Abstract
OBJECTIVE
Variation in the transcription factor 7-like 2 (TCF7L2) locus is associated with type 2 diabetes across multiple ethnicities. The aim of this study was to elucidate which variant in TCF7L2 confers diabetes susceptibility in African Americans.
RESEARCH DESIGN AND METHODS
Through the evaluation of tagging single nucleotide polymorphisms (SNPs), type 2 diabetes susceptibility was limited to a 4.3-kb interval, which contains the YRI (African) linkage disequilibrium (LD) block containing rs7903146. To better define the relationship between type 2 diabetes risk and genetic variation we resequenced this 4.3-kb region in 96 African American DNAs. Thirty-three novel and 13 known SNPs were identified: 20 with minor allele frequencies (MAF) >0.05 and 12 with MAF >0.10. These polymorphisms and the previously identified DG10S478 microsatellite were evaluated in African American type 2 diabetic cases (n = 1,033) and controls (n = 1,106).
RESULTS
Variants identified from direct sequencing and databases were genotyped or imputed. Fifteen SNPs showed association with type 2 diabetes (P < 0.05) with rs7903146 being the most significant (P = 6.32 × 10−6). Results of imputation, haplotype, and conditional analysis of SNPs were consistent with rs7903146 being the trait-defining SNP. Analysis of the DG10S478 microsatellite, which is outside the 4.3-kb LD block, revealed consistent association of risk allele 8 with type 2 diabetes (odds ratio [OR] = 1.33; P = 0.022) as reported in European populations; however, allele 16 (MAF = 0.016 cases and 0.032 controls) was strongly associated with reduced risk (OR = 0.39; P = 5.02 × 10−5) in contrast with previous studies.
CONCLUSIONS
In African Americans, these observations suggest that rs7903146 is the trait-defining polymorphism associated with type 2 diabetes risk. Collectively, these results support ethnic differences in type 2 diabetes associations.
Diabetes is estimated to affect nearly 24 million people in the United States. This significant disease burden translates to a major economic impact. Prevalence is observed disproportionately across ethnicities with the some of the highest rates observed in African Americans, i.e., 11.8% (1). Increased risk is likely to be multifactorial, resulting from the combination of shared cultural, environmental, and genetic factors. Although recent genome-wide association studies of type 2 diabetes in European-derived populations have revealed novel, reproducible susceptibility loci (2–11), few have been replicated in African Americans (12–14).
An exception to this observation is the association of the transcription factor 7-like 2 (TCF7L2) gene with type 2 diabetes in African (15) and African-derived populations, i.e., African Americans (12,16). TCF7L2 is a transcription factor involved in the Wnt signaling pathway (17). Although initial reports implicated TCF7L2 in the regulation of the glucagon gene in the L cells of the gut (18), more recent reports suggest involvement in insulin secretion (19) potentially through epigenetic mechanisms (20). The initial report of association between TCF7L2 and type 2 diabetes in an Icelandic cohort identified a 64-kb linkage disequilibrium (LD) block of strong LD encompassing the intron 3 to intron 4 region of the gene (21) in this European population. Refinement of this signal in expanded populations revealed the strongest evidence of association with the single nucleotide polymorphism (SNP) rs7903146 with a relative risk of 1.45–1.49 (15). Although it has been inferred from these studies that rs7903146 is most likely the causative variant, the large (64 kb) LD block in European-derived populations and the large number of variants in this region have made it challenging to definitively conclude that variation at this SNP confers susceptibility to type 2 diabetes based solely upon genetic studies.
We previously reported association of TCF7L2 variants and type 2 diabetes in a large African American case-control cohort (16). Of the SNPs evaluated, association was observed with rs7903146 and rs7901695 (admixture-adjusted additive P = 3.77 × 10−6 and 0.0030, respectively) in a collection of 577 type 2 diabetic case subjects enriched for nephropathy and 596 controls. Given the evidence of association in our African American cohort (12,16), we sought to refine the genomic interval of TCF7L2 associated with type 2 diabetes in African Americans and, using a comprehensive analysis of variation in TCF7L2, to define the genetic basis for type 2 diabetes susceptibility.
RESEARCH DESIGN AND METHODS
Study subjects.
This study was conducted under Institutional Review Board approval from Wake Forest University School of Medicine. Identification, clinical characteristics, and recruitment of African American patients and controls have been previously described in detail (22). Briefly, 1,033 unrelated African American patients with type 2 diabetes were recruited from dialysis facilities. Type 2 diabetes was diagnosed in African Americans who reported developing type 2 diabetes after the age of 25 years and who did not receive only insulin therapy since diagnosis. In addition, cases had to have at least one of the following three criteria for inclusion: 1) type 2 diabetes diagnosed at least 5 years before initiating renal replacement therapy, 2) background or greater diabetic retinopathy, and/or 3) ≥100 mg/dL proteinuria on urinalysis in the absence of other causes of nephropathy. An additional 1,106 unrelated African Americans without a current diagnosis of diabetes or renal disease were recruited from the community and internal medicine clinics as controls. All type 2 diabetic cases and nondiabetic controls were born in North Carolina, South Carolina, Georgia, Tennessee, or Virginia. DNA extraction was performed using the PureGene system (Gentra Systems, Minneapolis, MN).
Sequencing.
The DNA screening panel was composed of 96 African American subjects: 48 type 2 diabetic cases and 48 controls. PCR primers were designed to independently amplify a 4.3-kb region of TCF7L2. Primer sequences are available on request. DNA sequencing reactions were performed using BigDye Terminator v.1.1 Cycle Sequencing Kits and analyzed on the Applied Biosystems 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA). Sequencing reactions were performed on both DNA strands. Sequence alignment and polymorphism identification were performed using Sequencher 4.2 (Gene Codes, Ann Arbor, MI). All polymorphisms were validated through observation on both strands. A search of the region sequenced was performed at dbSNP (www.ncbi.nlm.nih.gov) to record all previously identified polymorphisms reported in the region.
Genotyping.
Genotyping of DG10S478 was performed by fragment length analysis on an ABI Prism DNA Analyzer 3700 with previously published primers (21) in a manner similar to that previously described (23). Fragment length was determined using ABI Prism GeneMapper software v3.0. Fifty-six duplicate samples were run for quality control purposes. SNP genotyping was performed on the iPlex MassARRAY genotyping platform (Sequenom, San Diego, CA). Blind duplicates and blanks were included for quality control and error checking. For all SNPs, the genotyping success rate was greater than 95%.
Imputation.
In preliminary analyses, SNPs in the TCF7L2 gene region ± 10 kb (C10:114689999–114926060) were imputed using data from HapMap phase II hybrid panel (1:1, YRI:CEU; 108 SNPs) and the 1000 Genomes YRI Pilot 1 dataset (497 SNPs) to circumvent limited coverage of the genetic diversity in this specific region by the Affymetrix 6.0 array. SNPs were imputed in 965 type 2 diabetic end-stage renal disease (ESRD) cases and 1,029 controls with high-quality score (rsq-hat ≥0.3) using the software MACH (www.sph.umich.edu/csg/abecasis) (24). The imputed most likely genotypes were then used for subsequent association tests.
Four common SNPs identified from direct sequence analysis had minor allele frequencies (MAF) ≥0.05 and failed assay design on the Sequenom platform. With the use of the resequencing genotype data obtained from the 96 African American samples as reference, these SNPs were imputed in the remaining 2,043 samples with high-quality score (rsq-hat ≥0.5) using the software MACH (www.sph.umich.edu/csg/abecasis) (24). The imputed most likely genotypes were then used for subsequent association tests.
Analysis.
SNPs were tested for departure from Hardy-Weinberg equilibrium (HWE) using an exact test of HWE proportions for the combined group of cases and controls and then for cases only and controls only (25). Those SNPs out of HWE were noted but still included for the genotypic analysis. Haplotype block structure was established using Haploview 4.1 (26), defining blocks using the method from Gabriel et al. (27).
Unadjusted measures of LD and association were assessed using the software SNPGWA (http://www.phs.wfubmc.edu) (28). SNPGWA computes LD statistics, D′ and r2, for each pair of tandem SNPs. SNPGWA also performs multiple tests of association including the overall two-degree of freedom test (genotype), dominant model, recessive model, additive model (Cochran-Armitage trend test), and the corresponding lack of fit to the additive model. Odds ratios, 95% confidence intervals, and P values were computed for each model of association. Population attributable risk (PAR) was calculated as (X − 1)/X. Assuming a log additive model, X = (1 − f)2 + 2f(1 − f)γ + f2γ2 where γ is the estimated odds ratio (OR) and f is the risk allele frequency. DG10S478 was converted to a biallelic marker for analysis.
Ancestry estimates were determined from 70 biallelic admixture informative markers (AIMs) as previously described (16,29). Briefly, AIMs were selected to maximize European and African allele frequency differences and sample all non-acrocentric arms of the autosomal genome. Reference population allele frequencies were derived by genotyping 44 African (Yoruba from Ibadan, Nigeria) and 39 European Americans. Individual ancestral proportions were generated for each subject using FRAPPE (30), an expectation-maximization algorithm, under a two-population model. The individual ancestral estimates were used as covariates in the association analyses.
Conditional haplotype analysis.
To test whether SNPs were independently associated with type 2 diabetes, we performed an omnibus test for the haplotype association using PLINK (31). We further adjusted the omnibus test by controlling for one of the SNPs at a time. An insignificant conditional test suggests that the conditioned SNP is sufficient to explain the haplotype association and there is a single, rather than multiple, association signals at the haplotype.
RESULTS
Population characteristics.
Characteristics of the African American case-control populations are shown in Table 1. Controls were significantly younger than type 2 diabetic cases (P < 0.0001), although they were significantly older than the mean age at type 2 diabetes diagnosis in the cases (P < 0.0001). BMI was not significantly different between cases and controls (P = 0.49). Similar proportions of women were present in cases and controls (61% and 58%, respectively). FRAPPE (30) analysis of AIM genotypes estimated the mean proportion of African ancestry overall was 0.79 ± 0.12 and differed significantly (P < 0.0001) between type 2 diabetic cases and controls (0.80 ± 0.12 and 0.78 ± 0.12, respectively). Therefore, all results are presented with adjustment for admixture.
TABLE 1.
Characteristics of African American study participants
| Trait | Type 2 diabetic ESRD cases |
Controls |
||
|---|---|---|---|---|
| n* | Mean ± SD | n* | Mean ± SD | |
| N | 1,033 | — | 1,106 | — |
| Women (%) | 626 | 60.6 | 639 | 57.8 |
| Age (years) | ||||
| At exam | 994 | 61.5 ± 10.4 | 881 | 49.1 ± 11.9 |
| At type 2 diabetes diagnosis | 965 | 41.4 ± 12.4 | — | — |
| At ESRD diagnosis | 960 | 57.9 ± 10.9 | — | — |
| BMI (kg/m2) | 996 | 29.8 ± 7.1 | 879 | 30.0 ± 7.1 |
*Number with data available.
Refinement of the type 2 diabetes associated genomic interval.
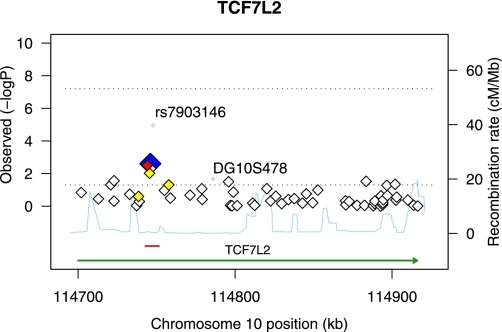
A preliminary analysis assessed association with type 2 diabetes for 59 SNPs across the entire 216-kb TCF7L2 gene ±10 kb from the Affymetrix 6.0 array-based analysis of 965 type 2 diabetic ESRD cases and 1,029 controls (Fig. 1). With the use of the Tagger program of Haploview (26), 43 of the 59 SNPs from the Affy array were available in the HapMap YRI dataset and captured common variation at 121 SNPs (MAF >0.05; aggressive tagging algorithm) with a mean r2 = 0.73. Notably, the SNP of greatest interest, rs7903146, is not typed on the Affymetrix 6.0 array. This SNP is located in a genomic interval that is not tagged well (max r2 = 0.45), resulting in only nominal evidence of association in the Affymetrix 6.0 analysis. To circumvent limited coverage of the genetic diversity in this specific region, imputation was used. Using data from HapMap phase II hybrid panel (1:1, YRI:CEU), 108 SNPs were imputed across the TCF7L2 gene ± 10 kb (Supplementary Fig. 1). As a result, no variants were identified with the same magnitude of significance as rs7903146. In a separate analysis, 497 SNPs were imputed from the 1000 Genomes YRI Pilot 1 dataset (Supplementary Fig. 2). As a result, two variants were identified (rs33998771, chr10:114740378) proximal to our region with significant P values (3.58 × 10−5 and 3.29 × 10−5, respectively) similar to that of rs7903146. To test whether these SNPs (rs33998771, chr10:114740378, rs7903146) were independently associated with type 2 diabetes, we performed an omnibus test for the haplotype association controlling for one SNP at a time. Four common haplotypes (ACT, ATT, TTT, and TTC) accounted for 99.9% of all haplotypes. These haplotype frequencies were significantly different between type 2 diabetic cases and controls (0.042, 0.019, 0.268, and 0.672, respectively for cases; 0.020, 0.016, 0.229, and 0.735, respectively for controls, omnibus test P = 5.4 × 10−6), with the haplotypes TTC and ACT strongly associated with protection and risk for type 2 diabetes, respectively (P < 0.0001). Omnibus analysis revealed that the haplotype association was significantly reduced after adjusting for rs7903146 (P = 0.023), whereas strong association remained by conditioning rs33998771 or chr10:114740378 (P = 0.0006 and 0.002, respectively), suggesting that rs7903146 alone explained most of the haplotype association.
FIG. 1.
Regional association plot for TCF7L2 ±10 kb (C10:114689999–114926060). All SNPs genotyped on the Affy 6.0 array are plotted with their −log10 P values of association with type 2 diabetes versus the genomic position (National Center for Biotechnology Information Build 36.1). The TCF7L2 gene position was taken from the University of California, Santa Cruz genome browser (green), and the core region of association (C10:114744078–114748339) analyzed by direct sequence analysis is depicted in red. The most significantly associated SNP from the array is depicted as a blue diamond with its correlated proxies (red = r2 ≥ 0.80; orange = 0.50 ≥ r2 > 0.80). SNP rs7903146 and microsatellite DG10S478, depicted as gray circles, were typed independently in the same set of samples. Estimated recombination rates from HapMap are plotted in the background to depict the LD structure in the region. (A high-quality color representation of this figure is available in the online issue.)
These results lead us to focus on a single region spanning ∼16.5 kb. Fourteen additional SNPs were genotyped in an expanded set of type 2 diabetic cases and controls within this interval and analyzed for association as summarized in Table 2. The core region of association was between SNPs rs4132115 and rs7903146 (admixture-adjusted additive P values ranging from 0.012 to 2.38 × 10−6). This region encompasses a 4.3-kb LD block in the YRI population (HapMap Phase II YRI data), which is bounded by the two SNPs rs7901695 and the previously associated rs7903146 (16).
TABLE 2.
Single SNP genotypic association results for SNPs in the TCF7L2 gene showing association with type 2 diabetes ESRD
| Marker | Chromosome position (bp) | Alleles | MAF |
OR (95% CI) | Additive P value | |
|---|---|---|---|---|---|---|
| Cases (n = 982) | Controls (n = 1,039) | |||||
| rs7079711* | 114735778 | G/A | 0.44 | 0.43 | 1.00 (0.88–1.13) | 0.99 |
| rs4074720 | 114738737 | T/C | 0.23 | 0.25 | 0.95 (0.82–1.11) | 0.53 |
| rs4074718 | 114738857 | A/G | 0.23 | 0.25 | 0.93 (0.80–1.08) | 0.35 |
| rs11196182* | 114740397 | C/T | 0.26 | 0.28 | 0.85 (0.74–0.98) | 0.025 |
| rs17747324† | 114742743 | T/C | 0.08 | 0.07 | 1.26 (0.99–1.60) | 0.058 |
| rs4132115 | 114745736 | G/T | 0.19 | 0.15 | 1.23 (1.04–1.46) | 0.014 |
| rs4506565 | 114746281 | A/T | 0.51 | 0.47 | 1.15 (1.01–1.30) | 0.030 |
| rs7068741 | 114746498 | C/T | 0.19 | 0.15 | 1.24 (1.05–1.46) | 0.012 |
| rs7069007 | 114746525 | G/C | 0.14 | 0.11 | 1.23 (1.02–1.48) | 0.033 |
| rs7903146 | 114748589 | C/T | 0.35 | 0.29 | 1.35 (1.18–1.54) | 1.76 × 10−5 |
| rs11196187 | 114749685 | G/A | 0.06 | 0.05 | 1.29 (0.97–1.71) | 0.082 |
| rs7092484 | 114751173 | G/A | 0.28 | 0.26 | 1.07 (0.93–1.23) | 0.35 |
| rs12098651* | 114751959 | G/A | 0.24 | 0.22 | 1.09 (0.94–1.26) | 0.25 |
| rs6585198 | 114752477 | A/G | 0.18 | 0.21 | 0.85 (0.73–1.00) | 0.051 |
*Inconsistent with HWE in cases.
†Inconsistent with HWE in controls.
In addition, the previously associated (21) microsatellite marker DG10S478, which lies 38 kb distal to and outside of this LD block, was typed, and the results of the analysis are presented in Table 3. Allele sizes and frequencies were consistent with prior genotyping in samples from African populations (15). Evidence of association was observed with the protective alleles −4 and 16 (P = 0.043 and 5.02 × 10−5, respectively) and the risk allele 8 (P = 0.022).
TABLE 3.
DG10S478 allelic association with type 2 diabetes ESRD in African Americans
| Allele | Frequency |
OR | P value | |
|---|---|---|---|---|
| Cases (n = 1,910) | Controls (n = 1,972) | |||
| −8 | 0.00052 | 0.0010 | 0.52 | 0.59 |
| −4 | 0.0063 | 0.012 | 0.47 | 0.043 |
| 0 | 0.72 | 0.73 | 0.91 | 0.21 |
| 4 | 0.11 | 0.11 | 1.14 | 0.20 |
| 8 | 0.081 | 0.064 | 1.33 | 0.022 |
| 12 | 0.059 | 0.049 | 1.27 | 0.10 |
| 16 | 0.016 | 0.032 | 0.39 | 5.02 × 10−5 |
| 20 | 0.0052 | 0.0056 | 0.94 | 0.89 |
Direct sequence analysis and association analysis of the associated genomic interval.
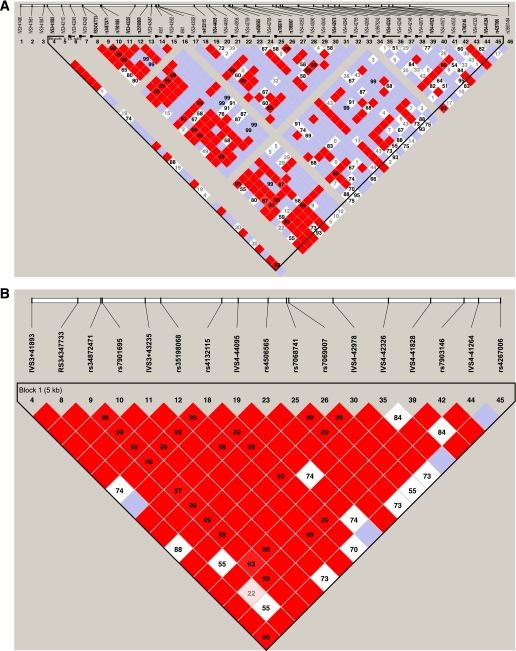
The core region of association (C10:114744078–114748339) ± 2 kb was analyzed by direct sequence analysis in 96 samples (48 type 2 diabetic cases and 48 controls). A total of 46 SNPs were identified, of which 72% (33/46) were novel and 17% (8/46) had a MAF >5%. When genotyped in the expanded cohort, five of the novel SNPs were found to be monomorphic and 10 could not be typed on the Sequenom platform because of the repetitive nature of the region. These 10 were genotyped via direct sequence analysis on a subset of 96 type 2 diabetic cases and 96 controls. Four SNPs were common, and these sequencing data were paired with existing data to be used as the known set of haplotypes for imputation in the remaining cohort. In addition, rs61875120 was common but yielded poor quality imputation (rsq = 0.31) and was therefore genotyped by direct sequence analysis. The remaining five SNPs had low MAF (MAF <0.05) and were not genotyped. Table 4 summarizes sequence variants and association analysis results. Of the 36 SNPs typed or imputed, 15 SNPs were found to be nominally associated with type 2 diabetes (admixture-adjusted additive P values ranging from 0.050 to 6.32 × 10−6). The most striking associations were observed at rs34872471, rs35198068 (imputed), and rs7903146, which were correlated (r2 > 0.74; Fig. 2) and associated with disease susceptibility (OR = 1.30–1.37) under an additive model. Three common haplotypes (CCT, CCC, and TTC) accounted for 99.6% of all haplotypes. These haplotype frequencies were significantly different between type 2 diabetic cases and controls (0.342, 0.056, and 0.602, respectively for cases; 0.278, 0.058, and 0.664, respectively for controls, omnibus test P = 3.7 × 10−5). Omnibus analysis revealed that the haplotype association was lost after adjusting for rs7903146 (P = 0.85), whereas modest association remained by conditioning rs34872471 or rs35198068 (P = 0.05), suggesting that rs7903146 alone is sufficient to explain the overall haplotype association.
TABLE 4.
Single SNP genotypic association results for SNPs identified by direct sequence analysis in the TCF7L2 gene showing association with type 2 diabetes ESRD
| Variant ID |
Position | Alleles | MAF |
OR (95% CI) | Additive P value | ||
|---|---|---|---|---|---|---|---|
| rs No. | Sequence ID | Cases (n = 1,033) | Controls (n = 1,106) | ||||
| IVS3 +41490 | 114742846 | C/− | 0.00 | 0.00 | — | — | |
| IVS3 +41661 | 114743017 | C/− | 0.00 | 0.00 | — | — | |
| rs61875120 | IVS3 +41847 | 114743203 | T/C | 0.08 | 0.07 | 1.29 (1.03–1.62) | 0.029 |
| IVS3 +41893† | 114743249 | T/C | 0.07 | 0.07 | 1.14 (0.90–1.44) | 0.28 | |
| IVS3 +42112 | 114743468 | C/− | 0.00 | 0.00 | — | — | |
| IVS3 +42245* | 114743601 | — | — | ||||
| IVS3 +42428* | 114743784 | — | — | ||||
| rs34347733† | IVS3 +42434 | 114743790 | C/T | 0.11 | 0.10 | 1.14 (0.94–1.39) | 0.19 |
| rs34872471 | IVS3 +42705 | 114744061 | T/C | 0.40 | 0.34 | 1.30 (1.14–1.48) | 8.34 × 10−5 |
| rs7901695 | IVS3 +42722 | 114744078 | T/C | 0.50 | 0.47 | 1.16 (1.03–1.32) | 0.017 |
| IVS3 +43235† | 114744591 | G/A | 0.18 | 0.15 | 1.26 (1.07–1.49) | 0.0053 | |
| rs35198068† | IVS3 +43418 | 114744774 | T/C | 0.40 | 0.34 | 1.31 (1.15–1.48) | 2.91 × 10−5 |
| IVS3 +43487* | 114744843 | — | — | ||||
| IVS3 +43552 | 114744908 | C/T | 0.02 | 0.01 | 1.80 (1.04–3.11) | 0.036 | |
| IVS3 +43592 | 114744948 | C/T | 0.01 | 0.01 | 1.64 (0.86–3.11) | 0.13 | |
| rs4132115 | IVS3 +44130 | 114745486 | G/T | 0.18 | 0.15 | 1.29 (1.09–1.53) | 0.0034 |
| IVS4 −44095 | 114745679 | C/G | 0.05 | 0.05 | 0.94 (0.71–1.25) | 0.68 | |
| IVS4 −44055 | 114745719 | C/G | 0.04 | 0.04 | 0.90 (0.66–1.23) | 0.51 | |
| IVS4 −43836 | 114745938 | A/G | 0.01 | 0.01 | 0.98 (0.48–1.99) | 0.95 | |
| IVS4 −43759 | 114746015 | T/− | 0.00 | 0.00 | — | — | |
| rs4506565 | IVS4 −43743 | 114746031 | A/T | 0.50 | 0.45 | 1.20 (1.06–1.36) | 0.0051 |
| IVS4 −43705 | 114746069 | A/G | 0.001 | 0.004 | 0.40 (0.11–1.49) | 0.17 | |
| rs7068741 | IVS4 −43526 | 114746248 | C/T | 0.18 | 0.15 | 1.25 (1.05–1.48) | 0.010 |
| IVS4 −43522 | 114746252 | — | — | ||||
| rs7069007 | IVS4 −43499 | 114746275 | G/C | 0.13 | 0.11 | 1.29 (1.07–1.57) | 0.0085 |
| IVS4 −43352 | 114746422 | T/C | 0.01 | 0.01 | 1.65 (0.87–3.13) | 0.12 | |
| IVS4 −43090 | 114746684 | A/G | 0.05 | 0.04 | 1.15 (0.84–1.56) | 0.38 | |
| IVS4 −43040§ | 114746734 | G/A | 0.001 | 0.004 | 0.39 (0.11–1.38) | 0.14 | |
| IVS4 −43007 | 114746767 | — | — | ||||
| IVS4 −42978 | 114746796 | C/T | 0.18 | 0.15 | 1.25 (1.04–1.50) | 0.019 | |
| IVS4 −42945 | 114746829 | G/T | 0.01 | 0.01 | 1.55 (0.82–2.95) | 0.18 | |
| IVS4 −42705 | 114747069 | A/T | 0.002 | 0.003 | 0.74 (0.21–2.64) | 0.64 | |
| IVS4 −42695 | 114747079 | T/− | 0.00 | 0.00 | — | — | |
| rs35676242 | IVS4 −42470 | 114747304 | C/A | 0.02 | 0.02 | 1.44 (0.90–2.30) | 0.13 |
| IVS4 −42326 | 114747448 | G/A | 0.07 | 0.07 | 0.95 (0.74–1.22) | 0.70 | |
| IVS4 −42248 | 114747526 | G/C | 0.03 | 0.03 | 0.92 (0.62–1.36) | 0.66 | |
| IVS4 −42148 | 114747626 | A/G | 0.02 | 0.01 | 1.75 (1.03–2.98) | 0.039 | |
| IVS4 −42079 | 114747695 | A/T | 0.01 | 0.01 | 1.41 (0.75–2.64) | 0.29 | |
| IVS4 −41828 | 114747946 | C/G | 0.13 | 0.13 | 0.93 (0.78–1.13) | 0.48 | |
| IVS4 −41672 | 114748102 | C/T | 0.01 | 0.01 | 1.11 (0.57–2.15) | 0.75 | |
| IVS4 −41633§ | 114748141 | G/C | 0.01 | 0.01 | 1.64 (0.72–3.77) | 0.24 | |
| rs7903146‡§ | IVS4 −41435 | 114748339 | C/T | 0.34 | 0.28 | 1.37 (1.19–1.57) | 6.32 × 10−6 |
| IVS4 −41323§ | 114748451 | G/A | 0.02 | 0.03 | 0.65 (0.43–0.99) | 0.043 | |
| IVS4 −41264 | 114748510 | C/T | 0.07 | 0.07 | 0.89 (0.69–1.14) | 0.36 | |
| rs4267006§ | IVS4 −41005 | 114748769 | G/T | 0.06 | 0.05 | 1.22 (0.93–1.60) | 0.14 |
| rs35801464 | IVS4 −39500 | 114750274 | C/T | 0.06 | 0.05 | 1.32 (1.00–1.75) | 0.050 |
*Low MAF in 96 samples resulting in poor imputation.
†Imputed genotypes.
‡Inconsistent with HWE in cases.
§Inconsistent with HWE in controls.
FIG. 2.
A: Haploview-generated LD map of the 40 SNPs identified by direct sequence analysis (C10:114742846–114750274) in African American controls (n = 1,106). Regions of high LD (D′ = 1 and logarithm of the odds [LOD] >2) are shown in dark red. Markers with lower LD (0.45 < D′ < 1 and LOD >2) are shown in dark through light red, with the color intensity decreasing with decreasing D′ value. Regions of low LD and low LOD scores (LOD <2) are shown in white. The number within each box indicates the r2 value. B: Haploview-generated LD map of the 17 common SNPs (MAF >0.05) identified by direct sequence analysis (C10:114742846–114750274) in African American controls (n = 1,106). Regions of high LD (D′ = 1 and LOD >2) are shown in dark red. Markers with lower LD (0.45 < D′ < 1 and LOD >2) are shown in light red, with the color intensity decreasing with decreasing D′ value. Regions of low LD and low LOD scores (LOD <2) are shown in white. The number within each box indicates the r2 value. (A high-quality color representation of this figure is available in the online issue.)
DISCUSSION
This study illustrates the power of genetic analyses in African-derived populations to facilitate identification of trait-defining variants. TCF7L2 has been identified as one of the strongest type 2 diabetes susceptibility genes to date with associations across multiple ethnically diverse populations (12,15,16,21,32). Our study is consistent with the initial association of SNP rs7903146 in an African American type 2 diabetic case-control population. By taking advantage of reduced LD in the African American population, we have been able to narrow the critical interval for association. This 4.3-kb region, flanked by SNPs rs4132115 and rs7903146, was the focus of resequencing in an effort to infer which sequence variant(s) are causally associated with type 2 diabetes.
Of the 46 SNPs identified by resequencing, 15 SNPs were nominally associated with type 2 diabetes with the most significant associations observed at rs34872471, rs35198068 (imputed), and rs7903146, which were highly correlated (r2 > 0.74; Fig. 1) and associated with disease susceptibility (OR = 1.30–1.37). Conditional omnibus haplotype analysis suggested that rs7903146 was sufficient to explain the haplotype association. This analysis suggests that association at rs34872471 and rs35198068 was the result of correlation with the true signal from rs7903146.
Although this study has eliminated the possibility of additional common variants (MAF >0.01) contributing to type 2 diabetes susceptibility within the fine-mapped interval (C10:114744078–114748339 ± 2 kb) of the TCF7L2 locus, four variants (IVS3 +42245, IVS3 +42428, IVS3 +43487, and IVS4 −43007) were found to have MAF <0.01. These variants, which were located in highly repetitive regions, were not evaluated. To date these variants have not been identified by other ongoing studies, i.e., the 1000 Genomes project, suggesting they are private mutations. Additionally, given the low MAF, these variants are not likely to explain the association observed at the TCF7L2 locus, but we cannot rule out the possibility that these and other unidentified rare variants contribute to disease susceptibility. If this were so, effect sizes of such rare variants would have to be in a range unprecedented for noncoding variants.
As a result of fine-mapping the TCF7L2 locus to determine the region most likely to harbor susceptibility variants, the microsatellite marker DG10S478 was excluded as the causal variant. DG10S478 is located 41 kb proximal to the critical interval defined in the African American population and is in weak LD with rs7903146 (D′ = 0.35, r2 = 0.07). Only a single common allele of DG10S478 is nominally associated with type 2 diabetes, with the strongest association, which is protective, seen with low MAF variants. These data suggest that the contribution to disease by DG10S478 is nominal.
This study represents the first comprehensive evaluation of variation within the TCF7L2 gene in a large African American population. Taking advantage of the LD structure in our African-derived sample of African Americans, we were able to reduce the genomic interval of association to ∼4.3 kb and exclude the possible contribution of the previously identified microsatellite marker to type 2 diabetes susceptibility. Our analysis identified three SNPs, rs34872471, rs35198068 (imputed), and rs7903146, which were highly associated with type 2 diabetes; all had P values that were two orders of magnitude stronger than other SNPs. Conditional omnibus haplotype analysis suggested that rs7903146 was sufficient to explain the haplotype association. SNP rs7903146 remains the most significantly associated variant within the TCF7L2 gene with a calculated PAR of 17.4%.
This investigation has used genetic approaches to focus on rs7903146. Alternative explanations can be proposed. For example, rs7903146 could be in LD with an unknown common variant. We cannot exclude this possibility with total confidence, but the assessment of markers in TCF7L2 by direct genotyping, imputation, and then through the use of the 1000 Genomes data using conditional analysis suggests the likelihood that such a common variant exists is low. Evaluation of long range LD on chromosome 10 shows little evidence for a remote variant (data not shown). An alternative is the possibility that a rare variant of large effect in LD with rs7903146 is the actual functional variant. This also seems unlikely. Although theoretically possible (33), we have recently shown empirically that it is easy to differentiate between a rare functional variant with large effect and a common variant in LD (34).
Thus, fine-mapping at the TCF7L2 locus using an African ancestry population has statistically implicated rs7903146 as the causal variant. It is noteworthy that Gaulton et al. (20) have implicated rs7903146 as a functional variant by mapping sequence variants to open chromatin sites. They found that rs7903146 is located in islet-selective open chromatin, and human islet samples heterozygous for rs7903146 showed allelic imbalance in islet enhancer activity. Thus genetic and functional studies make a consistent case for a functional role for rs7903146.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants K99-DK081350 (to N.D.P.), R01-DK066358 (to D.W.B.), R01-DK053591 (to D.W.B.), R01-HL56266 (to B.I.F.), and R01-DK070941 (to B.I.F.), and in part by the General Clinical Research Center of the Wake Forest University School of Medicine Grant M01-RR07122.
No potential conflicts of interest relevant to this article were reported.
N.D.P. researched data and wrote the article. J.M.H., S.S.A., A.A., and C.R. researched the data. C.D.L. provided analytical support, contributed to discussion, and reviewed and edited the article. B.I.F. contributed to discussion and reviewed and edited the article. M.C.Y.N. provided analytical support, contributed to discussion, and reviewed and edited the article. D.W.B. contributed to discussion and reviewed and edited the article.
The authors thank the patients, their relatives, and the staff of the Southeastern Kidney Council/ESRD Network 6 for their participation.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db10-0134/-/DC1.
REFERENCES
- 1.Diseases NIoDaDaK National Diabetes Statistics, 2007 fact sheet. Bethesda, MD, U.S. Department of Health and Human Services, National Institutes of Health, 2008 [Google Scholar]
- 2.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007;316:889–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet 2007;39:977–983 [DOI] [PubMed] [Google Scholar]
- 4.Prokopenko I, Langenberg C, Florez JC, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet 2009;41:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandhu MS, Weedon MN, Fawcett KA, et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet 2007;39:951–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxena R, Voight BF, Lyssenko V, et al. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 7.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007;316:1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881–885 [DOI] [PubMed] [Google Scholar]
- 9.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 2007;39:770–775 [DOI] [PubMed] [Google Scholar]
- 10.Zeggini E, Scott LJ, Saxena R, et al. Wellcome Trust Case Control Consortium Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008;40:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeggini E, Weedon MN, Lindgren CM, et al. Wellcome Trust Case Control Consortium (WTCCC) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis JP, Palmer ND, Hicks PJ, et al. Association analysis in African Americans of European-derived type 2 diabetes single nucleotide polymorphisms from whole-genome association studies. Diabetes 2008;57:2220–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore AF, Jablonski KA, McAteer JB, et al. Diabetes Prevention Program Research Group Extension of type 2 diabetes genome-wide association scan results in the diabetes prevention program. Diabetes 2008;57:2503–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmer ND, Goodarzi MO, Langefeld CD, et al. Quantitative trait analysis of type 2 diabetes susceptibility loci identified from whole genome association studies in the Insulin Resistance Atherosclerosis Family Study. Diabetes 2008;57:1093–1100 [DOI] [PubMed] [Google Scholar]
- 15.Helgason A, Pálsson S, Thorleifsson G, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 2007;39:218–225 [DOI] [PubMed] [Google Scholar]
- 16.Sale MM, Smith SG, Mychaleckyj JC, et al. Variants of the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in an African-American population enriched for nephropathy. Diabetes 2007;56:2638–2642 [DOI] [PubMed] [Google Scholar]
- 17.Prunier C, Hocevar BA, Howe PH. Wnt signaling: physiology and pathology. Growth Factors 2004;22:141–150 [DOI] [PubMed] [Google Scholar]
- 18.Yi F, Brubaker PL, Jin T. TCF-4 mediates cell type-specific regulation of proglucagon gene expression by beta-catenin and glycogen synthase kinase-3beta. J Biol Chem 2005;280:1457–1464 [DOI] [PubMed] [Google Scholar]
- 19.Lyssenko V, Lupi R, Marchetti P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest 2007;117:2155–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaulton KJ, Nammo T, Pasquali L, et al. A map of open chromatin in human pancreatic islets. Nat Genet 2010;42:255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006;38:320–323 [DOI] [PubMed] [Google Scholar]
- 22.Yu H, Bowden DW, Spray BJ, Rich SS, Freedman BI. Linkage analysis between loci in the renin-angiotensin axis and end-stage renal disease in African Americans. J Am Soc Nephrol 1996;7:2559–2564 [DOI] [PubMed] [Google Scholar]
- 23.Janssen B, Hohenadel D, Brinkkoetter P, et al. Carnosine as a protective factor in diabetic nephropathy: association with a leucine repeat of the carnosinase gene CNDP1. Diabetes 2005;54:2320–2327 [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Abecasis GR. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet 2006;S79:2290 [Google Scholar]
- 25.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet 2005;76:887–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265 [DOI] [PubMed] [Google Scholar]
- 27.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science 2002;296:2225–2229 [DOI] [PubMed] [Google Scholar]
- 28.Harley JB, Alarcón-Riquelme ME, Criswell LA, et al. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 2008;40:204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keene KL, Mychaleckyj JC, Leak TS, et al. Exploration of the utility of ancestry informative markers for genetic association studies of African Americans with type 2 diabetes and end stage renal disease. Hum Genet 2008;124:147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol 2005;28:289–301 [DOI] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang YC, Chang TJ, Jiang YD, et al. Association study of the genetic polymorphisms of the transcription factor 7-like 2 (TCF7L2) gene and type 2 diabetes in the Chinese population. Diabetes 2007;56:2631–2637 [DOI] [PubMed] [Google Scholar]
- 33.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol 2010;8:e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowden DW, An SS, Palmer ND, et al. Molecular basis of a linkage peak: exome sequencing and family-based analysis identify a rare genetic variant in the ADIPOQ gene in the IRAS Family Study. Hum Mol Genet 2010;19:4112–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]