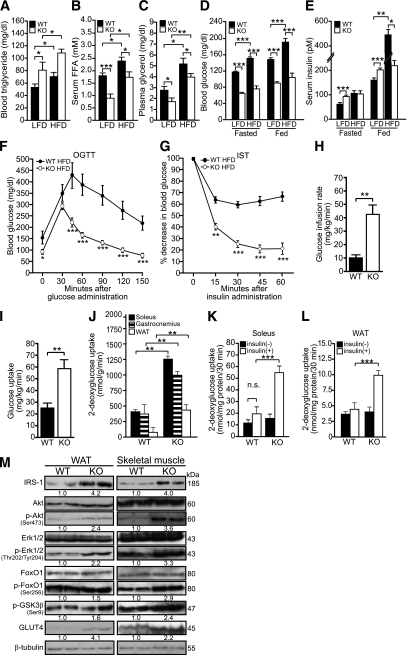
FIG. 7.
Smad3 deficiency ameliorates HFD-induced insulin resistance and glucose intolerance. A–E: The mean plasma triglyceride (A), FFA (B), glycerol (C), blood glucose (D), and blood insulin (E) in WT and KO mice on HFD or LFD. Blood glucose and insulin levels were measured at 6-h fasted and fed states. F: Changes in blood glucose concentration at the indicated time points after OGTT. G: Changes in blood glucose concentration at the indicated time points after IST. OGTT and IST were performed on mice fasted for 6 h on an 18-week LFD or HFD regimen. Data represent mean ± SEM, n= 10/group. H–J: Hyperinsulinemic-euglycemic clamp studies of Smad3-KO and WT mice. Glucose infusion rate required to maintained euglycemia in KO and WT mice (H); whole-body glucose uptake (peripheral insulin sensitivity) (I); 2-DG uptake by soleus, gastrocnemius muscles, and WAT of HFD-fed KO and WT mice during clamp studies (J). Data are represented as mean ± SEM, n= 5/group. K–L: In vitro insulin-stimulated glucose uptake in skeletal muscles and WAT. 2-DG uptake into soleus muscle (K) and WAT (L) was measured for 30 min. The tissues from WT and KO were incubated in the absence [insulin(-)] or presence of insulin [insulin(+), 14 nmol/L], followed by measurement of 2-DG uptake. Net uptake of glucose was determined by subtracting the amount of [14C]mannitol from that of 2-DG. Data represent mean ± SEM, n = 6/group. *P < 0.05, **P < 0.01, ***P < 0.001 vs. WT controls. M: Immunoblot analysis of indicated proteins from skeletal muscle and WAT of HFD-fed WT and KO mice. β-Tubulin was used as loading and transfer control. Representative blots are shown.
Glucose tolerance and insulin sensitivity tests (ISTs) were carried out to determine the effects of an HFD on the WT and KO mice. The blood glucose of WT mice on an HFD only started to decrease at 45 min after glucose administration and failed to return to the basal level even at the 120-min time point, indicative of impaired glucose tolerance in these diet-induced obese mice. By contrast, KO mice under an HFD retained the rapid glucose clearance ability as previously observed under the standard diet (Fig. 7F). In addition, HFD-induced obese WT mice also clearly displayed obesity-associated insulin resistance (Fig. 7G), which was absent in KO mice. In vivo insulin-stimulated glucose uptake by using hyperinsulinemic-euglycemic clamp studies (Fig. 7H–J) and in vitro tissue-specific insulin-stimulated 2-DG uptake (Fig. 7K and L) indicated that KO mice were more tolerant to HFD-induced peripheral insulin resistance compared with WT mice. In concordance, the protein expression levels of indicated key mediators of insulin-signaling cascades were elevated in the WAT and skeletal muscle of HFD-fed KO mice (Fig. 7M). Altogether, these observations indicated that KO mice are resistant to HFD-induced obesity and numerous obesity-linked pathologies.
DISCUSSION
Obesity increases risk for type 2 diabetes, hypertension, and cardiovascular diseases. Accordingly, much research effort currently targets identification of molecular targets and the development of drugs that reduce fat mass and improve insulin sensitivity. We examined the effect of Smad3 deficiency on adiposity in mice. By using Smad3-KO mice, we showed that Smad3 deficiency resulted in decreased adiposity associated with improved glucose tolerance and insulin responsiveness. Despite decreased physical activity, Smad3-KO mice were resistant to HFD-induced obesity. The deficiency in Smad3 also conferred resistance to the development of obesity-associated glucose intolerance, insulin resistance, and liver steatosis, thus providing an insight into the role of Smad3 in the pathogenesis of obesity and type 2 diabetes.
Previous in vitro studies showed that Smad3 inhibits the transactivation potential of C/EBPs and consequently abolishes the stimulation of the adipogenic master regulator PPARγ2 (17). In fact, an increase in adiposity might be anticipated in the absence of the inhibitory effect of Smad3 on C/EBPs, as in the case of Smad3 deficiency. We found that Smad3-KO mice displayed decreased adiposity resulting from reduced adipocyte number and size, suggesting defective adipogenesis and altered lipid accumulation. Gene expression profiling of the Smad3-KO WAT revealed reduced expression of PPARγ2 mRNA, with increased expression of the preadipocyte-specific marker Pref-1. PPARγ2 not only participates in adipogenesis and survival but also promotes lipid storage (33). Lipid accumulation relies on lipogenesis, which involves de novo synthesis of FA and glycerol, FA uptake, and synthesis of triglycerides. Notably, the expression of rate-limiting enzymes involved in these processes, such as FAS, PEPCK, and diacylglycerol acyltransferase, was downregulated in the Smad3-KO WAT (34). The reduced lipogenesis is exacerbated by concomitant downregulation of enzymes participating in the pentose phosphate pathway required for de novo synthesis of biomolecules. Lending support, knockout or adipose-specific deletion of some examined genes in mice has resulted in similar phenotypic features in Smad3-KO mice. Pref-1-null mice, for instance, display obesity and increased serum lipid metabolites (35). Adipose-specific knockout of PEPCK results in a fraction of mice developing lipodystrophy (36). As a central enzyme in lipogenesis, FAS has been identified as a candidate gene for determining body fat (37). Smad3 deficiency also resulted in increased FA β-oxidation as evidenced by the reduced plasma FFA level, offering another plausible explanation for the reduced adipocyte size in the Smad3-KO mice. Altogether, these findings indicate that Smad3 deficiency leads to impaired adipogenesis and lipogenesis in mice.
It has been proposed that adipose lipid storage functions to prevent peripheral lipotoxicity (5). Particularly evident in HFD-induced obesity, the chronic accumulation of FFA in skeletal muscles and liver eventually dampens their insulin responsiveness (5,7). Indeed, the ablation of PPARγ2 in obese mouse models results in reduced fat mass but leads to severe insulin resistance, β-cell failure, and dyslipidemia because of deposition of toxic reactive lipid species in the peripheral organs (38). However, this reported insulin resistance and β-cell failure were not observed in the Smad3-KO mice, which also exhibited reduced expression of adipose PPARγ2. This outcome may partly be the result of elevated plasma insulin levels and insulin hypersensitivity, which caused hypoglycemia in these mice. Indeed, our results also revealed enhanced insulin responsiveness in peripheral organs, such as skeletal muscle and WAT. Insulin activation of phosphatidylinositol-3-kinase/protein kinase B (PKB) (Akt) can inhibit TGF-β1 signaling via the formation of a PKB-Smad3 complex. This interaction does not inhibit PKB activity, but inhibits Smad3-mediated gene regulation (39,40). A recent study showed that Smad3 occupies the insulin gene promoter to repress its expression and increases glucose-stimulated insulin secretion because of enhanced insulin signaling in β-cell islets (21). Furthermore, the expression of adipocytokines, such as visfatin and resistin, can influence glucose uptake and metabolism (41). An increase in glucose uptake and glycolysis with the effects of these adipocytokines and a decreased plasma FFA level as observed in the Smad3-KO mice may partially contribute to the improved glucose tolerance and enhanced insulin sensitivity in these animals. Altogether, these findings place Smad3-PKB at a point of convergence in the crosstalk between TGF-β/Smad3 and insulin signaling pathways and provide insights into their roles in the development of obesity and insulin resistance. Our findings that Smad3-KO mice displayed increased insulin-stimulated whole-body (peripheral insulin sensitivity) and tissue-specific (skeletal muscle and WAT) glucose uptake underscored the relevance of this crosstalk in vivo.
The skeletal muscle is the largest energy consumer in mice and plays an important role in lowering plasma FFA and glycerol (42,43); however, we found that Smad3-KO mice exhibited reduced physical activity associated with muscular atrophy, in concordance with a role of Smad3 in β2-adrenergic–induced muscle hypertrophy (44). Thus, it is unlikely that increased energy expenditure in the skeletal muscles could account for the reduced adiposity in these mice. Although adipose tissue is not a major energy consumer compared with the skeletal muscle, increased β-oxidation in WAT can have a profound impact on adiposity and insulin sensitivity (41). Comparative gene expression analysis of WT and Smad3-KO WAT revealed upregulation of PPARβ/δ, UCP2, UCP3, and acyl-CoA oxidase 1, all of which are involved in energy dissipation and peroxisomal FA oxidation. The activation of PPARβ/δ in obese mice has been shown to selectively induce expression of an array of genes required for adipose FA catabolism and thermogenesis but not genes involved in lipogenesis and fat storage, which are controlled by PPARγ2 (14). Metabolic rate remained unchanged in Smad3-KO mice, reflecting a balance between reduced skeletal muscle activity and increased adipose lipid oxidation. Many regulatory mechanisms may influence the expression of PPARγ and PPARβ/δ in adipocytes. The relative proportion of homodimeric or heterodimeric C/EBP is likely to play important role. Although it is essential for PPARγ2 expression, C/EBPβ inhibits PPARβ/δ promoter activity. CHOP-10, which exhibited elevated expression in Smad3-KO WAT, acts as a dominant-negative inhibitor of C/EBP by preventing its binding to DNA (45); this function adds to the complexity of regulating the expression of these two PPAR isotypes with apparent opposing functions in the adipocyte. The reason for the differential regulation is unclear but may be attributed to different promoter context. The functional AP-1 site driving induction of the PPARβ/δ promoter is in close proximity to the identified C/EBP binding region. Smad3 may interact with c-JUN (AP-1), inhibiting c-JUN activity that is necessary for AP-1–stimulated PPARβ/δ expression, as reported in keratinocytes (46).
In summary, we have found that Smad3 is a multifaceted regulator in glucose and lipid metabolism, as well as in the pathogenesis of obesity and type 2 diabetes, thus identifying Smad3 as a potential target for the treatment of obesity and its associated disorders.
ACKNOWLEDGMENTS
This work was supported by grants from the Ministry of Education, Singapore (ARC 18/08), Nanyang Technological University, Singapore (RGD 127/05, 158/06) to N.S.T, and from the Swiss National Science Foundation and the National Research Center Frontiers in Genetics to W.W. All experimental protocols were approved by the Institutional Animal Care and Use Committee (ARF-SBS/NIE-A004 and -A0084).
No potential conflicts of interest relevant to this article were reported.
C.K.T. researched data, contributed to discussion, wrote the article, and reviewed and edited the article. N.L., M.J.T., Y.W.Y, Y.C., and R.K. researched data. W.W. researched data and reviewed and edited the article. N.S.T. contributed to discussion and reviewed and edited the article.
We acknowledge the Metabolic Evaluation platform of the Center for Integrative Genomics and Maude Husson (Center for Integrative Genomics) for help with the mice and for support.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db10-0801/-/DC1.
REFERENCES
- 1.Kopelman PG, Albon L. Obesity, non-insulin-dependent diabetes mellitus and the metabolic syndrome. Br Med Bull 1997;53:322–340 [DOI] [PubMed] [Google Scholar]
- 2.Wang P, Mariman E, Renes J, Keijer J. The secretory function of adipocytes in the physiology of white adipose tissue. J Cell Physiol 2008;216:3–13 [DOI] [PubMed] [Google Scholar]
- 3.Rabe K, Lehrke M, Parhofer KG, Broedl UC. Adipokines and insulin resistance. Mol Med 2008;14:741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol 2008;9:367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006;444:840–846 [DOI] [PubMed] [Google Scholar]
- 6.Xu H, Barnes GT, Yang Q, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 2003;112:1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu C, Chen Y, Cline GW, et al. Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J Biol Chem 2002;277:50230–50236 [DOI] [PubMed] [Google Scholar]
- 8.Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature 2000;405:421–424 [DOI] [PubMed] [Google Scholar]
- 9.Kersten S. Peroxisome proliferator activated receptors and obesity. Eur J Pharmacol 2002;440:223–234 [DOI] [PubMed] [Google Scholar]
- 10.Patsouris D, Müller M, Kersten S. Peroxisome proliferator activated receptor ligands for the treatment of insulin resistance. Curr Opin Investig Drugs 2004;5:1045–1050 [PubMed] [Google Scholar]
- 11.Yoon M. The role of PPARalpha in lipid metabolism and obesity: focusing on the effects of estrogen on PPARalpha actions. Pharmacol Res 2009;60:151–159 [DOI] [PubMed] [Google Scholar]
- 12.Guerre-Millo M, Gervois P, Raspé E, et al. Peroxisome proliferator-activated receptor alpha activators improve insulin sensitivity and reduce adiposity. J Biol Chem 2000;275:16638–16642 [DOI] [PubMed] [Google Scholar]
- 13.Fajas L, Fruchart JC, Auwerx J. Transcriptional control of adipogenesis. Curr Opin Cell Biol 1998;10:165–173 [DOI] [PubMed] [Google Scholar]
- 14.Wang YX, Lee CH, Tiep S, et al. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell 2003;113:159–170 [DOI] [PubMed] [Google Scholar]
- 15.Barish GD, Narkar VA, Evans RM. PPAR delta: a dagger in the heart of the metabolic syndrome. J Clin Invest 2006;116:590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massagué J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 2000;103:295–309 [DOI] [PubMed] [Google Scholar]
- 17.Choy L, Derynck R. Transforming growth factor-beta inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J Biol Chem 2003;278:9609–9619 [DOI] [PubMed] [Google Scholar]
- 18.Suzawa M, Takada I, Yanagisawa J, et al. Cytokines suppress adipogenesis and PPAR-gamma function through the TAK1/TAB1/NIK cascade. Nat Cell Biol 2003;5:224–230 [DOI] [PubMed] [Google Scholar]
- 19.Samad F, Yamamoto K, Pandey M, Loskutoff DJ. Elevated expression of transforming growth factor-beta in adipose tissue from obese mice. Mol Med 1997;3:37–48 [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeiffer A, Middelberg-Bisping K, Drewes C, Schatz H. Elevated plasma levels of transforming growth factor-beta 1 in NIDDM. Diabetes Care 1996;19:1113–1117 [DOI] [PubMed] [Google Scholar]
- 21.Lin HM, Lee JH, Yadav H, et al. Transforming growth factor-beta/Smad3 signaling regulates insulin gene transcription and pancreatic islet beta-cell function. J Biol Chem 2009;284:12246–12257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Letterio JJ, Lechleider RJ, et al. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J 1999;18:1280–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hevener AL, He W, Barak Y, et al. Muscle-specific Pparg deletion causes insulin resistance. Nat Med 2003;9:1491–1497 [DOI] [PubMed] [Google Scholar]
- 24.Chong HC, Tan MJ, Philippe V, et al. Regulation of epithelial-mesenchymal IL-1 signaling by PPARbeta/delta is essential for skin homeostasis and wound healing. J Cell Biol 2009;184:817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wensaas AJ, Rustan AC, Lövstedt K, et al. Cell-based multiwell assays for the detection of substrate accumulation and oxidation. J Lipid Res 2007;48:961–967 [DOI] [PubMed] [Google Scholar]
- 26.Listenberger LL, Han X, Lewis SE, et al. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA 2003;100:3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh W, Abu-Elheiga L, Kordari P, et al. Glucose and fat metabolism in adipose tissue of acetyl-CoA carboxylase 2 knockout mice. Proc Natl Acad Sci USA 2005;102:1384–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denton RM, Randle PJ. Concentrations of glycerides and phospholipids in rat heart and gastrocnemius muscles. Effects of alloxan-diabetes and perfusion. Biochem J 1967;104:416–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frayn KN, Maycock PF. Skeletal muscle triacylglycerol in the rat: methods for sampling and measurement, and studies of biological variability. J Lipid Res 1980;21:139–144 [PubMed] [Google Scholar]
- 30.Di-Poï N, Desvergne B, Michalik L, Wahli W. Transcriptional repression of peroxisome proliferator-activated receptor beta/delta in murine keratinocytes by CCAAT/enhancer-binding proteins. J Biol Chem 2005;280:38700–38710 [DOI] [PubMed] [Google Scholar]
- 31.Rosen ED, Hsu CH, Wang X, et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev 2002;16:22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Surwit RS, Kuhn CM, Cochrane C, McCubbin JA, Feinglos MN. Diet-induced type II diabetes in C57BL/6J mice. Diabetes 1988;37:1163–1167 [DOI] [PubMed] [Google Scholar]
- 33.Imai T, Takakuwa R, Marchand S, et al. Peroxisome proliferator-activated receptor gamma is required in mature white and brown adipocytes for their survival in the mouse. Proc Natl Acad Sci USA 2004;101:4543–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tontonoz P, Hu E, Devine J, Beale EG, Spiegelman BM. PPAR gamma 2 regulates adipose expression of the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol 1995;15:351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moon YS, Smas CM, Lee K, et al. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol 2002;22:5585–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beale EG, Harvey BJ, Forest C. PCK1 and PCK2 as candidate diabetes and obesity genes. Cell Biochem Biophys 2007;48:89–95 [DOI] [PubMed] [Google Scholar]
- 37.Mobbs CV, Makimura H. Block the FAS, lose the fat. Nat Med 2002;8:335–336 [DOI] [PubMed] [Google Scholar]
- 38.Medina-Gomez G, Gray SL, Yetukuri L, et al. PPAR gamma 2 prevents lipotoxicity by controlling adipose tissue expandability and peripheral lipid metabolism. PLoS Genet 2007;3:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Remy I, Montmarquette A, Michnick SW. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat Cell Biol 2004;6:358–365 [DOI] [PubMed] [Google Scholar]
- 40.Conery AR, Cao Y, Thompson EA, Townsend CM, Jr, Ko TC, Luo K. Akt interacts directly with Smad3 to regulate the sensitivity to TGF-beta induced apoptosis. Nat Cell Biol 2004;6:366–372 [DOI] [PubMed] [Google Scholar]
- 41.Ahima RS, Flier JS. Adipose tissue as an endocrine organ. Trends Endocrinol Metab 2000;11:327–332 [DOI] [PubMed] [Google Scholar]
- 42.Baron AD, Laakso M, Brechtel G, Edelman SV. Reduced capacity and affinity of skeletal muscle for insulin-mediated glucose uptake in noninsulin-dependent diabetic subjects. Effects of insulin therapy. J Clin Invest 1991;87:1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perseghin G. Muscle lipid metabolism in the metabolic syndrome. Curr Opin Lipidol 2005;16:416–420 [DOI] [PubMed] [Google Scholar]
- 44.Pearen MA, Ryall JG, Lynch GS, Muscat GE. Expression profiling of skeletal muscle following acute and chronic beta2-adrenergic stimulation: implications for hypertrophy, metabolism and circadian rhythm. BMC Genomics 2009;10:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev 1992;6:439–453 [DOI] [PubMed] [Google Scholar]
- 46.Tan NS, Michalik L, Di-Poï N, et al. Essential role of Smad3 in the inhibition of inflammation-induced PPARbeta/delta expression. EMBO J 2004;23:4211–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]